Down-regulation of CaMET1-like1 in pepper leads to DNA hypomethylation and premature fruit ripening, and interactions between DNA methylation and phytohormones appear to fine-tune the ripening process.
Keywords: Carotenoids, DNA methylation, fruit ripening, gene expression, pepper, plant hormone
Abstract
There is growing evidence to suggest that epigenetic tags, especially DNA methylation, are critical regulators of fruit ripening. To examine whether this is the case in sweet pepper (Capsicum annuum) we conducted experiments at the transcriptional, epigenetic, and physiological levels. McrBC PCR, bisulfite sequencing, and real-time PCR demonstrated that DNA hypomethylation occurred in the upstream region of the transcription start site of some genes related to pepper ripening at the turning stage, which may be attributed to up-regulation of CaDML2-like and down-regulation of CaMET1-like1, CaMET1-like2, CaCMT2-like, and CaCMT4-like. Silencing of CaMET1-like1 by virus-induced gene silencing led to DNA hypomethylation, increased content of soluble solids, and accumulation of carotenoids in the fruit, which was accompanied by changes in expression of genes involved in capsanthin/capsorubin biosynthesis, cell wall degradation, and phytohormone metabolism and signaling. Endogenous ABA increased during fruit ripening, whereas endogenous IAA showed an opposite trend. No ethylene signal was detected during ripening. DNA hypomethylation repressed the expression of auxin and gibberellin biosynthesis genes as well as cytokinin degradation genes, but induced the expression of ABA biosynthesis genes. In mature-green pericarp, exogenous ABA induced expression of CaDML2-like but repressed that of CaCMT4-like. IAA treatment promoted the transcription of CaMET1-like1 and CaCMT3-like. Ethephon significantly up-regulated the expression of CaDML2-like. Treatment with GA3 and 6-BA showed indistinct effects on DNA methylation at the transcriptional level. On the basis of the results, a model is proposed that suggests a high likelihood of a role for DNA methylation in the regulation of ripening in the non-climacteric pepper fruit.
Introduction
The ripening of fleshy fruit involves an elaborate process that is regulated by many factors at multiple levels. There is growing evidence to suggest that epigenetic tags are a critical regulator of fruit ripening, especially DNA methylation (Zhong et al., 2013; Liu et al., 2015; Lang et al., 2017; Cheng et al., 2018).
DNA methylation mainly refers to the methyl at the 5´ position of cytosine (5mC) (Bender, 2004). Based on the neighboring nucleotides, it can be classified into symmetric (CG or CHG) and asymmetric (CHH) types (Deleris et al., 2016). The formation of asymmetric DNA methylation is catalysed by CHROMOMETHYLASE 2 (CMT2) and DOMAIN REARRANGED METHYLTRANSFERASES (DRM1 and DRM2) through the RNA-directed DNA methylation (RdDM) pathway, whereas the symmetric type is catalysed by METHYLTRANSFERASE 1 (MET1) and CHROMOMETHYLASE 3 (CMT3) (Finnegan and Kovac, 2000; Lindroth et al., 2001; Bender, 2004; Zhang et al., 2010; Deleris et al., 2016). DNA methylation can be passively lost after DNA replication in the absence of methyltransferase activity or it can be actively eliminated by the DNA demethylases REPRESSOR OF SILENCING 1 (ROS1), DEMETER (DME), DME-LIKE 2 (DML2), and DME-LIKE 3 (DML3), which remove the 5mC base and then fill the single-nucleotide gap with a non-methylated cytosine (Gong et al., 2002; Hsieh et al., 2009; Wu and Zhang, 2010; Liu et al., 2015; Lang et al., 2017).
DNA methylation has been demonstrated to regulate fruit ripening in tomato, strawberry, and sweet orange. A global decrease of DNA methylation, especially at the 5´- and 3´-regulatory regions of the genes, has been associated with tomato and strawberry fruit ripening, and artificially accelerating this process by application of the methyltransferase inhibitor 5‐azacytidine induces premature ripening (Zhong et al., 2013; Cheng et al., 2018). In contrast, inhibition of ripening-associated hypomethylation through knockdown of SlDML2 disturbs the natural dynamic expression patterns of many tomato genes related to ripening and ultimately leads to retarded ripening (Liu et al., 2015; Lang et al., 2017). In the case of sweet orange, a global hypermethylation of DNA is found in the peel, which is critical for the ripening-associated process of de-greening (Huang et al., 2019). Differentially expressed genes regulated by ripening-associated changes in DNA methylation are usually enriched in the processes of pigment biosynthesis (such as anthocyanin biosynthesis in strawberry and carotenoid biosynthesis in tomato), aromatic compound biosynthesis, cell wall degradation, and ripening-related phytohormone biosynthesis (such as ABA biosynthesis in strawberry and sweet orange, and ethylene biosynthesis in tomato) (Lang et al., 2017; Cheng et al., 2018; Huang et al., 2019). The ripening-associated variation in DNA methylation is controlled by DNA methyltransferase and demethylase genes at the transcriptional level. In tomato, the ripening-associated DNA hypomethylation is attributed to both the down-regulation of DNA methyltransferase genes and the up-regulation of DNA demethylase genes (Cao et al., 2014). On the other hand, in strawberry this epigenetic change is mainly caused by the down-regulated of genes involving in RdDM (Cheng et al., 2018). In the peel of sweet orange, ripening-associated DNA hypermethylation is primarily caused by the down-regulation of DNA demethylase genes (Huang et al., 2019).
In contrast with the recent evidence for the involvement of epigenetic tags, the role of phytohormones in fruit ripening is long established. Ethylene plays a critical role in triggering ripening in climacteric fruit, and suppression of its biosynthesis and/or blocking of its signal pathway severely delays the normal ripening process (Oeller et al., 1991; Lanahan et al., 1994; Li et al., 2007; Lee et al., 2012; Sekeli et al., 2014). ABA is well accepted as an important positive regulator of ripening in both climacteric and non-climacteric fruit (Jia et al., 2011; Sun et al., 2012a, 2012b; Ji et al., 2014; Wang et al., 2013, 2017). The accumulation of ABA is associated with ripening in many fleshy fruits, such as tomato (Sun et al., 2012a, 2012b), cucumber (Wang et al., 2013), watermelon (Wang et al., 2017), grape (Sun et al., 2010), and strawberry (Jia et al., 2011). Artificially inhibiting ABA accumulation and/or blocking its perception results in retarded fruit ripening (Chai et al., 2011; Jia et al., 2011; Sun et al., 2012a, 2012b; Ji et al., 2014). Auxin has been found to have contrasting functions in the regulation fruit ripening. In most cases, it inhibits ripening by antagonizing the effects of ethylene or ABA (Manning et al., 1994; Davies et al., 1997; Liu et al., 2005; Böttcher et al., 2011; Symons et al., 2012; Ziliotto et al., 2012; Hao et al., 2015; Su et al., 2015; Li et al., 2016, 2017; Liao et al., 2018; Sravankumar et al., 2018); however, in some cases, especially in climacteric fruits such as tomato, peach, and fig, auxin promotes ripening by inducing the expression of ethylene biosynthesis genes (Gillaspy et al., 1993; Jones et al., 2002; Owino et al., 2006; Trainotti et al., 2007). Gibberellin generally plays a repressive role in the regulation of fleshy fruit ripening through antagonizing the effects of ethylene (Dostal and Leopold, 1967; Biale, 1978; Martínez et al., 1994; Martínez-Romero et al., 2000; Alós et al., 2006; Kader, 2008). Our knowledge about the role of cytokinin in the regulation of fleshy fruit ripening is very limited, although it has been reported to repress ripening in grape berries (Peppi and Fidelibus, 2008).
Pepper (Capsicum annuum) is a fruit that is consumed worldwide and it is sold at the mature and ripening stages. Ripening determines pepper fruit quality because, on one hand, it promotes the accumulation of pigments and other nutrients that increase the quality of the ripe fruit, and on the other hand, post-harvest ripening and coloration severely reduce fruit quality and shelf-life of mature-green peppers. To date, our understanding of pepper fruit ripening is very limited. Pepper is currently considered as a medium type between climacteric and non-climacteric fruits (Villavicencio et al., 2001) since some varieties, especially hot peppers, show an ethylene burst during ripening (Villavicencio et al., 1999; Hou et al., 2018) whereas some varieties, such as sweet peppers, do not (Lurie and Ben-Yehoshua 1986; Pretel et al., 1995; Watkins, 2002). The regulatory mechanism of pepper fruit ripening remains unclear, although a few studies have found that ethylene and ABA seem to promote ripening whilst auxin, gibberellin, and cytokinin are likely to play opposite roles (Deruère et al., 1994; Hou et al., 2018). Whether DNA methylation affects the regulation of ripening and interacts with phytohormones in pepper is even more unclear. In this study, we therefore examined the effects of DNA methylation on pepper fruit ripening and investigated its interactions with phytohormones. Our results demonstrated that the degree of DNA methylation in the fruit may be controlled by DNA methyltransferase and demethylase genes at the transcriptional level. DNA hypomethylation caused by silencing of CaMET1-like1 led to premature ripening. DNA methylation interacted with phytohormones in a way that may fine-tune the ripening of the fruit.
Materials and methods
Plant material
The sweet pepper (Capsicum annuum L.) inbred line 16C391 was developed in our lab and bears non-climacteric fruit. Plants used for developmental studies were grown in a greenhouse under natural daylight at the Experimental Station of China Agricultural University in 2017 and 2018. Plants were supplied with adequate water and nutrients according to standard horticultural practice. Pericarps were collected at the following stages: immature-green (IM, 15 d post anthesis, DPA), mature-green (MG, 38 DPA), breaker (B, 44 DPA), turning (T, 50 DPA), and red-ripening (R, 55 DPA). They were immediately frozen with liquid nitrogen and stored at –80 °C.
McrBC-PCR analysis
McrBC-PCR was conducted to test the DNA methylation levels in the upstream regions of the transcriptional start site (UROT) of 12 ripening-related genes, namely CaPSY1 (Capana04g002519, encoding phytoene synthase), CaPDS (Capana03g000054, encoding phytoene desaturase), CaCCS (Capana06g000615, encoding capsanthin-capsorubin synthase), CaPG (Capana10g002229, encoding polygalacturonase), CaPME2.2 (Capana00g004152, encoding a pectin methylesterase), CaCEL1 (Capana01g000166, encoding an endo-1,4-beta-glucanase), CaCEL2 (Capana08g002622, encoding an endo-1,4-beta-glucanase), CaNCED1 (Capana00g003114, encoding a 9-cis-epoxycarotenoid dioxygenase, a key enzyme in ABA biosynthesis), CaCYP707A1 (Capana04g001637, encoding an 8′‐hydroxylase, a key enzyme in ABA catabolism), CaRIN (Capana11g002005, encoding a MADS-box transcription factor), CaCNR (Capana02g001917, encoding a MADS-box transcription factor), and CaNOR (Capana07g002220, encoding a NAC domain transcription factor). Genomic DNA was extracted using the CTAB method according to Healey et al. (2014) from ~3 g pericarp tissue collected at the IM and T stages. McrBC digestion was performed with 1 µg of genomic DNA using a McrBC kit (NEB Beijing, China) following the manufacturer’s instructions. The digestion system without GTP was used as a negative control. The tested regions (~800–1000 bp) with relatively higher GC level were selected from the 2-kb regions upstream the putative transcriptional start sites (TSSs) (Supplementary Fig. S1 at JXB online). The GC level was calculated using the MethPrimer software (Li and Dahiya, 2002). Primers were designed using Primer5 and are listed in Supplementary Table S1. PCR was performed with 50 ng of DNA as the template and the products were analysed using 1.5% agarose gel electrophoresis.
