Spatiotemporal localization of Arabidopsis sucrose synthase isoforms implicates these proteins in phloem loading and unloading.
Keywords: Arabidopsis, carbon metabolism, cellulose, phloem loading, sucrose synthase
Abstract
Sucrose synthase (SuSy) is one of two enzyme families capable of catalyzing the first degradative step in sucrose utilization. Several earlier studies examining SuSy mutants in Arabidopsis failed to identify obvious phenotypic abnormalities compared with wild-type plants in normal growth environments, and as such a functional role for SuSy in the previously proposed cellulose biosynthetic process remains unclear. Our study systematically evaluated the precise subcellular localization of all six isoforms of Arabidopsis SuSy via live-cell imaging. We showed that yellow fluorescent protein (YFP)-labeled SuSy1 and SuSy4 were expressed exclusively in phloem companion cells, and the sus1/sus4 double mutant accumulated sucrose under hypoxic conditions. SuSy5 and SuSy6 were found to be parietally localized in sieve elements and restricted only to the cytoplasm. SuSy2 was present in the endosperm and embryo of developing seeds, and SuSy3 was localized to the embryo and leaf stomata. No single isoform of SuSy was detected in developing xylem tissue of elongating stem, the primary site of cellulose deposition in plants. SuSy1 and SuSy4 were also undetectable in the protoxylem tracheary elements, which were induced by the vascular-related transcription factor VND7 during secondary cell wall formation. These findings implicate SuSy in the biological events related to sucrose translocation in phloem.
Introduction
In the vast majority of terrestrial plants, assimilated carbon from photosynthesis is primarily transported as sucrose to non-photosynthetic tissues, where it serves an integral role as a carbon and energy source. In sink tissues, sucrose is essential for the maintenance of cellular metabolism and cell wall biosynthesis, and can be converted to starch for storage and later use. Two enzymes catalyze the entry of sucrose into metabolic pathways: sucrose synthase (SuSy) and invertase (INV). While INV irreversibly cleaves sucrose into glucose and fructose, SuSy reversibly catalyzes the formation of fructose and UDP-glucose (Koch, 2004). SuSy predominantly occurs as a soluble form in the cytoplasm, but has also been found associated with the plasma membrane (Amor et al., 1995).
Sucrose synthases have also been shown to play an important role in modulating sink strength (D’Aoust et al., 1999). Phloem loading and sucrose breakdown efficiency in sink organs are thought to be important for defining sink strength. SuSy localization in phloem tissue has been observed through β-glucuronidase (GUS) histochemical staining or immunolabeling in many plant species, including maize (Nolte and Koch, 1993; Regmi et al., 2016), potato (Fu and Park, 1995), rice (Shi et al., 1994), citrus (Nolte and Koch, 1993), and Arabidopsis (Martin et al., 1993; Fallahi et al., 2008). In addition, several lines of evidence implicate SuSy in the development of sink organs. For example, inhibition of SuSy activity in potato led to decreased total tuber dry weight (Zrenner et al., 1995), while in carrot plants suppression of SuSy expression resulted in shorter and thinner roots (Tang and Sturm, 1999). Along with being involved in phloem loading, SuSy has also been shown to participate in starch biosynthesis (Chourey et al., 1998; Craig et al., 1999), as cellular starch concentrations have been reported to be correlated with altered SuSy activity in potato leaves and tubers (Munoz et al., 2005; Baroja-Fernandez et al., 2009), tomato fruit (D’Aoust et al., 1999), and maize seed endosperm (Li et al., 2013).
In sink tissues, such as secondary xylem, SuSy has been proposed to have a specific role in the synthesis of cell wall polymers, including cellulose and callose (Amor et al., 1995). A prevailing model of cellulose production suggests that a plasma membrane-localized SuSy interacts directly with the cellulose synthase complex to channel UDP-glucose derived from sucrose cleavage directly to cellulose synthesis (Koch, 2004; Verbancic et al., 2018). In developing cotton fibers, which deposit extremely pure cellulose in the secondary cell wall, Amor et al. (1995) showed via western blot analysis that SuSy was tightly bound to the plasma membrane. Consistent with this model, immunolabeling demonstrated that SuSy was abundant in sites where cellulose was rapidly synthesized and was distributed in a pattern paralleling the cellulose microfibrils (Amor et al., 1995; Haigler et al., 2001; Salnikov et al., 2003). In addition, co-immunoprecipitation experiments conducted with developing Populus xylem showed that SuSy was pulled down with the cellulose synthase complex, among other known and putative cell wall-related proteins (Song et al., 2010).
However, the essential role of SuSy in cellulose production has been questioned by Barratt et al. (2009), who reported that Arabidopsis sus1/sus2/sus3/sus4 quadruple mutants lacking the majority of SuSy activity displayed, via Fourier transform infrared spectroscopy analysis, normal cell wall structure and cellulose content. This conclusion was later challenged by Baroja-Fernández et al. (2012), as they showed that the remaining SuSy5 and SuSy6 activity in the quadruple mutant was sufficient to support normal rates of starch and cellulose synthesis in Arabidopsis when measured under optimum conditions (Baroja-Fernandez et al., 2012). These results suggest that a high functional redundancy exists within the SuSy gene family. Moreover, mutant studies could not exclude the possibility that SuSy may directly facilitate cellulose biosynthesis, via the association with cellulose synthase complex. Therefore, this study employed high-resolution imaging of each SuSy isoform to provide additional information to understand the potential functionalities of SuSy in Arabidopsis development.
In Arabidopsis, xylem vessels and fiber cells form a thick secondary cell wall and serve as a major sink for cellulose deposition. If SuSy supplies the substrate to the cellulose synthase complex, one would expect to see SuSy isoforms in developing xylem cells as they are actively depositing cellulose polymers into their secondary cell walls. To investigate the potential role of SuSy in Arabidopsis growth and development, yellow fluorescent protein (YFP) fusion constructs were generated under the control of native SuSy promoters, and live-cell imaging was used to determine their precise intracellular localization. Both SuSy1 and SuSy4 were specifically localized to phloem companion cells, but were not detectable in the developing xylem of the organs examined, including stems, roots, petioles, and siliques. In addition, none of the other four SuSy isoforms was found in the developing xylem of elongating stem, indicating that Arabidopsis SuSy could not directly facilitate cellulose biosynthesis. Our data support a functional role for SuSy1 and SuSy4 in energy production in companion cells that could contribute to the intricacies of sucrose translocation.
Materials and methods
Plant material, growth conditions, and flooding treatment
The sus1/sus4 double knock out mutant line was previously described in Bieniawska et al. (2007). Seeds of wild-type (WT) Arabidopsis thaliana ecotype Columbia-0 and sus1/sus4 were sterilized with 70% (v/v) ethanol for 5 min, followed by 10% (v/v) sodium hypochlorite (bleach) for 15 min, and finally rinsed several times with sterile distilled water. After 2–4 d in the dark at 4 °C, seedlings were germinated on half-strength Murashige and Skoog (MS) medium without sucrose and transferred to soil 7 d post-germination. All plants were grown in a growth chamber maintained at 21 °C, 50% humidity, 16 h light, 8 h dark, and a photosynthetic photon flux density of 150–180 μmol m–2 s–1. After 4 weeks of growth, a subset of plants was subjected to flooding treatment by adding and maintaining degassed water in the growth trays at a level just above the surface of the soil for 5 d. Leaf diameters were measured at the widest part of the rosettes following the 5 d of flooding.
Chlorophyll content analysis
The chlorophyll content index was measured on excised leaves, at the leaf tips where chlorosis was visually apparent by determining the absorbance at two wavelengths (931 nm and 653 nm) using a Chlorophyll Content Meter CCM-200 (OPTISCIENCE, USA). The output, the ratio of transmission of radiation from a light-emitting diode centered at 931 nm to transmission of radiation from a light-emitting diode centered at 653 nm (CCM-200 user manual), was used to establish the chlorophyll content index.
Soluble carbohydrate analysis
Soluble sucrose was extracted as previously described (Coleman et al., 2006) and analyzed by anion-exchange HPLC (ICS-5000; Dionex, Sunnyvale, CA, USA) that was fitted with a Dionex™ CarboPac™ PA1 column and a pulsed amperometric detector containing a gold electrode. Sugars were eluted with 16 mM NaOH and 2 mM NaOAc at a flow rate of 0.8 ml min–1. The concentration of sucrose was determined using an external calibration standard curve generated with known sucrose concentrations.
