Inducible expression of AvrRps4 activates RRS1/RPS4-mediated effector-triggered immunity without the presence of pathogens, allowing us to characterize downstream immune responses triggered by TIR-NLRs without cell surface receptor-mediated immunity.
Keywords: Cell death, defence gene expression, estradiol-inducible expression system, Golden Gate modular cloning, hypersensitive response, MAP kinase, NLR activation, plant innate immunity, protein complex
Abstract
Plant nucleotide-binding domain, leucine-rich repeat receptor (NLR) proteins play important roles in recognition of pathogen-derived effectors. However, the mechanism by which plant NLRs activate immunity is still largely unknown. The paired Arabidopsis NLRs RRS1-R and RPS4, that confer recognition of bacterial effectors AvrRps4 and PopP2, are well studied, but how the RRS1/RPS4 complex activates early immediate downstream responses upon effector detection is still poorly understood. To study RRS1/RPS4 responses without the influence of cell surface receptor immune pathways, we generated an Arabidopsis line with inducible expression of the effector AvrRps4. Induction does not lead to hypersensitive cell death response (HR) but can induce electrolyte leakage, which often correlates with plant cell death. Activation of RRS1 and RPS4 without pathogens cannot activate mitogen-associated protein kinase cascades, but still activates up-regulation of defence genes, and therefore resistance against bacteria.
Introduction
To investigate plant immunity, researchers routinely conduct pathogen inoculations on plants in a controlled environment. Upon pathogen attack, plants activate innate immune responses via both membrane-associated and intracellular receptors, which makes it difficult to unravel the distinct contribution of each component. Most plasma membrane-localized receptors perceive conserved pathogen-associated molecular patterns (PAMPs) or host-cell-derived damage-associated molecular patterns (DAMPs) and activate PAMP-triggered immunity (PTI) or DAMP-triggered immunity (DTI). Plant intracellular immune receptors belong to a family of nucleotide-binding leucine-rich repeat (NB-LRR) proteins, also known as NLRs. NLRs recognize pathogen effectors and activate effector-triggered immunity (ETI), which often leads to accumulation of reactive oxygen species (ROS) and a hypersensitive cell death response (HR). Most plant NLRs carry either coiled-coil (CC) or Toll/interleukin-1 receptor (TIR) N-terminal domains. Both CC and TIR domains are believed to function in signalling upon activation of NLRs, but the detailed mechanisms are unknown. Many CC-NLRs localize at and function in association with the plasma membrane, whereas TIR-NLRs can function in diverse locations, including the nucleus. Regardless of the distinct localization patterns between CC- and TIR-NLRs, their downstream outputs culminate in elevated resistance, but have never been directly compared side-by-side. To study the specific immune outputs generated by ETI, inducible expression tools have been applied (McNellis et al., 1998; Tornero et al., 2002; Allen et al., 2004; Porter et al., 2012).
In Arabidopsis, functionally paired NLRs RRS1-R and RPS4 confer resistance against a soil-borne bacterial pathogen Ralstonia solanacearum through the recognition of an effector PopP2 secreted via a Type III secretion system and a hemibiotrophic ascomycetous fungal pathogen Colletotrichum higginsianum (Narusaka et al., 2009). They can also confer resistance against the bacterium Pseudomonas syringae pv. tomato (Pst) DC3000 carrying AvrRps4, an effector protein from Pseudomonas syringae pv. pisi, causing bacterial blight in Pisum sativum (pea) (Narusaka et al., 2009; Sohn et al., 2009). Previously, it was reported that residues 135–138 of AvrRps4, lysine–arginine–valine–tyrosine (KRVY), are required for the recognition of AvrRps4 by RRS1 and RPS4 (Sohn et al., 2009). The crystal structure of the C-terminus of AvrRps4 revealed that residue 187 (glutamate; E187) is also required for HR and immunity (Sohn et al., 2012). PopP2 recognition by RRS1 occurs by the integrated WRKY domain at the C-terminus of a resistant allele of RRS1-R [from the Wassilewskija-2 (Ws-2) ecotype of Arabidopsis] but not the susceptible allele of RRS1-S [from the Columbia-0 (Col-0) ecotype of Arabidopsis] (Sarris et al., 2015). Crystal structure information of RRS1 and RPS4 on the TIR domains and the co-crystal structures between the RRS1-R WRKY domain and effector PopP2 have indicated some structural basis for how RRS1/RPS4 have been activated (Williams et al., 2014; Zhang et al., 2017). However, it is still unknown how the protein complex assembles and functions.
Here we report tools for studying the immune complex of RRS1-R and RPS4 in vivo. We established a set of transgenic Arabidopsis lines to study RRS1/RPS4-mediated ETI in the absence of pathogens. Using these lines, we show that some, but not all, immune outputs induced by the conditionally expressed AvrRps4 resemble other reported effector-inducible lines.
Materials and methods
Plant material and growth conditions
Arabidopsis thaliana accessions Ws-2 and Col-0 were used as the wild type in this study. The eds1-2 mutant used has been described previously (Falk et al., 1999). Seeds were sown on compost, and plants were grown at 21 °C with 10 h under light and 14 h in dark, and at 70% humidity. Tobacco plants were grown at 22 °C with 16 h light/8 h dark, and at 80% constant humidity. The light level is ~180–200 µmol with fluorescent tubes.
FastRed selection for transgenic Arabidopsis
Seeds harvested from Agrobacterium-transformed Arabidopsis were resuspended in 0.1% agarose and exposed under a fluorescence microscope with a DsRed (red fluorescent protein) filter. Seeds with bright red fluorescence are selected as the positive transformants.
β-Glucuronidase (GUS) staining
Nicotiana benthamiana leaves were infiltrated with agrobacteria carrying constructs with the GUS reporter gene expressed under selected Arabidopsis promoters (Supplementary Table S1 at JXB online). Leaves were collected at 2 days post-infiltration (dpi), and vacuum-infiltrated with GUS staining buffer [0.1 M sodium phosphate pH 7.0, 10 mM EDTA pH 7.0, 0.5 mM K3Fe(CN)6, 0.5 mM K4Fe(CN)6, 0.76 mM 5-bromo-4-chloro-3-indolyl-β-d-glucuronide cyclohexylamine salt or X-Gluc, and 0.04% Triton X-100]. After vacuum infiltration, the leaves were incubated at 37 °C overnight in the dark. The leaves were rinsed with 70% ethanol until the whole leaf de-stained to a clear white.
Immunoblotting
Nicotiana benthamiana leaves were infiltrated with agrobacteria carrying our stacking constructs (Supplementary Table S2). At 2 dpi, the same leaves were infiltrated with either DMSO or 50 µM β-estradiol (E2) diluted in water. Samples were collected at 6 hours post-infiltration (hpi) of DMSO or E2 treatment, and snap-frozen in liquid nitrogen. Proteins were extracted using GTEN buffer (10% glycerol, 25 mM Tris pH 7.5, 1 mM EDTA, 150 mM NaCl) with 10 mM DTT, 1% NP-40, and protease inhibitor cocktail (cOmplete™, EDTA-free; Merck). For Arabidopsis seedlings, seedlings grown for 8 d after germination were treated with DMSO or E2 at the indicated time points and snap-frozen in liquid nitrogen. After centrifugation at 13 000 rpm for 15 min to remove cell debris, the protein concentration of each sample was measured using the Bradford assay (Protein Assay Dye Reagent Concentrate; Bio-Rad). After normalization, extracts were incubated with 3× SDS sample buffer at 95 °C for 5 min; 6% SDS–PAGE gels were used to run the protein samples. After transferring proteins from gels to polyvinylidene fluoride (PVDF) membranes (Merck-Millipore) using the Trans-Blot Turbo System (Bio-Rad), membranes were immunoblotted with horseradish peroxidase (HRP)-conjugated Flag antibodies (Monoclonal ANTI-FLAG® M2-Peroxidase HRP antibody produced in mouse, A5892; Merck-Millipore), HRP-conjugated HA antibodies (12013819001; Merck-Roche), or Phospho-p44/42 mitogen-acivated protein kinase (MAPK; Erk1/2) (Thr202/Tyr204) (D13.14.4E) XP® rabbit monoclonal antibody (4370; Cell Signalling Technology). Anti-rabbit IgG (whole molecule)–peroxidase antibody produced in goat (A0545; Merck-Sigma-Aldrich) was used as secondary antibody following the use of Phospho-p44/42 MAPK antibody.