Bisulfite sequencing
Bisulfite sequencing was performed to further confirm the DNA methylation level. DNA bisulfite conversion was conducted with 1 µg of genomic DNA using the a DNA Bisulfite Conversion Kit (Tiangen, China) following the manufacturer’s instructions. Methylation-specific PCR was set up with ~100 ng converted or non-converted genomic DNA as the template using a Methylation-specific PCR Kit (Tiangen, China) according to the user’s manual. Primers for the methylation-specific PCR were designed within the McrBC-PCR tested regions and are listed in Supplementary Table S2. The length of the amplification fragments ranged from 150–300 bp (Supplementary Fig. S1). Since every cytosine can be potentially methylated, to avoid any sequence selection bias during the PCR, the ‘C’ and ‘G’ nucleotides were replaced by ‘Y’ and ‘R’ in the forward and reverse primers, respectively. Ten single colonies for each PCR fragment were sequenced. The methylation ratio for each C-G site was calculated by dividing the number of non-changed nucleotides by the sequencing depth.
Identification, phylogenetic analysis, and prediction of conserved functional regions of DNA methyltransferase and demethylase genes
BLAST searches were performed with the Arabidopsis and tomato orthologs as queries against the pepper genome in the NCBI (https://www.ncbi.nlm.nih.gov/), SOL (https://www.solgenomics.net), and The Pepper Genome (http://peppersequence.genomics.cn/page/species/index.jsp) databases. A Neighbor-joining phylogenetic tree was constructed using the ClustalX2.0.12 and MEGA4.0.2 software (bootstrap =1000 replicates). Conserved functional regions were predicted using a MOTIF Search (https://www.genome.jp/tools/motif/) with the default settings.
Quantitative real-time PCR analysis
Total RNA extraction, first strand cDNA synthesis, and real-time PCR were conducted according to Sun et al. (2011). UBI-3 (Capana06g002873) was selected as the internal control (Wan et al., 2011) and relative expression values were calculated using the 2–∆∆CT method (Livak and Schmittgen, 2001). For each gene, three biological replicates were performed, and for each biological replicate at least three technical replicates were performed. Primers used in this analysis are listed in Supplementary Table S3.
Subcellular localization of CaMET1-like1, CaCMT3-like, and CaDML2-like
The coding sequence fragments of CaMET1-like1, CaCMT3-like, and CaDML2-like without the stop codon were PCR-amplified and subcloned into the Super1300 vector in frame with green fluorescence protein (GFP) driven by the CaMV 35S promoter. Plasmids were transferred into onion epidermal cells using particle bombardment according to Lee et al. (2006). GFP fluorescence was detected and captured using a Carl Zeiss LSM 510 system. Each assay was repeated three times. The primers used are listed in Supplementary Table S4.
Determination of IAA, ABA, and ethylene levels
Levels of auxin (IAA) and ABA were determined in the pericarp of normally developing fruits at the IM, M, T, and R stages and also in premature-ripe and green pericarps of the CaMET1-like1-silenced fruit using UPLC-MS (Waters Corporation and ThermoFisher Scientific) according to Wang et al. (2017). Ethylene levels of normally developing fruit were measured at the IM, M, T, and R stages using gas chromatography (GC 17A, Shimadzu) according to Xue et al., (2008).
Virus-induced gene silencing
A VIGS system based on the tobacco rattle virus (TRV) was used to investigate gene function (Liu et al., 2002). Fragments of CaMET1-like1 (423 bp) and CaPDS (452 bp) were PCR-amplified from pepper cDNA and cloned into the pTRV2 vector to generate the plasmids pTRV2-CaMET1-like1 and pTRV2-CaPDS, respectively. pTRV1, pTRV2, pTRV2-CaPDS, and pTRV2-CaMET1-like1 were introduced into Agrobacterium tumefaciens strain GV3101 by electroporation. The transformed cells were grown for 8–10 h at 28 °C in Luria–Bertani (LB) medium containing the appropriate antibiotics, and were then collected by centrifugation (5000 g for 15 min), re-suspended in infiltration buffer (10 mM MgCl2, 10 mM MES, and 100 µM acetosyringone), and incubated at 28 °C for 3 h. pTRV1 and pTRV2 or its derivatives were mixed at a 1:1 (v/v) ratio and infiltrated into cotyledons of 1-week-old pepper seedlings using a 1-cm3 syringe without a needle (Liu et al. 2002). The infiltrated plants were grown in a culture room at 22 °C, 16/8 h light/dark, 150 μmol m–2 s–1. Since silencing of CaPDS leads to light bleaching of green organs, plants infiltrated with pTRV1+pTRV2-CaPDS were used as the positive control to check the activity of pTRV1 and to indicate the spatio-temporal pattern of gene silencing (Guo et al., 2018). When light-bleaching appeared in the positive control, reverse-transcription PCR was conducted to detect pTRV1, pTRV2 and its derivatives in all the infiltrated plants. Plants carrying pTRV1+pTRV2-CaMET1-like1 and pTRV1+pTRV2 (negative control) were used for further analysis. Flowers at anthesis were hand-pollinated and tagged, and the fruit ripening time was evaluated as the number of days from anthesis to the breaker stage. At least 100 plants were infiltrated with pTRV1+pTRV2-CaMET1-like1 and at least 30 plants were injected with negative or positive control plasmid mixtures. The whole experiment was repeated twice. The primers used are listed in Supplementary Table S5.
RNA-seq analysis
Premature-ripe and green pericarps of the CaMET1-like1-silenced fruit and red-ripe pericarps of the negative control were collected from the two biological repeats as well as from a preliminary experiment. RNA extraction and library construction were conducted according to Zhong et al. (2011). Sequencing was conducted by the Biomarker Technologies Corporation (Beijing, China) on a GAII platform (Illumina) and paired-end reads were generated. Filtration of the reads, alignment, and expression calculations were carried out according to Huang et al. (2013). Differentially expressed genes (DEGs) were calculated using DESeq2 in R and were then screened according the following conditions: the average expression value of each gene was greater than 2 fragments per kilobase of transcript per million fragments mapped reads in at least one of the three samples, and the expected false positive rate value was less than 0.05. The KEGG statistics of pathway enrichment were analysed and plotted using BMKCloud (http://www.biocloud.net/) with the default settings. Principal component analysis (PCA) was conducted with expression profiles of all the samples using the stats::prcomp() function in R. Prior to that, data were mean centered and Z-scored. The PCA results were plotted and edited using ggplot2::ggplot() (Wickham, 2016) in R and Photoshop CS5, respectively.
Ripening-related parameters
Soluble solids content (SSC) and titratable acidity (TA) were measured using 50–100 g pericarp tissue taken from 3–5 fruit according to Kong et al. (2013). Carotenoids were extracted from 50 mg freeze-dried pericarp samples and measured using aLC-APCI-MS/MS system (AB SCIEX, USA) according to Giuffrida et al. (2013). Standard substances examined in study are listed in Supplementary Table S6. Three or more replicates of each assay were performed.
Phytohormone treatments
Fruit were harvested at the MG stage, surface-sterilized with 2.5% (w/v) NaClO aqueous solution for 15 min, and then washed three times with sterile water. Pericarp discs (radius 0.6 cm) were cut from the equatorial region of the fruit using a hole puncher. The discs were soaked in MES buffer (pH=5.5) containing 5 mmol l–1 CaCl2, 5 mmol/l–1 MgCl2, 5 mmol/l–1 EDTA, 5 mmol/l–1 vitamin C, and 200 mmol/l–1 mannitol for 30 mins (Télef et al., 2006). Phytohormones were then added into the buffer to give final concentrations of 25 μM for auxin (IAA; Sigma-Aldrich I5148), 100 μM for ABA (Sigma-Aldrich A1049), 50 μM for gibberellic acid (GA3; Sigma-Aldrich G7645), 25 μM for cytokinin (6-BA; Sigma-Aldrich B3408), and 25 μM for ethephon (Sigma-Aldrich 45473). The optimal concentration for each hormone was determined in preliminary experiments. At 4 h after adding the hormones the pericarp discs were collected, frozen with liquid nitrogen, and stored at –80 °C. For each treatment, at least 15 discs were used and the whole experiment was repeated three times. Expression of the DNA methyltransferase and demethylases genes, ripening-related genes (CaPSY1, CaPDS, CaPG, CaNCED1, and CaCYP707A1) together with several hormone-responsive genes (CaARF2 for IAA, CaABI2 for ABA, CaDELLA for GA3, CaARR5 for 6-BA, and CaETR1 for ethylene) were analysed using real-time PCR. Primers used in this experiment are listed in Supplementary Table S3.
Statistical analyses
Significant differences between two groups of samples were determined using Student’s t-test via the t.test function in Excel. Significant differences among multiple groups of samples were tested by ANOVA and post hoc Tukey’s HSD test using stats::anova() and agricolae::HSD.test(), respectively, in R (https://CRAN.R-project.org/package=agricolae). For real-time PCR, a significant difference between two groups of samples had to satisfy the following conditions: statistically significant difference at the 0.05 level, and the log2-transformed expression fold-change had to be either greater than 1 or less than 0.5.
