Illumination conditions and acquisition of source status by winter oilseed rape leaves jointly impact on proline levels, expression levels of proline metabolism-related genes, and associated metabolic fluxes.
Keywords: Brassica napus, fluxes, osmotic stress, Δ1-PYRROLINE-5-CARBOXYLATE SYNTHASE (P5CS), PROLINE DEHYDROGENASE (ProDH), proline, regulation, senescence
Abstract
Proline metabolism is an essential component of plant adaptation to multiple environmental stress conditions that is also known to participate in specific developmental phases, particularly in reproductive organs. Recent evidence suggested a possible role for proline catabolism in Brassica napus for nitrogen remobilization processes from source leaves at the vegetative stage. Here, we investigate transcript levels of Δ1-PYRROLINE-5-CARBOXYLATE SYNTHASE (P5CS) and PROLINE DEHYDROGENASE (ProDH) genes at the vegetative stage with respect to net proline biosynthesis and degradation fluxes in leaves having a different sink/source balance. We showed that the underexpression of three P5CS1 genes in source leaves was accompanied by a reduced commitment of de novo assimilated 15N towards proline biosynthesis and an overall depletion of free proline content. We found that the expression of ProDH genes was strongly induced by carbon starvation conditions (dark-induced senescence) compared with early senescing leaves. Our results suggested a role for proline catabolism in B. napus, but acting only at a late stage of senescence. In addition, we also identified some P5CS and ProDH genes that were differentially expressed during multiple processes (leaf status, dark to light transition, and stress response).
Introduction
Winter oilseed rape (Brassica napus L.) is a major oleaginous crop ranked the third most essential source of plant oil in the world. Considering the agroecological transition and climate change contexts, there is a need to maintain crop seed yield and quality under fluctuating environments and to reduce chemical inputs. However, oilseed rape is highly demanding for mineral nitrogen (N) inputs (150–250 kg N ha–1) and characterized by a poor N use efficiency, with only 50% of the N absorbed by plants being present in seeds (Rossato et al., 2001; Rathke et al., 2005). Therefore, optimization of N management by oilseed rape under reduced N input conditions and/or during abiotic stress is a promising target for future crop improvement (Masclaux-Daubresse et al., 2010; Bouchet et al., 2016). In plants, N is taken up first from the soil by roots through nitrate and ammonia transporters. Nitrate is preferentially transported in the xylem to be assimilated in source leaves and then further reallocated to sink tissues in the form of amino acids (Tegeder and Masclaux-Daubresse, 2018). Although oilseed rape is characterized by a strong N uptake efficiency at the vegetative stage, it has a low N use efficiency, mainly due to a low N remobilization efficiency (Malagoli et al., 2005). At the vegetative stage, N remobilization in oilseed rape occurs essentially between leaves according to their specific sink/source balance, through regulated sequential senescence processes, and involves a complex proteolytic degradation system (Girondé et al., 2015). Indeed, a complete metabolic reprogramming is triggered to help protein degradation and successive steps of proteinogenic transamination of amino acids into glutamine for N recycling and reallocation (Chrobok et al., 2016). The specific overexpression of glutamine synthetase in midveins during natural leaf senescence and under N limitation conditions suggests that glutamine is a major vector of N for N remobilization through the phloem in oilseed rape (Orsel et al., 2014). However, others amino acids may also contribute to N remobilization, depending on the physiological leaf status and environmental agroclimatic conditions. In B. napus, previous studies pointed out the importance of proline metabolism for stress response under N and water depletion, as well as a potential role in N remobilization from source tissues at the vegetative stage (Albert et al., 2012; Faës et al., 2015; Zhang and Becker, 2015; Clément et al., 2018).
Proline is considered as a compatible osmolyte that accumulates widely in the plant kingdom during varying abiotic stress conditions (i.e. drought, high salinity, high light, heavy metals, and phosphate starvation, and in response to biotic challenges) (Trotel et al., 1996; Slama et al., 2015; Aleksza et al., 2017). It can act as an osmoprotective and osmoregulating compound by chaperoning macromolecules under low water environment and readjusting the internal cellular osmotic potential (Verslues and Sharma, 2010). In plants, proline biosynthesis from glutamate takes place in the cytosol and the chloroplasts, and is initiated first by the NADPH-dependent delta-1-pyrroline-5-carboxylate synthase (P5CS) (Szabados and Savoure, 2010). This enzyme produces a glutamate semialdehyde that is spontaneously converted to pyrroline-5-carboxylate (P5C). Then P5C is further reduced to proline by the NADPH-dependent P5C reductase (P5CR). Proline biosynthesis is directly connected to the NADPH/NADP+ balance and is often seen as a sink for reducing power under stress-disturbed redox status, thereby lowering reactive oxygen species (ROS) production (Ben Rejeb et al., 2014). In Arabidopsis, two P5CS genes were identified with specific functions: P5CS1 is induced by drought and osmotic stress while P5CS2 is required during seed maturation and expressed in response to biotic stress (Savouré et al., 1995; Yoshiba et al., 1995; Fabro et al., 2004; Székely et al., 2008). In B. napus, a total of six P5CS1 genes have been identified (Wang et al., 2014). Previous analysis revealed that a P5CS1 (BnaA05g05760D) and a P5CS2 (BnaA09g35230D) were involved in proline production during water shortage, salt stress, or polyethyleneglycol (PEG) stress (Xue et al., 2009; Albert et al., 2012). Proline catabolism occurs in mitochondria via the sequential action of proline dehydrogenase (ProDH) and P5C dehydrogenase, which convert proline successively to P5C then to glutamate (Schertl et al., 2014). Two ProDH genes exist in Arabidopsis, with ProDH1 playing an essential role for proline-dependent oxidation/respiration in Arabidopsis mitochondria (Cabassa-Hourton et al., 2016). Recently, the response of the single mutants prodh1 and prodh2 and the double mutant prodh1prodh2 to a prolonged darkness treatment was analysed. The results showed that ProDH1 and ProDH2 were both involved in the proline-dependent mitochondrial respiration during dark-induced senescence (Launay et al., 2019). In B. napus, six ProDH1 and two ProDH2 genes were identified by phylogenetic analysis with parental ancestors (B. rapa and B. oleracea) (Faës et al., 2015). BnaA&C.ProDH1a and BnaA&C.ProDH1b genes were very highly expressed in leaves and roots, whereas BnaA&C.ProDH1c genes were almost exclusively expressed in reproductive organs. BnaProDH2 genes were mainly expressed in flowers, roots, and senescent leaves. Analysis of promoter–β-glucuronidase (GUS) lines showed a strong expression of BnaProDH2 genes in vascular tissues, proposing a role for proline degradation, which would provide energy for phloem transport (Faës et al., 2015). Proline biosynthesis can also derive from ornithine through the action of ornithine δ- or α-aminotransferase, but these alternative pathways were shown to be irrelevant for proline biosynthesis during stress conditions (Funck et al., 2008).
Regulation of proline metabolism under optimal or stress conditions is well documented and usually involves the modulation of the P5CS/ProDH balance at the transcriptional level, through either an abscisic acid (ABA)-dependent or an ABA-independent pathway (Zarattini and Forlani, 2017). Under osmotic/salt/drought/and multiple other abiotic stress conditions, induction of P5CS gene transcription allows plants to acclimate to the stress by accumulating proline and scavenging reducing power (Yoshiba et al., 1995; Signorelli and Monza, 2017). During the post-stress metabolic recovery phase, induction of ProDH gene transcription plays a key role in proline recycling, as it is a readily available source of N and reducing power for mitochondrial metabolism (Sharma and Verslues, 2010; Dobrá et al., 2011). Additionally, light and dark conditions can affect proline metabolism in Arabidopsis. P5CS gene transcription is promoted during the light period and repressed during dark periods, whereas ProDH transcript levels show the opposite behaviour (Hayashi et al., 2000; Abrahám et al., 2003). Prolonged exposure to darkness (dark-induced senescence) was also reported to cause an activation of ProDH transcription and a depletion of proline content, mediated by activationof sucrose-dependent bZIP factors (Dietrich et al., 2011; Launay et al., 2019). At the reproductive stage, decreasing water potential during flower senescence in Rosa hybrida L. can trigger the accumulation of proline through increasing P5CS transcription (Kumar et al., 2008; Zhang and Becker, 2015).
Considering the role of ProDH genes during dark-induced senescence in Arabidopsis (Launay et al., 2019), and the specific expression of ProDH2 genes in the midveins of source tissues in B. napus (Faës et al., 2015), proline catabolism might contribute to phloem-dependent N remobilization processes in source leaves of B. napus. Such a role would demonstrate that proline metabolism is a novel target for the improvement of N remobilization efficiency in crops. In this work, we addressed the question of the role of proline metabolism in the source leaves at the vegetative stage in the economically important plant crop B. napus. We systematically analysed the expression of the P5CS and ProDH genes related to the biosynthesis and degradation of proline, and measured the associated net proline fluxes (synthesis versus degradation) using 15N labelling experiments. Considering that the allopolyploid nature of oilseed rape can confer multiple possibilities of sub-/neofunctionnalization of proline metabolism-associated gene copies, we also analysed their expressions during stress conditions (osmotic stress and dark-induced senescence), in order to delimit the regulatory genes of interest. The analysis of the BnaP5CS/BnaProDH transcript balance with respect to the modifications of net proline metabolic fluxes revealed that proline catabolism was weakly activated according to the sink/source balance of the leaves. The depletion of proline content in source leaves was associated with a decrease in the net proline biosynthesis flux and was correlated with decreased expression of five BnaP5CS1 genes. In addition, we identified some BnaP5CS and BnaProDH genes that were differentially expressed during multiple processes (leaf status, dark to light transition, and stress response).