Bacterial growth assay
Pst strain DC3000 carrying pVSP61 empty vector was grown on selective King’s B (KB) medium plates containing 15% (w/v) agar, 25 µg ml–1 rifampicin, and 50 µg ml–1 kanamycin for 48 h at 28 °C. Bacteria were harvested from the plates, resuspended in infiltration buffer (10 mM MgCl2), and the concentration was adjusted to an optical density of 0.001 at 600 nm [OD600=0.001, representing ~5×105 colony-forming units (CFU) ml–1]. Bacteria were infiltrated into abaxial surfaces of 5-week-old Arabidopsis leaves with a 1 ml needleless syringe. For quantification, leaf samples were harvested with a 6 mm diameter cork borer (Z165220; Merck-Sigma-Aldrich), resulting in leaf discs with an area of 0.283 cm2. Two leaf discs per leaf were harvested as a single sample. For each condition, four samples were collected immediately after infiltration as ‘day 0’ samples to ensure no significant difference introduced by unequal infiltrations, and six samples were collected at 3 dpi as ‘day 3’ samples to compare the bacterial growth between different genotypes, conditions, and treatments. For ‘day 0’, samples were ground in 200 μl of infiltration buffer and spotted (10 μl per spot) on selective KB medium agar plates to grow for 48 h at 28 °C. For ‘day 3’, samples were ground in 200 μl of infiltration buffer, serially diluted (5, 50, 500, 5000, and 50 000 times), and spotted (6 μl per spot) on selective KB medium agar plates to grow for 48 h at 28 °C. The number of colonies (CFU per drop) was monitored and bacterial growth was represented as CFU cm–2 of leaf tissue. All results are plotted using ggplot2 in R (Wickham, 2016), and a detailed statistical summary can be found in Supplementary Table S5.
Hypersensitive cell death response phenotyping in Arabidopsis
Pseudomonas fluorescens engineered with a Type III secretion system (Pf0-1 ‘EtHAn’ strains) expressing one of the wild-type or mutant effectors, AvrRps4, AvrRps4KRVY135-138AAAA, PopP2, PopP2C321A, AvrRpt2, or pVSP61 empty vector were grown on selective KB plates for 24 h at 28 °C (Thomas et al., 2009; Sohn et al., 2014). Bacteria were harvested from the plates, resuspended in infiltration buffer (10 mM MgCl2), and the concentration was adjusted to OD600=0.2 (108 CFU ml–1). The abaxial surfaces of 5-week-old Arabidopsis leaves were hand infiltrated with a 1 ml needleless syringe. Cell death was monitored 24 h after infiltration.
Electrolyte leakage assay
Either 50 μM E2 or DMSO was hand infiltrated into 5-week-old Arabidopsis leaves with a 1 ml needleless syringe for electrolyte leakage assay. Leaf discs were taken with a 4 mm diameter cork borer from infiltrated leaves. Discs were dried and washed in deionized water for 1 h before being floated on deionized water (15 discs per sample, three samples per biological replicate). Electrolyte leakage was measured as water conductivity with a Pocket Water Quality Meters (LAQUAtwin-EC-33; Horiba) at the indicated time points. All results are plotted using ggplot2 in R (Wickham, 2016), and a detailed statistics summary can be found in Supplementary Table S5.
Trypan blue staining
Either 50 μM E2 or DMSO was hand infiltrated in 5-week-old Arabidopsis leaves with a 1 ml needleless syringe for trypan blue staining. Six leaves per sample were collected 24 hpi. Leaves were boiled in trypan blue solution (1.25 mg ml–1 trypan blue dissolved in 12.5% glycerol, 12.5% phenol, 12.5% lactic acid, and 50% ethanol) in a boiling water bath for 1 min and de-stained by chloral hydrate solution (2.5 g ml–1). De-stained leaves were mounted, and pictures were taken under on a Leica M165FC fluorescent stereomicroscope. All images were taken with identical settings at ×2.5 magnification. Scale bars=0.5 mm.
Reverse transcription–quantitative PCR (RT–qPCR) for measuring relative gene expression
For gene expression analysis, RNA was isolated from 5-week-old Arabidopsis leaves and used for subsequent RT–qPCR analysis. RNA was extracted with a Quick-RNA Plant Kit (R2024; Zymo Research) and treated with RNase-free DNase (4716728001; Merck-Roche). Reverse transcription was carried out using SuperScript IV Reverse Transcriptase (18090050; ThermoFisher Scientific). qPCR was performed using a CFX96 Touch™ Real-Time PCR Detection System. Primers for qPCR analysis of Isochorismate Synthase1 (ICS1), Pathogenesis-Related1 (PR1), AvrRps4, and Elongation Factor 1 Alpha (EF1α) are listed in Supplementary Table S4. Data were analysed using the double delta Ct method (Livak and Schmittgen, 2001). All results are plotted using ggplot2 in R (Wickham, 2016), and a detailed statistical summary can be found in Supplementary Table S5.
Confocal laser scanning microscopy (CLSM) imaging
Transgenic plant materials were imaged with the Leica DM6000/TCS SP5 confocal microscope (Leica Microsystems) for confirmation of expression of inducible AvrRps4 fused with monomeric yellow–green fluorescent protein, mNeonGreen, or mNeon (Shaner et al., 2013). Roots from 3-week-old Arabidopsis seedlings were sprayed with 50 μM E2 and imaged at 1 d post-spray. Fluorescence of mNeon was excited at 500 nm and detected at between 520 nm and 540 nm. CLSM images of root cells from Arabidopsis seedlings are recorded via the camera. The images were analysed with the Leica application Suite and Fiji software (Schindelin et al., 2012).
Co-immunoprecipitation (co-IP)
Arabidopsis transgenic seedlings, and the background ecotype Col-0 grown for 7 days after germination (DAG) were treated with 0.1% DMSO or 50 μM E2 for 3 h. Proteins from seedlings were extracted using IP buffer (10% glycerol, 50 mM Tris–HCl pH 6.8, 50 mM KCl, 1 mM EDTA, 5 mM MgCl2, 1% NP-40, 10 mM DTT, 1 mM dATP). Crude extract of the seedlings was centrifuged, and supernatants were incubated with anti-HA-conjugated beads (EZviewTM Red Anti-HA Affinity Gel; E6779; Sigma). A small portion of supernatants were taken for input samples. At 2 h after incubation of the extract with beads, the beads were washed three times with IP buffer containing 0.1% NP-40. Proteins bound to beads were eluted by boiling the beads with SDS sample buffer. Immunoblotting of the input and eluted samples was performed as described above.