Results
DNA hypomethylation occurs in the UROT of several ripening-related genes during fruit ripening
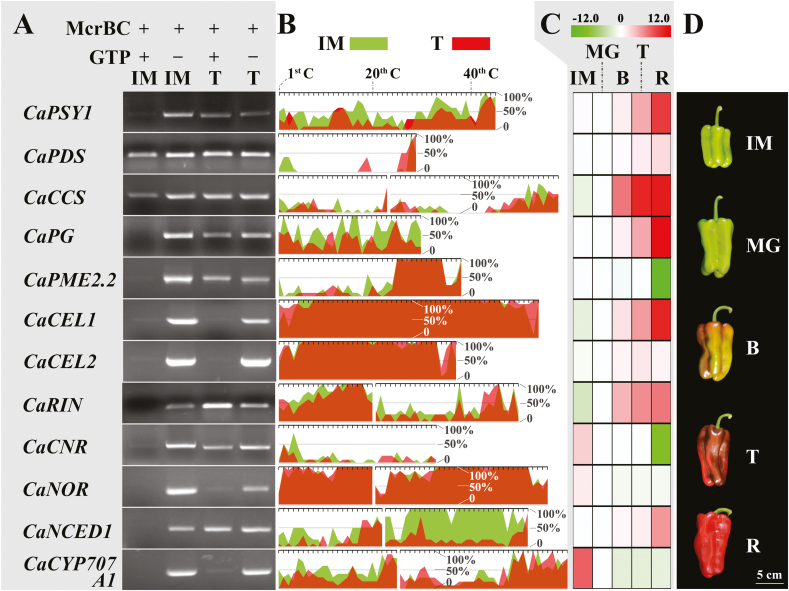
To explore the link between DNA methylation and pepper fruit ripening, the DNA methylation levels of the upstream region of the transcriptional start site (UROT) of several ripening-related genes were determined in the pericarp at the IM and T stages, namely those involved in capsanthin/capsorubin biosynthesis (CaPSY1, CaPDS, and CaCCS), cell wall degradation (CaPG, CaPME2.2, CaCEL1, and CaCEL2), ABA metabolism (CaNCED1 and CaCYP707A1) together with genes encoding ripening-related transcription factors (CaRIN, CaCNR, and CaNOR) (Fig. 1A, B). The expression of CaPSY1, CaPDS, CaCCS, CaPG, CaCEL1, CaCEL2, CaRIN, and CaNCED1 was up-regulated during ripening, whereas the expression of the other genes was down-regulated (Fig. 1C, D). According to the results of the McrBC-PCR, DNA hypomethylation was most likely to occur in the UROT of CaPSY1, CaPG, CaPME2.2, CaRIN, CaCNR, and CaNCED1 at the T stage (Fig. 1A). In contrast, low levels of DNA methylation were detected in the UROT of CaPDS and CaCCS at the IM and T stages, whilst consistently high levels of DNA methylation were recorded in the tested regions of CaCEL1, CaCEL2, CaNOR, and CaCYP707A1 (Fig. 1A). Although the bisulfite-sequencing results were not completely consistent with those of McrBC-PCR, various degrees (~12.42–61.84%) of DNA hypomethylation were recorded in the UROT of CaPSY1, CaCCS, CaPG, CaPME2.2, CaRIN, CaCNR, CaNCED1, and CaCYP707A1 at the T stage (Fig. 1B).
Fig. 1.
Dynamic changes of DNA methylation and expression levels of ripening-related genes during ripening of pepper fruit. (A) McrBC-PCR analysis of the upstream region of the transcriptional start site (UROT) of ripening-related genes in the pericarp at the immature-green (IM) and turning (T) stages. + and – indicate the presence or absence, respectively, of McrBC and GTP. (B) Bisulfite sequencing of the UROT of ripening-related genes in the pericarp at the IM and T stages. The scale at the top of each chart represents the position of each cytosine in the tested region. (C) Relative expression of the tested genes during pepper fruit ripening. Data were generated by real-time PCR and are log2-transformed. For each gene, expression at the MG stage was set as 1. (D) Pepper fruit at five different developmental stages (MG, mature-green; B, breaker; R, red-ripening).
Identification and expression analysis of DNA methyltransferase and demethylase genes
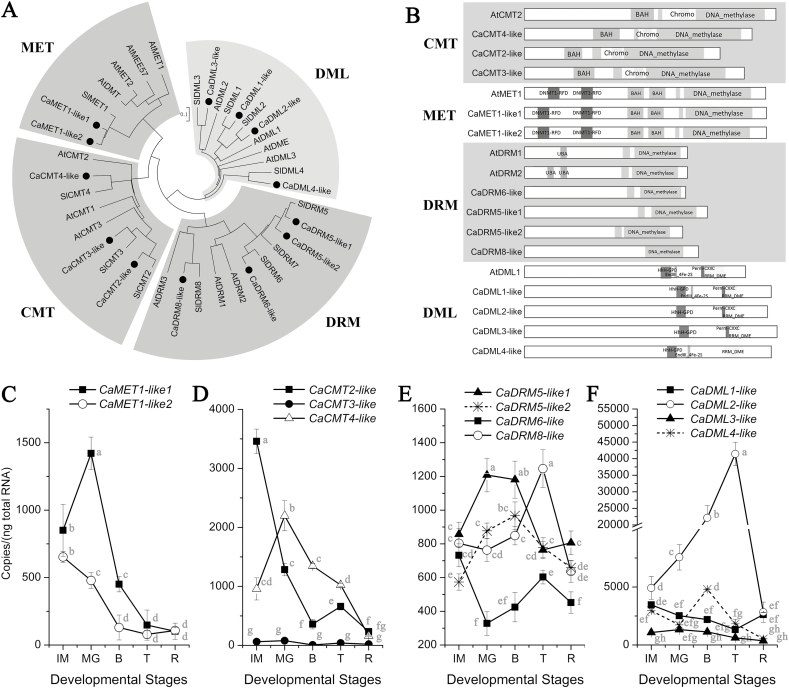
Orthologs of Arabidopsis and tomato DNA methyltransferase and demethylase genes were identified in the pepper genome. Based on the phylogenetic tree (Fig. 2A), they were renamed as CaMET1-like1 (Capana04g000012), CaMET1-like2 (Capana12g001109), CaCMT2-like (Capana12g000016), CaCMT3-like (Capana01g001654), CaCMT4-like (Capana01g004297), CaDRM5-like1 (Capana02g000460), CaDRM5-like2 (Capana07g000549), CaDRM6-like (Capana10g002486), CaDRM8-like (Capana05g002234), CaDML1-like (Capana09g002341), CaDML2-like (Capana10g001947), CaDML3-like (Capana12g002335), and CaDML4-like (Capana03g000092). Motif analysis showed that CaCMTs, CaMETs, CaDRMs, and CaDMLs shared conserved functional domains with their Arabidopsis orthologs (Fig. 2B; Supplementary Tables S7, S8), suggesting that they may play similar roles. CaMET1-like1, CaCMT3-like, and CaDML2-like were all localized in the nucleus, which further suggested their potential roles in the regulation of DNA methylation (Supplementary Fig. S2). Expression analysis indicated that both CaMET1-like1 and CaMET1-like2 followed a similar pattern of decreased expression after the MG stage; however, at most stages CaMET1-like1 was expressed at a higher level than CaMET1-like2 (Fig. 2C). In the CaCMT family, expression of CaCMT2-like dramatically decreased from the IM to B stage, whereas expression of CaCMT4-like peaked at the MG stage and then decreased (Fig. 2D). The expression of CaCMT3-like was relatively low during the whole ripening period (Fig. 2D). In the CaDRM family, expression of CaDRM5-like1, CaDRM5-like2, and CaDRM8-like was significantly higher than that of CaDRM6-like at most stages (Fig. 2E). In particular, CaDRM5-like1 and CaDRM5-like2 were highly expressed at the MG and B stages, while expression of CaDRM8-like peaked at the T stage (Fig. 2E). In the CaDML family, expression of CaDML2-like, which peaked at the T stage, was significantly higher than that of the other members at most stages (Fig. 2F). The expression of CaDML1-like, CaDML3-like, and CaDML4-like peaked at the IM, MG, and B stages, respectively. It can be assumed that CaMET1-like1, CaMET1-like2, CaCMT2-like, CaCMT4-like, CaDRM5-like1, CaDRM5-like2, CaDRM8-like, and CaDML2-like may play important roles in the regulation of DNA methylation during pepper fruit development and ripening.
Fig. 2.
Identification of DNA methyltransferase and demethylase genes in pepper. (A) Phylogenetic tree of DNA methyltransferase and demethylase genes in pepper (Ca), tomato (Sl), and Arabidopsis (At). (B) Conserved domains of the DNA methyltransferase and demethylase genes. (C–F) Dynamic expression changes of CaMETs (C), CaCMTs (D), CaDRMs (E), and CaDMLs (F) during pepper fruit development and ripening. Significant differences were determined using ANOVA followed by Tukey’s HSD test (P<0.05).
Silencing of CaMET1-like1 promotes fruit ripening and leads to DNA hypomethylation
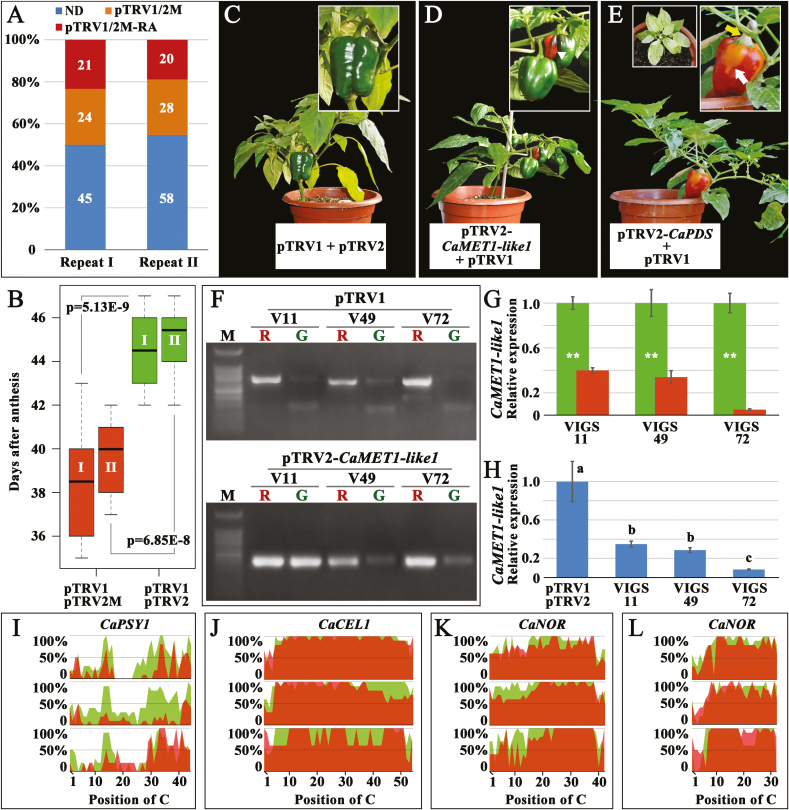
In order to further explore the effect of DNA methylation on ripening, CaMET1-like1 was silenced using VIGS. In replicates I and II, accelerated fruit ripening was observed in 21 and 20 plants, respectively, which was 4–8 d earlier than that of the negative control (Fig. 3A, B). The color transition in each precocious fruit was restricted to certain areas of the pericarp for several days and led to heterogeneous fruit color (Fig. 3C, D), which was different from that of the untreated fruit (Fig. 1D) and the negative controls (Fig. 3C). Heterogenous coloration was also observed in the CaPDS-silenced fruit (Fig. 3E). Later, the premature-ripe and green pericarps were sampled from the precocious fruits produced by the CaMET1-like1-silenced plants V11, V49, and V72. RT-PCR indicated that the premature-ripe pericarp simultaneously carried the pTRV1 and pTRV2-CaMET1-like1 plasmids, whereas the green pericarp only harbored one of them (Fig. 3F). Consistent with this, expression of CaMET1-like1 in the premature-ripe pericarp decreased to less than 40% of that of the green pericarp (Fig. 3G). Besides the fruit, reduced CaMET1-like1 expression was also observed in the leaves of the three tested plants (Fig. 3H). Bisulfite sequencing demonstrated that the DNA methylation levels of the UROT of CaPSY1, CaCEL1, and CaNOR were down-regulated in most premature-ripe pericarps (Fig. 3I–L). In accordance with the molecular data, premature-ripe pericarps had a higher level of soluble solids content (SSC) and accumulated more phytoene, β-carotene, zeaxanthin, antheraxanthin, capsanthin, capsorubin, and α-carotene than green pericarps (Tables 1, 2). However, when compared with red-ripe pericarps of the negative control, premature-ripe pericarps accumulated higher levels of phytoene, β-carotene, zeaxanthin, antheraxanthin, violaxanthin, α-carotene, and lutein (Table 2). In contrast, the SSC and carotenoid levels of green pericarps in silenced fruit were similar to those in mature-green fruit of the negative control (Tables 1, 2). With regards to phytohormones, premature-ripe pericarps accumulated more ABA and less IAA than green pericarps (Supplementary Table S9). It can therefore be concluded that silencing of CaMET1-like1 resulted in premature ripening and DNA hypomethylation in the pepper fruit.