Real-time quantitative PCR analysis
RNA was extracted from Arabidopsis stems and leaves using the TRIzol® reagent (Invitrogen) according to the manufacturer’s instructions, and treated with TURBO DNase™ (Ambion) to remove residual DNA. cDNA was synthesized using the OneScript® Plus cDNA Synthesis Kit (Applied Biological Materials), and real-time quantitative PCR was performed using a Bio-Rad CFX96 Touch Real-Time PCR Detection System following the manufacturer’s protocol. Three biological replicates were harvested for each treatment, and samples were run in triplicate with BrightGreen Express 2× qPCR MasterMix-No Dye (Applied Biological Materials) using the primers listed in Supplementary Table S1 at JXB online. The Arabidopsis ubiquitin5 gene (AtUBQ5; At3g62250) was employed as a reference gene. The conditions for real-time analyses were: 95 °C for 20 s, followed by 40 cycles of 95 °C for 3 s, 57 °C for 30 s. Relative expression was determined according to Levy et al. (2004) using the equation: ΔCt=2–(CtSuSy–CtUBQ5).
Plasmid constructs
SuSy coding sequences were amplified from Arabidopsis stem and silique cDNA employing the iProof™ High-Fidelity PCR kit (Bio-Rad) using the primers listed in Supplementary Table S2. SuSy promoter fragments were amplified from genomic DNA, and the full details are listed in Supplementary Table S3. SuSy1 and SuSy4 coding sequences and individual promoters were first cloned into pDONR/Zeo using Gateway™ technology, and subsequently cloned into a modified pBin19 vector in which kanamycin resistance was replaced by a sulfadiazine resistance gene. C-terminal YFP fusions (SuSy1pro::SuSy1::YFP and SuSy4pro::SuSy4::YFP) were then independently transformed into sus1/sus4 double mutant plants using Agrobacterium tumefaciens (strain GV3101). In the case of SuSy2, SuSy3, SuSy5, and SuSy6, all DNA sequences were cloned into pDONR/Zeo vectors and subcloned into pH7WG2 vectors containing YFP at the C-terminus and were driven by their respective native SuSy promoters (~2000 bp upstream of the start codon). All constructs were subsequently transformed into WT Col-0 plants.
VND7 induction system for secondary cell wall formation
Seedlings containing both 35Spro::VND7::VP16::GR and SuSy–YFP fusions (SuSy1pro::SuSy1::YFP and SuSy4pro::SuSy4::YFP) were crossed, selected, and germinated on germination medium (1× MS, 1% sucrose, 1× Gamborg’s Vitamin mix, 0.05% MES, 0.8% agar at pH 5.8) for 5 d. Dexamethasone at 10 μM (diluted in half-strength MS liquid solution, Sigma) was supplemented to the plates for 12 h (the earliest stage of secondary cell wall deposition) to 24 h to induce the differentiation of protoxylem tracheary elements (Yamaguchi et al., 2010; Watanabe et al., 2015). Induction time for each cell in the individual seedling was slightly different between plants and lines, so the stage of development was defined based on the morphology of secondary cell wall banding pattern, as per Watanabe et al. (2015, 2018).
Western blotting
Approximately 200 mg of 6-week-old Arabidopsis stems were ground in liquid nitrogen and used for protein extraction following a previously described method (Wang et al., 2006). Proteins were quantified using DC™ Protein Assay (Bio-Rad) and diluted in phosphate-buffered saline (PBS) containing 1% SDS. A 15–25 μg aliquot of total protein was loaded onto an 8% SDS–PAGE gel with a 5% stacking gel and separated for 120 min at 110 V using the Mini Protean® Tetra Cell system (Bio-Rad). Proteins were subsequently transferred to a 0.1 μm pore nitrocellulose membrane and used for western blot analysis performed as previously described (Schilmiller et al., 2007). Primary antibodies (Hill et al., 2014; Watanabe et al., 2018) were used at the dilutions of 1:2000 for anti-CesA4, 1:500 for anti-CesA8, and 1:250 for anti-CesA7 in Tris-buffered saline with 0.01% Tween-20 containing 1% non-fat dry milk. Anti-SuSy immune serum raised against soybean root nodule SuSy (Zhang et al., 1999; Fedosejevs et al., 2014) was used at a 1:5000 dilution. Blots were washed four times with Tris-buffered saline with 0.01% Tween-20 and then incubated with a Superclonal™ ECL horseradish peroxidase- (HRP) conjugated secondary antibody (goat anti-rabbit IgG, Thermo Fisher) at a concentration of 1:15 000. Protein–antibody complexes were visualized with the SuperSignal West Pico Chemiluminescent Substrate kit (Thermo Fisher) following the manufacturer’s instructions.
Microscopy
Live-cell imaging was performed on a Leica DMI 6000 B inverted microscope with a Perkin Elmer Ultraview VoX Spinning Disk scan head with excitation/emission wavelengths of 514 nm/540 nm for visualization of all SuSy–YFP fusions. Propidium iodide and FM4-64 were detected using the 561 nm laser line and the 595 nm emission filter, respectively, while aniline blue was excited by a 405 nm laser and observed with the 540 nm detection filter. Images were captured on a Hamamatsu 9100-02 CCD camera using Volocity 6.3 software (Improvision) and processed using Image J software (National Institutes of Health; https://rsb.info.nih.gov/ij/). Background correction was performed by adjusting brightness or contrast of the images. For visualizing SuSy localization in mature plants, thin cross-sections were hand-sectioned from the base to the top of elongating stems and from leaves which were still expanding. In addition, 60–100 μm thick longitudinal sections of developing petioles and siliques were generated using a sliding benchtop microtome (American optical, Model #860). Cell wall staining with propidium iodide was performed by incubating 7-day-old seedlings in 10 μg ml–1 propidium iodide (in water) for 5 min, followed by thorough washing in water prior to imaging. The plasma membrane was labeled with a 10 μM FM4-64 dye (in water, Invitrogen) for 1 min and imaged directly, while the sieve plates were labeled using 0.01% aniline blue (in water) for 10 min in the dark and then rinsed, prior to imaging.
Results
Phenotype of sus1/sus4 under hypoxic conditions
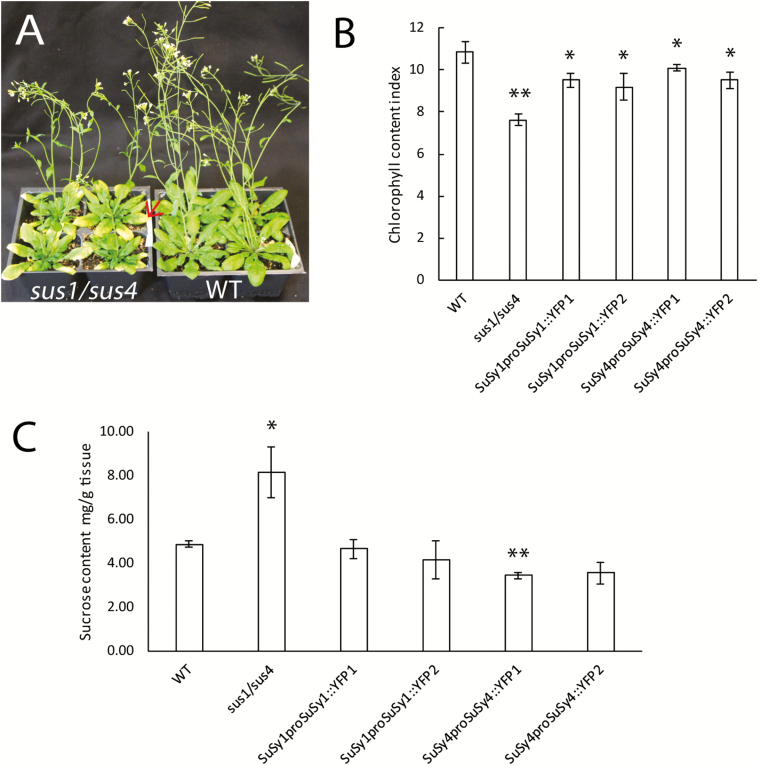
The Arabidopsis stem is considered a model system to study cell wall synthesis and, more specifically, cellulose biosynthesis, as it is largely composed of xylem vessels and fiber cells, which in turn are dominated by the cellulose polymer. In addition, SuSy1 and SuSy4 have been identified as the most highly expressed stem isoforms (Bieniawska et al., 2007), and as such were selected to examine their putative contribution to cellulose synthesis and deposition. Consistent with Bieniawska et al. (2007), under normal growing conditions, the sus1/sus4 double mutant exhibited no obvious growth phenotypes compared with WT plants, except for the prevalence of chlorotic leaves (Fig. 1A). In contrast, when subjected to 5 d of flooding, these lines showed reduced growth rates and significantly elevated soluble sucrose content in the rosette leaves (Fig. 1C; Supplementary Table S4).