Results
RRS1 overexpression can compromise RPS1/RPS4 function
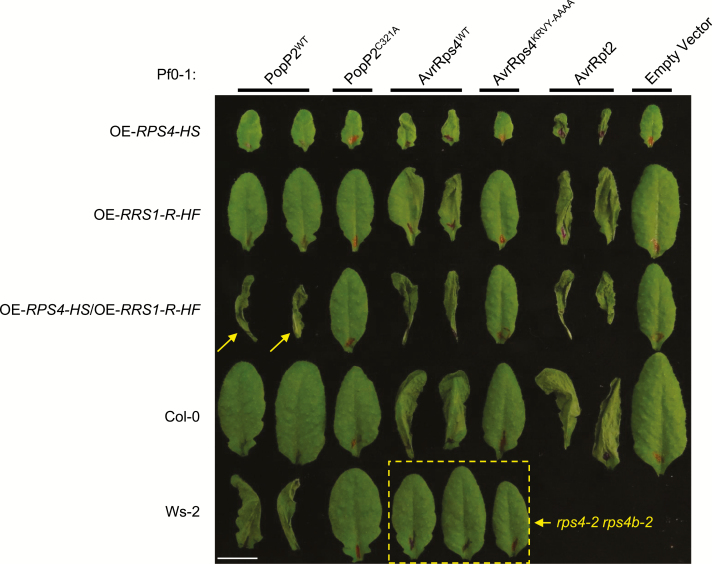
Overexpression of RPS4 leads to autoimmunity and dwarfism under standard growth conditions (see the Materials and methods) (Heidrich et al., 2013). This autoimmunity is both temperature and RRS1 dependent. In contrast, elevated expression of RRS1-R from ecotype Ws-2 in Col-0, an ecotype expressing a dominant allele of RRS1-S, does not trigger autoimmunity (Huh et al., 2017). Furthermore, high level RRS1-R expression does not confer recognition of effector PopP2 (Fig. 1). Overexpression of RRS1 in an RPS4 overexpression line attenuates dwarfism and autoimmunity (Huh et al., 2017). We infiltrated non-pathogenic strains of P. fluorescens Pf0-1 engineered with the Type III secretion system from the Pst DC3000 strain expressing the effectors PopP2, mutant PopP2C321A, AvrRps4, mutant AvrRps4KRVY-AAAA, AvrRpt2, and empty vector, respectively (Sohn et al., 2009; Thomas et al., 2009; Saucet et al., 2015). This enabled the assessment of HR activated by individual effectors with their corresponding NLR proteins without artefactual tissue damage from the carrier. The Ws-2 ecotype containing RRS1-R recognizes wild-type PopP2 (PopP2WT), whereas the RRS1-S-containing Col-0 ecotype shows no HR with PopP2WT (Fig. 1). Mutant PopP2 (PopP2C321A), mutant AvrRps4 (AvrRps4KRVY-AAAA), and empty vector served as non-recognition negative controls which do not activate HR (Fig. 1). AvrRpt2 is known to be recognized by CC-NLR RPS2 (Axtell and Staskawicz, 2003; Mackey et al., 2003), and therefore HR was observed in all tested lines. We found that only simultaneously overexpressing RRS1-R and RPS4 can lead to the gain of recognition of PopP2 in the susceptible ecotype Col-0 (Fig. 1). No HR was observed in the rps4-2 rps4b-2 double mutant when infiltrated with Pf0-1:AvrRps4 (Fig. 1). Thus, we propose that a balanced protein expression of RRS1 and RPS4 is required for both suppressing autoimmunity and functional recognition of the corresponding effectors.
Fig. 1.
Overexpression of RPS4 and RRS1-R reconstructs the recognition of PopP2 in Col-0. Arabidopsis transgenic lines overexpressing RPS4 (OE-RPS4-HS), RRS1-R (OE-RRS1-R-HF), or both generated by crossing (OE-RPS4-HS/OE-RRS1-R-HF) in the Col-0 background, with Col-0 and Ws-2 accessions were tested for hypersensitive response (HR). Five-week-old leaves were infiltrated with Pseudomonas fluorescens Pf0-1 strains carrying empty vector (EV), wild-type (WT) AvrRps4, mutant AvrRps4KRVY-AAAA, WT PopP2, mutant PopP2C321A, and WT AvrRpt2. Leaves were collected 1 day post-infiltration (dpi) for imaging. Scale bar=1 cm. Arrows indicate reconstructed PopP2 recognition of Col-0 background overexpressing RRS1-R and RPS4. The dashed box highlights loss of AvrRps4 recognition in the double mutant rps4-2 rps4b-2. Infiltration of EV and AvrRpt2 serves as a negative and positive control of HR, respectively. (This figure is available in colour at JXB online.)
A survey of leaf-expressed genes reveals promoters for moderate and balanced expression levels of RRS1 and RPS4
Genome-wide expression profiling has revealed numerous genes altered by PTI alone or PTI plus ETI at early time points of RRS1/RPS4-mediated immune activation (Sohn et al., 2014). This analysis also enabled the discovery of genes that are moderately and constitutively expressed without changing their transcript abundance during immune activation. In plants, gene expression patterns and levels are usually specified by their promoters. Based on the endogenous expression relative transcript abundance in the ‘stable gene set’, we selected six promoters with ‘moderate’ expression (Supplementary Table S1). We define the ‘moderate’ expression based on two criteria: (i) the gene transcript abundance with those promoters is at least 100 times more than the endogenous transcript abundance of RRS1 and RPS4; and (ii) the gene transcript abundance with those promoters is lower than that with the 35S promoter. The selected genes encode proteins that are involved in essential biological processes that we expect to be expressed in most mesophyll cells, including a δ-tonoplast intrinsic protein (our name: At1, locus identifier AT3G16240, protein symbol name TIP2-1), a ribosomal protein S16 (At2, AT4G34620, RPS16-1), a cysteine synthase isomer CysC1 (At3, AT3G61440, CYSC1), a PSII subunit Q (At4, AT4G21280, PSBQ1), a xyloglucan endotransglucosylase/hydrolase 6 (At5, AT5G65730, XTH6), and a ubiquitin-like protein 5 (At6, AT5G42300, UBL5) (Supplementary Table S1).
To test the strength of the selected Arabidopsis promoters (pAt1–pAt6) for driving gene expression in planta, constructs were designed and generated to express GUS (pAt:GUS). Agrobacterium strains carrying each pAt:GUS construct were infiltrated in N. benthamiana leaves with the infiltration buffer as negative control and GUS expressed under the Cauliflower mosaic virus (CaMV) 35S promoter (35S:GUS) as positive control. GUS expressed under pAt4 shows a similar level of activity to that with 35S, whereas GUS activities detected from other pAt promoters are significantly weaker (Supplementary Fig. S1).
A T-DNA construct expresses RPS4, RRS1, and inducible AvrRps4
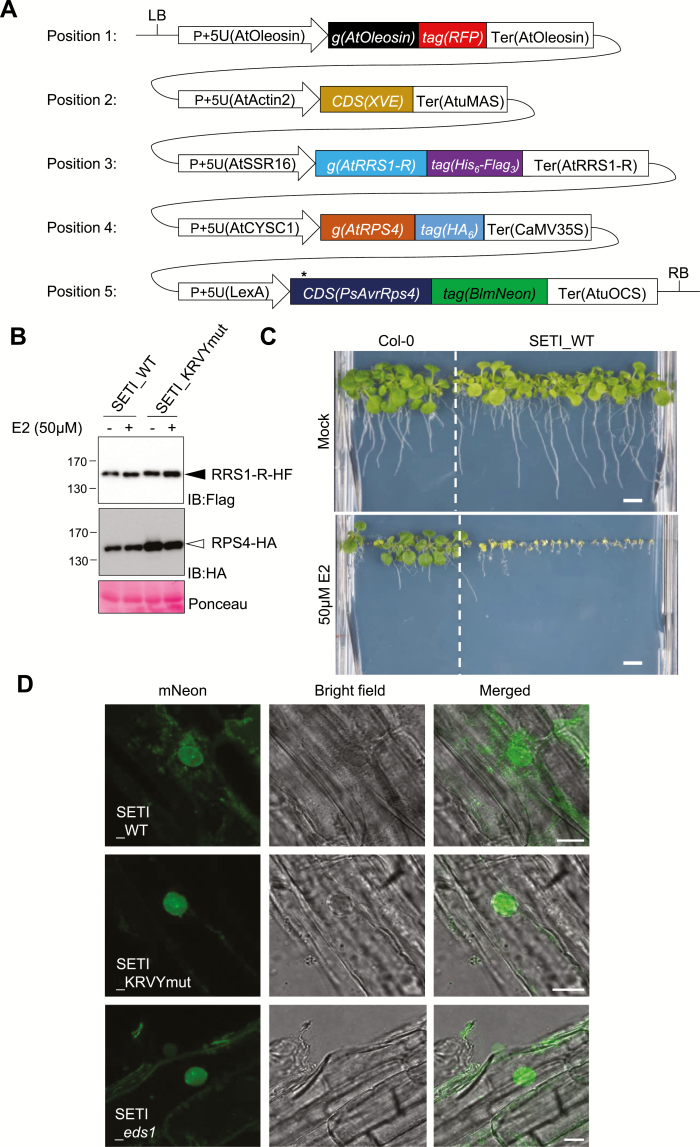
We designed a binary vector to reconstruct the effector ligand AvrRps4 and its receptors RRS1 and RPS4, using the Golden Gate Modular Cloning Toolbox (Fig. 2A) (Engler et al., 2014). We chose moderate and balanced promoters pAt2 and pAt3 from our promoter survey experiment for expressing RRS1 and RPS4, respectively. We have also cloned the RRS1-R full-length coding sequence (CDS) from Ws-2 and the RPS4 full-length CDS from Col-0 for the expression of RRS1-R and RPS4 proteins. We chose synthetic C-terminal fusion epitope tags His6-TEV-FLAG3 (HF) and HA6 for detecting RRS1 and RPS4 protein expression, respectively (Fig. 2A; Supplementary Table S2) (Gauss et al., 2005; Soleimani et al., 2013). We have used an E2-inducible system for AvrRps4 expression (Zuo et al., 2000). We named this multigene stacking binary construct ‘Super ETI’, or SETI. We have also generated constructs inducing mutant AvrRps4KRVY-AAAA or mutant AvrRps4E187A as negative controls, and named them SETI_KRVYmut and SETI_E187A, respectively. SETI_KRVYmut and SETI_E187A can induce the expression of mutant AvrRps4 alleles, but no induction of immunity because these two mutant AvrRps4 alleles cannot be recognized by RRS1 and RPS4 (Sohn et al., 2009, 2014). All restriction enzyme sites for BsaI and BpiI in modules for promoters, CDSs for genes or epitope tags, and the terminators were eliminated (Fig. 2A; Supplementary Table S4). More detailed information on the cloning can be found in Supplementary Table S3. To verify the SETI construct, we used a transient expression system in N. benthamiana by infiltrating agrobacteria that deliver the SETI T-DNA. Protein accumulation of RRS1-R-HF and RPS4-HA was detected (Supplementary Fig. S2).