Fig. 3.
Silencing of CaMET1-like1 in pepper fruit leads to premature ripening and DNA hypomethylation. (A) Infection efficiency of the virus-induced gene silencing (VIGS). ‘pTRV1/2M-RA’ indicates injected plants that carried both the pTRV1 and pTRV2-CaMET1-like1 plasmids and which showed accelerated fruit ripening. ‘pTRV1/2M’ indicates injected plants that carried both the pTRV1 and pTRV2-CaMET1-like1 plasmids and which showed no effect on fruit ripening. ‘ND’ indicates injected plants that only carried one of the above plasmids. (B) Boxplot of the time of onset of ripening of fruit injected with pTRV1 + pTRV2-CaMET1-like1 (pTRV2M) and pTRV1 + pTRV2. Data are shown from two replicates (I, II). Student’s t-test was used to determine the P-values. (C–E) Plants injected with pTRV1 + pTRV2 (C), pTRV1 + pTRV2-CaMET1-like1 (D), and pTRV1 + pTRV2-CaPDS (E). The white arrowhead in (D) indicates the coloration on a premature-ripe fruit. The yellow and white arrows in (E) indicate a light-bleached peduncle and pericarp, respectively. Light-bleached leaves are also shown in (E). (F) PCR-based detection of pTRV1 and pTRV2-CaMET1-like1 plasmids in three heterogeneously colored precocious fruits, which were collected from three replicate injected plants (V11, V49, and V72). ‘R’ and ‘G’ indicate premature-ripe and green pericarps, respectively, collected from the same precocious fruit. (G) Relative expression of CaMET1-like1 in the premature-ripe (red bars) and green pericarps (green bars). Significant differences were determined using Student’s t-test: **P<0.01. (H) Relative expression of CaMET1-like1 in leaves collected from three replicate plants injected with pTRV1 + pTRV2 and pTRV1 + pTRV2-CaMET1-like1 (VIGS11, VIGS49, and VIGS72). Significant differences were determined using ANOVA followed by Tukey’s HSD test (P<0.01). (I–L) 5mC levels of the upstream region of the transcriptional start site (UROT) of CaPSY1 (I), CaCEL1 (J), and CaNOR (K, L) in pericarps of premature-ripe (shaded red) and green fruit (shaded green) of V11 (top), V49 (middle), and V72 (bottom) plants. (K) and (L) are two different regions of the UROT of CaNOR.
Table 1.
Soluble solids concentration (SSC) and titratable acids (TA) in pepper fruit samples at different stages of development
| Pericarp sample | SSC (%) | TA (ml 1 M NaOH) |
|---|---|---|
| Control-MG | 1.65±0.15b | 6.03±1.38a |
| Control-R | 6.00±0.50a | 10.4±2.15a |
| Silenced-MG | 1.85±0.63b | 7.13±0.55a |
| Silenced-R | 4.83±1.31a | 10.5±2.33a |
Control-MG, negative control mature-green fruit; Control-R, negative control red-ripening fruit; Silenced-MG, CaMET1-like1-silenced mature-green fruit; Silenced-R, CaMET1-like1-silenced premature-ripe.
Different letters indicate significant differences as determined using ANOVA followed by Tukey’s HSD test (P<0.01).
Table 2.
Carotenoid concentrations in pepper fruit samples at different stages of development
| Pericarp sample | Phytoene | β-carotene | Zeaxanthin | Antheraxanthin | Violaxanthin | Capsanthin | Capsorubin | α-carotene | Lutein | Capsanthin |
|---|---|---|---|---|---|---|---|---|---|---|
| Control-MG | 0.54±0.15c | 26.0±4.59b | 5.49±1.14c | 7.15±1.32a | 56.1±5.20a | 0.16±0.02b | ND | 1.02±0.24c | 107±25.7a | 0.16±0.02b |
| Control-R | 4.83±2.76b | 18.9±4.57b | 15.4±2.09b | ND | 7.27±1.54c | 26.9±6.20a | 5.25±1.09a | 0.73±0.05c | 0.78±0.03c | 26.9±6.20a |
| Silenced-MG | 0.22±0.02c | 20.7±1.44b | 2.65±0.65c | 3.33±0.58b | 52.6±14.4ab | 0.15±0.01b | ND | 1.84±0.16b | 93.6±4.20a | 0.15±0.01b |
| Silenced-R | 10.8±0.51a | 62.2±16.1a | 48.5±3.43a | 9.63±1.81a | 32.2±5.07b | 37.0±4.00a | 5.61±1.32a | 3.82±0.18a | 9.25±1.50b | 37.0±4.00a |
All data are μg g–1 fresh weight. ND, not detected.
Control-MG, negative control mature-green fruit; Control-R, negative control red-ripening fruit; Silenced-MG, CaMET1-like1-silenced mature-green fruit; Silenced-R, CaMET1-like1-silenced premature-ripe.
Different letters indicate significant differences as determined using ANOVA followed by Tukey’s HSD test (P<0.01).
Silencing of CaMET1-like1 affects gene expression in the fruit
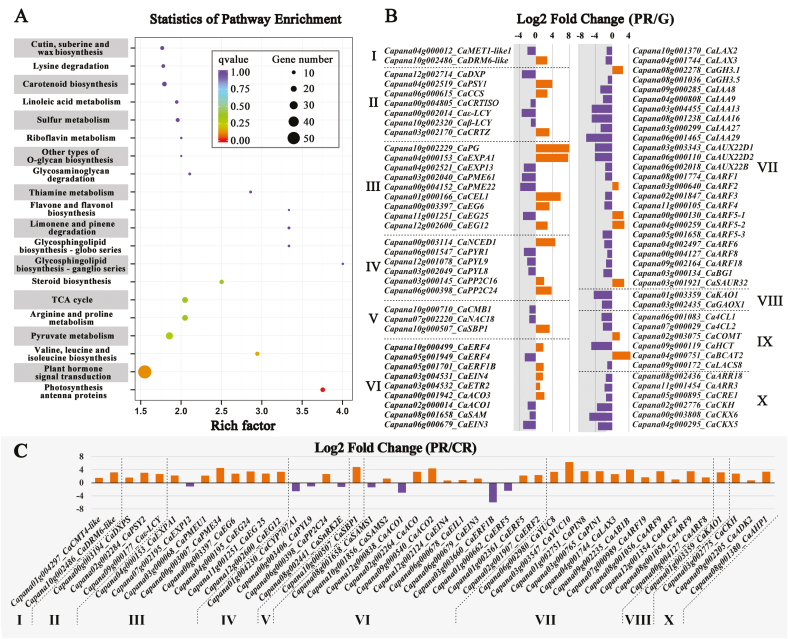
Comparative transcriptional analysis was conducted in order to investigate the effects of CaMET1-like1 on pepper fruit ripening. PCA showed that samples collected from red-ripe pericarps of the negative control and from premature-ripe and green pericarps of the silenced fruit were clearly separated in the plot defined by PC1 and PC2, which together explained ~80% of the variation (Supplementary Fig. S3), suggesting that gene silencing affected the pepper fruit transcriptome. DEGs between the premature-ripe and green pericarp were mainly enriched in the categories of plant hormone signal transduction, pyruvate metabolism, arginine and proline metabolism, the TCA cycle, and steroid and cutin biosynthesis (Fig. 4A).DEGs between the premature-ripe pericarp and the red-ripe negative control pericarp were mainly enriched in phenylpropanoid biosynthesis, photosynthesis, DNA replication, porphyrin, and chlorophyll metabolism (Supplementary Fig. S4). Putative ripening-related DEGs were extracted from the two comparisons, and included those involved in DNA methylation, carotenoid biosynthesis, cell wall degradation, phytohormone (ABA, ethylene, auxin, GA, and cytokinin) metabolism and signaling, auxin polar transport, transcriptional regulation, and capsaicin biosynthesis (Fig. 4B, C). In particular, compared with the genes in the green pericarp of the silenced fruit, in premature-ripe pericarps CaMET1-like1 was down-regulated while CaDRM6-like was up-regulated; CaPSY1 and CaCCS, which are involved in capsanthin/capsorubin biosynthesis, were up-regulated while Caε-LCY and Caβ-LCY were down-regulated; and CaPG, CaEXPA1, and CaCEL1, which are involved in cell wall degradation, were up-regulated. With regards to ethylene biosynthesis genes, CaACS10 and CaACO1 were down-regulated while CaACO3 was up-regulated. For auxin-related genes, most of the CaLAXs, CaIAAs, CaAUX, and CaARFs were down-regulated, except for CaGH3.1, CaARF2, CaARF5-1, and CaARF5-2, which were up-regulated; CaKAO1 and CaGAOX,1, which are involved in GA metabolism, were down-regulated. For capsaicin biosynthesis genes, expression of CaCOMT and CaBCAT2 was increased, while expression of Ca4CL1/2, CaHCT, and CaLACS8 was decreased. CaARR3/18, CaCRE1, CaCKH, and CaCKX5/6, which are involved in cytokinin metabolism, were down-regulated. Transcription factors CaCMB1 and CaNAC18 were down-regulated, whereas CaSBP1 was up-regulated (Fig. 4B). In another comparison, CaDRM6-like and CaCMT4-like were up-regulated in premature-ripe pericarps relative to red-ripe pericarps, whilst CaMET1-like1 was not identified as a DEG. CaDXPS, CaPSY2, and Caε-LCY, which are involved in carotenoid metabolism, were up-regulated. With regard to cell wall-related genes, expression of CaEXPA1, CaPMEU1/34, and CaEG6/12/24/25 was increased, and expression of CaEXP12 was decreased. CaCYP707A1 (involved in ABA degradation), CaPYL9, and CaSnRK2E (both participants in ABA signaling) were all down-regulated, while CaPP2C24 was up-regulated. Regarding ethylene biosynthesis genes, expression of CaSAMS2 and CaACO/2 was increased, whilst expression of CaSAMS1 and CaACO1 was decreased; CaEIN3/4, CaEIL1, and CAERFID2/5, which are involved in ethylene signaling, were up-regulated, while expression of CaERF1B/5 was down-regulated. Genes participating in auxin metabolism (CaYUC8/10), signaling (CaARF2/8/9/19), and polar transport (CaPIN1/8 and CaLAX3), GA (CaKAO1) and cytokinin metabolism (CaCKH, CaADK2, and CaAHP1), and transcriptional regulation (CaSPB1) were all up-regulated (Fig. 4C).