Materials and methods
Plant material and growth conditions
Seeds of oilseed rape B. napus genotype Aviso were obtained from our ‘Bracysol’ biological resource centre. After a 3 d germination step on soaked blotting paper, seedling were transferred in 4 litre pots filled with a non-fertilized commercial substrate (Falienor, reference 922016F3). Plants were grown in a 6 m3 growth chamber under a 14 h light/10 h dark cycle (22 °C/16 °C) in ambient air (65% and 80% humidity) with a photosynthetically active radiation of 100 µmol photons m–2 s–1 at the top of the canopy. A commercial fertilized solution (Liquoplant Bleu, 2.5% N, 5% P, 2.5% K) was applied twice a week on plants. All experiments were performed on plants grown for 60 days after sowing (60 DAS), possessing 14 leaf ranks, annotated from the bottom to the top (L3 to L16). The two oldest leaves (L1 and L2) had already fallen off, confirming that the remobilization processes between leaves holding a different sink/source balance were operating.
Chlorophyll and soluble protein contents
Relative chlorophyll quantification in SPAD units was achieved using a non-destructive chlorophyll SPAD-502 meter (Minolta). For protein extraction, samples were freeze-dried for 72 h then ground to a fine powder. Soluble proteins were extracted in a buffer containing 20 mM citrate, 160 mM Na2HPO4 (pH 6.8), and a pinch of polyvinylpyrrolidone by a 15 min incubation step at 1500 rpm. After a 30 min centrifugation step at 4 °C, proteins from the supernatant were quantified using the Bradford reagent with BSA as the standard.
Nitrogen and carbon percentage
N% and C% were determined on 1 mg of lyophilized plant powder and the Dumas combustion method with an NA1500CN Fisons instrument (Thermoquest, Runcorn, Cheshire, UK) analyser.
Leaf discs assays
Leaf discs (0.8 cm2) were randomly punched with a cork-borer in both laminas of the leaves. For 15N labelling experiments, leaf discs were floated for 4 h in a 6-well microplate filled with a buffer containing 10 mM MES-KOH (pH 6.5), 10 mM 15NH4Cl (99%), or [15N]l-proline (98%). From each leaf, two time points were sampled (0 and 4 h) in order to calculate 15N incorporation into amino acids before and after the experiment. For hyperosmotic experiments, leaf discs were floated on a buffer containing 5 mM HEPES (pH 6), 1.5 mM CaCl2, and 10 mM KCl supplemented or not with 400 g of PEG 6000 per kg of H2O. This PEG concentration was calculated accordingly to Michel and Kaufman’s equation to provide a –1.88 MPa osmotic shock to leaf discs (Michel and Kaufmann, 1973). After 18 h of stress, leaf discs were transferred to a hypo-osmotic medium (the same medium without PEG) for 6 h to allow tissue rehydration. All incubations were performed under continuous light in an orbital shaker at 70 rpm. For each harvested time point, leaf discs were rinsed three times with a neutral buffer (without PEG, 15NH4Cl, or [15N]l-proline) and dried for 30 s on a clean tissue before being frozen in liquid nitrogen and stored at –80 °C. For dark-induced senescence experiments, leaf discs were floated on water and the control experiment was done by incubating the microplate under a classical light/dark cycle (14 h/10 h). Complete dark conditions were mimicked by covering the microplate with a thick black plastic bag.
Amino acid content and 15N labelling
Prior to metabolite extraction, samples were freeze-dried for 72 h, allowing measurement of the dry weight. Metabolites were extracted with a mixture of MTBE/MeOH/H2O, except that dl-3-aminobutyric acid (111 µM final concentration in a 0.9 ml MeOH/H2O fraction) was used as internal standard for amino acid quantification (Salem et al., 2016). A 200 µl aliquot of the remaining MeOH/H2O fraction was evaporated using a SpeedVac concentrator at 35 °C for 2 h. Amino acids were resuspended in 50 µl of ultrapure water, and 5 µl were derivatized using an AccQTag derivatization kit (Waters) following the manufacturer’s procedures. Separation and UV detection of amino acids was performed on an Acquity UPLC-DAD (Renault et al., 2010). Amino acids were quantified using external calibration curves for each amino acid and normalized with the internal standard. For 15N experiments, isotopologues (M+0, M+1, M+2, M+3, M+4) from AccQTag-derivatized amino acid were analysed by MS by coupling a triple quadrupole spectrometer (Waters) to the analytical system. The electrospray source was settled in positive mode and each isotopologue was tracked with the Selected Ion Recording function. The settings for the mass spectrometer electrospray source were as follow: 4,4 kV capillary voltage, 35 V cone voltage, 150 °C cone temperature, 450 °C desolvatation temperature, and 900 l h–1 N flow for desolvatation. Derivatization with AccQTag added ~20% of artificial M+1 enrichment for each amino acid, which was corrected for each sample using the M+1 enrichment observed in 15N-free external standards. Considering an amino acid with n atoms of N, true 15N labelling (%) for each isotopologue x was calculated as:
Net 15N labelling for each isotopologue was calculated by subtracting the M+x labelling (%) from T0 of the experiment (natural abundancy). Net 15N incorporation for each amino acid was defined as the absolute quantity of net 15N detected in each amino acid from leaf discs after 4 h of 15N labelling. The commitment of [15N]amino acids in the protein fraction was too low to be detected in our experiments.
Identification of P5CS genes and phylogenetic analysis
P5CS genes were identified by blasting Arabidopsis thaliana P5CS1 (AT2G39800) and P5CS2 (AT3G55610) coding sequences retrieved from TAIR database (Berardini et al., 2015), respectively, on the available genomes from ENSEMBL or Phytozome database [Arabidopsis lyrata (v.1.0), B. napus var Darmor (AST_PRJEB5043 v1, assembly 2014), B. rapa (Brapa_1.0), B. oleracea (BOL), Oryza sativa (IRGSP-1.0), Eutrema salsugineum (v1.0), and human (GRCh38.p12)]. Sequence alignments and phylogenetic analysis were performed with MEGAX software (Tamura et al., 2013). The evolutionary phylogenetic gene tree was generated with the maximum likelihood method and the nucleotide substitution model of Tamura–Nei, assuming uniform rates among sites (1000 bootstraps).
Quantitative analysis of transcript levels
Total RNAs were extracted with the NucleoSpin RNA Plus Kit (Macherey Nagel) from frozen or lyophilized tissue ground into a fine powder, following the manufacturer’s instructions. Potential DNA traces were removed with DNase using the RQ1 RNase-free DNase (Promega) according to the manufacturer’s protocol. The absence of DNA contamination in samples was controlled by PCR amplification of the BnaEF1α reference gene. RNA integrity was verified by running samples on 1.2% agarose gels. A 1 μg aliquot of total RNA treated with the DNase was reverse-transcribed using oligo(dT)15 primer and Goscript reverse transcriptase (Promega) according to the manufacturer’s instructions. For quantuitative PCR (qPCR) experiments, 2 μl of diluted cDNA were mixed with the LightCycler 480 SYBR Green I Master mix (Roche) and primers pairs (see Supplementary Table S2 at JXB online). qPCRs were achieved on a Light Cycler LC480 (Roche) under the following conditions: an initial step at 95 °C for 5 min, then 45 cycles of 95 °C for 15 s and 60 °C for 40 s. Two reference genes [BnaC.UBQ11 (BnaC04g09150D) and BnaC.RibS3 (BnaC04g13040D)] were selected for their expression stability in our samples harvested from different conditions (leaf ranks, light/dark conditions, and stress conditions) using the GeNorm method (Vandesompele et al., 2002). Gene expression was normalized using the geometrical mean expression of two reference genes and the efficiency of qPCR primers (Supplementary Table S2). The efficiency of qPCR primers was calculated from a serial dilution of mixed samples from our experiments. Fold change (FC) values were calculated as the ratio of the relative gene expression between two consecutive time points for treated (FC treatment) or control (FC control) leaf discs (T0 and PEG, PEG and PEG+rehydration for osmotic stress; T2d and T4d for dark-induced senescence). Log2 (FC treatment/FC control) corresponded to the log2 value of the ratio (FC treatment/FC control).
Statistical analysis
The variability of the results is expressed as the mean ±SD of n independent biological replicates (n=3–6). Means of different leaf ranks were compared together with one-way ANOVA followed by a post-hoc Tukey’s HSD test for multiple pairwise comparison. Means of two treatments (light versus dark; control versus dark-induced senescence; control versus osmotic stress) for each leaf rank were compared using a Student t-test (bilateral distribution, equal variance). For statistical analysis of qPCR experiments, relative expression values were log-transformed to obtain normally distributed values. For each test, a P-value <0.05 was applied. All statistical analyses were achieved with “R” software.