Fig. 1.
Phenotypes of sus1/sus4 mutants and complemented Arabidopsis lines. (A) Representative photos of 6-week-old WT and sus1/sus4 mutants grown in a growth chamber maintained at 21 °C, 50% humidity, 16 h light, 8 h dark, and a photosynthetic photon flux density of 150–180 μmol m–2 s–1, showing the leaf chlorosis (red arrow) in double mutant plants. (B) Chlorophyll content index of the rosette leaves of sus1/sus4, WT, and SuSy transgenic plants. Results represent the means ±SE of three replicates from each of four individual plants (n=4). (C) Leaf soluble sucrose content after 5 d of flooding. Data are means ±SE calculated from four individual plants per line. Asterisks indicate a significant difference (*P<0.05, **P<0.01) comparable with WT plants using a Benjamini–Hochberg-corrected Student t-test.
SuSy1 and SuSy4 are detected in phloem, but not in developing xylem
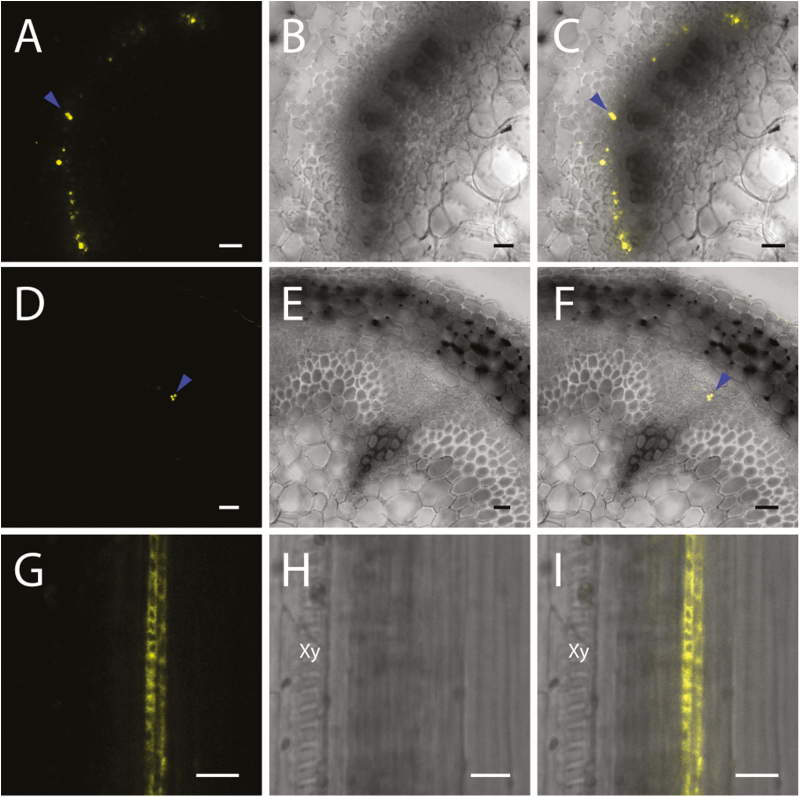
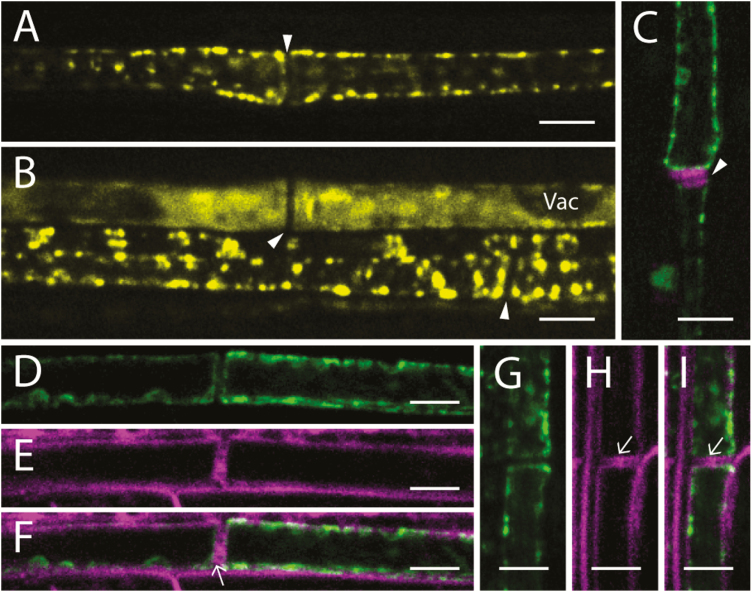
To investigate the spatiotemporal localization of these isoforms, Arabidopsis SuSy1 (At5g20830) and SuSy4 (At3g43190) coding sequences were cloned and subsequently fused to a C-terminal YFP fluorescence tag. The constructs were placed under the regulation of their respective native AtSuSy1 or AtSuSy4 promoters, and the functionality of the SuSy–YFP proteins was tested by complementing the sus1/sus4 loss-of-function mutant. In general, individual SuSy–YFP fusions partially rescued the sus1/sus4 phenotypes, demonstrating partial recovery of leaf chlorosis (Fig. 1B) and a reduced growth rate (Supplementary Table S4), and complete recovery of the soluble sucrose concentrations under hypoxic conditions (Fig. 1C). Subsequently, live-cell imaging was performed to visualize the cellular distribution of SuSy1 and SuSy4 proteins via spinning disk confocal microscopy. By checking the cross-sections from the base to the top of an elongating stem, YFP signal was not detectable in the xylem, but was consistently shown to be abundantly present in phloem tissue (Fig. 2A–C; Supplementary Fig. S1A–C). The same distribution pattern was also observed in the petioles of expanding leaves and developing siliques (Fig. 2D–I; Supplementary Fig. S1D–I). In the roots of 7-day-old seedlings, protoxylem cells were stained with propidium iodide and the fluorescently tagged SuSy signal was observed specifically in the phloem poles (Supplementary Fig. S2). SuSy1 and SuSy4 were also found in the funiculus, and only SuSy1 was present in the unloading domain of seeds at the end of the funiculus (Supplementary Fig. S3).
Fig. 2.
Confocal images of AtSuSy1–YFP in aerial tissues. (A–C) SuSy1–YFP localization in the phloem of stem cross-sections. (D–F) Cross-sections of leaf petioles showing YFP signal of AtSuSy1 (blue arrowheads) in the phloem. (G–I) Longitudinal sections of silique walls showing SuSy1–YFP in phloem tissue, not in protoxylem. Xy, protoxylem; (A), (D), and (G) are YFP fluorescence panels, while (B), (E), and (H) represent bright field images, and (C), (F), and (I) are merged images of YFP fluorescence and bright field images. Scale bars=50 μm in (A–F) and 20 μm in (G–I).
SuSy1 and SuSy4 are confined to companion cells
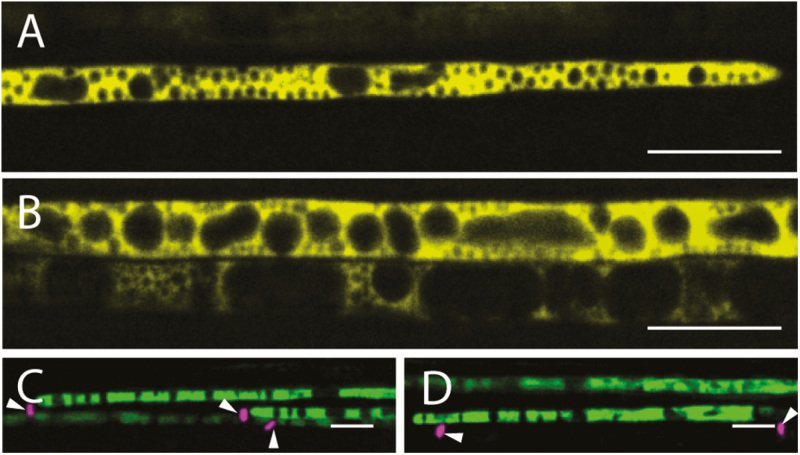
To further examine the precise localization of SuSy1 and SuSy4 proteins, thin longitudinal sections of leaf petioles that allowed single-cell imaging were employed. SuSy1 and SuSy4 were detected extensively in cells showing the typical morphology of companion cells (Fig. 3A, B), which differ from sieve elements as they inherently contain nuclei and vacuoles. To support this observation, the identity of these cells was confirmed by aniline blue staining, which specifically labels the callose-enriched sieve plates of sieve elements (Fig. 3C, D). Interestingly, both SuSy–YFP fusion proteins were consistently observed streaming within the cytoplasm of the companion cells, as shown by time-lapse video (Supplementary Movie S1).