Fig. 4.
Transcriptome analysis of premature-ripe (PR) and green (G) pericarps of CaMET1-like1-silenced pepper fruit and of red-ripe pericarp (CR) of the negative control. (A) Statistics of pathway enrichment of the differentially expressed genes (DEGs) between the premature-ripe and green pericarps. (B) DEGs of interest between the premature-ripe and green pericarps of the CaMET1-like1-silenced fruits. (C) DEGs of interest between the premature-ripe and the red-ripe pericarps. The numerals indicate genes involved in (I) ‘DNA methylation’, (II) ‘carotenoids biosynthesis’, (III) ‘cell wall degradation’, (IV) ‘ABA metabolism and signaling’, (V) ‘transcriptional regulation’, (VI) ‘ethylene biosynthesis and signaling’, (VII) ‘auxin biosynthesis and signaling’, (VIII) ‘gibberellin biosynthesis and signaling’, (IX) ‘capsaicin biosynthesis’, and (X) ‘cytokinin biosynthesis and signaling’.
Phytohormones affect the expression of genes related to DNA methylation and ripening in the pericarp
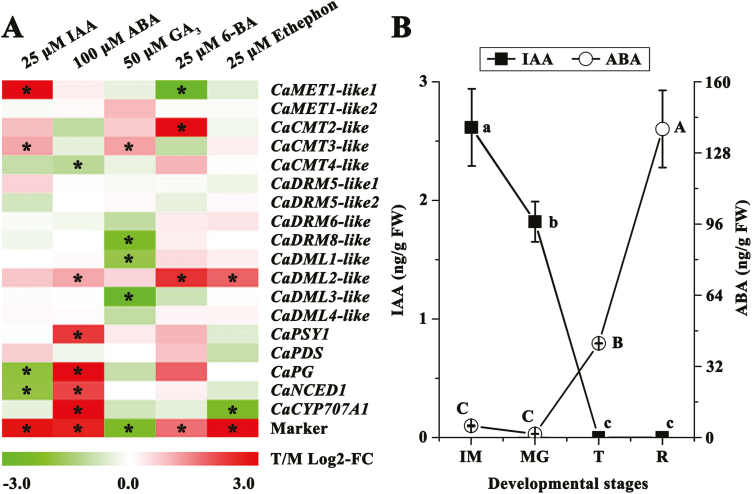
In order to investigate the relationships between phytohormones and genes related to DNA methylation during pepper fruit ripening, phytohormones were applied exogenously to discs of mature-green pericarp. Exogenous IAA clearly up-regulated the expression of CaMET1-like1 and CaCMT3-like, but down-regulated the expression of CaPG and CaNCED1 (Fig. 5A). Exogenous ABA repressed the expression of CaCMT4-like, but induced the accumulation of transcripts of CaDML2-like, CaPSY1, CaPG, CaNCED1, and CaCYP707A1. Gibberellin down-regulated the expression of CaDRM8-like, CaDML1-like, and CaDML3-like, but up-regulated the expression of CaCMT3-like. Application of 6-BA, a synthetic cytokinin, resulted in up-regulation of CaCMT2-like, CaDML2-like, and CaPG, and in down-regulation of CaMET1-like1. Ethephon clearly induced the expression of CaDML2-like, but repressed the expression of CaCYP707A1.
Fig. 5.
Responses of genes to phytohormones in mature-green (MG) pericarps and dynamic changes in endogenous ABA and IAA during pepper fruit ripening. (A) Changes in gene expression in response to exogenous auxin (IAA), ABA, gibberellin (GA3), cytokinin (6-BA), and ethephon in pericarp discs at the MG stage. Data are log2-transformed. ‘T/M’ indicates the treatment value divided by the mock control value. ‘*’ indicates that the log2 fold-change (FC) value is either greater than 1 or less than –1. (B) Dynamic changes of ABA and IAA levels during pepper fruit ripening. ‘IM’, immature-green; ‘M’, mature-green; ‘T’, turning; ‘R’, red-ripening. Different letters indicate significant differences as determined using ANOVA followed by Tukey’s HSD test (P<0.01): IAA, lowercase letters; ABA, uppercase letters.
Dynamic changes of IAA, ABA, and ethylene levels during ripening
In order to examine the effects of IAA and ABA on pepper fruit ripening, their concentrations were measured at different developmental stages. A decrease in endogenous IAA was observed from the IM to T stage (Fig. 5B). In contrast, the levels of endogenous ABA decreased slightly from the IM to MG stage and then increased and peaked at the R stage. No ethylene signal was detected at any of the tested stages (Supplementary Fig. S5).
Discussion
DNA methylation may be involved in the regulation of ripening in pepper fruit
DNA methylation has been recently been discovered to regulate fruit ripening in tomato, strawberry, and sweet orange (Liu et al., 2015; Lang et al., 2017; Cheng et al., 2018; Huang et al., 2019). In our present study, DNA hypomethylation was observed in the UROT of some ripening-related genes during pepper fruit ripening (Fig. 1A, B). Consistent with this, dynamic changes in the expression of DNA methyltransferase and demethylase genes associated with ripening were also observed (Fig. 2C–F), suggesting that there was very likely a link between DNA methylation and fruit ripening. Moreover, silencing of CaMET1-like1 using VIGS resulted in DNA hypomethylation (Fig. 3I–L), premature accumulation of carotenoids (Table 2), increased levels of SSCs and ABA (Table 1, Supplementary Table S9), and decreased levels of IAA (Supplementary Table S9). The gene expression profile of premature-ripe pericarps of the CaMET1-like1-silenced fruit was also shifted towards to that of the red-ripe pericarps of the negative control (Supplementary Fig. S3). In particular, capsanthin/capsorubin biosynthesis (up-regulation of CaPSY1 and CaCCS), cell wall degradation (up-regulation of CaPG, CaEXPA1, and CaCEL1; down-regulation of CaEXP13, CaPME22/61, and CaEG25), and ABA biosynthesis (up-regulation of CaNCED1) were promoted, whereas IAA signaling (down-regulation of CaIAA8/9/13/16/27/29 and CaAUX22B/D1/D2) and gibberellin biosynthesis (down-regulation of CaKAO and CaGAOX1) were repressed (Fig. 4B). Therefore, it can be deduced that CaMET1-like1, which is involved in the regulation of DNA methylation and hypomethylation in pepper fruit, may promote ripening. In accordance with this, ripening-associated DNA hypomethylation is also observed in tomato and strawberry, and its disturbance leads to abnormal fruit ripening (Liu et al., 2015; Lang et al., 2017; Cheng et al., 2018). This suggests that the regulation of DNA methylation in fleshy fruit ripening may be common in some species. DNA hypomethylation occurs in the putative promoter region of PSY1 in tomato and pepper and it enhances the expression of the gene (Liu et al., 2015; Lang et al., 2017; Fig. 1A, B), suggesting that PSY1 may be a key point in the regulation of fruit ripening by DNA methylation.
The comparison between premature-ripe and red-ripe pericarps was also informative. Compared with red-ripe pericarps, the premature-ripe ones accumulated more phytoene, β-carotene, zeaxanthin, antheraxanthin, violaxanthin, α-carotene, and lutein (Table 2), whereas the levels of capsanthin and capsorubin levels were not changed significantly. A possible reason for this was the higher expression of CaDXPS, CaPSY2, and Caε-LCY in premature-ripe pericarps. The up-regulation of the first two genes may have led to the increase of total carotenoids, while the increased expression of Caε-LCY may have enhanced the δ-carotene–α-carotene–lutein pathway and ultimately led to high levels of α-carotene and lutein. However, since the expression of CaCCS and CaVDE was not included in the list of DEGs (Fig. 4B), the capsanthin and capsorubin levels may not have been significantly changed.
Transcriptional regulation of DNA methylation during ripening
The DNA methylation level is controlled by DNA methyltransferases (MET1, CMT2, CMT3, and DRMs) and demethylases (ROS1, DME, and DMLs) (Deleris et al., 2016; Gallusci et al., 2016). In our study, members of these families were identified in the pepper genome (Fig. 2A), which is consistent with the hypothesis that DNA methylation is conserved in all flowering plants (Goll and Bestor, 2005). At the transcriptional level, expression of CaMETs and CaCMTs showed dynamic changes associated with ripening (fold-change >2), whereas expression of CaDRMs was barely altered (fold-change <2), especially after the MG stage (Fig. 2C–F), suggesting the CaMETs and CaCMTs may mainly control the maintenance and de novo methylation of cytosine, respectively, during pepper fruit ripening. Moreover, most of the CaMETs and CaCMTs were down-regulated after the MG stage (Fig. 2C, D), suggesting decreased DNA methylation activity. However, similar to tomato, pepper fruit generally undergo genome endoreduplication after the MG stage (Cheniclet et al., 2005; Chevalier et al., 2007; Ogawa et al., 2010, 2012). Therefore, passive DNA demethylation is highly likely to occur during pepper fruit ripening. On the other hand, expression of CaDML2, an ortholog of ROS1 (AtDML1) and the highest expressed member of the CaDML family, peaked at the T stage (Fig. 2F), suggesting that an active demethylation process may be occurring during pepper fruit ripening. Thus, it seems that the ripening-associated DNA hypomethylation in pepper may be mainly controlled by CaMETs, CaCMTs, and CaDML2 at the transcriptional level. Similar phenomena have been observed in tomato, in which the DNA hypomethylation associated with ripening is controlled by the expression of both DNA methyltransferase and demethylase genes (Teyssier et al., 2008; Liu et al., 2015; Lang et al., 2017; Yang et al., 2019). However, in strawberry and sweet orange, changes in ripening-associated DNA methylation are only attributed to variations in the expression of RdDM-related and DNA demethylase genes, respectively (Cheng et al., 2018; Huang et al., 2019). Therefore, it can be concluded that the transcriptional regulatory mechanism of the variations in DNA methylation that are associated with ripening may be not the same among different species.