Results
Source status of B. napus leaves is associated with a decrease of proline content
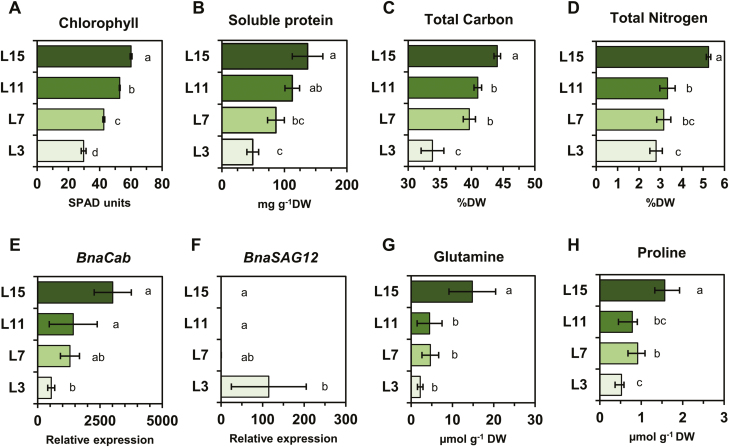
In order to obtain B. napus plants with a gradient of leaf ranks having different sink/source balance, plants were grown for 2 months at the vegetative stage in growth chambers to reach the 16-leaf stage (the two oldest leaves had already fallen off). From this stage, we decided to work on four leaf ranks: a young growing leaf (L15), an immature leaf (L11), a post-mature leaf (L7), and a senescent leaf (L3), essentially functioning as a source tissue (Albert et al., 2012). The status of the four different leaf ranks was checked by quantifying different parameters: chlorophyll and soluble protein contents, total carbon (C) and N contents, the expression of the Cab/SAG12 reporter genes, and free amino acid contents (Gombert et al., 2006). First, dry weight per leaf area was decreased according to the sink/source balance of the leaves whereas the fresh weight per leaf area was relatively maintained, leading to an increase of the FW/DW ratio between the different leaf ranks (Supplementary Fig. S2). Therefore, all results were expressed on a dry weight basis to compare the different leaf ranks, except for chlorophyll content which was measured per leaf area. Nevertheless, the observed decreased of chlorophyll content per leaf area for the different leaf ranks might still be relevant by normalizing with dry weight (Fig. 1A; Supplementary Fig. S1). Along the leaf nodal ranks, we observed a significant decrease of the content of total soluble proteins and the total C and N contents (Fig. 1B–D). Since amino acids (and therefore proteins) have a high C/N ratio, the decrease of C and N% per dry weight seemed to be attributed to an activation of protein degradation. These results suggest that both C and N remobilization are strongly induced from the L11 leaf. Those observations were also confirmed by the expression levels of Cab and SAG12 genes. Cab gene expression was significantly and gradually repressed according to the source status of the different leaf ranks, whereas the SAG12 gene was significantly expressed in the L3 leaf, mirroring its actual senescent status (Fig. 1E, F). From the overall free amino acid profiles, glutamine content showed a significant decrease, except for L11 and L7 (Fig. 1G; Supplementary Table S1). Such variations of glutamine content between the different leaf ranks is often attributed to the activation of the remobilization processes (proteolytic degradation and amino acids exported as glutamine) (Orsel et al., 2014; Tegeder and Masclaux-Daubresse, 2018). Consequently, the L15 leaf may have a higher ‘sink’ demand compared with its ‘source’ export. Interestingly, although at a lower amplitude, proline content showed statistically significant variations between the different leaf ranks, similarly to glutamine content variations, thereby suggesting a potential role for proline metabolism in nutrient recycling processes in the sources leaves of B. napus (Fig. 1H). To summarize, we have identified four leaf ranks with gradually increased levels of source status and gradually decreased levels of proline content from the top to the bottom of the plant axis.
Fig. 1.
Proline is a potential source of nitrogen remobilized from source leaves at the vegetative stage in B. napus. (A) Chlorophyll and (B) soluble protein contents. (C) Total carbon and (D) total nitrogen contents. (E, F) qPCR analysis of BnaCab and BnaSAG12 genes. (G, H) Glutamine and proline contents from four leaf ranks having a different sink/source balance at the vegetative stage (60 DAS). Leaves were harvested 3 h after the beginning of the illumination period. Values are the means ±SD of 3–6 independent biological replicates. Different letters indicate that mean values are significantly different (P-value <0.05) between the different leaf ranks. Overall amino acid analysis of the four leaf ranks is listed in Supplementary Table S1. (This figure is available in colour at JXB online.)
Decrease of BnaP5CS1 gene expression in source leaves correlates with a decrease of net proline biosynthesis flux
Next, we decided to analyse the P5CS and ProDH transcript balance according to the sink/source balance of the leaves and during a dark to light transition. These different conditions may solicit proline metabolism for N reallocation, respiration feeding, and/or redox status adjustment. In order to design gene-specific primers for each copy of BnaP5CS (Supplementary Table S2), we performed a genome-wide analysis of P5CS2 genes in B. napus and its progenitors B. rapa and B. oleracea, using known P5CS2 coding sequences from the Arabidopsis genome as a query. Our analysis identified four AtP5CS2 (Bna.P5CS2) orthologues, corresponding to two P5CS2 orthologues in each progenitor genome (B. rapa and B. oleracea) (Supplementary Fig. S1). We also performed similar work for P5CS1 in order to localize the genomic region of the six P5CS1 genes previously identified (Wang et al., 2014). The identified P5CS1 and P5CS2 genes from B. napus, B. rapa, and B. oleracea were correspondingly renamed in this study based on the genome to which they belonged or inherited (A for B. rapa and C for B. oleracea), their chromosome location, and their phylogenetic relationship (Supplementary Fig. S2). Considering ProDH genes, a previous work already identified six BnaProDH1 and two BnaProDH2 genes and designed specific primers for each gene (Faës et al., 2015).
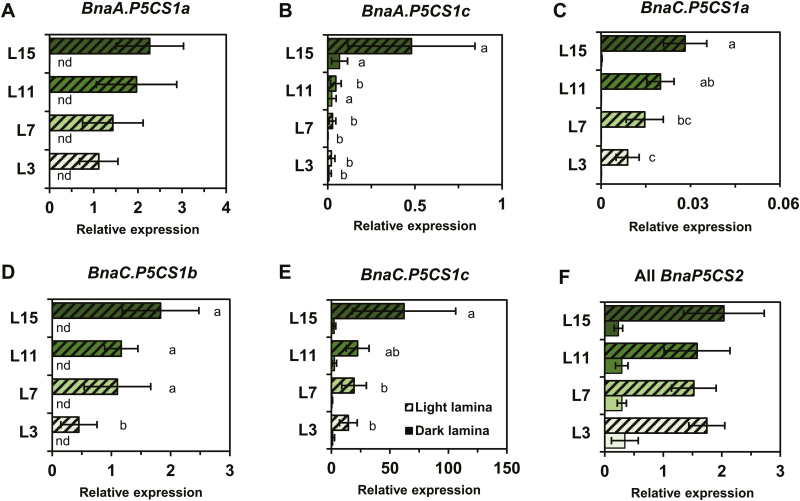
First, P5CS transcript levels were quantified relative to the reference genes in the four leaf ranks sampled 3 h before or 3 h after the beginning of the illumination period, in order to evaluate the possibly combined effects of the sink/source balance of the leaves and the natural dark to light transition. Transcript levels for four BnaP5CS1 genes were significantly decreased in leaves having a very strong source leaf status compared with the L15 leaf, in accordance with the previously observed decrease in proline content (Figs 1F, 2A–E). The expression levels of BnaA.P5CS1b are not shown since they were too weak to be repeatedly detected in our samples. BnaC.P5CS1c showed the strongest expression levels compared with other BnaP5CS1 genes, suggesting a preponderant role in controlling endogenous proline levels (Fig. 2B). All BnaP5CS2 transcript levels were not significantly changed between the different leaf ranks (Fig. 2F; expression levels for each BnaP5CS2 gene are available in Supplementary Fig. S3). However, all BnaP5CS genes (except BnaA.P5CS1b) were significantly overexpressed in light conditions compared with dark conditions for all the leaf ranks.
Fig. 2.
Four P5CS1 genes are underexpressed in source leaves in B. napus. Laminae from four leaf ranks having a different sink/source balance at the vegetative stage (60 DAS) were sampled either 3 h before (dark) or 3 h after (light) the beginning of the illumination period. Expression of each gene copy was normalized relative to two reference genes UBQ11 and RibS3. Relative expression of genes showing no significantly different behaviour during leaf development were pooled together (all BnaP5CS2 genes). The complete data set for each BnaP5CS2 gene is available in Supplementary (Fig. S3). Values are the means ±SD of five independent biological replicates. Different letters indicate that mean values are significantly different (P-value <0.05) between the different leaf ranks. Comparison of mean values from light and dark conditions were always significantly different for each leaf rank and each gene (P-value ≤0.05). (This figure is available in colour at JXB online.)