Fig. 2.
Single T-DNA expresses RRS1-R-HF, RPS4-HA, and inducible wild-type AvrRps4 or AvrRps4 mutant variants. (A) Illustrative layout of the Super-ETI (SETI) construct. There are five individual expression units or Golden Gate Level 2 positional components listed, which are indicated as positions 1–5. Position 1, expression unit of the FastRed selection marker (Shimada et al., 2010). Positions 2 and 5, chimeric transactivator XVE (LexA-VP16-ER) and the corresponding LexA-inducible system to express AvrRps4 or its mutant variants under the control of β-estradiol (E2) treatment. Positions 3 and 4, full-length RRS1-R and RPS4 proteins with epitope tags His6-Flag3 and HA6, respectively. All individual units used for construct assembly can be found in Supplementary Tables S2 and S3. (B) Protein accumulation of RRS1-R-HF (IB:Flag, black arrowhead) and RPS4-HA (IB:HA, white arrowhead) of SETI lines expressing AvrRps4 (SETI_WT) or mutant AvrRps4 KRVY-AAAA (SETI_KRVYmut). Seedlings were grown in liquid culture and induced with 50 μM E2 for 2 h at 7 days after germination (DAG). Ponceau staining of Rubisco large subunits was used as a loading control. (C) Seedling phenotype of the SETI Arabidopsis transgenic line at 14 DAG in GM medium containing mock (0.1% DMSO) or 50 μM E2. Col-0 was sown as control for the effect of E2 on seedling growth. Scale bar=0.5 cm. (D) Confocal images of SETI_WT, SETI_KRVYmut, SETI_eds1 root cells expressing AvrRps4–mNeon, and AvrRps4KRVY-AAAA–mNeon induced by 50 μM E2 for 24 h. The mNeon channel shows nucleocytoplasmic localization of AvrRps4–mNeon and AvrRps4KRVY-AAAA–mNeon. Bright field channel and a merged image of mNeon and the bright field channel are shown together. Scale bars=10 μm. (This figure is available in colour at JXB online.)
The single-locus lines carrying the SETI T-DNA show inducible growth arrest
We generated transgenic Arabidopsis lines using the SETI, SETI_KRVYmut, and SETI_E187A construct expressing AvrRps4 (SETI_WT), AvrRps4KRVY-AAAA, and AvrRps4E187A, respectively. With the FastRed selection module, we have selected ~20 positive SETI_WT T1 lines. The seedlings from the T2 generation of three T1 lines were further tested for response to E2 treatment (see the Materials and methods; Supplementary Table S2). On E2-containing growth medium, SETI_WT transgenic lines display severe growth arrest (Supplementary Fig. S3). We selected one of the lines (T1-#8_T2-#4; SETI_WT) for subsequent experiments (Fig. 2C; Supplementary Fig. S3). We confirmed the protein expression of RRS1-R-HF and RPS4-HA (Fig. 2B). We also tested the expression of inducible AvrRps4–mNeon under a fluorescence microscope upon treatment with E2. mNeonGreen signal was detected at 24 h post-spray on transgenic seedlings, consistent with the mRNA accumulation of AvrRps4 at 4 h post E2 infiltration in leaves (Fig. 2D; Supplementary Fig. S2C).
RRS1-R and RPS4 form pre-activation complexes in Arabidopsis
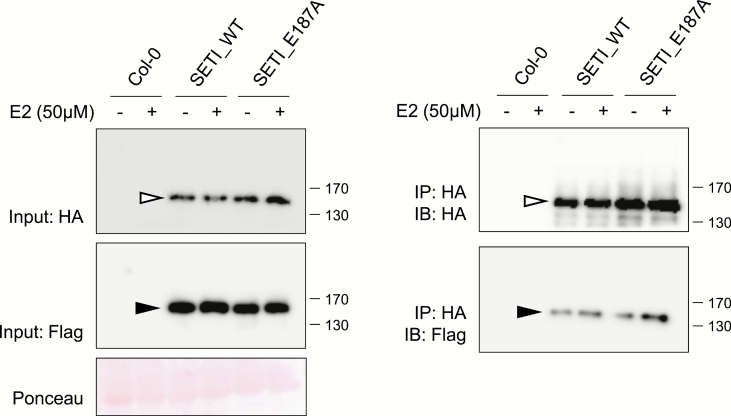
The SETI lines enable detection of epitope-tagged RRS1-R and RPS4 (Fig. 2B). We investigated in vivo interaction of tagged RRS1-R and RPS4 by co-IP with SETI_WT and SETI_E187A seedling extracts with or without E2 induction. When RPS4-HA was immunoprecipitated using HA beads, we found that RRS1-R and RPS4 stay in association with each other both before and 3 h after the induction of AvrRps4 expression (Fig. 3). There were no significant differences of RRS1-R and RPS4 association upon AvrRps4 induction. Induction of AvrRps4 E187A also had no effect on RRS1-R and RPS4 association. While all previous studies on interactions of RRS1-R and RPS4 only used the N. benthamiana transient expression system (Huh et al., 2017), generation of the SETI lines enabled the detection of RRS1-R and RPS4 interaction in their native system in Arabidopsis.
Fig. 3.
RRS1-R and RPS4 interact in vivo. Co-immunoprecipitation of RRS1-R-HF with RPS4-HA. Col-0, SETI_WT, and SETI_E187A seedlings at 7 DAG were treated with 50 μM E2 for 3 h. Crude extracts were centrifuged and RPS4-HA proteins were immunoprecipitated with anti-HA-conjugated beads. Immunoprecipitation of RPS4-HA, and co-immunoprecipitation of RRS1-R-HF were determined by immunoblot analysis with HA (IB:HA) or Flag (IB:Flag). Ponceau staining indicates equal loading of the input samples. RRS1-R-HF (black arrowhead) and RPS4-HA (white arrowhead) are indicated.