Fig. 3.
AtSuSy1–YFP and AtSuSy4–YFP were found specifically in the companion cells of petioles. (A) Longitudinal sections of leaf petioles showing AtSuSy1 in companion cells containing numerous intracellular organelles. (B) AtSuSy4–YFP was confined to companion cells of leaf petioles. (C) Associated sieve plates were stained with aniline blue (magenta; white arrowheads) in panels showing YFP signal of AtSuSy1 (green). (D) Sieve plates were labeled with aniline blue (magenta; white arrowheads) in panels showing YFP signal of AtSuSy4 (green). Scale bars=10 μm.
Secondary cell wall CesAs are expressed concurrently with the SuSy isoforms
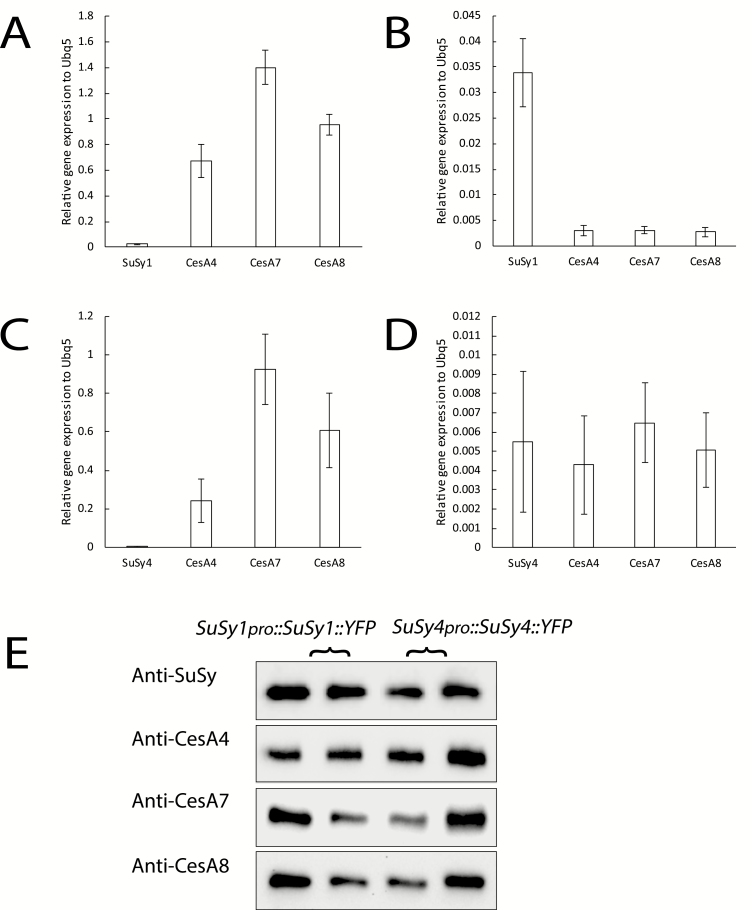
Cellulose synthesis will slow as the xylem matures. Should a SuSy–cellulose synthase complex interaction exist, SuSy–YFP fusion proteins may go undetected if xylem development has transitioned to later phases of development. To exclude this possibility, transcript levels and protein abundances of secondary cell wall-specific CesAs (CesA4, CesA7, and CesA8) were examined in the same transgenic SuSy1 and SuSy4 lines used for live-cell imaging. In general, CesA4, CesA7, and CesA8 displayed extremely high expression levels in elongating stem tissues but not in developing rosette leaves (Fig. 4A–D), which is consistent with their biological roles in secondary cell wall cellulose synthesis. Moreover, the transcript abundance of the secondary cell wall CesAs was much higher than that of SuSy in the stem tissue of both SuSy1–YFP and SuSy4–YFP fusion lines (Fig. 4A, C). Furthermore, western blot analysis showed that the CesA4, CesA7, and CesA8 proteins were present in the examined elongating stem tissues (Fig. 4E), indicating that live-cell imaging was carried out at an appropriate developmental window when secondary cell walls were still being actively deposited.
Fig. 4.
Transcript level and protein abundance of Arabidopsis secondary cell wall CesAs in 6-week-old SuSy1 and SuSy4 transgenic plant lines. (A, B) Expression of Arabidopsis AtSuSy1, AtCesA4, AtCesA7, and AtCesA8 relative to AtUBQ5 in the stems (A) and rosette leaves (B) of a SuSy1 transgenic line. (C, D) Expression of Arabidopsis AtSuSy4, AtCesA4, AtCesA7, and AtCesA8 relative to AtUBQ5 in the stems (C) and rosette leaves (D) of a SuSy4 transgenic line. (E) Western blots showing that SuSy–YFP and secondary cell wall CesA (CesA4, CesA7, and CesA8) proteins were all present in the stems of transgenic lines containing SuSy1pro::SuSy1::YFP or SuSy4pro::SuSy4::YFP. Data in expression bar graphs are means ±SE calculated from three biological replicates (n=3). For western blots, two biological replicates were used (n=2).
SuSy1 and SuSy4 are not present in the VND7-induced tracheary element system
In 2005, Kubo et al. (2005) demonstrated that the overexpression of Arabidopsis VND7 transcription factor could induce the trans-differentiation of seedling cells into protoxylem tracheary elements. The VND7 transcription factor was later paired with an inducible transcriptional activator, the viral protein 16 (VP16) coupled with a glucocorticoid receptor (GR), and, upon induction with dexamethasone, even the epidermal cells in Arabidopsis seedlings were shown to differentiate into tracheary elements (Yamaguchi et al., 2010). This system has subsequently been used to successfully perform high-resolution imaging of proteins expressed during secondary cell wall deposition (Watanabe et al., 2015, 2018; Takenaka et al., 2018). SuSy1–YFP and SuSy4–YFP fusions were transformed into an Arabidopsis line carrying the VND7–VP16–GR insertion and the ensuing plants showed YFP signal confined to the vasculature of seedling roots (Supplementary Fig. S2). However, the YFP signal of either SuSy1 or SuSy4 was undetectable in the induced tracheary element cells over the course of secondary cell wall development (Supplementary Fig. S4), indicating that SuSy1 and SuSy4 are not concurrently up-regulated during secondary cell wall formation in protoxylem tracheary elements.
SuSy5 and SuSy6 are restricted to sieve elements
In order to exclude the possibility that the four remaining SuSy proteins may be localized to the xylem and facilitate the production of UDP-glucose directly from sucrose to permit cellulose biosynthesis, C-terminal YFP fusion constructs of SuSy2, SuSy3, SuSy5, and SuSy6 were generated (SuSy2pro:: SuSy2::YFP, SuSy3pro::SuSy3::YFP, SuSy5pro::SuSy5::YFP, and SuSy6pro::SuSy6::YFP), independently transformed into WT Arabidopsis, and analyzed by confocal microscopy. Similarly, cross-sections from the base to the top of an elongating stem clearly showed a phloem-specific localization of SuSy5 and SuSy6 (Supplementary Fig. S5A–F). Again, longitudinal sections of leaf petioles were examined to determine the subcellular localization of SuSy5 and SuSy6. Consistent with Barratt et al. (2009), SuSy5 and SuSy6 were only apparent in the sieve elements, including both immature sieve elements containing vacuoles and mature sieve elements that are fully functional (Fig. 5A–C). These observations were further confirmed by aniline blue staining of sieve plates (Fig. 5C). Interestingly, in the translocating mature sieve elements, YFP fusions appeared to be parietally positioned and largely accumulate as puncta, and were distributed along the entire sieve element (Fig. 5A, C). Moreover, both the SuSy5–YFP and SuSy6–YFP fusions were immobile, as shown by time-lapse video (Supplementary Movie S2). In contrast, fluorescence occurred uniformly in the cytoplasm when the sieve element was immature (Fig. 5B). Employing the styryl dye FM4-64, which specifically labels the plasma membrane (Bolte et al., 2004), the precise localization of SuSy5 and SuSy6 was shown to be restricted to the inner side of the plasma membrane, indicating a cytoplasmic localization of these two proteins (Fig. 5D–I). This observation was most evident at the sieve plates, where the plasmodesmata were also stained by FM4-64. These observations provide additional evidence that SuSy proteins are not present in the developing xylem of Arabidopsis plants.
Fig. 5.