Interactions between phytohormones and DNA methylation during ripening
Ethylene is considered to be a positive regulator of climacteric fruit ripening (Oeller et al., 1991; Lanahan et al., 1994; Li et al., 2007; Lee et al., 2012; Sekeli et al., 2014). However, in this study, no endogenous ethylene burst was detected during fruit ripening (Supplementary Fig. S5), suggesting that our pepper inbred line harbors non-climacteric fruit. Consistent with this, compared with green pericarps of the silenced fruit, CaACS was not up-regulated in premature-ripe pericarps (Fig. 4B). A similar phenomenon was also observed by Aizat et al. (2013). This suggests that an endogenous ethylene burst is not indispensable for the non-climacteric ripening of pepper fruit. In accordance with this, expression of CaPSY1, CaPDS, and CaPG did not clearly respond to short-term treatment with ethephon (Fig. 5A), suggesting that these genes may not be directly regulated by exogenous ethylene. However, a previous study determined that continuous treatment with exogenous ethylene promoted non-climacteric fruit ripening (Fox et al., 2005), suggesting that exogenous ethylene may have some slow or indirect effects on the ripening of this type of fruit. In our study, expression of CaDML2-like was clearly induced by short-term ethephon treatment (Figs 5A, Fig. 6), suggesting that this gene and even DNA methylation may be involved in the process.
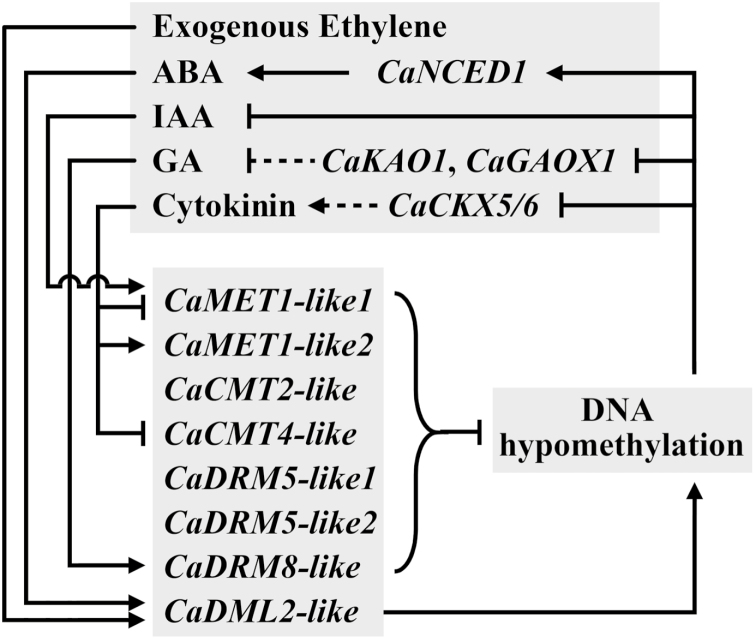
Fig. 6.
A proposed model of the interactions between DNA methylation and phytohormones during ripening of pepper fruit. Arrows indicate promotion, blocked lines indicate inhibition. Solid lines indicate that the interaction is supported by data obtained from this study. Dashed lines indicate that the interaction is deduced from the gene function.
ABA has been widely accepted as an important regulator of non-climacteric fruit ripening (Deruère et al., 1994; Jia et al., 2011; Leng et al., 2014; Wang et al., 2013, 2017). Consistent with this, we found that exogenous ABA promoted carotenoid biosynthesis, cell wall degradation, and DNA hypomethylation through altering the expression of CaPYS1, CaPG, and genes related to DNA methylation (CaDML2-like and CaCMT4-like), respectively (Figs 5A, Fig. 6). Endogenous ABA dramatically increased from the MG to R stage (Fig. 5B), suggesting a positive role in the regulation of pepper fruit ripening. ABA levels as well as the expression of CaNCED1 and two CaPP2Cs were up-regulated in premature-ripe pericarps (Supplementary Table S9, Figs 4B, Fig. 6). CaNCED1 encodes a key enzyme in ABA biosynthesis (Schwartz et al., 1997; Tan et al., 1997; Burbidge et al., 1999). CaPP2C encodes a negative component of ABA signaling and is usually up-regulated by ABA (Ma et al., 2009; Sun et al., 2010; Wang et al., 2015). Therefore, it can be deduced that ABA biosynthesis and signaling were activated by DNA hypomethylation in the non-climacteric pepper fruit. Similar phenomena have also been observed in strawberry, in which ABA biosynthesis genes are up-regulated during fruit ripening whilst at the same time undergoing a loss of DNA methylation in their 5´- and 3´-regulatory regions (Liao et al., 2018; Cheng et al., 2018). Hence, it can be concluded that ABA and DNA methylation may interact with each other at the transcriptional level during pepper fruit ripening.
Auxin is often considered as a repressor of non-climacteric fruit ripening that acts fully or partially through antagonizing the effects of ABA (Manning et al., 1994; Davies et al., 1997; Liu et al., 2005; Böttcher et al., 2011; Symons et al., 2012; Ziliotto et al., 2012; Liao et al., 2018). In accordance with this, we found that exogenous IAA inhibited cell wall degradation, ABA biosynthesis, and DNA hypomethylation through affecting the expression of CaPG, CaNCED1, and DNA methyltransferase genes (CaMET1-like1 and CaCMT3-like), respectively (Figs 5A, Fig. 6). Moreover, endogenous IAA levels decreased from the IM to T stage, suggesting that it may be involved in the regulation of ripening of the non-climacteric pepper fruit. In premature-ripe pericarps, the endogenous IAA level as well as most of the differentially expressed AUX/IAA genes were down-regulated (Supplementary Table S9, Fig. 4B). AUX/IAA genes are important elements of auxin signaling and positively respond to auxin treatment (Waseem et al., 2018). Therefore, it can be concluded that DNA hypomethylation caused by the silencing of CaMET1-like1 led to the inhibition of auxin signaling and a decreased level of auxin.
Generally, gibberellin (GA) and cytokinin are considered to be suppressors of fruit ripening (Dostal and Leopold, 1967; Biale, 1978; Deruère et al., 1994; Martínez et al., 1994; Martínez-Romero et al., 2000; Alós et al., 2006; Kader, 2008; Peppi and Fidelibus, 2008). However, in our study, exogenous GA3 and 6-BA did not significantly affect the expression of CaPDS, CaPSY1, CaPG, CaNCED, or CaCYP707A1 (Fig. 5A), indicating that under our experimental condition these two hormones had weak effects on pepper fruit ripening. In tobacco leaves, GA is considered as a determinant for global DNA hypomethylation (Manoharlal et al., 2018a, 2018b). However, in our study, exogenous GA3 and 6-BA did not show clear effects on DNA methylation at the transcriptional level (Figs 5A, Fig. 6). DNA hypomethylation seemed to inhibit GA accumulation at the transcriptional level, since the expression levels of CaKAO1 and CaGAOX1 were lower in premature-ripe pericarps than in green pericarps of CaMET1-like1-silenced fruit (Fig. 4B, Fig. 6). With regard to cytokinin, DNA hypomethylation was likely to inhibit its degradation by down-regulating the expression of CaCKX5/6 (Figs 4B, Fig. 6).
Conclusions
A hypothetical model is proposed (Fig. 6) that indicates a high likelihood of a role for DNA methylation in the regulation of fruit ripening in non-climacteric pepper, which is mainly controlled by CaMET1-like1, CaMET1-like2, CaCMT2-like, CaCMT4-like, and CaDML2-like at the transcriptional level. Silencing of CaMET1-like1 by VIGS results in DNA hypomethylation and premature fruit ripening, which includes the accumulation of carotenoids, increased levels of SSC and ABA. and decreased levels of IAA. Under in vitro conditions, ABA promotes DNA hypomethylation in the pericarp at the MG stage at the transcriptional level, whereas IAA inhibits this process. Exogenous gibberellin and cytokinin show indistinct roles in DNA methylation in MG pericarps of non-climacteric pepper fruit.
Supplementary data
Supplementary data are available at JXB online.
Table S1. Primers used for McrBC-PCR.
Table S2. Primers used for bisulfite sequencing.
Table S3. Primers used for real-time PCR.
Table S4. Primers used for subcellular localization plasmid construction.
Table S5. Primers used for VIGS construction and detection.
Table S6. Standard substances used in study.
Table S7. IDs and conserved domains of pepper DNA methyltransferase genes.
Table S8. IDs and conserved domains of pepper DNA demethylases genes.
Table S9. ABA and IAA levels in premature-ripe and green pericarps of CaMET1-like1-silenced fruit.
Fig. S1. Upstream regions of transcriptional start sites for McrBC-PCR and bisulfite sequencing.
Fig. S2. Subcellular localization of CaDML2-like, CaMET1-like1, and CaCMT3-like in onion epidermal cells.
Fig. S3. PCA of the RNA-seq data.
Fig. S4. Enrichment of DEGs between premature-ripe pericarps of CaMET1-like1-silenced fruit and red-ripe pericarps of negative control fruits.
Fig. S5. Ethylene production of fruit at different developmental stages.
Acknowledgments
We thank Prof. Yule Liu of the School of Life Sciences, Tsinghua University, for kindly sending us the pTRV1 and pTRV2 vectors. We thank Dr Yanjie Xu of the Department of Ornamental Horticulture, College of Horticulture, China Agricultural University, for help with the measurement of ethylene. We thank Dr Nathan King Taitano of the Institute for Plant Breeding, Genetics & Genomics, University of Georgia, for help with the modification of language. This work was supported by the National Key Research and Development Program of China (2017YFD0101903 to LS and HS), the Beijing Fruit Vegetables Innovation Team of Modern Agricultural Industry Technology System (BAIC01-2018 to HS), and The Construction of Beijing Science and Technology Innovation and Service Capacity in Top Subjects (CEFF-PXM2019_014207_000032 to LS and HS).