Some but not all defence responses are induced by E2 in SETI lines
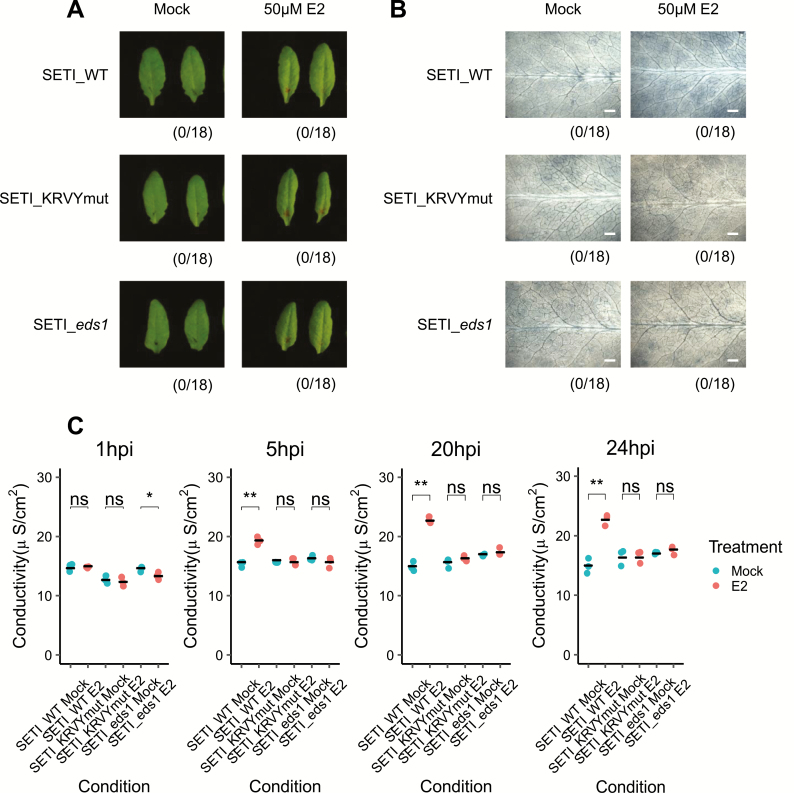
The induced expression of multiple effectors, such as AvrRpt2, AvrRpm1, and ATR13, can induce cell death or specific macroscopic HR in Arabidopsis leaves (McNellis et al., 1998; Tornero et al., 2002; Allen et al., 2004). We therefore tested whether induced expression of AvrRps4 can trigger macroscopic HR in Arabidopsis. We used SETI_eds1 as control, in which SETI_WT was crossed with the mutant eds1. EDS1 is a downstream genetic component of TIR-NLR-mediated ETI (Aarts et al., 1998; Falk et al., 1999). As seen in Fig. 4A, no HR can be observed after AvrRps4 expression is induced in the SETI leaves. However, only the expression of AvrRps4 but not that of AvrRps4_KRVYmut leads to electrolyte leakage (Fig. 4C). We also observed slightly stronger trypan blue stains in the SETI leaves treated with E2 compared with mock treatment; suggesting that the expression of AvrRps4 causes microscopic or weak but not macroscopic or strong HR in contrast to other known inducible effector-expressing lines (Fig. 4B).
Fig. 4.
Induced expression of AvrRps4 in Arabidopsis causes microscopic but not macroscopic cell death. (A) HR phenotype assay in Arabidopsis. Five-week-old SETI_WT, SETI_KRVYmut, and SETI_eds1 leaves were mock infiltrated (1% DMSO) or infiltrated with 50 μM E2. Images were taken at 1 dpi. Numbers indicate the number of leaves displaying cell death from the total number of infiltrated leaves (18 for each genotype and treatment). (B) Trypan blue staining. Five-week-old SETI_WT, SETI_KRVYmut, and SETI_eds1 were mock infiltrated (1% DMSO) or infiltrated with 50 μM E2. Leaves were stained with trypan blue solution at 1 dpi. After destaining, leaves were imaged using a stereoscopic microscope. Scale bar=0.5 mm. (C) Electrolyte leakage assay. Five-week old SETI_WT, SETI_KRVYmut, and SETI_eds1 leaves were mock infiltrated (1% DMSO) or infiltrated with 50 μM E2. Fifteen leaf discs were collected for each data point. Conductivity was measured at 1, 5, 20, and 24 hours post-infiltration (hpi). Each data point represents one technical replicate, and three technical replicates are included per treatment and genotype for one biological replicate. The black line represents the mean of the technical replicates. This experiment was repeated three times independently with similar results (Supplementary Fig. S2). Significant differences relative to the mock treatment in each genotype were calculated with t-test, and the P-values are indicated as ns (non-significant), P>0.05; *P<0.05; **P<0.01; ***P<0.001. (This figure is available in colour at JXB online.)
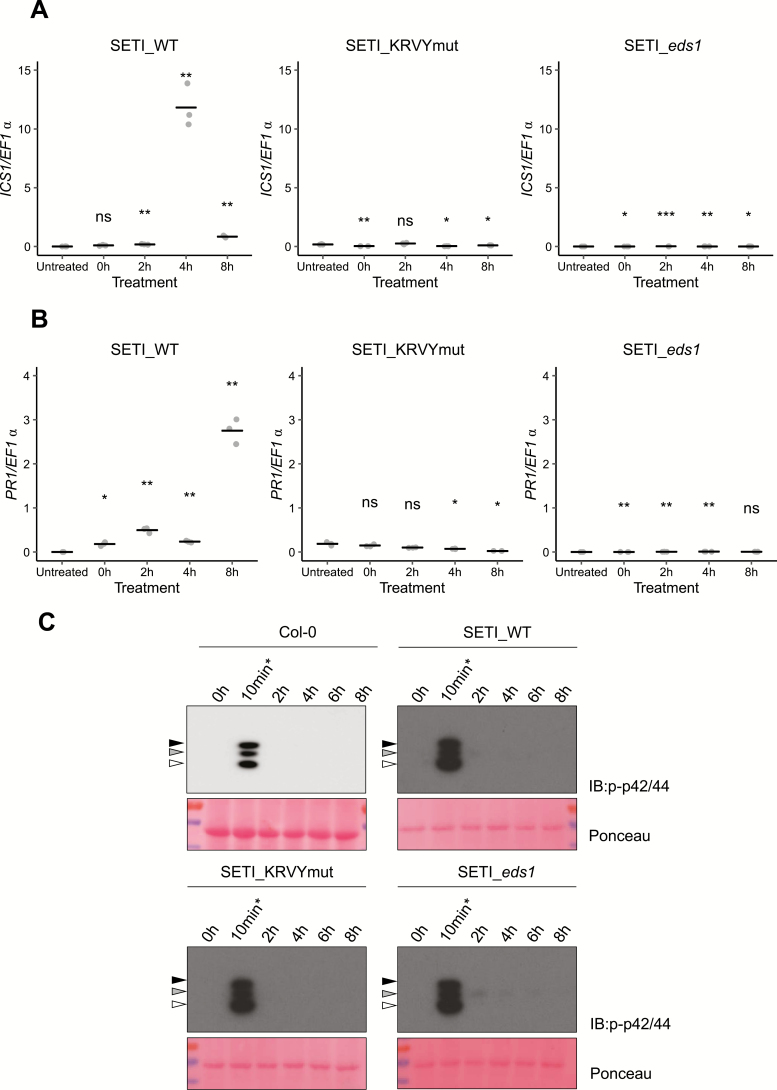
Salicylic acid induction is another hallmark of ETI (Castel et al., 2019). Enzymes such as isochorismate synthase 1 (ICS1), enhanced disease susceptibility 5 (EDS5), and AvrPphB susceptible 3 (PBS3) are involved in the biosynthesis of salicylic acid (Rekhter et al., 2019; Torrens-Spence et al., 2019), and the expression of these genes is also highly induced during ETI (Sohn et al., 2014). The expression of ICS1 after AvrRps4 induction was tested by quantitative real-time PCR. ICS1 was highly induced 4 h after the induction of AvrRps4 by E2 but not in the negative controls of SETI_KRVYmut or SETI_eds1 (Fig. 5A). In contrast, the Pathogenesis-Related protein 1 (PR1) gene was highly induced only 8 h after the induction of AvrRps4 (Fig. 5B). This shows that ETI triggered by RRS1/RPS4 is sufficient for the induction of ICS1 and the biosynthesis of salicylic acid, which subsequently leads to expression of PR1.
Fig. 5.
Induced expression of AvrRps4 in Arabidopsis leads to ICS1 and PR1 expression, but not MAPK activation. (A and B) ICS1 (A) and PR1 (B) expression after induction with E2 for 2, 4, and 8 h in SETI (left panel), SETI_KRVYmut (middle panel), and SETI_eds1 (right panel) leaf samples. Five-week-old SETI and SETI_KRVYmut leaves were infiltrated with 50 μM E2. Samples were collected at 0, 2, 4, and 8 hpi for RNA extraction and subsequent qPCR. The expression level is presented as relative to EF1α expression. Each data point represents one technical replicate. The black line represents the mean of the technical replicates. This experiment was repeated three times independently with similar results. Significant differences relative to the untreated samples were calculated with t-test, and the P-values are indicated as ns (non-significant), P>0.05; *P<0.05; **P<0.01; ***P<0.001. (C) Activation of MAPKs in Col-0, SETI_WT, SETI_KRVYmut, and SETI_eds1 seedlings by E2 induction of effector AvrRps4 or mutant AvrRps4KRVY-AAAA. Seedlings grown in liquid culture at 7 DAG were treated with 50 μM E2 for the indicated times (0, 2, 4, 6, or 8 h) and samples were collected. Col-0, SETI_WT, SETI_KRVYmut, and SETI_eds1 seedlings treated with 100 nM flg22 for 10 min (10 min*) were used as positive control. Proteins were extracted from these seedlings and phosphorylated MAPKs were detected using p-p42/44 antibodies. Arrowheads indicate phosphorylated MAPKs (black, pMPK6; grey, pMPK3; white, pMPK4/11). Ponceau staining was used as loading control. (This figure is available in colour at JXB online.)