AtSuSy5–YFP and AtSuSy6–YFP were confined to the sieve elements of petioles. (A, B) Longitudinal sections of leaf petioles showing AtSuSy5 in mature sieve elements (A) and in immature sieve elements (B) containing vacuoles (Vac). (C) YFP labeling of AtSuSy6 (green) in mature sieve elements; the magenta signal indicates the sieve plates (white arrowhead) stained with aniline blue. (D–F) Longitudinal sections of leaf petioles showing the cytoplasmic localization of AtSuSy5. YFP signal of AtSuSy5 (green) is shown in (D), while the plasma membrane stained with FM4-64 (magenta) is shown in (E). Merged images of SuSy5–YFP and plasma membrane stained with FM4-64 are shown in (F). Plasmodesmata are indicated by an arrow. (G–I) Longitudinal sections of leaf petioles showing AtSuSy6–YFP (green) in (G) while the plasma membrane is labeled with FM4-64 (magenta) in (H). Merged images of SuSy6–YFP and plasma membrane stained with FM4-64 are shown in (I). Scale bars=5 μm.
SuSy2 and SuSy3 are highly expressed in developing seeds and leaf stomata
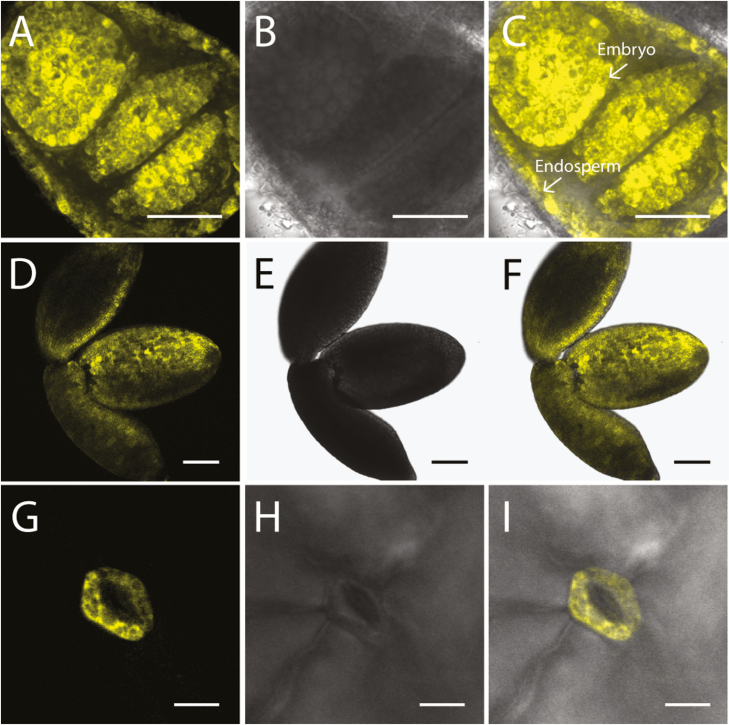
SuSy2 and SuSy3 were absent or undetectable in the vasculature of elongating stem (Supplementary Fig. S5G–L). Previous studies have shown that Arabidopsis SuSy2 and SuSy3 were induced in seeds, but barely detectable in other tissues (Bieniawska et al., 2007; Nunez et al., 2008). Therefore, live-cell imaging was performed on the cross-sections of transgenic seed, and SuSy2 was apparent in the embryo and endosperm (Fig. 6A–C). YFP signal of SuSy2 was widely detected in embryo cells from 10 to 17 days post-anthesis (DPA), and then the signal faded quickly after 17 DPA (Supplementary Fig. S6). Consistent with previous data, our current work shows that SuSy2 activity was not detectable in the outer integuments of the seed coat, when the mucilage was secreted and secondary cell wall was deposited (Supplementary Fig. S7 C,D). In contrast, SuSy3 was highly expressed in embryo cells at 17 DPA and was also observed in the guard cells of leaf stomata (Fig. 6D–I). Images in higher resolution showed that SuSy2–YFP and SuSy3–YFP were confined to the cytoplasm of embryo cells (Supplementary Fig. S8).
Fig. 6.
AtSuSy2–YFP and AtSuSy3–YFP were highly expressed in developing seeds and leaf stomata. (A–C) Cross-sections of Arabidopsis seeds at 13 DPA showing SuSy2 in the embryo and endosperm (arrows). (D–F) Intact embryo at 17 DPA showing that SuSy3–YFP fusions were present in abundance in embryo cells. (G–I) Leaf imaging showing that SuSy3 was detected in the guard cells of stomata. (A), (D), and (G) are YFP fluorescence panels, while (B), (E), and (H) represent bright field images, and (C), (F), and (I) are merged images of YFP fluorescence and bright field images. Scale bars=50 μm in (A–C), 100 μm in (D–F), and 10 μm in (G–I).
Discussion
Although the SuSy pathway has been generally considered the dominant route of sucrose catabolism in many plants, its essential role in Arabidopsis growth and development has been questioned by several studies (Bieniawska et al., 2007; Barratt et al., 2009). Thus, to gain further insights into the potential functional role(s) of SuSy in Arabidopsis, as well as to investigate its direct role in cellulose biosynthesis, we used live-cell imaging to localize all six SuSy isoforms under the control of their respective native promoters. High-resolution imaging of fluorescently tagged SuSy was carried out using a confocal microscope in several xylem-producing tissues including stems, leaves, siliques, and roots. Our results clearly demonstrated that most SuSy isoforms were exclusively expressed in the phloem, and none of the six SuSy was detectable in developing Arabidopsis xylem. SuSy1 and SuSy4 were specifically localized to phloem companion cells, where they are most probably involved in sucrose translocation.
Consistent with Bieniawska et al. (2007), sus1/sus4 double mutant plants exhibited no obvious growth phenotype when grown in well-aerated conditions, with the exception of mild chlorosis, but showed significantly reduced growth rates and accumulation of soluble carbohydrate when subjected to flooding (Fig. 1; Supplementary Table S4). Similar phenomena were also apparent in maize mutants lacking SuSy activity, which showed root death only during anoxic growth conditions (Ricard et al., 1998). It has been suggested that both SuSy1 and SuSy4 are necessary for plants to cope with anaerobic stress, as the transcript abundance of SuSy1 and SuSy4 dramatically increased when plants were subject to anaerobic environments (Baud et al., 2004; Bieniawska et al., 2007). Elevation of SuSy transcript abundance and activity in response to hypoxic conditions has also been reported in wheat (Albrecht and Mustroph, 2003) and potato (Biemelt et al., 1999). This specific response has been proposed to be biologically beneficial under these conditions, as it offers an energetically more efficient means to supply UDP-glucose to biological processes: requiring a single enzymatic step (via SuSy), while sucrose hydrolysis via invertases requires several additional biochemical conversions for the generation of UDP-glucose (Canam et al., 2006). Thus, under anaerobic stress and with limited ATP supply, the SuSy pathway may be favored in some plants. Evidence supporting this claim includes the rapid decrease in the INV:SuSy activity ratio when maize roots (Zeng et al., 1999) and rice seedlings (Guglielminetti et al., 1995) were exposed to low levels of oxygen. Moreover, leaf chlorosis has been reported to be associated with the inhibition of phloem loading (Zhang et al., 2014). Consistent with these observations, in this study, leaf chlorosis was obvious in 6-week-old sus1/sus4 plants and was more severe after flooding treatment, supporting a putative role for these two SuSy isoforms in phloem loading in Arabidopsis.