References
- Aizat WM, Able JA, Stangoulis JC, Able AJ. 2013. Characterisation of ethylene pathway components in non-climacteric capsicum. BMC Plant Biology 13, 191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alós E, Cercós M, Rodrigo MJ, Zacarías L, Talón M. 2006. Regulation of color break in citrus fruits. Changes in pigment profiling and gene expression induced by gibberellins and nitrate, two ripening retardants. Journal of Agricultural and Food Chemistry 54, 4888–4895. [DOI] [PubMed] [Google Scholar]
- Bender J. 2004. DNA methylation and epigenetics. Annual Review of Plant Biology 55, 41–68. [DOI] [PubMed] [Google Scholar]
- Biale JB. 1978. On the interface of horticulture and plant physiology. Annual Review of Plant Biology 29, 1–24. [Google Scholar]
- Böttcher C, Harvey KE, Forde CG, Boss PK, Davies C. 2011. Auxin treatment of pre-veraison grape (Vitis vinifera L.) berries both delays ripening and increases the synchronicity of sugar accumulation. Australian Journal of Grape and Wine Research 17, 1–8. [Google Scholar]
- Burbidge A, Grieve TM, Jackson A, Thompson A, McCarty DR, Taylor IB. 1999. Characterization of the ABA-deficient tomato mutant notabilis and its relationship with maize Vp14. The Plant Journal 17, 427–431. [DOI] [PubMed] [Google Scholar]
- Cao D, Ju Z, Gao C, Mei X, Fu D, Zhu H, Luo Y, Zhu B. 2014. Genome-wide identification of cytosine-5 DNA methyltransferases and demethylases in Solanum lycopersicum. Gene 550, 230–237. [DOI] [PubMed] [Google Scholar]
- Chai YM, Jia HF, Li CL, Dong QH, Shen YY. 2011. FaPYR1 is involved in strawberry fruit ripening. Journal of Experimental Botany 62, 5079–5089. [DOI] [PubMed] [Google Scholar]
- Cheng J, Niu Q, Zhang B, Chen K, Yang R, Zhu JK, Zhang Y, Lang Z. 2018. Downregulation of RdDM during strawberry fruit ripening. Genome Biology 19, 212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheniclet C, Rong WY, Causse M, Frangne N, Bolling L, Carde JP, Renaudin JP. 2005. Cell expansion and endoreduplication show a large genetic variability in pericarp and contribute strongly to tomato fruit growth. Plant Physiology 139, 1984–1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevalier C. 2007. Cell cycle control and fruit development, in cell cycle control and plant development. In: Inze D, ed. Cell cycle control and plant development. Annual Plant Reviews, vol. 32. Oxford: Blackwell Publishing, 269–293. [Google Scholar]
- Davies C, Boss PK, Robinson SP. 1997. Treatment of grape berries, a nonclimacteric fruit with a synthetic auxin, retards ripening and alters the expression of developmentally regulated genes. Plant Physiology 115, 1155–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deleris A, Halter T, Navarro L. 2016. DNA methylation and demethylation in plant immunity. Annual Review of Phytopathology 54, 579–603. [DOI] [PubMed] [Google Scholar]
- Deruère J, Römer S, d’Harlingue A, Backhaus RA, Kuntz M, Camara B. 1994. Fibril assembly and carotenoid overaccumulation in chromoplasts: a model for supramolecular lipoprotein structures. The Plant Cell 6, 119–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dostal HC, Leopold AC. 1967. Gibberellin delays ripening of tomatoes. Science 158, 1579–1580. [DOI] [PubMed] [Google Scholar]
- Finnegan EJ, Kovac KA. 2000. Plant DNA methyltransferases. Plant Molecular Biology 43, 189–201. [DOI] [PubMed] [Google Scholar]
- Fox AJ, Del Pozo-Insfran D, Lee JH, Sargent SA, Talcott ST. 2005. Ripening-induced chemical and antioxidant changes in bell peppers as affected by harvest maturity and postharvest ethylene exposure. HortScience 40, 732–736. [Google Scholar]
- Gallusci P, Hodgman C, Teyssier E, Seymour GB. 2016. DNA methylation and chromatin regulation during fleshy fruit development and ripening. Frontiers in Plant Science 7, 807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillaspy G, Ben-David H, Gruissem W. 1993. Fruits: a developmental perspective. The Plant Cell 5, 1439–1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giuffrida D, Dugo P, Torre G, Bignardi C, Cavazza A, Corradini C, Dugo G. 2013. Characterization of 12 Capsicum varieties by evaluation of their carotenoid profile and pungency determination. Food Chemistry 140, 794–802. [DOI] [PubMed] [Google Scholar]
- Goll MG, Bestor TH. 2005. Eukaryotic cytosine methyltransferases. Annual Review of Biochemistry 74, 481–514. [DOI] [PubMed] [Google Scholar]
- Gong Z, Morales-Ruiz T, Ariza RR, Roldán-Arjona T, David L, Zhu JK. 2002. ROS1, a repressor of transcriptional gene silencing in Arabidopsis, encodes a DNA glycosylase/lyase. Cell 111, 803–814. [DOI] [PubMed] [Google Scholar]
- Guo JJ, Liu C, Wang P, Cheng Q, Sun L, Yang WC, Shen HL. 2018. The aborted microspores (AMS)-like gene is required for anther and microspore development in pepper (Capsicum annuum L.). International Journal of Molecular Sciences 19, 1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao Y, Hu G, Breitel D, Liu M, Mila I, Frasse P, Fu Y, Aharoni A, Bouzayen M, Zouine M. 2015. Auxin response factor SlARF2 is an essential component of the regulatory mechanism controlling fruit ripening in tomato. PLoS Genetics 11, e1005649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Healey A, Furtado A, Cooper T, Henry RJ. 2014. Protocol: a simple method for extracting next-generation sequencing quality genomic DNA from recalcitrant plant species. Plant Methods 10, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou BZ, Li CL, Han YY, Shen YY. 2018. Characterization of the hot pepper (Capsicum frutescens) fruit ripening regulated by ethylene and ABA. BMC Plant Biology 18, 162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh TF, Ibarra CA, Silva P, Zemach A, Eshed-Williams L, Fischer RL, Zilberman D. 2009. Genome-wide demethylation of Arabidopsis endosperm. Science 324, 1451–1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Liu R, Niu Q, Tang K, Zhang B, Zhang H, Chen K, Zhu JK, Lang Z. 2019. Global increase in DNA methylation during orange fruit development and ripening. Proceedings of the National Academy of Sciences, USA 116, 1430–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang ZJ, Van Houten J, Gonzalez G, Xiao H, van der Knaap E. 2013. Genome-wide identification, phylogeny, and expression analysis of SUN, OFP and YABBY gene family in tomato. Molecular Genetics and Genomics 288, 111–129. [DOI] [PubMed] [Google Scholar]
- Ji K, Kai W, Zhao B, et al. 2014. SlNCED1 and SlCYP707A2: key genes involved in ABA metabolism during tomato fruit ripening. Journal of Experimental Botany 65, 5243–5255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia HF, Chai YM, Li CL, Lu D, Luo JJ, Qin L, Shen YY. 2011. Abscisic acid plays an important role in the regulation of strawberry fruit ripening. Plant Physiology 157, 188–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones B, Frasse P, Olmos E, Zegzouti H, Li ZG, Latché A, Pech JC, Bouzayen M. 2002. Down-regulation of DR12, an auxin-response-factor homolog, in the tomato results in a pleiotropic phenotype including dark green and blotchy ripening fruit. The Plant Journal 32, 603–613. [DOI] [PubMed] [Google Scholar]
- Kader AA. 2008. Flavor quality of fruits and vegetables. Journal of the Science of Food and Agriculture 88, 1863–1868. [Google Scholar]
- Kong Y, Avraham L, Perzelan Y, Alkalai-Tuvia S, Ratner K, Shahak Y, Fallik E. 2013. Pearl netting affects postharvest fruit quality in ‘Vergasa’ sweet pepper via light environment manipulation. Scientia Horticulturae 150, 290–298. [Google Scholar]
- Lanahan MB, Yen HC, Giovannoni JJ, Klee HJ. 1994. The never ripe mutation blocks ethylene perception in tomato. The Plant Cell 6, 521–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang ZB, Wang YH, Tang K, Tang DG, Datsenka T, Cheng JF, Zhang YJ, Handa AK, Zhu JK. 2017. Critical roles of DNA demethylation in the activation of ripening-induced genes and inhibition of ripening-repressed genes in tomato fruit. Proceedings of the National Academy of Sciences, USA 114, 4511–4519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JM, Joung JG, McQuinn R, Chung MY, Fei Z, Tieman D, Klee H, Giovannoni J. 2012. Combined transcriptome, genetic diversity and metabolite profiling in tomato fruit reveals that the ethylene response factor SlERF6 plays an important role in ripening and carotenoid accumulation. The Plant Journal 70, 191–204. [DOI] [PubMed] [Google Scholar]
- Lee SC, Kim SH, An SH, Yi SY, Hwang BK. 2006. Identification and functional expression of the pepper pathogen-induced gene, CAPIP2, involved in disease resistance and drought and salt stress tolerance. Plant Molecular Biology 62, 151–164. [DOI] [PubMed] [Google Scholar]
- Leng P, Yuan B, Guo Y. 2014. The role of abscisic acid in fruit ripening and responses to abiotic stress. Journal of Experimental Botany 65, 4577–4588. [DOI] [PubMed] [Google Scholar]
- Li J, Tao X, Bu J, Ying T, Mao L, Luo Z. 2017. Global transcriptome profiling analysis of ethylene-auxin interaction during tomato fruit ripening. Postharvest Biology and Technology 130, 28–38. [Google Scholar]
- Li J, Tao X, Li L, Mao L, Luo Z, Khan ZU, Ying T. 2016. Comprehensive RNA-Seq analysis on the regulation of tomato ripening by exogenous auxin. PLoS ONE 11, e0156453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li LC, Dahiya R. 2002. MethPrimer: designing primers for methylation PCRs. Bioinformatics 18, 1427–1431. [DOI] [PubMed] [Google Scholar]
- Li Y, Zhu B, Xu W, Zhu H, Chen A, Xie Y, Shao Y, Luo Y. 2007. LeERF1 positively modulated ethylene triple response on etiolated seedling, plant development and fruit ripening and softening in tomato. Plant Cell Reports 26, 1999–2008. [DOI] [PubMed] [Google Scholar]
- Liao X, Li M, Liu B, Yan M, Yu X, Zi H, Liu R, Yamamuro C. 2018. Interlinked regulatory loops of ABA catabolism and biosynthesis coordinate fruit growth and ripening in woodland strawberry. Proceedings of the National Academy of Sciences, USA 115, E11542–E11550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindroth AM, Cao X, Jackson JP, Zilberman D, McCallum CM, Henikoff S, Jacobsen SE. 2001. Requirement of CHROMOMETHYLASE3 for maintenance of CpXpG methylation. Science 292, 2077–2080. [DOI] [PubMed] [Google Scholar]
- Liu K, Kang BC, Jiang H, Moore SL, Li H, Watkins CB, Setter TL, Jahn MM. 2005. A GH3-like gene, CcGH3, isolated from Capsicum chinense L. fruit is regulated by auxin and ethylene. Plant Molecular Biology 58, 447–464. [DOI] [PubMed] [Google Scholar]