Activation of MAPKs by PTI has been reported in many cases, and happens within a few minutes of the activation of PTI. However, the activation of MAPKs by ETI is slower and lasts longer than PTI-induced MAPK activation (Tsuda et al., 2013). We tested whether the induced expression of AvrRps4 can lead to MAPK activation in SETI_WT and control lines Col-0, SETI_KRVYmut, and SETI_eds1. Treatment with flg22 for 10 min triggered phosphorylation of MAPKs (Fig. 5C). However, in contrast to AvrRpt2-inducible transgenic Arabidopsis plants (Tsuda et al., 2013), induced expression of AvrRps4 does not activate MAPKs (Fig. 5C).
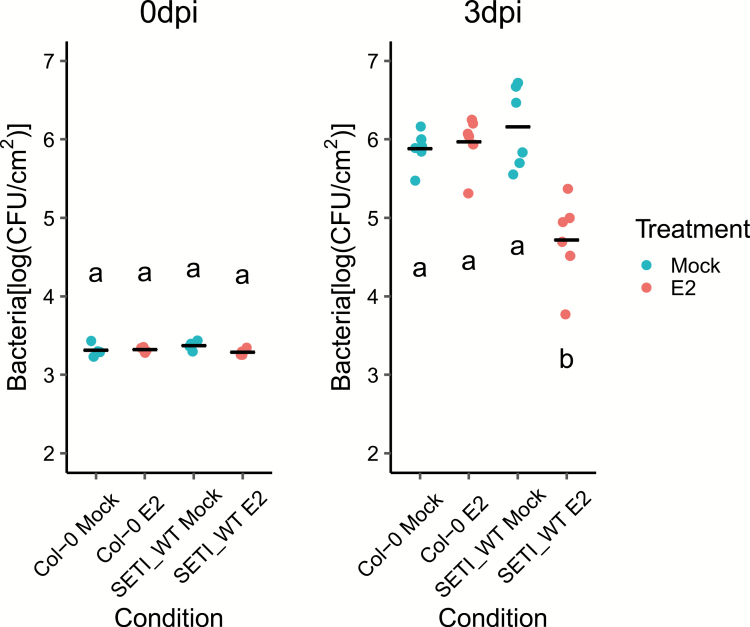
We further tested if the induction of ETI would elevate resistance. We infiltrated the leaves with E2 or mock solution 1 d before we infiltrated plants with Pst DC3000 (see the Materials and methods). SETI_WT plants pre-treated with E2 are more resistant to the bacteria than those mock pre-treated, while there was no significant difference between E2 and mock pre-treatment in Col-0 (Fig. 6).
Fig. 6.
Effector-triggered immunity triggered by the expression of AvrRps4 leads to resistance against Pseudomonas syringae pv. tomato strain DC3000. Five-week-old SETI_WT and Col-0 leaves were mock infiltrated (1% DMSO) or infiltrated with 50 μM E2. At 1 dpi, leaves were inoculated with Pst DC3000 (OD600=0.001). Bacteria in the leaves were then quantified as colony-forming units (CFU) at 0 and 3 dpi. Each data point represents two leaves collected from one individual plant. Samples from four individual plants were collected for 0 dpi and samples from six individual plants were collected for 3 dpi. The black line represents the mean of the technical replicates. This experiment was repeated three times independently with similar results. Biological significance of the values was determined by one-way ANOVA followed by post-hoc Tukey HSD analysis. Letters above the data points indicate significant differences (P<0.05). (This figure is available in colour at JXB online.)
Discussion
To facilitate studying the functional complex of RRS1 and RPS4 in vivo, we generated an expression construct of E2-inducible AvrRps4 stacked with epitope-tagged RRS1 and RPS4. To achieve balanced expression levels higher than endogenous expression of RRS1 and RPS4, we surveyed constitutively expressed gene promoters. Here, we report six new and tested promoter modules that are compatible with the Golden Gate Modular Cloning Toolbox. We used two of the promoters to express RRS1 and RPS4, and we avoided autoimmunity induced by excessive expression of RPS4 or non-recognition of PopP2 caused by excessive expression of RRS1-R. We were also able to generate inducible AvrRps4 expression to activate RRS1/RPS4-mediated ETI under the control of E2 treatment. We thus were able to stack genes for inducible expression of a pathogen effector and its NLR receptors in one construct. In addition, with the epitope tags, we are able to monitor effector-dependent changes in the NLR proteins without interference from using a pathogen effector delivery system. We could thus express any effectors or pathogen ligands that will trigger immunity in plant cells with the E2-inducible module, and their immune receptors using the same gene stacking strategy.
There are multiple advantages to enabling investigation of ETI without the complication of co-activating PTI. First, we could test the contribution of other genes to ETI activation by introducing mutants into the SETI background, either using conventional crossing or using genome editing such as CRISPR/Cas9 [clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9]. These lines can also help in investigating downstream signalling from plant NLRs. Multiple forward genetic screens have been conducted, but few novel components have been found, and most mutations are in either the NLRs or regulatory elements rather than signalling components (van Wersch et al., 2016). Another plausible explanation is that the signalling path downstream of plant NLRs is very short, but this is debatable because several significant steps are required for immunity. EDS1, PAD4, and SAG101 are required for TIR–NLR signalling (Falk et al., 1999; Gantner et al., 2019). NRC family proteins in Solanaceae species are required for many NLRs, NRG1/ADR1s in Arabidopsis are required for TIR-NLRs, and ADR1s are required for some CC-NLRs (Bonardi et al., 2011; Dong et al., 2016; Wu et al., 2017; Castel et al., 2019; Wu et al., 2019). NRG1s and ADR1s seem to function downstream of EDS1 and may function distinctly with SAG101 and PAD4, respectively (Lapin et al., 2019). SETI lines carry heterologously expressed RRS1-R/RPS4 and also endogenous RRS1-S/RPS4 and RRS1B/RPS4B, which together provide three redundant copies of NLR pairs that can recognize AvrRps4. In theory, in an ethyl methanesulfonate (EMS) mutagenesis forward genetic screen to identify suppressors of immunity induced by AvrRps4, there should be a reduced background of mutations in the receptor(s), improving prospects to reveal mutations in genes that are functionally important in NLR signalling and regulation.
With SETI, we are able to assess the pure ETI response mediated by the TIR-NLRs, RRS1 and RPS4. E2 induction provoked rapid transcriptional changes in activation of defence genes and also ion leakage. AvrRps4-induced ETI-enhanced resistance against bacterial pathogens; but neither MAPK activation nor macroscopic HR, in contrast to other inducible ETI examples (Tornero et al., 2002; Tsuda et al., 2013). This indicates that outputs of plant NLRs might differ. Both TIR and CC domains alone are sufficient to activate plant immunity. However, whether they signal through similar or different downstream components is still unknown.
In diverse multicellular eukaryotes, immune complexes are assembled into oligomeric complexes to signal downstream. The mammalian inflammasome, assembled in response to bacterial peptide recognition by NAIP proteins and subsequent activation and binding of NLRC4 proteins, is a classic example (Zhang et al., 2015). The plant CC-NLR ZAR1 forms an effector-dependent resistosome, which is a pentamer of ZAR1 assembled together with cofactors PBL2 and RKS1 (Wang et al., 2019). The structure of TIR domains implies that activation might require the disassociation of the RRS1 and RPS4 TIR domains and the oligomerization of RPS4 TIR domains (Williams et al., 2014). In SETI lines, RRS1 and RPS4 form a pre-activation complex in the absence of pathogen effector. However, co-IP data cannot distinguish the ratio with which RRS1 and RPS4 bind to each other. It will be interesting to check via various non-denaturing methods if RRS1-R and RPS4 form a dimer or a higher order oligomer in vivo, or whether there is a conformational change in the complex upon effector recognition. Furthermore, with the SETI lines generated in this study, we can ask what other cofactors are required for the activation of RRS1-R and RPS4 under native conditions.