In support of this claim, our live-cell imaging data clearly show that SuSy1 and SuSy4 were exclusively localized to phloem companion cells, and were not present in the developing xylem of all the tissues examined (Figs 2, 3; Supplementary Fig. S1). Transcript results and western blot analysis indicate that secondary cell wall cellulose was still actively being synthesized in the elongating stem tissues employed for imaging (Fig. 4). In addition, our results show that neither the SuSy1–YFP nor the SuSy4–YFP fusion was detectable during secondary cell wall deposition in VND7-induced protoxylem tracheary elements (Supplementary Fig. S4). Instead, it was apparent that both SuSy1 and SuSy4 isoforms were actively streaming in the cytoplasm of companion cells of mature leaf petioles (Supplementary Movie S1). The current findings are consistent with the observed SuSy1 activity within phloem cells shown by promoter–GUS fusion studies (Martin et al., 1993). Furthermore, companion cell-specific localization of SuSy has been reported in several species (Nolte and Koch, 1993; Fallahi et al., 2008; Regmi et al., 2016). Our current study uniquely examined the tissue and subcellular localization of all SuSy isoforms. These findings, therefore, provide insight into the potential role(s) of SuSy in energy-dependent sucrose loading. Companion cells are very metabolically active and distinguished by a dense cytoplasm containing numerous mitochondria, plastids, and free ribosomes (Cayla et al., 2015), and communicate with sieve elements via plasmodesmata to supply energy and macromolecules. In Arabidopsis, sucrose is believed to be actively loaded into the phloem from the apoplast via a plasma membrane-localized AtSUC2 H+–sucrose symporter in the companion cells (Stadler and Sauer, 1996). This symporter has been proposed to be important not only for phloem loading in the sink tissues, but also for sucrose retrieval during long-distance transport (Truernit and Sauer, 1995; Stadler and Sauer, 1996; Gottwald et al., 2000). Our data show that the distribution pattern of SuSy isoforms is similar to that of the AtSUC2 H+–sucrose symporter. In addition, we show that SuSy1 was highly expressed in the symplastic unloading zone at the end of the funiculus (Supplementary Fig. S3A–C). In Arabidopsis, sucrose unloading at the terminal ends of phloem has been reported to be mainly symplastic, as observed in anthers (Imlau et al., 1999), developing seeds (Imlau et al., 1999; Stadler et al., 2005a), and root tips (Stadler et al., 2005b). Though the exact function of SuSy in this passive diffusion mechanism is not clear, one possibility is that the sucrose concentration is modulated by SuSy, and may regulate the efficiency of symplastic unloading.
In contrast to the companion cell specificity of SuSy1 and SuSy4, we show that SuSy5 and SuSy6 were confined to phloem sieve elements (Fig. 5A–C), consistent with Barratt et al. (2009). However, our high-resolution imaging expands these findings to show that SuSy5 and SuSy6 occurred predominantly as immobile puncta at the margin of sieve tubes, and were not co-localized with the plasma membrane (Fig. 5D–I). These unique observations may be explained by the ultrastructure of a sieve element, which has lost its nuclei, ribosomes, and vacuole during maturation, but still contains smooth endoplasmic reticulum, plastids, and P-protein (Knoblauch and van Bel, 1998; Cayla et al., 2015). These organelles are usually embedded in an amorphous ground matrix that attaches to the plasma membrane or P-protein filaments (Froelich et al., 2011). Thus, we propose that SuSy5 and SuSy6 are likely to be trapped in this parietal protein layer (see lack of movement in Supplementary Movie S2) and play a role in cellular metabolism of sieve elements, such as controlling osmolyte levels. Sieve plates provide connections among adjacent sieve elements, and the callose lining of sieve plate pores is essential for normal phloem transport (Barratt et al., 2011). SuSy5 and SuSy6 may therefore also be involved in callose synthesis, as double mutants displayed defects in callose lining of the sieve plates (Barratt et al., 2009). Our subcellular localization supports these claims. In addition, via immunofluorescence, SuSy has been shown to co-localize with H+-ATPases in sieve elements of castor bean tumor (Wächter et al., 2003). A sucrose transporter, AtSUC3, has previously been identified in the phloem sieve elements and proposed to be responsible for the retrieval of sucrose during phloem transport (Meyer et al., 2004). Therefore, we postulate that SuSy5 and SuSy6 could be involved in providing energy for this sucrose retrieval.
Consistent with previous studies, SuSy2 and SuSy3 were not present or detectable in the vasculature of elongating stems, but were specifically and strongly expressed in developing seeds and leaf stomata (Fig. 6; Supplementary Figs S5G–L, S6; Bieniawska et al., 2007; Fallahi et al., 2008; Nunez et al., 2008). Our results show that SuSy2 activity was highly induced in the cytoplasm of embryo cells from 10 to 17 DPA (Supplementary Fig. S6), while SuSy3 was widely expressed throughout the embryo at 17 DPA (Supplementary Fig. S8). In addition, transiently decreased starch content and sucrose accumulation has been reported in the single knock outs of SuSy2 and SuSy3 (Angeles-Nunez and Tiessen, 2010), suggesting that SuSy2 and SuSy3 may have a specific role in embryo development. Furthermore, SuSy3 was shown to be present in the guard cells of leaf stomata. Daloso et al. (2016) have shown that the up-regulation of AtSuSy3 in tobacco plants led to increased stomatal aperture and conductance, transpiration rate, net photosynthesis rate, and plant growth. These findings suggest that SuSy3 may be specifically involved in guard cell metabolism/function. Moreover, based on the collective observations, we suggest that SuSy2 and SuSy3 are not likely to be associated with cellulose synthesis, but are important to seed development and stomatal metabolism.
No single fluorescently tagged Arabidopsis SuSy was detectable in the xylem of the organs examined, while most SuSy proteins were restricted to the phloem. These findings suggest that the bulk of cellulose biosynthesis in Arabidopsis does not use UDP-glucose channelled directly from particulate SuSy, but instead relies on the free pool of cytoplasmic UDP-glucose. Free UDP-glucose may be generated partially by the activity of SuSy, but is more likely to be formed via UDP-glucose pyrophosphorylase from glucose released from sucrose via INVs. This is consistent with the recent findings of Barnes and Anderson (2018), who showed that an Arabidopsis mutant lacking two cytosolic INV isoforms exhibited abnormal cellulose biosynthesis and significantly reduced UDP-glucose content. However, SuSy could still impact cellulose deposition indirectly, by modulating carbon allocation and sink strength. Thus, the current model in Arabidopsis, at the very least, should be reconsidered, and similar studies should be initiated in other species reporting the phloem specificity of SuSy. However, it is still possible that in some plant species the proposed association between SuSy and cellulose synthase exists in certain cell types, such as cotton fibers and developing poplar xylem, which have extremely high rates of cellulose synthesis. Consistent with this, in these systems, overexpression of SuSy genes has manifested in improved cell wall biosynthesis, resulting in elevated cellulose production (Coleman et al., 2009; Xu et al., 2012). However, this does not necessarily have to come from a direct interaction between SuSy and the cellulose synthase complex. In addition, Gerber et al. (2014) showed that deficient SuSy activity in developing wood of aspen (Populus) did not affect cellulose biosynthesis, but led to a decrease in wood density, which again suggests that SuSy might play a role in defining carbon partitioning into wood. Therefore, our results highlight the need to re-examine the intricacies of secondary cell wall development and more specifically cellulose biosynthesis in other species, especially those generating significant quantities of cellulose in a relatively short period of time.
In summary, we examined the precise subcellular localization of three phylogenetically related pairs of Arabidopsis SuSy proteins. Despite the high expression level of SuSy1 and SuSy4 in stem tissues as previous studies suggest, they were specifically localized to phloem companion cells, not to developing xylem, which implicates a predominant role in sucrose translocation, not direct cellulose biosynthesis. Furthermore, SuSy5 and SuSy6 were confined to the cytoplasm of sieve elements, and SuSy2 and SuSy3 were highly induced during seed development and only SuSy3 was found in leaf stomata. Our data contribute to a better understanding of the potential functionality of SuSy, and permit a more refined understanding of the potential roles for SuSy isoforms in cell wall development, and general growth and development in Arabidopsis.
Supplementary data
Supplementary data are available at JXB online.
Fig. S1. Spatial localization of AtSuSy4.
Fig. S2. Phloem localization of AtSuSy1–YFP and AtSuSy4–YFP in the roots.
Fig. S3. Different spatial localization of AtSuSy1–YFP and AtSuSy4–YFP in developing seeds.
Fig. S4. YFP signal of SuSy1 and SuSy4 was not detected in the VND7-induced tracheary elements.
Fig. S5. Confocal images of AtSuSy2–YFP, AtSuSy3–YFP, AtSuSy5–YFP, and AtSuSy6–YFP in elongating inflorescence stems.
Fig. S6. Localization of AtSuSy2 in developing embryo of Arabidopsis.
Fig. S7. AtSuSy2–YFP was not detected in the epidermis of Arabidopsis seed coat.
Fig. S8. Subcellular localization of AtSuSy2–YFP and AtSuSy3–YFP in embryo cells.
Movie S1. Motility of SuSy4–YFP in the companion cells.
Movie S2. SuSy5–YFP was immobile in the sieve elements of petiole longitudinal sections.
Table S1. Primers employed for real-time PCR analysis.
Table S2. Primers employed for the amplification of SuSy-coding sequences.
Table S3. Primers used for amplification of SuSy promoter fragments.
Table S4. Growth phenotype of sus1/sus4 mutant plants subjected to flooding.