- Liu R, How-Kit A, Stammitti L, et al. 2015. A DEMETER-like DNA demethylase governs tomato fruit ripening. Proceedings of the National Academy of Sciences, USA 112, 10804–10809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Schiff M, Dinesh-Kumar SP. 2002. Virus-induced gene silencing in tomato. The Plant Journal 31, 777–786. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Lurie S, Ben-Yehoshua S. 1986. Changes in membrane properties and abscisic acid during senescence of harvested bell pepper fruit. Journal of The American Society for Horticultural Science 111, 886–889. [Google Scholar]
- Ma Y, Szostkiewicz I, Korte A, Moes D, Yang Y, Christmann A, Grill E. 2009. Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 324, 1064–1068. [DOI] [PubMed] [Google Scholar]
- Manning K. 1994. Changes in gene expression during strawberry fruit ripening and their regulation by auxin. Planta 194, 62–68. [Google Scholar]
- Manoharlal R, Saiprasad GV, Kaikala V, Kumar RS, Kovařik A. 2018a Analysis of DNA methylome and transcriptome profiling following gibberellin A3 (GA3) foliar application in Nicotiana tabacum L. Indian Journal of Plant Physiology 23, 543–556. [Google Scholar]
- Manoharlal R, Saiprasad GV, Ullagaddi C, Kovařik A. 2018b Gibberellin A3 as an epigenetic determinant of global DNA hypo-methylation in tobacco. Biologia Plantarum 62, 11–23. [Google Scholar]
- Martínez GA, Chaves AR, Anon MC. 1994. Effect of gibberellic acid on ripening of strawberry fruits (Fragaria annanassa Duch.). Journal of Plant Growth Regulation 13, 87–91. [Google Scholar]
- Martínez-Romero D, Valero D, Serrano M, Burlo F, Carbonell A, Burgos L, Riquelme F. 2000. Exogenous polyamines and gibberellic acid effects on peach (Prunus persica L.) storability improvement. Journal of Food Science 65, 288–294. [Google Scholar]
- Oeller PW, Lu MW, Taylor LP, Pike DA, Theologis A. 1991. Reversible inhibition of tomato fruit senescence by antisense RNA. Science 254, 437–439. [DOI] [PubMed] [Google Scholar]
- Ogawa D, Ishikawa K, Mii M. 2012. Difference in the polysomaty degree during fruit development among plants with different ploidy levels produced by artificial chromosome doubling of a pepper (Capsicum annuum) cultivar ‘Shishitou No. 562’. Scientia Horticulturae 134, 121–126. [Google Scholar]
- Ogawa D, Ishikawa K, Nunomura O, Mii M. 2010. Correlation between fruit characters and degree of polysomaty in fruit tissues of Capsicum. Journal of the Japanese Society for Horticultural Science 79, 168–173. [Google Scholar]
- Owino WO, Manabe Y, Mathooko FM, Kubo Y, Inaba A. 2006. Regulatory mechanisms of ethylene biosynthesis in response to various stimuli during maturation and ripening in fig fruit (Ficus carica L.). Plant Physiology and Biochemistry 44, 335–342. [DOI] [PubMed] [Google Scholar]
- Peppi MC, Fidelibus MW. 2008. Effects of forchlorfenuron and abscisic acid on the quality of ‘flame seedless’ grapes. Hortscience 43, 173–176. [Google Scholar]
- Pretel MT, Serrano M, Amoros A, Riquelme F, Romojaro F. 1995. Non-involvement of ACC and ACC oxidase activity in pepper fruit ripening. Postharvest Biology and Technology 5, 295–302. [Google Scholar]
- Schwartz SH, Tan BC, Gage DA, Zeevaart JA, McCarty DR. 1997. Specific oxidative cleavage of carotenoids by VP14 of maize. Science 276, 1872–1874. [DOI] [PubMed] [Google Scholar]
- Sekeli R, Abdullah J, Namasivayam P, Muda P, Baka U, Yeong W, Pillai V. 2014. RNA interference of 1-aminocyclopropane-1-carboxylic acid oxidase (ACO1 and ACO2) genes expression prolongs the shelf life of Eksotika (Carica papaya L.) papaya fruit. Molecules 19, 8350–8362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sravankumar T, Akash, Naik N, Kumar R. 2018. A ripening-induced SlGH3-2 gene regulates fruit ripening via adjusting auxin-ethylene levels in tomato (Solanum lycopersicum L.). Plant Molecular Biology 98, 455–469. [DOI] [PubMed] [Google Scholar]
- Su L, Diretto G, Purgatto E, Danoun S, Zouine M, Li Z, Roustan JP, Bouzayen M, Giuliano G, Chervin C. 2015. Carotenoid accumulation during tomato fruit ripening is modulated by the auxin–ethylene balance. BMC Plant Biology 15, 114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Sun Y, Zhang M, et al. 2012. a Suppression of 9-cis-epoxycarotenoid dioxygenase, which encodes a key enzyme in abscisic acid biosynthesis, alters fruit texture in transgenic tomato. Plant Physiology 158, 283–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Wang YP, Chen P, Ren J, Ji K, Li Q, Li P, Dai SJ, Leng P. 2011. Transcriptional regulation of SlPYL, SlPP2C, and SlSnRK2 gene families encoding ABA signal core components during tomato fruit development and drought stress. Journal of Experimental Botany 62, 5659–5669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Yuan B, Zhang M, Wang L, Cui M, Wang Q, Leng P. 2012b Fruit-specific RNAi-mediated suppression of SlNCED1 increases both lycopene and β-carotene contents in tomato fruit. Journal of Experimental Botany 63, 3097–3108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Zhang M, Ren J, Qi J, Zhang G, Leng P. 2010. Reciprocity between abscisic acid and ethylene at the onset of berry ripening and after harvest. BMC Plant Biology 10, 257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symons GM, Chua YJ, Ross JJ, Quittenden LJ, Davies NW, Reid JB. 2012. Hormonal changes during non-climacteric ripening in strawberry. Journal of Experimental Botany 63, 4741–4750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan BC, Schwartz SH, Zeevaart JA, McCarty DR. 1997. Genetic control of abscisic acid biosynthesis in maize. Proceedings of the National Academy of Sciences, USA 94, 12235–12240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Télef N, Stammitti-Bert L, Mortain-Bertrand A, Maucourt M, Carde JP, Rolin D, Gallusci P. 2006. Sucrose deficiency delays lycopene accumulation in tomato fruit pericarp discs. Plant Molecular Biology 62, 453–469. [DOI] [PubMed] [Google Scholar]
- Teyssier E, Bernacchia G, Maury S, How Kit A, Stammitti-Bert L, Rolin D, Gallusci P. 2008. Tissue dependent variations of DNA methylation and endoreduplication levels during tomato fruit development and ripening. Planta 228, 391–399. [DOI] [PubMed] [Google Scholar]
- Trainotti L, Tadiello A, Casadoro G. 2007. The involvement of auxin in the ripening of climacteric fruits comes of age: the hormone plays a role of its own and has an intense interplay with ethylene in ripening peaches. Journal of Experimental Botany 58, 3299–3308. [DOI] [PubMed] [Google Scholar]
- Villavicencio LE, Blankenship SM, Sanders DC, Swallow WH. 1999. Ethylene and carbon dioxide production in detached fruit of selected pepper cultivars. Journal of the American Society for Horticultural Science 124, 402–406. [Google Scholar]
- Villavicencio LE, Blankenship SM, Sanders DC, Swallow WH. 2001. Ethylene and carbon dioxide concentrations in attached fruits of pepper cultivars during ripening. Scientia Horticulturae 91, 17–24. [Google Scholar]
- Wan H, Yuan W, Ruan M, et al. 2011. Identification of reference genes for reverse transcription quantitative real-time PCR normalization in pepper (Capsicum annuum L.). Biochemical and Biophysical Research Communications 416, 24–30. [DOI] [PubMed] [Google Scholar]
- Wang YP, Chen P, Sun L, Li Q, Dai SJ, Sun YF, Kai WB, Zhang YS, Liang B, Leng P. 2015. Transcriptional regulation of PaPYLs, PaPP2Cs and PaSnRK2s during sweet cherry fruit development and in response to abscisic acid and auxin at onset of fruit ripening. Plant Growth Regulation 75, 455–464. [Google Scholar]
- Wang Y, Guo S, Tian S, Zhang J, Ren Y, Sun H, Gong G, Zhang H, Xu Y. 2017. Abscisic acid pathway involved in the regulation of watermelon fruit ripening and quality trait evolution. PLoS ONE 12, e0179944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Wang Y, Ji K, et al. 2013. The role of abscisic acid in regulating cucumber fruit development and ripening and its transcriptional regulation. Plant Physiology and Biochemistry 64, 70–79. [DOI] [PubMed] [Google Scholar]
- Waseem M, Ahmad F, Habib S, Li Z. 2018. Genome-wide identification of the auxin/indole-3-acetic acid (Aux/IAA) gene family in pepper, its characterisation, and comprehensive expression profiling under environmental and phytohormones stress. Scientific Reports 8, 12008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins CB. 2002. Ethylene synthesis, mode of action, consequences and control. In Knee M, ed, Fruit quality and its biological basis. Boca Raton, FL: CRC Press, 180–224. [Google Scholar]
- Wickham H. 2016. ggplot2: elegant graphics for data analysis. New York: Springer-Verlag. [Google Scholar]
- Wu SC, Zhang Y. 2010. Active DNA demethylation: many roads lead to Rome. Nature Reviews Molecular Cell Biology 11, 607–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue J, Li Y, Tan H, Yang F, Ma N, Gao J. 2008. Expression of ethylene biosynthetic and receptor genes in rose floral tissues during ethylene-enhanced flower opening. Journal of Experimental Botany 59, 2161–2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Tang K, Datsenka TU. 2019. Critical function of DNA methyltransferase 1 in tomato development and regulation of the DNA methylome and transcriptome. Journal of Integrative Plant Biology 61, 1224–1242. [DOI] [PubMed] [Google Scholar]
- Zhang MS, Kimatu JN, Xu K, Liu B. 2010. DNA cytosine methylation in plant development. Journal of Genetics and Genomics 37, 1–12. [DOI] [PubMed] [Google Scholar]
- Zhong S, Fei Z, Chen YR, et al. 2013. Single-base resolution methylomes of tomato fruit development reveal epigenome modifications associated with ripening. Nature Biotechnology 31, 154–159. [DOI] [PubMed] [Google Scholar]
- Zhong S, Joung JG, Zheng Y, Chen YR, Liu B, Shao Y, Xiang JZ, Fei Z, Giovannoni JJ. 2011. High-throughput Illumina strand-specific RNA sequencing library preparation. Cold Spring Harbor Protocols 2011, 940–949. [DOI] [PubMed] [Google Scholar]
- Ziliotto F, Corso M, Rizzini FM, Rasori A, Botton A, Bonghi C. 2012. Grape berry ripening delay induced by a pre-véraison NAA treatment is paralleled by a shift in the expression pattern of auxin- and ethylene-related genes. BMC Plant Biology 12, 185. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.