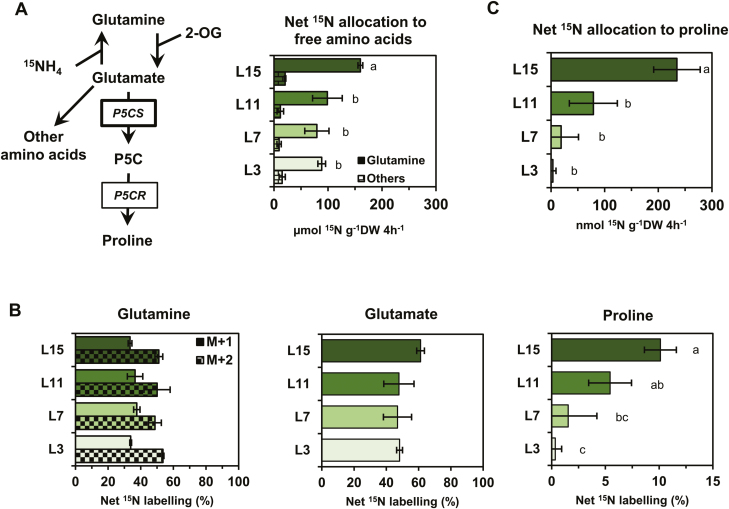
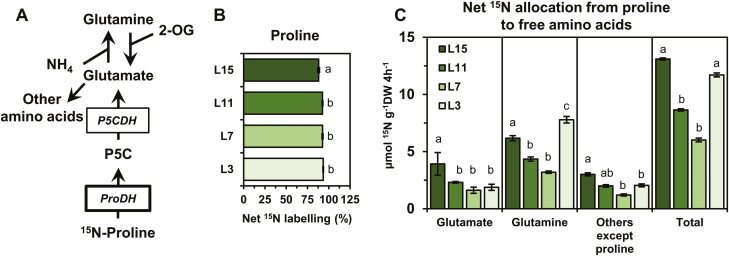
In order to connect the variations of BnaP5CS1 transcript levels with the variations of proline content observed in the different leaf ranks, we performed 15N labelling experiments on leaf discs isolated from the four leaf ranks. This experiment aimed at comparing in vivo the capacity for net proline biosynthesis between the different leaf ranks using the repartition of the 15N as a proxy. We took advantage of the leaf capacity to assimilate de novo N using 15NH4 to label glutamate, the major precursor for proline biosynthesis (Fig. 3A). We analysed net 15N enrichment and contents of amino acids using an UPLC-TQD system. After 4 h of 15N labelling in isolated leaf discs, we found that net 15N was in great part allocated to glutamine (Fig. 3A). This allocation was significantly higher in L15 compared with other leaves, showing that L15 has a higher capacity for free ammonia assimilation compared with leaves having a very strong source status. Analysis of 15N labelling in glutamine showed similar values for each leaf rank (no statistical differences), with ~30–35% for the M+1 isotopologue and 50–55% for the M+2 isotopologue (Fig. 3B). The 15N labelling in the glutamate molecule was also statistically identical between each leaf rank, with values ranging between 48% and 60%. Interestingly, the net 15N labelling in the proline molecule was significantly lower between the different leaf ranks (~10% in L15, 7% in L11, and 2–0.5 in L7 and L3). Since the 15N labelling in glutamate was similar between the different leaf ranks, our results suggested that the commitment of de novo assimilated 15N toward proline biosynthesis was lowered in leaves having a very strong source status. To quantitatively evaluate this commitment, we calculated the net 15N allocation to proline by assuming that proline commitment toward the protein fraction between the different leaf ranks was negligible (15N labelling was undetectable in the protein fraction) (Fig. 3A). Although the net 15N allocation to proline underestimates the true net proline biosynthesis flux, it is still a reliable tool to compare net flux variations. We found that net 15N allocation to proline was significantly lowered from L15 to L11, L7, and L3, showing values of ~200 nmol 15N g–1 DW 4 h–1 in L15 to drop to 75 in L11 and then nearly 0 in L7/L3 (Fig. 3C). The results were consistent with a decrease of the net proline biosynthesis flux in source leaves. Such a decrease was strongly correlated with the decrease of BnaP5CS1 transcript levels.
Fig. 3.
Partitioning of de novo assimilated 15N towards proline biosynthesis in source leaves correlates with the down-regulation of P5CS genes. Leaf discs from four leaves having a different sink/source balance at the vegetative stage (60 DAS) were incubated for 4 h under continuous light with 15NH4Cl (99%). (A) Metabolic route for 15NH4 to proline and net 15N allocation to free amino acids. (B) Glutamine, glutamate, and proline net 15N labelling. (C) Net 15N allocation to proline.15N labelling detection, and absolute quantification of amino acids were performed by MS and UV detection. Values are the means ±SD of three independent biological replicates. Different letters indicate that mean values are significantly different between the different leaf ranks (P-value <0.05). (This figure is available in colour at JXB online.)
Increase of BnaA.ProDH2a transcript levels in source leaves of B. napus does not correlate with variations of maximal proline degradation capacity
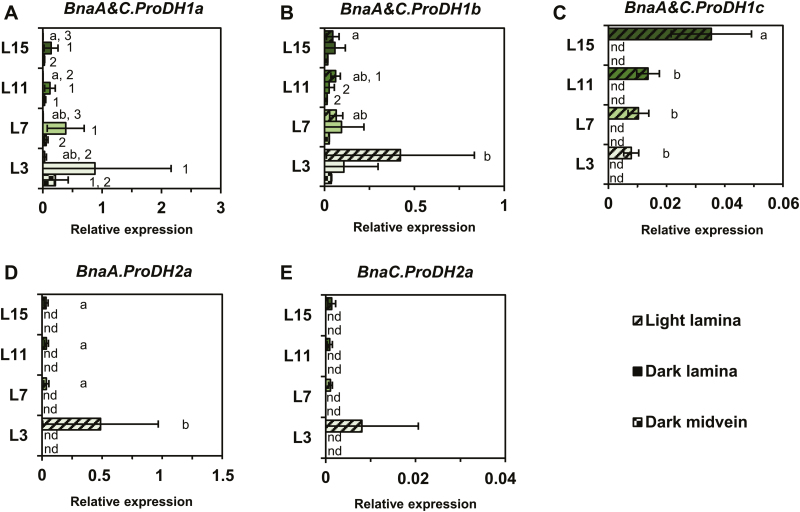
After analysing the relationship between the net proline biosynthesis capacity and the transcriptional regulation of P5CS genes, we focused our work on the proline degradation pathway. Consequently, we quantified ProDH transcript levels with respect to dark–light transition and the sink/source balance of the leaves. Since the BnaC.ProDH2a gene has already been reported to be expressed in the midveins of senescent leaves (Faës et al., 2015) and as ProDH genes were overexpressed during dark periods in Arabidopsis (Hayashi et al., 2000), we also analysed BnaProDH gene expression in the midveins of leaves sampled in the dark. Out of the six BnaProDH1 genes, BnaA&C.ProDH1a and BnaA&C.ProDH1b genes were the most expressed and were significantly overexpressed in the lamina of leaves L7 and L3 compared with L15 (Fig. 4A–C). This overexpression was dependent on the light/dark condition (BnaA&C.ProDH1a genes for light, BnaA&C.ProDH1b genes for dark). BnaA&C.ProDH1c gene expression was significantly increased in leaves having a very low source status. However, those last genes were only detected in the lamina sampled in the light and were poorly expressed. Considering the two BnaProDH2 genes, their expression levels were only detected in the lamina sampled in the light. The BnaA.ProDH2a gene was the most expressed and was significantly overexpressed in the lamina of the L3 senescing leaves (Fig. 4D, E). No BnaProDH gene was overexpressed in the midveins of leaves sampled in the dark compared with other conditions/tissues (Fig. 4). To summarize, BnaA&C.ProDH1b and BnaA.ProDH2a genes were overexpressed in leaves having a very strong source status, and showed a maximal expression value in the L3 senescent leaves. Since the net 15N allocation to proline was close to 0 in the L3 leaf rank (Fig. 3C), our results suggested that proline degradation could be activated in these tissues. Therefore, we also performed 15N labelling experiments on leaf discs isolated from the four leaf ranks using [15N]proline. This experiment aimed at comparing in vivo the maximal proline degradation capacity between the different leaf ranks, by measuring the incorporation of the 15N (resulting from proline) into other amino acids (Fig. 5A). After 4 h of [15N]proline labelling in isolated leaf discs, net 15N labelling in the proline molecule (M+1 isotopologue) was ~87–93% in the different leaf ranks (Fig. 5B). Although this enrichment was significantly lower in the L15 leaf rank compared with the other leaf ranks, the values were relatively similar. This suggested that [15N]proline might have been similarly incorporated in the leaf discs during the labelling experiment. From the net 15N labelling and quantification of amino acids, we calculated the net 15N allocation from proline to free amino acids (Fig. 5C). We found that net 15N allocation to glutamate and others amino acids (except proline) was significantly reduced from the leaf ranks L15 to L7. However, net 15N allocation to glutamine was significantly higher in L3 compared with other leaf ranks. Considering the total net 15N allocation from proline to free amino acids, our results showed that the maximal proline degradation capacity was decreased from L15 to L7, but was restored in L3 leaves (similarly to L15 leaves). Overall, our results suggest that proline catabolism is weakly activated in leaves having a very strong source status and may not be specifically controlled by variations of BnaProDH transcript levels.
Fig. 4.
Two ProDH genes are overexpressed in source leaves of B. napus. Laminae from four leaf ranks having a different sink/source balance at the vegetative stage (60 DAS) were sampled either 3 h before (dark) or 3 h after (light) the beginning of the illumination period. In addition, the midveins sampled in dark conditions were also analysed. Expression of each gene copy was normalized relative to two reference genes UBQ11 and RibS3. Values are the means ±SD of five independent biological replicates. Different letters indicate that mean values are significantly different between the different leaf ranks (P-value <0.05). Different numbers indicate that mean values are significantly different between the three conditions (light lamina, dark lamina, and dark midvein) for each leaf rank (P-value <0.05). (This figure is available in colour at JXB online.)
Fig. 5.
The variation of the maximal proline degradation capacity in leaves having a different sink/source balance poorly correlates with the variations of BnaProDH gene transcript levels. Leaf discs from four leaves having a different sink/source balance at the vegetative stage (60 DAS) were incubated for 4 h under continuous light with [15N]l-proline (98%). (A) Metabolic route for [15N]l-proline to amino acids. (B) Proline net 15N labelling. (C) Net 15N allocation from proline to free amino acids. 15N labelling detection and absolute quantification of proline were performed by MS and UV detection. Values are the means ±SD of three independent biological replicates. Different letters indicate that mean values are significantly different between the different leaf ranks (P-value <0.05). (This figure is available in colour at JXB online.)