Acknowledgements
DY was funded by the China Scholarship Council (CSC). SDM acknowledges funding from the Natural Sciences and Engineering Research Council (NSERC) of Canada Discovery Program (RGPIN-2017–04566). The authors are grateful to Dr Ming Tien (Pennsylvania State University) for the gift of secondary cell wall CesA antibodies, and to Dr Taku Demura (Nara Institute of Science and Technology) for kindly providing the seeds containing 35Spro::VND7::VP16::GR. The authors also appreciate the professional advice on imaging and microscopy from Dr Anne Lacey Samuels (University of British Columbia).
References
- Albrecht G, Mustroph A. 2003. Localization of sucrose synthase in wheat roots: increased in situ activity of sucrose synthase correlates with cell wall thickening by cellulose deposition under hypoxia. Planta 217, 252–260. [DOI] [PubMed] [Google Scholar]
- Amor Y, Haigler CH, Johnson S, Wainscott M, Delmer DP. 1995. A membrane-associated form of sucrose synthase and its potential role in synthesis of cellulose and callose in plants. Proceedings of the National Academy of Sciences, USA 92, 9353–9357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angeles-Núñez JG, Tiessen A. 2010. Arabidopsis sucrose synthase 2 and 3 modulate metabolic homeostasis and direct carbon towards starch synthesis in developing seeds. Planta 232, 701–718. [DOI] [PubMed] [Google Scholar]
- Barnes WJ, Anderson CT. 2018. Cytosolic invertases contribute to cellulose biosynthesis and influence carbon partitioning in seedlings of Arabidopsis thaliana. The Plant Journal 94, 956–974. [DOI] [PubMed] [Google Scholar]
- Baroja-Fernández E, Muñoz FJ, Li J, Bahaji A, Almagro G, Montero M, Etxeberria E, Hidalgo M, Sesma MT, Pozueta-Romero J. 2012. Sucrose synthase activity in the sus1/sus2/sus3/sus4 Arabidopsis mutant is sufficient to support normal cellulose and starch production. Proceedings of the National Academy of Sciences, USA 109, 321–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baroja-Fernández E, Muñoz FJ, Montero M, et al. 2009. Enhancing sucrose synthase activity in transgenic potato (Solanum tuberosum L.) tubers results in increased levels of starch, ADPglucose and UDPglucose and total yield. Plant & Cell Physiology 50, 1651–1662. [DOI] [PubMed] [Google Scholar]
- Barratt DH, Derbyshire P, Findlay K, Pike M, Wellner N, Lunn J, Feil R, Simpson C, Maule AJ, Smith AM. 2009. Normal growth of Arabidopsis requires cytosolic invertase but not sucrose synthase. Proceedings of the National Academy of Sciences, USA 106, 13124–13129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barratt DH, Kölling K, Graf A, Pike M, Calder G, Findlay K, Zeeman SC, Smith AM. 2011. Callose synthase GSL7 is necessary for normal phloem transport and inflorescence growth in Arabidopsis. Plant Physiology 155, 328–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baud S, Vaultier MN, Rochat C. 2004. Structure and expression profile of the sucrose synthase multigene family in Arabidopsis. Journal of Experimental Botany 55, 397–409. [DOI] [PubMed] [Google Scholar]
- Biemelt S, Hajirezaei MR, Melzer M, Albrecht G, Sonnewald U. 1999. Sucrose synthase activity does not restrict glycolysis in roots of transgenic potato plants under hypoxic conditions. Planta 210, 41–49. [DOI] [PubMed] [Google Scholar]
- Bieniawska Z, Paul Barratt DH, Garlick AP, Thole V, Kruger NJ, Martin C, Zrenner R, Smith AM. 2007. Analysis of the sucrose synthase gene family in Arabidopsis. The Plant Journal 49, 810–828. [DOI] [PubMed] [Google Scholar]
- Bolte S, Talbot C, Boutte Y, Catrice O, Read ND, Satiat-Jeunemaitre B. 2004. FM-dyes as experimental probes for dissecting vesicle trafficking in living plant cells. Journal of Microscopy 214, 159–173. [DOI] [PubMed] [Google Scholar]
- Canam T, Park JY, Yu KY, Campbell MM, Ellis DD, Mansfield SD. 2006. Varied growth, biomass and cellulose content in tobacco expressing yeast-derived invertases. Planta 224, 1315–1327. [DOI] [PubMed] [Google Scholar]
- Cayla T, Batailler B, Le Hir R, Revers F, Anstead JA, Thompson GA, Grandjean O, Dinant S. 2015. Live imaging of companion cells and sieve elements in Arabidopsis leaves. PLoS One 10, e0118122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chourey PS, Taliercio EW, Carlson SJ, Ruan YL. 1998. Genetic evidence that the two isozymes of sucrose synthase present in developing maize endosperm are critical, one for cell wall integrity and the other for starch biosynthesis. Molecular & General Genetics 259, 88–96. [DOI] [PubMed] [Google Scholar]
- Coleman HD, Ellis DD, Gilbert M, Mansfield SD. 2006. Up-regulation of sucrose synthase and UDP-glucose pyrophosphorylase impacts plant growth and metabolism. Plant Biotechnology Journal 4, 87–101. [DOI] [PubMed] [Google Scholar]
- Coleman HD, Yan J, Mansfield SD. 2009. Sucrose synthase affects carbon partitioning to increase cellulose production and altered cell wall ultrastructure. Proceedings of the National Academy of Sciences, USA 106, 13118–13123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig J, Barratt P, Tatge H, Dejardin A, Handley L, Gardner CD, Barber L, Wang T, Hedley C, Martin C, Smith AM. 1999. Mutations at the rug4 locus alter the carbon and nitrogen metabolism of pea plants through an effect on sucrose synthase. The Plant Journal 17, 353–362. [Google Scholar]
- Daloso DM, Williams TC, Antunes WC, Pinheiro DP, Müller C, Loureiro ME, Fernie AR. 2016. Guard cell-specific upregulation of sucrose synthase 3 reveals that the role of sucrose in stomatal function is primarily energetic. New Phytologist 209, 1470–1483. [DOI] [PubMed] [Google Scholar]
- D’Aoust MA, Yelle S, Nguyen-Quoc B. 1999. Antisense inhibition of tomato fruit sucrose synthase decreases fruit setting and the sucrose unloading capacity of young fruit. The Plant Cell 11, 2407–2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fallahi H, Scofield GN, Badger MR, Chow WS, Furbank RT, Ruan YL. 2008. Localization of sucrose synthase in developing seed and siliques of Arabidopsis thaliana reveals diverse roles for SUS during development. Journal of Experimental Botany 59, 3283–3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedosejevs ET, Ying S, Park J, Anderson EM, Mullen RT, She YM, Plaxton WC. 2014. Biochemical and molecular characterization of RcSUS1, a cytosolic sucrose synthase phosphorylated in vivo at serine 11 in developing castor oil seeds. Journal of Biological Chemistry 289, 33412–33424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froelich DR, Mullendore DL, Jensen KH, Ross-Elliott TJ, Anstead JA, Thompson GA, Pélissier HC, Knoblauch M. 2011. Phloem ultrastructure and pressure flow: sieve-element-occlusion-related agglomerations do not affect translocation. The Plant Cell 23, 4428–4445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu H, Park WD. 1995. Sink- and vascular-associated sucrose synthase functions are encoded by different gene classes in potato. The Plant Cell 7, 1369–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber L, Zhang B, Roach M, Rende U, Gorzsás A, Kumar M, Burgert I, Niittylä T, Sundberg B. 2014. Deficient sucrose synthase activity in developing wood does not specifically affect cellulose biosynthesis, but causes an overall decrease in cell wall polymers. New Phytologist 203, 1220–1230. [DOI] [PubMed] [Google Scholar]
- Gottwald JR, Krysan PJ, Young JC, Evert RF, Sussman MR. 2000. Genetic evidence for the in planta role of phloem-specific plasma membrane sucrose transporters. Proceedings of the National Academy of Sciences of the USA 97, 13979–13984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guglielminetti L, Perata P, Alpi A. 1995. Effect of anoxia on carbohydrate metabolism in rice seedlings. Plant Physiology 108, 735–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haigler CH, Ivanova-Datcheva M, Hogan PS, Salnikov VV, Hwang S, Martin K, Delmer DP. 2001. Carbon partitioning to cellulose synthesis. Plant Molecular Biology 47, 29–51. [PubMed] [Google Scholar]