The availability of SETI lines will also enable us to study how PTI and ETI interact with each other, especially in the context of RRS1- and RPS4-mediated immunity. Some models have been proposed in discussing this topic (Tsuda et al., 2009; Cui et al., 2015). From the zig–zag model, PTI and ETI have different thresholds for activation of immunity (Jones and Dangl, 2006). With the SETI line, we could specifically ask how PTI and ETI can physically influence each other. A lot of evidence shows that the PTI receptors PRRs usually have very specific post-translational modification (PTM) events at early time points; there is also some evidence showing that ETI can activate somewhat overlapping but different PTMs on immune-related proteins (Withers and Dong, 2017; Kadota et al., 2019). It will be interesting to know how the activation of RRS1/PRS4 leads to the changes of PTMs and how those changes contribute to the robustness of immunity. In addition, transcriptional changes are not the only process reported as the early changes of ETI, but also changes in translation (Meteignier et al., 2017; Yoo et al., 2020). Work using inducible AvrRpm1 or AvrRpt2 reveals interesting observations on the trade-off between defence and growth, and the specific regulatory element in the genome (Meteignier et al., 2017; Yoo et al., 2020). Both effectors are recognized by CC-type NLRs, so it will be interesting to know what changes in translation will be induced by TIR-NLRs using the SETI line. One can also use proteomics tools to generate complex information using inducible SETI to fish for ETI-specific interaction networks.
Recently it has been shown that plant NLRs can also form higher order protein complexes, similar to the inflammasome in the mammalian immune system. However, it is unknown if all plant NLRs form the same kind of complex or use the same mechanism to activate defence. It was noted that NLRs have evolved to partner with other NLRs to function genetically, but whether this model is also true biochemically is still unknown (Adachi et al., 2019). Unlike ZAR1, RRS1 and RPS4 require each other to function, and they localize and function exclusively in the nuclei but not the cell membrane, so it will be interesting to compare them once the cryo-EM structure of the RRS1 and RPS4 complex is resolved. The SETI line could be a very good toolkit for mutagenesis to verify the function based on the structural information.
We have observed that the activation of ETI alone in the absence of pathogens is sufficient to prime the resistance against bacterial pathogens in Arabidopsis (Fig. 6). Previously, we have reported that a group of up-regulated genes at the early time point of activation of RRS1-R/RPS4 are related to the salicylic acid pathway, so it will be interesting to determine whether the elevated or primed resistance against bacteria induced in SETI lines is due to the activation of the salicylate pathway (Sohn et al., 2014).
Another major question regarding the signalling pathways is that SAG101 and PAD4 seem to be redundant but functionally equivalent to EDS1 (Wagner et al., 2013; Lapin et al., 2019). They also have been shown to be genetically linked to helper NLRs, NRG1s and/or ADR1s, to function (Castel et al., 2019; Wu et al., 2019). Using the SETI line, one can test their function more specifically in ETI in the absence of PTI and interference by many other unwanted pathogens.
Supplementary data
Supplementary data are available at JXB online.
Table S1. Information on synthetic promoters used in this study.
Table S2. Golden Gate stacking construct of R genes and effectors.
Table S3. Golden Gate cloning modules used in this work.
Table S4. Primers used in this study.
Table S5. Statistical analysis results.
Fig. S1. GUS-staining activity of synthetic promoters in N. benthamiana.
Fig. S2. Transient expression of Super-ETI (SETI) constructs in N. benthamiana.
Fig. S3. SETI T2 lines grown under E2 treatment.
Acknowledgements
We thank the Gatsby Foundation (UK) for funding to the JDGJ laboratory. BN was supported by the Norwich Research Park (NRP) Biosciences Doctoral Training Partnership (DTP) from the Biotechnology and Biological Sciences Research Council (BBSRC) (grant agreement: BB/M011216/1). HA were supported by a European Research Council Advanced Grant ‘ImmunitybyPairDesign’ (grant agreement: 669926). PD acknowledges support from the European Union’s Horizon 2020 Research and Innovation Program under Marie Skłodowska-Curie Actions (grant agreement: 656243) and a Future Leader Fellowship from BBSRC (grant agreement: BB/R012172/1). AR acknowledges support from an EMBO Long Term Fellowship (grant agreement: ALTF-842-2015). HB was supported by the NRP DTP funding from BBSRC (grant agreement: BB/M011216/1). YM was supported by a BBSRC grant (grant agreement: BB/M008193/1). LT, MY, and JDGJ were supported by the Gatsby Foundation funding to the Sainsbury Laboratory.
References
- Aarts N, Metz M, Holub E, Staskawicz BJ, Daniels MJ, Parker JE. 1998. Different requirements for EDS1 and NDR1 by disease resistance genes define at least two R gene-mediated signaling pathways in Arabidopsis. Proceedings of the National Academy of Sciences, USA 95, 10306–10311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adachi H, Derevnina L, Kamoun S. 2019. NLR singletons, pairs, and networks: evolution, assembly, and regulation of the intracellular immunoreceptor circuitry of plants. Current Opinion in Plant Biology 50, 121–131. [DOI] [PubMed] [Google Scholar]
- Allen RL, Bittner-Eddy PD, Grenville-Briggs LJ, Meitz JC, Rehmany AP, Rose LE, Beynon JL. 2004. Host–parasite coevolutionary conflict between Arabidopsis and downy mildew. Science 306, 1957–1960. [DOI] [PubMed] [Google Scholar]
- Axtell MJ, Staskawicz BJ. 2003. Initiation of RPS2-specified disease resistance in Arabidopsis is coupled to the AvrRpt2-directed elimination of RIN4. Cell 112, 369–377. [DOI] [PubMed] [Google Scholar]
- Bonardi V, Tang S, Stallmann A, Roberts M, Cherkis K, Dangl JL. 2011. Expanded functions for a family of plant intracellular immune receptors beyond specific recognition of pathogen effectors. Proceedings of the National Academy of Sciences, USA 108, 16463–16468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castel B, Ngou PM, Cevik V, Redkar A, Kim DS, Yang Y, Ding P, Jones JDG. 2019. Diverse NLR immune receptors activate defence via the RPW8-NLR NRG1. New Phytologist 222, 966–980. [DOI] [PubMed] [Google Scholar]
- Cui H, Tsuda K, Parker JE. 2015. Effector-triggered immunity: from pathogen perception to robust defense. Annual Review of Plant Biology 66, 487–511. [DOI] [PubMed] [Google Scholar]
- Dong OX, Tong M, Bonardi V, El Kasmi F, Woloshen V, Wünsch LK, Dangl JL, Li X. 2016. TNL-mediated immunity in Arabidopsis requires complex regulation of the redundant ADR1 gene family. New Phytologist 210, 960–973. [DOI] [PubMed] [Google Scholar]
- Engler C, Youles M, Gruetzner R, Ehnert TM, Werner S, Jones JD, Patron NJ, Marillonnet S. 2014. A golden gate modular cloning toolbox for plants. ACS Synthetic Biology 3, 839–843. [DOI] [PubMed] [Google Scholar]
- Falk A, Feys BJ, Frost LN, Jones JD, Daniels MJ, Parker JE. 1999. EDS1, an essential component of R gene-mediated disease resistance in Arabidopsis has homology to eukaryotic lipases. Proceedings of the National Academy of Sciences, USA 96, 3292–3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gantner J, Ordon J, Kretschmer C, Guerois R, Stuttmann J. 2019. An EDS1–SAG101 complex is essential for TNL-mediated immunity in Nicotiana benthamiana. The Plant Cell 31, 2456–2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauss R, Trautwein M, Sommer T, Spang A. 2005. New modules for the repeated internal and N-terminal epitope tagging of genes in Saccharomyces cerevisiae. Yeast 22, 1–12. [DOI] [PubMed] [Google Scholar]