Some BnaP5CS and BnaProDH genes are specifically involved in development and stress response processes
We have shown that multiple copies of P5CS and ProDH genes were differentially expressed according to the sink/source status of the leaves in B. napus. Since the variation of P5CS and ProDH gene expression can control proline contents during some stress conditions, we decided to test whether specific BnaP5CS and BnaProDH genes were differentially expressed during multiple processes (sink/source status, dark to light transition, and stress response). Consequently, we analysed the gene-specific expression of BnaP5CS and BnaProDH genes during two stressing conditions known to regulate proline metabolism: an osmotic shock followed by a recovery period and a dark-induced senescence kinetic experiment (Trotel et al., 1996; Dietrich et al., 2011; Launay et al., 2019). We decided to perform this experiment on the L7 leaf, since its source but not senescing leaf status allows a rapid entry into senescence during the dark-induced senescence experiment.
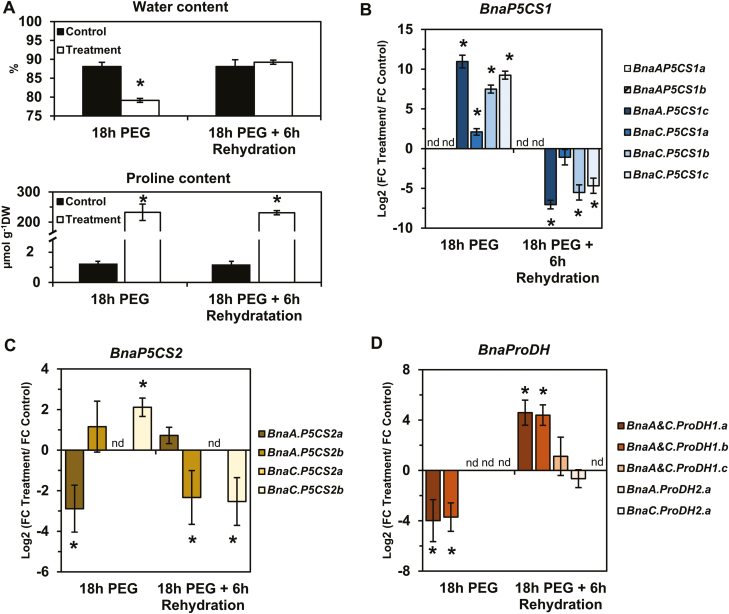
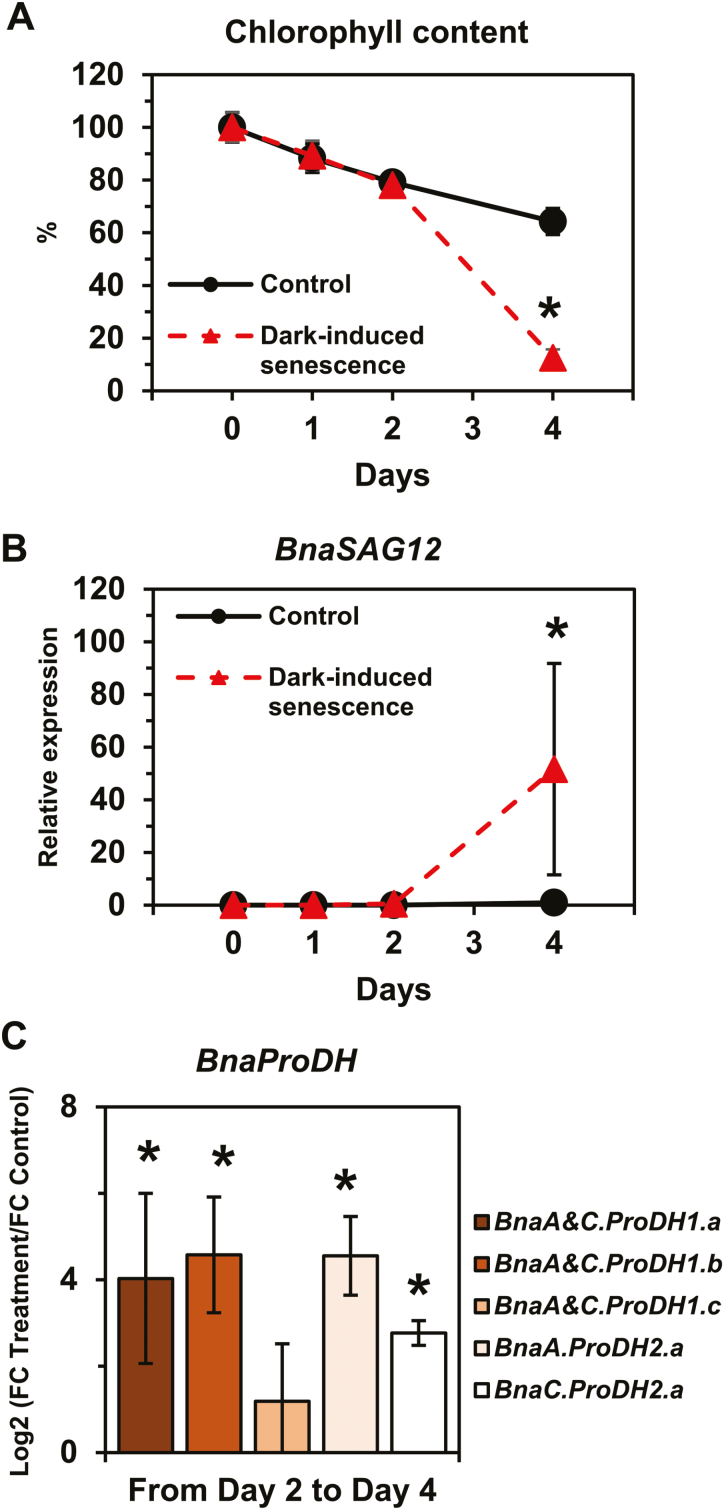
First, leaf discs from L7 were subjected to a PEG-induced hyperosmotic stress (–1.88 MPa) for 18 h under light conditions, followed by a 6 h recovery phase in a hypo-osmotic medium. The evolution of the leaf disc water content in the PEG treatment condition compared with the control condition confirmed that tissue dehydration occurred during hyperosmotic treatment whereas full tissue rehydration was restored after the recovery phase (Fig. 6A). A strong accumulation of proline was observed in the stressed tissues compared with the control after 18 h of PEG treatment. However, after 6 h of recovery, there was no significant decrease in proline content. Out of the four BnaP5CS1 genes detected, the expression of the three genes BnaA.P5CS1c, BnaC.P5CS1b, and BnaC.P5CS1c was significantly highly induced (log2 FC variation >7) during the osmotic shock and was significantly repressed (log2 FC variation less than –4) during the post-stress recovery phase (Fig. 6B). BnaC.P5CS1a gene expression was moderately induced during the stress phase and repressed during the post-stress recovery phase. Surprisingly, we found that BnaC.P5CS2b gene expression was also moderately induced by the osmotic shock, whereas BnaA.P5CS2a gene expression was moderately repressed (Fig. 6C). During the recovery from hyperosmotic stress, BnaA.P5CS2b and BnaC.P5CS2b gene expression was significantly repressed. Regarding BnaProDH genes, only BnaA&C.ProDH1a and BnaA&C.ProDH1b gene expression was repressed during osmotic shock then induced during the recovery phase (Fig. 6D). Overall, our results demonstrated that three BnaPCS1 genes were specifically involved in the control of proline content according to the sink/source balance of the leaves and during a dark–light transition and an osmotic stress and recovery. Next, we performed a time-course dark-induced senescence experiment on leaf discs from L7 to explore the differential expression of BnaProDH genes during stress-induced senescence processes. After 2 d of incubation, chlorophyll content was not significantly different in the control compared with the prolonged darkness condition (Fig. 7A). After 4 d, a significant decrease in chlorophyll content was observed in the prolonged darkness condition compared with the control. Concomitantly, transcription of the senescence marker genes BnaA&C.SAG12-1&2 was strongly induced by prolonged darkness from day 2 to day 4 compared with the control (Fig. 7B). Those results confirmed that dark-induced senescence only operated from day 2 to day 4 in our experimental set-up. As a result, except for BnaA&C.ProDH1c, all BnaProDH genes were significantly induced (log2 FC variation >3) by dark-induced senescence compared with the control (Fig. 7C). Therefore, our results demonstrated that BnaA&C.ProDH1a gene expression was specifically controlled during a dark–light transition, an osmotic shock, recovery, and a dark-induced senescence, whereas BnaA&C.ProDH1b and BnaA.ProDH2a gene expression was specifically controlled by senescence (natural or dark induced).
Fig. 6.
Transcript level variations of the BnaP5CS and BnaProDH genes during osmotic stress and rehydration in B. napus. For the ‘treatment’ condition, leaf discs from L7 leaves were subjected to a hyperosmotic stress (–1.88 MPa) for 18 h and then transferred to a hypo-osmotic medium for 6 h of rehydration under continuous light. For the ‘control’ condition, leaf discs from L7 leaves were incubated in the hypo-osmotic medium (same medium without PEG) for 18 h and then transferred to a new fresh medium for 6 h, under continuous light. (A) Water and proline content and log2 values for the ratio of fold change (FC) expression levels in the treated leaf discs over the control leaf discs for BnaP5CS1 (B), BnaP5CS2 (C), and BnaProDH genes (D). Values are the mean ±SD of three independent biological replicates. Asterisks indicate that mean values are significantly different between the two conditions (treatment versus control) (P-value <0.05). (This figure is available in colour at JXB online.)
Fig. 7.
Dark-induced senescence triggers the expression of four BnaProDH genes in B. napus. Leaf discs from L7 were floated on water for 4 d and incubated under a light/dark cycle (14 h/10 h) (control) or under complete darkness (dark-induced senescence). (A) Chlorophyll content and (B) SAG12 expression levels during the experiment. (C) Log2 values for the ratio of fold change (FC) expression levels in the treated leaf discs over the control leaf discs for BnaProDH genes from day 2 to day 4. Values are the mean ±SD of three independent biological replicates. Asterisks indicate that mean values are significantly different between the two conditions (treatment versus control) (P-value <0.05). (This figure is available in colour at JXB online.)