- Hill JL Jr, Hammudi MB, Tien M. 2014. The Arabidopsis cellulose synthase complex: a proposed hexamer of CESA trimers in an equimolar stoichiometry. The Plant Cell 26, 4834–4842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imlau A, Truernit E, Sauer N. 1999. Cell-to-cell and long-distance trafficking of the green fluorescent protein in the phloem and symplastic unloading of the protein into sink tissues. The Plant Cell 11, 309–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoblauch M, van Bel AJ. 1998. Sieve tubes in action. The Plant Cell 10, 35–50. [Google Scholar]
- Koch K. 2004. Sucrose metabolism: regulatory mechanisms and pivotal roles in sugar sensing and plant development. Current Opinion in Plant Biology 7, 235–246. [DOI] [PubMed] [Google Scholar]
- Kubo M, Udagawa M, Nishikubo N, Horiguchi G, Yamaguchi M, Ito J, Mimura T, Fukuda H, Demura T. 2005. Transcription switches for protoxylem and metaxylem vessel formation. Genes & Development 19, 1855–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy M, Edelbaum O, Sela I. 2004. Tobacco mosaic virus regulates the expression of its own resistance gene N. Plant Physiology 135, 2392–2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Baroja-Fernández E, Bahaji A, et al. 2013. Enhancing sucrose synthase activity results in increased levels of starch and ADP-glucose in maize (Zea mays L.) seed endosperms. Plant & Cell Physiology 54, 282–294. [DOI] [PubMed] [Google Scholar]
- Martin T, Frommer WB, Salanoubat M, Willmitzer L. 1993. Expression of an Arabidopsis sucrose synthase gene indicates a role in metabolization of sucrose both during phloem loading and in sink organs. The Plant Journal 4, 367–377. [DOI] [PubMed] [Google Scholar]
- Meyer S, Lauterbach C, Niedermeier M, Barth I, Sjolund RD, Sauer N. 2004. Wounding enhances expression of AtSUC3, a sucrose transporter from Arabidopsis sieve elements and sink tissues. Plant Physiology 134, 684–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz FJ, Baroja-Fernández E, Morán-Zorzano MT, Viale AM, Etxeberria E, Alonso-Casajús N, Pozueta-Romero J. 2005. Sucrose synthase controls both intracellular ADP glucose levels and transitory starch biosynthesis in source leaves. Plant & Cell Physiology 46, 1366–1376. [DOI] [PubMed] [Google Scholar]
- Nolte KD, Koch KE. 1993. Companion-cell specific localization of sucrose synthase in zones of phloem loading and unloading. Plant Physiology 101, 899–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Núñez JG, Kronenberger J, Wuillème S, Lepiniec L, Rochat C. 2008. Study of AtSUS2 localization in seeds reveals a strong association with plastids. Plant & Cell Physiology 49, 1621–1626. [DOI] [PubMed] [Google Scholar]
- Regmi KC, Zhang S, Gaxiola RA. 2016. Apoplasmic loading in the rice phloem supported by the presence of sucrose synthase and plasma membrane-localized proton pyrophosphatase. Annals of Botany 117, 257–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricard B, Toai TV, Chourey P, Saglio P. 1998. Evidence for the critical role of sucrose synthase for anoxic tolerance of maize roots using a double mutant. Plant Physiology 116, 1323–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salnikov VV, Grimson MJ, Seagull RW, Haigler CH. 2003. Localization of sucrose synthase and callose in freeze-substituted secondary-wall-stage cotton fibers. Protoplasma 221, 175–184. [DOI] [PubMed] [Google Scholar]
- Schilmiller AL, Koo AJ, Howe GA. 2007. Functional diversification of acyl-coenzyme A oxidases in jasmonic acid biosynthesis and action. Plant Physiology 143, 812–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Wang M, Powell K, Van Damme E, Hilder V, Gatehouse A, Boulter D, Gatehouse J. 1994. Use of the rice sucrose synthase-1 promoter to direct phloem-specific expression of β-glucuronidase and snowdrop lectin genes in transgenic tobacco plants. Journal of Experimental Botany 45, 623–631. [Google Scholar]
- Song D, Shen J, Li L. 2010. Characterization of cellulose synthase complexes in Populus xylem differentiation. New Phytologist 187, 777–790. [DOI] [PubMed] [Google Scholar]
- Stadler R, Lauterbach C, Sauer N. 2005a Cell-to-cell movement of green fluorescent protein reveals post-phloem transport in the outer integument and identifies symplastic domains in Arabidopsis seeds and embryos. Plant Physiology 139, 701–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler R, Sauer N. 1996. The Arabidopsis thaliana AtSUC2 gene is specifically expressed in companion cells. Botanica Acta 109, 299–306. [Google Scholar]
- Stadler R, Wright KM, Lauterbach C, Amon G, Gahrtz M, Feuerstein A, Oparka KJ, Sauer N. 2005b Expression of GFP-fusions in Arabidopsis companion cells reveals non-specific protein trafficking into sieve elements and identifies a novel post-phloem domain in roots. The Plant Journal 41, 319–331. [DOI] [PubMed] [Google Scholar]
- Takenaka Y, Watanabe Y, Schuetz M, et al. 2018. Patterned deposition of xylan and lignin is independent from that of the secondary wall cellulose of Arabidopsis xylem vessels. The Plant Cell 30, 2663–2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang GQ, Sturm A. 1999. Antisense repression of sucrose synthase in carrot (Daucus carota L.) affects growth rather than sucrose partitioning. Plant Molecular Biology 41, 465–479. [DOI] [PubMed] [Google Scholar]
- Truernit E, Sauer N. 1995. The promoter of the Arabidopsis thaliana SUC2 sucrose–H+ symporter gene directs expression of beta-glucuronidase to the phloem: evidence for phloem loading and unloading by SUC2. Planta 196, 564–570. [DOI] [PubMed] [Google Scholar]
- Verbančič J, Lunn JE, Stitt M, Persson S. 2018. Carbon supply and the regulation of cell wall synthesis. Molecular Plant 11, 75–94. [DOI] [PubMed] [Google Scholar]
- Wächter R, Langhans M, Aloni R, et al. 2003. Vascularization, high-volume solution flow, and localized roles for enzymes of sucrose metabolism during tumorigenesis by Agrobacterium tumefaciens. Plant Physiology 133, 1024–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Vignani R, Scali M, Cresti M. 2006. A universal and rapid protocol for protein extraction from recalcitrant plant tissues for proteomic analysis. Electrophoresis 27, 2782–2786. [DOI] [PubMed] [Google Scholar]
- Watanabe Y, Meents MJ, McDonnell LM, Barkwill S, Sampathkumar A, Cartwright HN, Demura T, Ehrhardt DW, Samuels AL, Mansfield SD. 2015. Visualization of cellulose synthases in Arabidopsis secondary cell walls. Science 350, 198–203. [DOI] [PubMed] [Google Scholar]
- Watanabe Y, Schneider R, Barkwill S, Gonzales-Vigil E, Hill JL Jr, Samuels AL, Persson S, Mansfield SD. 2018. Cellulose synthase complexes display distinct dynamic behaviors during xylem transdifferentiation. Proceedings of the National Academy of Sciences, USA 115, E6366–E6374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu SM, Brill E, Llewellyn DJ, Furbank RT, Ruan YL. 2012. Overexpression of a potato sucrose synthase gene in cotton accelerates leaf expansion, reduces seed abortion, and enhances fiber production. Molecular Plant 5, 430–441. [DOI] [PubMed] [Google Scholar]
- Yamaguchi M, Goué N, Igarashi H, et al. 2010. VASCULAR-RELATED NAC-DOMAIN6 and VASCULAR-RELATED NAC-DOMAIN7 effectively induce transdifferentiation into xylem vessel elements under control of an induction system. Plant Physiology 153, 906–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Y, Wu Y, Avigne WT, Koch KE. 1999. Rapid repression of maize invertases by low oxygen. Invertase/sucrose synthase balance, sugar signaling potential, and seedling survival. Plant Physiology 121, 599–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Han L, Slewinski TL, Sun J, Zhang J, Wang ZY, Turgeon R. 2014. Symplastic phloem loading in poplar. Plant Physiology 166, 306–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XQ, Lund AA, Sarath G, Cerny RL, Roberts DM, Chollet R. 1999. Soybean nodule sucrose synthase (nodulin-100): further analysis of its phosphorylation using recombinant and authentic root-nodule enzymes. Archives of Biochemistry and Biophysics 371, 70–82. [DOI] [PubMed] [Google Scholar]
- Zrenner R, Salanoubat M, Willmitzer L, Sonnewald U. 1995. Evidence of the crucial role of sucrose synthase for sink strength using transgenic potato plants (Solanum tuberosum L.). The Plant Journal 7, 97–107. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.