- Heidrich K, Tsuda K, Blanvillain-Baufumé S, Wirthmueller L, Bautor J, Parker JE. 2013. Arabidopsis TNL-WRKY domain receptor RRS1 contributes to temperature-conditioned RPS4 auto-immunity. Frontiers in Plant Science 4, 403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh SU, Cevik V, Ding P, Duxbury Z, Ma Y, Tomlinson L, Sarris PF, Jones JDG. 2017. Protein–protein interactions in the RPS4/RRS1 immune receptor complex. PLoS Pathogens 13, e1006376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JD, Dangl JL. 2006. The plant immune system. Nature 444, 323–329. [DOI] [PubMed] [Google Scholar]
- Kadota Y, Liebrand TWH, Goto Y, et al. 2019. Quantitative phosphoproteomic analysis reveals common regulatory mechanisms between effector- and PAMP-triggered immunity in plants. New Phytologist 221, 2160–2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapin D, Kovacova V, Sun X, et al. 2019. A coevolved EDS1–SAG101–NRG1 module mediates cell death signaling by TIR-domain immune receptors. The Plant Cell 31, 2430–2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Mackey D, Belkhadir Y, Alonso JM, Ecker JR, Dangl JL. 2003. Arabidopsis RIN4 is a target of the type III virulence effector AvrRpt2 and modulates RPS2-mediated resistance. Cell 112, 379–389. [DOI] [PubMed] [Google Scholar]
- McNellis TW, Mudgett MB, Li K, Aoyama T, Horvath D, Chua NH, Staskawicz BJ. 1998. Glucocorticoid-inducible expression of a bacterial avirulence gene in transgenic Arabidopsis induces hypersensitive cell death. The Plant Journal 14, 247–257. [DOI] [PubMed] [Google Scholar]
- Meteignier LV, El Oirdi M, Cohen M, Barff T, Matteau D, Lucier JF, Rodrigue S, Jacques PE, Yoshioka K, Moffett P. 2017. Translatome analysis of an NB-LRR immune response identifies important contributors to plant immunity in Arabidopsis. Journal of Experimental Botany 68, 2333–2344. [DOI] [PubMed] [Google Scholar]
- Narusaka M, Shirasu K, Noutoshi Y, Kubo Y, Shiraishi T, Iwabuchi M, Narusaka Y. 2009. RRS1 and RPS4 provide a dual Resistance-gene system against fungal and bacterial pathogens. The Plant Journal 60, 218–226. [DOI] [PubMed] [Google Scholar]
- Porter K, Shimono M, Tian M, Day B. 2012. Arabidopsis actin-depolymerizing factor-4 links pathogen perception, defense activation and transcription to cytoskeletal dynamics. PLoS Pathogens 8, e1003006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rekhter D, Lüdke D, Ding Y, Feussner K, Zienkiewicz K, Lipka V, Wiermer M, Zhang Y, Feussner I. 2019. Isochorismate-derived biosynthesis of the plant stress hormone salicylic acid. Science 365, 498–502. [DOI] [PubMed] [Google Scholar]
- Sarris PF, Duxbury Z, Huh SU, et al. 2015. A plant immune receptor detects pathogen effectors that target WRKY transcription factors. Cell 161, 1089–1100. [DOI] [PubMed] [Google Scholar]
- Saucet SB, Ma Y, Sarris PF, Furzer OJ, Sohn KH, Jones JD. 2015. Two linked pairs of Arabidopsis TNL resistance genes independently confer recognition of bacterial effector AvrRps4. Nature Communications 6, 6338. [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, et al. 2012. Fiji: an open-source platform for biological-image analysis. Nature Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner NC, Lambert GG, Chammas A, et al. 2013. A bright monomeric green fluorescent protein derived from Branchiostoma lanceolatum. Nature Methods 10, 407–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada TL, Shimada T Hara-Nishimura I. 2010. A rapid and non‐destructive screenable marker, FAST, for identifying transformed seeds of Arabidopsis thaliana. The Plant Journal 61, 519–528. [DOI] [PubMed] [Google Scholar]
- Sohn KH, Hughes RK, Piquerez SJ, Jones JD, Banfield MJ. 2012. Distinct regions of the Pseudomonas syringae coiled-coil effector AvrRps4 are required for activation of immunity. Proceedings of the National Academy of Sciences, USA 109, 16371–16376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohn KH, Segonzac C, Rallapalli G, Sarris PF, Woo JY, Williams SJ, Newman TE, Paek KH, Kobe B, Jones JD. 2014. The nuclear immune receptor RPS4 is required for RRS1SLH1-dependent constitutive defense activation in Arabidopsis thaliana. PLoS Genetics 10, e1004655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohn KH, Zhang Y, Jones JD. 2009. The Pseudomonas syringae effector protein, AvrRPS4, requires in planta processing and the KRVY domain to function. The Plant Journal 57, 1079–1091. [DOI] [PubMed] [Google Scholar]
- Soleimani VD, Palidwor GA, Ramachandran P, Perkins TJ, Rudnicki MA. 2013. Chromatin tandem affinity purification sequencing. Nature Protocols 8, 1525–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas WJ, Thireault CA, Kimbrel JA, Chang JH. 2009. Recombineering and stable integration of the Pseudomonas syringae pv. syringae 61 hrp/hrc cluster into the genome of the soil bacterium Pseudomonas fluorescens Pf0-1. The Plant Journal 60, 919–928. [DOI] [PubMed] [Google Scholar]
- Tornero P, Chao RA, Luthin WN, Goff SA, Dangl JL. 2002. Large-scale structure–function analysis of the Arabidopsis RPM1 disease resistance protein. The Plant Cell 14, 435–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torrens-Spence MP, Bobokalonova A, Carballo V, Glinkerman CM, Pluskal T, Shen A, Weng JK. 2019. PBS3 and EPS1 complete salicylic acid biosynthesis from isochorismate in Arabidopsis. Molecular Plant 12, P1577–P1586. [DOI] [PubMed] [Google Scholar]
- Tsuda K, Mine A, Bethke G, Igarashi D, Botanga CJ, Tsuda Y, Glazebrook J, Sato M, Katagiri F. 2013. Dual regulation of gene expression mediated by extended MAPK activation and salicylic acid contributes to robust innate immunity in Arabidopsis thaliana. PLoS Genetics 9, e1004015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda K, Sato M, Stoddard T, Glazebrook J, Katagiri F. 2009. Network properties of robust immunity in plants. PLoS Genetics 5, e1000772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wersch R, Li X, Zhang Y. 2016. Mighty dwarfs: Arabidopsis autoimmune mutants and their usages in genetic dissection of plant immunity. Frontiers in Plant Science 7, 1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner S, Stuttmann J, Rietz S, Guerois R, Brunstein E, Bautor J, Niefind K, Parker JE. 2013. Structural basis for signaling by exclusive EDS1 heteromeric complexes with SAG101 or PAD4 in plant innate immunity. Cell Host & Microbe 14, 619–630. [DOI] [PubMed] [Google Scholar]
- Wang J, Hu M, Wang J, et al. 2019. Reconstitution and structure of a plant NLR resistosome conferring immunity. Science 364, eaav5870. [DOI] [PubMed] [Google Scholar]
- Wickham H. 2016. ggplot2—elegant graphics for data analysis. New York: Springer-Verlag New York. [Google Scholar]
- Williams SJ, Sohn KH, Wan L, et al. 2014. Structural basis for assembly and function of a heterodimeric plant immune receptor. Science 344, 299–303. [DOI] [PubMed] [Google Scholar]
- Withers J, Dong X. 2017. Post-translational regulation of plant immunity. Current Opinion in Plant Biology 38, 124–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CH, Abd-El-Haliem A, Bozkurt TO, Belhaj K, Terauchi R, Vossen JH, Kamoun S. 2017. NLR network mediates immunity to diverse plant pathogens. Proceedings of the National Academy of Sciences, USA 114, 8113–8118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Li M, Dong OX, Xia S, Liang W, Bao Y, Wasteneys G, Li X. 2019. Differential regulation of TNL-mediated immune signaling by redundant helper CNLs. New Phytologist 222, 938–953. [DOI] [PubMed] [Google Scholar]
- Yoo H, Greene GH, Yuan M, Xu G, Burton D, Liu L, Marqués J, Dong X. 2020. Translational regulation of metabolic dynamics during effector-triggered immunity. Molecular Plant 13, 88–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Chen S, Ruan J, et al. 2015. Cryo-EM structure of the activated NAIP2–NLRC4 inflammasome reveals nucleated polymerization. Science 350, 404–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang ZM, Ma KW, Gao L, Hu Z, Schwizer S, Ma W, Song J. 2017. Mechanism of host substrate acetylation by a YopJ family effector. Nature Plants 3, 17115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo J, Niu QW, Chua NH. 2000. An estrogen receptor-based transactivator XVE mediates highly inducible gene expression in transgenic plants. The Plant Journal 24, 265–273. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.