Discussion
Proline metabolism plays an essential role in plant adaptation to multiple environmental stress conditions, specifically when they are facing biotic and abiotic stresses (Szabados and Savouré, 2010). The tight control of proline content, generally achieved by regulating the P5CS/ProDH balance at the transcriptional level, allows the plants to tolerate the stress condition (Slama et al., 2015). However, proline metabolism can also participate in plant developmental events, by contributing to proline accumulation and/or degradation during specific growth phases. In particular in reproductive organs and tissues with actively dividing cells, proline accumulation was shown to be stimulated (Mattioli et al., 2009; Biancucci et al., 2015). Conversely, at the vegetative stage, several piesces of evidence suggested that proline catabolism could be activated during leaf senescence and source leaf status acquisition in B. napus and could fuel mitochondrial respiration (Faës et al., 2015; Clément et al., 2018; Launay et al., 2019). Such a phenomenon of metabolic reconversion of proline can be of functional interest for N recycling and nutrient remobilization processes through the phloem only if it is adequately fed with proline arising from neosynthesis or proteolysis.
In this study, we have shown that four BnaP5CS1 genes were underexpressed in leaves having a very strong source status during the vegetative growth phase, whereas transcript levels of two BnaProDH genes were moderately overexpressed in the L3 senescent leaves (Figs 2B–E, 4B, D). These changes in transcript levels were strongly correlated with the depletion in proline content and with the reduction in commitment of de novo assimilated N towards proline biosynthesis, whereas maximal proline degradation capacity was not different between L15 (very low source status) and L3 (very strong source status) leaves (Figs 1H, 3C, 5C). We also found that transcription of three BnaP5CS1 genes was strongly induced during an osmotic stress, and was repressed during the post-recovery phase (Fig. 6B). In particular, these changes were associated with strong variations of proline contents (Fig. 6A). In Arabidopsis, during an osmotic stress and the post-recovery phase, the modulation of proline content was associated with the overexpression of the P5CS1 gene, whereas expression of the P5CR gene was not changed at all (Sharma and Verslues, 2010). Indeed, P5CR from Arabidopsis has a higher protein stability compared with P5CR from Escherichia coli, which may explain why this gene is less transcribed (Funck et al., 2012). Therefore, our results suggest that the variations of P5CS transcript levels in B. napus may actively affect the biosynthesis of proline.
The specific control of proline content by the transcript levels of BnaP5CS1 genes according to the sink/source status of the leaves may be explained by different physiological processes. First, growth and development of young leaves requires protein biosynthesis whereas old leaves having a very strong source status degrade their proteins and recycle their macromolecules. Increasing proline biosynthesis in young leaves compared with old source leaves could support the demand for proline to synthesize proteins in young leaves and to fuel high-energy-consuming cell division processes (Kavi Kishor and Sreenivasulu, 2014). Secondly, old leaves having a very strong source status are more hydrated than young leaves (Musse et al., 2013; Sorin et al., 2015). We have shown that the transcription of BnaP5CS1 genes was decreased during tissue hydration after a hyperosmotic shock. Therefore, the decrease of net proline biosynthesis flux in source leaves may be a side effect of the tissue hydration. In addition to these factors, BnaP5CS1 may also be down-regulated by other factors, including a crosstalk between senescence and the redox balance, regarding its up-regulation by light (our work; Hayashi et al., 2000) and by the redox status (Aswani et al., 2019).
We have identified that BnaA&C.ProDH1a genes were specifically overexpressed in dark conditions compared with light conditions (Fig. 4A), in good agreement with previous work performed in Arabidopsis (Hayashi et al., 2000). ProDH1 and ProDH2 are located in mitochondria and can provide electrons to the mitochondrial electron transfer chain through proline degradation (Cabassa-Hourton et al., 2016; Launay et al., 2019). Since mitochondrial respiration is inhibited up to 95% by light in plants (Tcherkez et al., 2005), we propose that BnaA&C.ProDH1a genes are involved in proline-dependent mitochondrial respiration. However, other BnaProDH genes are overexpressed in dark-induced senescence conditions and therefore could be involved in this proline-dependent respiration (Fig. 7C). Indeed, in Arabidopsis, some C starvation conditions {dark-induced senescence or [3-(3,4-dichlorophenyl)-1,1-dimethylurea] (DCMU)} can promote the expression of ProDH1 (Dietrich et al., 2011). Recently, the analysis of the double mutant prodh1prodh2 in Arabidopsis confirmed that proline was effectively used as a substrate for mitochondrial respiration after 5 d of dark-induced senescence (Launay et al., 2019). Our [15N]proline labelling experiments showed that the proline catabolism may be weakly activated between L7 and L3 leaves (i.e. between a source leaf and a source-to-senescent leaf) (Fig. 5C). In plants, many amino acids can be used as alternative respiratory substrates when C is missing (dark-induced senescence), such as leucine, isoleucine, valine, and tyrosine (Araujo et al., 2008, 2010, 2012, 2013; Chrobok et al., 2016; Wang et al., 2019). In our conditions, the overexpression of six BnaProDH genes during dark-induced senescence was observed in leaf discs containing very low chlorophyll contents (equivalent to 4 SPAD units) (Fig. 7C). Conversely, at an intermediary chlorophyll content in the L3 leaves (~20 SPAD units), only three BnaProDH genes were moderately overexpressed (Fig. 4A, B, D). Those observations suggest that the activation of proline catabolism in B. napus may be effective in leaves having a late stage of senescence, namely when the major part of the N content has been remobilized and other respiratory sources have already been depleted.
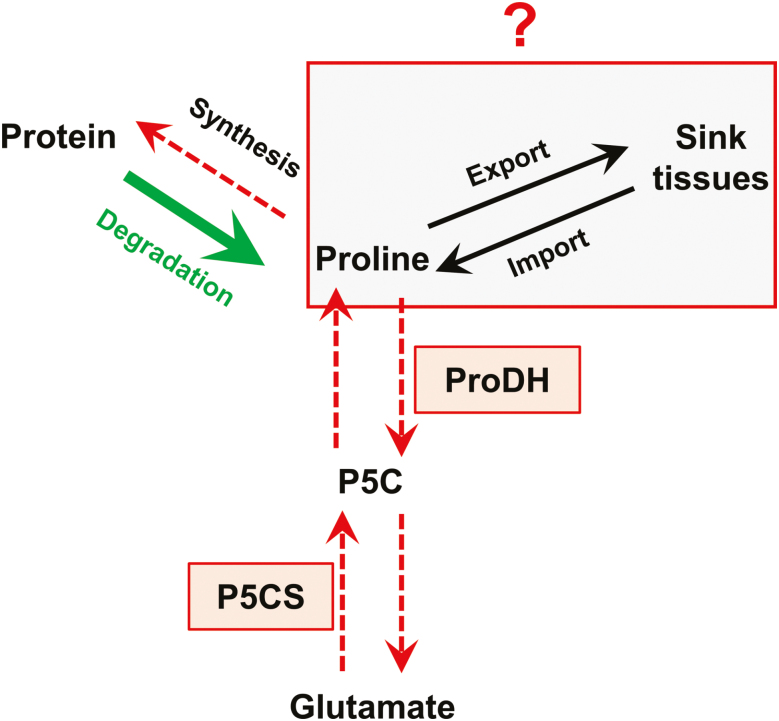
Our results showed that proline was not accumulated in source leaves and that proline catabolism was weakly activated according to the sink/source status of the leaves (Figs 1H, 5C). Consequently, proline arising from protein degradation in source leaves could be transported to young leaves and sink tissues through the phloem (Fig. 8). The source–sink apoplastic transport of amino acid through the phloem represents a key determinant of the N utilization efficiency for crops (Perchlik and Tegeder, 2017). In white clover, 15N tracing experiments showed that proline can be actively loaded to the phloem for transport between roots and shoots, suggesting that an equivalent transport between leaves having a different sink/source balance may also occur at the vegetative stage (Lee et al., 2009). The differential proline enrichment found in phloem sap along the leaf ranks during drought stress also showed that an active source–sink proline transport can take place under stress conditions (Girousse et al., 1996; Albert et al., 2012). To date, no cell to cell proline transporter has been identified to play a role in proline remobilization at the vegetative stage in Brassica species. However, one proline-specific transporter (AtProT2) and one amino acid permease (AAP1) can both contribute to proline uptake under normal or stress conditions (Lehmann et al., 2011; Wang et al., 2017).
Fig. 8.
The regulation of proline content in source leaves of B. napus at the vegetative growth phase. Protein degradation in source leaves produces proline, which is not accumulated compared with leaves having a low sink/source balance. Our results showed that net proline biosynthesis flux was reduced in source leaves, whereas the maximal proline degradation capacity was weakly affected by the sink/source balance of the leaves. Consequently, proline arising from protein degradation could be exported from source leaves to organs having a low sink/source balance. Overall, our results suggest that proline catabolism plays a minor role in nitrogen remobilization processes in source leaves. (This figure is available in colour at JXB online.)
Concerning the sub-/neofunctionnalization of proline metabolism-associated gene copies, we have shown that some BnaP5CS1 and BnaProDH genes shared conserved multiple functions in B. napus. The expression of three out of six BnaP5CS1 genes was controlled by both the sink/source status of the leaf and osmotic stress (Figs 2B, D, E, 6B), and the expression of four out of six ProDH1 genes was controlled by both osmotic stress and dark-induced senescence (Figs 6D, 7C). Since parental ancestors of oilseed rape have undergone a whole-genome triplication event, the presence of multiple copies was expected to relax natural and artificial selective pressures on any individual gene copy, thereby facilitating the emergence of novel gene function (Conant and Wolfe, 2008). In the oilseed rape genome, duplicated flowering time genes were shown to be preferentially retained compared with other genes, and ~70% of them had a divergent expression patterns relative to each other across development, suggesting neo- or subfunctionalization (Jones et al., 2018). Such conserved multifunctionalization of P5CS and ProDH genes in B. napus clearly reflects the importance of finely controlling both phases of proline metabolism (biosynthesis and degradation) to face issues related to both development and stress conditions.
In conclusion, we have shown that the depletion of proline content in source leaves in B. napus was associated with a decrease in the net proline biosynthesis flux rather than an activation of proline catabolism. Such flux variations were correlated with the strong underexpression of four BnaP5CS1 genes, whereas the expression of three BnaDH genes was moderately increased in source leaves to senescent leaves. Since BnaProDH genes were strongly overexpressed during dark-induced senescence (C starvation condition), our results suggested that proline catabolism may played a role in B. napus leaves but during late stages of senescence, perhaps to contribute to proline-dependent mitochondrial respiration. Consequently, the proline arising from protein degradation in source leaves might be remobilized toward young leaves and sink tissues through the phloem. Recent works showed that N use efficiency and seed yields could be dramatically increased in crops by enhancing the expression of the amino acid transporter AAP1, which can contribute to proline uptake (Perchlik and Tegeder, 2017; Wang et al., 2017). Future research should focus on the possible source–sink proline transport at the vegetative stage to identify potential new targets for improvement of N use efficiency.
Supplementary data
Supplementary data are available at JXB online.
Fig. S1. FW and DW per leaf area and FW/DW ratio of the four leaf ranks having a different sink/source balance (L15, L11, L7, L3).
Fig. S2. Molecular phylogenetic analysis of the Brassica napus P5CS coding sequences.
Fig. S3. Relative expression for each BnaP5CS2 gene in four leaf ranks having a different sink/source balance (L15, L11, L7, L3) harvested under either dark or light conditions.
Table S1. Amino acid contents in the laminae of four leaf ranks having a different sink/source balance (L15, L11, L7, L3) sampled 3 h after the beginning of the illumination period.
Table S2. List of primers used in this study for qPCR analysis and primer efficiency.
Acknowledgements
We are grateful to Dr. Anne Marmagne for performing C and N analyses. We thank Dr. Laurent Leport for helpful discussions, Françoise Leprince, Sophie Rolland and the ‘molecular biology group’ for access to facilities, and Patrick Leconte and the ‘experiment group’ for help with growing B. napus plants (IGEPP, INRA Rennes). We also thank members of the ‘Plateau de Profilage Métabolique et Métabolique’ (P2M2) for access to the analytical systems, especially Solenne Berardocco. This work was supported by a ‘Young Researcher’ grant from the Institut National de la Recherche Agromomique (FLUCOLSA project), and dedicated funding from the University of Rennes 1 (TOPIC project).
References
- Abrahám E, Rigó G, Székely G, Nagy R, Koncz C, Szabados L. 2003. Light-dependent induction of proline biosynthesis by abscisic acid and salt stress is inhibited by brassinosteroid in Arabidopsis. Plant Molecular Biology 51, 363–372. [DOI] [PubMed] [Google Scholar]
- Albert B, Le Cahérec F, Niogret MF, Faes P, Avice JC, Leport L, Bouchereau A. 2012. Nitrogen availability impacts oilseed rape (Brassica napus L.) plant water status and proline production efficiency under water-limited conditions. Planta 236, 659–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aleksza D, Horváth GV, Sándor G, Szabados L. 2017. Proline accumulation is regulated by transcription factors associated with phosphate starvation. Plant Physiology 175, 555–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araújo WL, Ishizaki K, Nunes-Nesi A, et al. 2010. Identification of the 2-hydroxyglutarate and isovaleryl-CoA dehydrogenases as alternative electron donors linking lysine catabolism to the electron transport chain of Arabidopsis mitochondria. The Plant Cell 22, 1549–1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araújo WL, Nunes-Nesi A, Trenkamp S, Bunik VI, Fernie AR. 2008. Inhibition of 2-oxoglutarate dehydrogenase in potato tuber suggests the enzyme is limiting for respiration and confirms its importance in nitrogen assimilation,. Plant Physiology 148, 1782–1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araújo WL, Tohge T, Osorio S, Lohse M, Balbo I, Krahnert I, Sienkiewicz-Porzucek A, Usadel B, Nunes-Nesi A, Fernie AR. 2012. Antisense inhibition of the 2-oxoglutarate dehydrogenase complex in tomato demonstrates its importance for plant respiration and during leaf senescence and fruit maturation. The Plant Cell 24, 2328–2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araújo WL, Trofimova L, Mkrtchyan G, Steinhauser D, Krall L, Graf A, Fernie AR, Bunik VI. 2013. On the role of the mitochondrial 2-oxoglutarate dehydrogenase complex in amino acid metabolism. Amino Acids 44, 683–700. [DOI] [PubMed] [Google Scholar]
- Aswani V, Rajsheel P, Bapatla RB, Sunil B, Raghavendra AS. 2019. Oxidative stress induced in chloroplasts or mitochondria promotes proline accumulation in leaves of pea (Pisum sativum): another example of chloroplast–mitochondria interactions. Protoplasma 256, 449–457. [DOI] [PubMed] [Google Scholar]
- Ben Rejeb K, Abdelly C, Savouré A. 2014. How reactive oxygen species and proline face stress together. Plant Physiology and Biochemistry 80, 278–284. [DOI] [PubMed] [Google Scholar]
- Berardini TZ, Reiser L, Li D, Mezheritsky Y, Muller R, Strait E, Huala E. 2015. The Arabidopsis information resource: making and mining the ‘gold standard’ annotated reference plant genome. Genesis 53, 474–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biancucci M, Mattioli R, Forlani G, Funck D, Costantino P, Trovato M. 2015. Role of proline and GABA in sexual reproduction of angiosperms. Frontiers in Plant Science 6, 680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchet AS, Laperche A, Bissuel-Belaygue C, et al. 2016. Genetic basis of nitrogen use efficiency and yield stability across environments in winter rapeseed. BMC Genetics 17, 131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabassa-Hourton C, Schertl P, Bordenave-Jacquemin M, et al. 2016. Proteomic and functional analysis of proline dehydrogenase 1 link proline catabolism to mitochondrial electron transport in Arabidopsis thaliana. The Biochemical Journal 473, 2623–2634. [DOI] [PubMed] [Google Scholar]
- Chrobok D, Law SR, Brouwer B, et al. 2016. Dissecting the metabolic role of mitochondria during developmental leaf senescence. Plant Physiology 172, 2132–2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clément G, Moison M, Soulay F, Reisdorf-Cren M, Masclaux-Daubresse C. 2018. Metabolomics of laminae and midvein during leaf senescence and source–sink metabolite management in Brassica napus L. leaves. Journal of Experimental Botany 69, 891–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conant GC, Wolfe KH. 2008. Turning a hobby into a job: how duplicated genes find new functions. Nature Reviews. Genetics 9, 938–950. [DOI] [PubMed] [Google Scholar]
- Dietrich K, Weltmeier F, Ehlert A, Weiste C, Stahl M, Harter K, Dröge-Laser W. 2011. Heterodimers of the Arabidopsis transcription factors bZIP1 and bZIP53 reprogram amino acid metabolism during low energy stress. The Plant Cell 23, 381–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrá J, Vanková R, Havlová M, Burman AJ, Libus J, Storchová H. 2011. Tobacco leaves and roots differ in the expression of proline metabolism-related genes in the course of drought stress and subsequent recovery. Journal of Plant Physiology 168, 1588–1597. [DOI] [PubMed] [Google Scholar]
- Fabro G, Kovács I, Pavet V, Szabados L, Alvarez ME. 2004. Proline accumulation and AtP5CS2 gene activation are induced by plant–pathogen incompatible interactions in Arabidopsis. Molecular Plant-Microbe Interactions 17, 343–350. [DOI] [PubMed] [Google Scholar]
- Faës P, Deleu C, Aïnouche A, et al. 2015. Molecular evolution and transcriptional regulation of the oilseed rape proline dehydrogenase genes suggest distinct roles of proline catabolism during development. Planta 241, 403–419. [DOI] [PubMed] [Google Scholar]
- Funck D, Stadelhofer B, Koch W. 2008. Ornithine-delta-aminotransferase is essential for arginine catabolism but not for proline biosynthesis. BMC Plant Biology 8, 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funck D, Winter G, Baumgarten L, Forlani G. 2012. Requirement of proline synthesis during Arabidopsis reproductive development. BMC Plant Biology 12, 191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girondé A, Etienne P, Trouverie J, et al. 2015. The contrasting N management of two oilseed rape genotypes reveals the mechanisms of proteolysis associated with leaf N remobilization and the respective contributions of leaves and stems to N storage and remobilization during seed filling. BMC Plant Biology 15, 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girousse C, Bournoville R, Bonnemain JL. 1996. Water deficit-induced changes in concentrations in proline and some other amino acids in the phloem sap of alfalfa. Plant Physiology 111, 109–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gombert J, Etienne P, Ourry A, Le Dily F. 2006. The expression patterns of SAG12/Cab genes reveal the spatial and temporal progression of leaf senescence in Brassica napus L. with sensitivity to the environment. Journal of Experimental Botany 57, 1949–1956. [DOI] [PubMed] [Google Scholar]
- Hayashi F, Ichino T, Osanai M, Wada K. 2000. Oscillation and regulation of proline content by P5CS and ProDH gene expressions in the light/dark cycles in Arabidopsis thaliana L. Plant & Cell Physiology 41, 1096–1101. [DOI] [PubMed] [Google Scholar]
- Jones DM, Wells R, Pullen N, Trick M, Irwin JA, Morris RJ. 2018. Spatio-temporal expression dynamics differ between homologues of flowering time genes in the allopolyploid Brassica napus. The Plant Journal 96, 103–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavi Kishor PB, Sreenivasulu N. 2014. Is proline accumulation per se correlated with stress tolerance or is proline homeostasis a more critical issue? Plant, Cell & Environment 37, 300–311. [DOI] [PubMed] [Google Scholar]
- Kumar N, Srivastava GC, Dixit K. 2008. Flower bud opening and senescence in roses (Rosa hybrida L.). Plant Growth Regulation 55, 81–99. [Google Scholar]
- Launay A, Cabassa-Hourton C, Eubel H, et al. 2019. Proline oxidation fuels mitochondrial respiration during dark-induced leaf senescence in Arabidopsis thaliana. Journal of Experimental Botany 70, 6203–6214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee BR, Jin YL, Avice JC, Cliquet JB, Ourry A, Kim TH. 2009. Increased proline loading to phloem and its effects on nitrogen uptake and assimilation in water-stressed white clover (Trifolium repens). New Phytologist 182, 654–663. [DOI] [PubMed] [Google Scholar]
- Lehmann S, Gumy C, Blatter E, Boeffel S, Fricke W, Rentsch D. 2011. In planta function of compatible solute transporters of the AtProT family. Journal of Experimental Botany 62, 787–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malagoli P, Laine P, Rossato L, Ourry A. 2005. Dynamics of nitrogen uptake and mobilization in field-grown winter oilseed rape (Brassica napus) from stem extension to harvest. II. An 15N-labelling-based simulation model of N partitioning between vegetative and reproductive tissues. Annals of Botany 95, 1187–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masclaux-Daubresse C, Daniel-Vedele F, Dechorgnat J, Chardon F, Gaufichon L, Suzuki A. 2010. Nitrogen uptake, assimilation and remobilization in plants: challenges for sustainable and productive agriculture. Annals of Botany 105, 1141–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattioli R, Falasca G, Sabatini S, Altamura MM, Costantino P, Trovato M. 2009. The proline biosynthetic genes P5CS1 and P5CS2 play overlapping roles in Arabidopsis flower transition but not in embryo development. Physiologia Plantarum 137, 72–85. [DOI] [PubMed] [Google Scholar]
- Michel BE, Kaufmann MR. 1973. The osmotic potential of polyethylene glycol 6000. Plant Physiology 51, 914–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musse M, De Franceschi L, Cambert M, Sorin C, Le Caherec F, Burel A, Bouchereau A, Mariette F Leport L. 2013. Structural changes in senescing oilseed rape leaves at tissue and subcellular levels monitored by nuclear magnetic resonance relaxometry through water status. Plant Physiology 63, 392–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orsel M, Moison M, Clouet V, Thomas J, Leprince F, Canoy AS, Just J, Chalhoub B, Masclaux-Daubresse C. 2014. Sixteen cytosolic glutamine synthetase genes identified in the Brassica napus L. genome are differentially regulated depending on nitrogen regimes and leaf senescence. Journal of Experimental Botany 65, 3927–3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perchlik M, Tegeder M. 2017. Improving plant nitrogen use efficiency through alteration of amino acid transport processes. Plant Physiology 175, 235–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathke GW, Christen O, Diepenbrock W. 2005. Effects of nitrogen source and rate on productivity and quality of winter oilseed rape (Brassica napus L.) grown in different crop rotations. Field Crops Research 94, 103–113. [Google Scholar]
- Renault H, Roussel V, El Amrani A, Arzel M, Renault D, Bouchereau A, Deleu C. 2010. The Arabidopsis pop2-1 mutant reveals the involvement of GABA transaminase in salt stress tolerance. BMC Plant Biology 10, 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossato L, Lainé P, Ourry A. 2001. Nitrogen storage and remobilization in Brassica napus L. during the growth cycle: nitrogen fluxes within the plant and changes in soluble protein patterns. Journal of Experimental Botany 52, 1655–1663. [PubMed] [Google Scholar]
- Salem MA, Jüppner J, Bajdzienko K, Giavalisco P. 2016. Protocol: a fast, comprehensive and reproducible one-step extraction method for the rapid preparation of polar and semi-polar metabolites, lipids, proteins, starch and cell wall polymers from a single sample. Plant Methods 12, 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savouré A, Jaoua S, Hua XJ, Ardiles W, Van Montagu M, Verbruggen N. 1995. Isolation, characterization, and chromosomal location of a gene encoding the delta 1-pyrroline-5-carboxylate synthetase in Arabidopsis thaliana. FEBS Letters 372, 13–19. [DOI] [PubMed] [Google Scholar]
- Schertl P, Cabassa C, Saadallah K, Bordenave M, Savouré A, Braun HP. 2014. Biochemical characterization of proline dehydrogenase in Arabidopsis mitochondria. The FEBS Journal 281, 2794–2804. [DOI] [PubMed] [Google Scholar]
- Sharma S, Verslues PE. 2010. Mechanisms independent of abscisic acid (ABA) or proline feedback have a predominant role in transcriptional regulation of proline metabolism during low water potential and stress recovery. Plant, Cell & Environment 33, 1838–1851. [DOI] [PubMed] [Google Scholar]
- Signorelli S, Monza J. 2017. Identification of delta(1)-pyrroline 5-carboxylate synthase (P5CS) genes involved in the synthesis of proline in Lotus japonicus. Plant Signaling & Behavior 12, e1367464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slama I, Abdelly C, Bouchereau A, Flowers T, Savouré A. 2015. Diversity, distribution and roles of osmoprotective compounds accumulated in halophytes under abiotic stress. Annals of Botany 115, 433–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorin E, Etienne P, Maillard A, Zamarreño AM, Garcia-Mina JM, Arkoun M, Jamois F Cruz F, Yvin JC, Ourry A. 2015. Effect of sulphur deprivation on osmotic potential components and nitrogen metabolism in oilseed rape leaves: identification of a new early indicator. Journal of Experimetal Botany 66, 6175–6189. [DOI] [PubMed] [Google Scholar]
- Szabados L, Savouré A. 2010. Proline: a multifunctional amino acid. Trends in Plant Science 15, 89–97. [DOI] [PubMed] [Google Scholar]
- Székely G, Abrahám E, Cséplo A, et al. 2008. Duplicated P5CS genes of Arabidopsis play distinct roles in stress regulation and developmental control of proline biosynthesis. The Plant Journal 53, 11–28. [DOI] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Molecular Biology and Evolution 30, 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tcherkez G, Cornic G, Bligny R, Gout E, Ghashghaie J. 2005. In vivo respiratory metabolism of illuminated leaves. Plant Physiology 138, 1596–1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tegeder M, Masclaux-Daubresse C. 2018. Source and sink mechanisms of nitrogen transport and use. New Phytologist 217, 35–53. [DOI] [PubMed] [Google Scholar]
- Trotel P, Bouchereau A, Niogret MF, Larher F. 1996. The fate of osmo-accumulated proline in leaf discs of rape (Brassica napus L.) incubated in a medium of low osmolarity. Plant Science 118, 31–45. [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. 2002. Accurate normalization of real-time quantitative RT–PCR data by geometric averaging of multiple internal control genes. Genome Biology 3, RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verslues PE, Sharma S. 2010. Proline metabolism and its implications for plant–environment interaction. The Arabidopsis Book 8, e0140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C-P, Lin B, Zhang Y-Q, Lin Y-H, Liu A-H, Hua X-J. 2014. The evolutionary fate of Δ1-pyrroline-5-carboxylate synthetase 1 (P5CS1) genes in allotetraploid Brassica napus. Journal of Systematics and Evolution 52, 566–579. [Google Scholar]
- Wang M, Toda K, Block A, Maeda HA. 2019. TAT1 and TAT2 tyrosine aminotransferases have both distinct and shared functions in tyrosine metabolism and degradation in Arabidopsis thaliana. Journal of Biological Chemistry 294, 3563–3576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Chen Y, Zhang M, Chen J, Liu J, Han H, Hua X. 2017. Arabidopsis AMINO ACID PERMEASE1 contributes to salt stress-induced proline uptake from exogenous sources. Frontiers in Plant Science 8, 2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue X, Liu A, Hua X. 2009. Proline accumulation and transcriptional regulation of proline biosynthesis and degradation in Brassica napus. BMB Reports 42, 28–34. [DOI] [PubMed] [Google Scholar]
- Yoshiba Y, Kiyosue T, Katagiri T, Ueda H, Mizoguchi T, Yamaguchi-Shinozaki K, Wada K, Harada Y, Shinozaki K. 1995. Correlation between the induction of a gene for delta 1-pyrroline-5-carboxylate synthetase and the accumulation of proline in Arabidopsis thaliana under osmotic stress. The Plant Journal 7, 751–760. [DOI] [PubMed] [Google Scholar]
- Zarattini M, Forlani G. 2017. Toward unveiling the mechanisms for transcriptional regulation of proline biosynthesis in the plant cell response to biotic and abiotic stress conditions. Frontiers in Plant Science 8, 927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Becker DF. 2015. Connecting proline metabolism and signaling pathways in plant senescence. Frontiers in Plant Science 6, 552. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.