Abstract
Genes that provide resistance to fungi and/or bacteria usually reduce plant growth and ultimately affect grain yield. Thus, crop breeding programs need to find genetic resources that balance disease resistance with growth. The receptor kinase FERONIA regulates cell growth and survival in Arabidopsis. Here, we investigate, in rice, the role of members of the FERONIA-like receptor (FLR) gene family in the balance between growth and the response to the fungal pathogen Magnaporthe oryzae (Pyricularia oryzae), which causes the most devastating disease in rice. We carried out genome-wide gene expression and functional screenings in rice via a gene knockout strategy, and we successfully knocked out 14 FLR genes in rice. Using these genetic resources, we found that mutations in the FLR2 and FLR11 genes provide resistance to rice blast without a profound growth penalty. Detailed analyses revealed that FLR2 mutation increased both defense-related gene expression and M. oryzae-triggered production of reactive oxygen species. Thus, our results highlight novel genetic tools for studying the underlying molecular mechanisms of enhancing disease resistance without growth penalty.
Keywords: FERONIA, FLR, Magnaporthe oryzae, rice, ROS), stress responses
The FERONIA-like receptor (FLR) is essential for the response to Magnaporthe oryzae in rice, and mutations in FLR2 and FLR11 enhance resistance in the absence of significantly growth penalty.
Introduction
Genetic immunity to disease is usually accompanied by unexpected reductions in growth and productivity (Ning et al., 2017). Promising new crop varieties (for example in rice) should have a sufficient balance between plant disease resistance and economic characteristics. Thus, finding genetic resources that balance disease resistance with growth is needed for crop breeding.
Plants perceive and process external and internal information to form coordinated responses, which are critical to adapt to environmental conditions. Most of the information is processed by plasma membrane (PM)-localized receptor-like kinases (RLKs), which perceive signals via an N-terminal extracellular domain and activate downstream signaling cascades via a C-terminal intracellular kinase domain (Liao et al., 2017; Franck et al., 2018). Members of the RLK1-like (CrRLK1Ls) subfamily, which are also known as malectin RLKs, were first discovered in Catharanthus roseus (Schulze-Muth et al., 1996) and found to be conserved in green plants. Malectin, first described in Xenopus laevis, plays a role in recognizing Glc2-N-glycan (Schallus et al., 2008). There are 17 CrRLK1L members in Arabidopsis, each of which contains an extracellular malectin-like domain (MLD), a transmembrane domain, and a cytoplasmic kinase domain (Lindner et al., 2012). FERONIA (FER), a member of the CrRLK1L subfamily, and its relatives are promising candidates that are capable of balancing disease resistance with yield and growth. FER and other CrRLK1L members play essential role in cell growth, seed yield, and stress responses in plants (Liao et al., 2017; Franck et al., 2018). FER loss-of-function mutants present distorted trichomes, relatively short root hairs (Duan et al., 2010), and box-shaped epidermal cells (Li et al., 2015) in Arabidopsis, and exhibit semi-dwarf plant height (Li et al., 2016; Pu et al., 2017) in rice. FER is involved in the crosstalk between several hormone pathways that regulate plant growth. For example, FER regulates auxin-stimulated root hair elongation (Duan et al., 2010) and mediates the crosstalk between abscisic acid (ABA) and rapid alkalinization factor1 (RALF1, a ligand of FER) to modulate root growth under stress conditions (Yu et al., 2012). FER has also been implicated in ethylene- (Mao et al., 2015) and brassinosteroid-mediated (Guo et al., 2009) responses that control cell growth. One of the downstream responses of the RALF1–FER pathway is the recruitment of RIPK, which inhibits H+ pump activity to increase the pH and inhibit cell growth in roots (Pearce et al., 2001; Haruta et al., 2008, 2014; Du et al., 2016). Recently, EBP1 was identified as a downstream factor in the RALF1–FER pathway and was shown to regulate nuclear gene expression and cell size (Li et al., 2018). Relatives of the FER gene family also affect Arabidopsis growth; THESEUS1 (THE1) and HERCULES1 (HERK1) act redundantly to control cell elongation, as the1 and herk1 single mutants exhibit no defective vegetative phenotype, while the leaves and stems of the1/herk1 double mutants and the1/herk1/herk2 triple mutants exhibit cell elongation defects (Guo et al., 2009). ERULUS/[Ca2+]cyt-associated protein kinase (ERU/CAP1) and CURVY1 (CVY1) mutants have short root hairs and box-shaped pavement cells, respectively (Bai et al., 2014; Gachomo et al., 2014). ANXUR1 (ANX1) and ANX2 are preferentially expressed in pollen, and control pollen tube growth and rupture by interacting with BUDDHA PAPER SEAL1 (BUPS1) and BUPS2 via ectodomains and via the formation of complexes (Boisson-Dernier et al., 2009; Miyazaki et al., 2009; Ge et al., 2017). During plant reproductive development, RALF4 and RALF19 directly interact with the ANX1/2–BUPS1/2 complex to regulate pollen tube integrity and growth, and RALF34 can subsequently replace RALF4 and RALF19 to bind with ANX1/2–BUPS1/2 and induce pollen tube rupture and sperm release when the pollen tube enters the ovule (Ge et al., 2017; Mecchia et al., 2017). In rice, the FER homologs FLR1 and FLR2 redundantly control plant height, tillering, branching, and fertility (Li et al., 2016; Pu et al., 2017); the last three are critical factors that control grain yield (Gravois and McNew, 1993). Ruptured pollen tube (RUPO), a relative of the FLR1/2 gene family, is expressed specifically in pollen and regulates pollen tube growth and integrity (Liu et al., 2016), indicating that FER and its homologs are highly conserved signaling molecules that are involved in plant cell growth and yield.
FER also affects the immune response of Arabidopsis. FER mutations have conferred increased resistance to the pathogens Golovinomyces (syn. Erysiphe) orontii (Kessler et al., 2010) and Fusarium oxysporum (Masachis et al., 2016), but also increased susceptibility to Hyaloperonospora arabidopsidis and Colletotrichum higginsianum (Kessler et al., 2010). Recently, FER was confirmed to be essential for pathogen-associated molecular pattern (PAMP)-triggered immunity (PTI). FER is required for flg22-, elf18-, and chitin-triggered reactive oxygen species (ROS) production. Moreover, Arabidopsis RALF23 inhibits plant immunity via recruiting FER and inhibiting pattern recognition receptor (PRR) complex formation (Stegmann et al., 2017). Fungal RALF mimics have been identified in a wide range of plant pathogens and in some bacteria (Masachis et al., 2016; Thynne et al., 2017). The pathogenic fungus F. oxysporum can secrete RALF1-like proteins (F-RALFs), which can bind with FER to negatively regulate H+ pump activity and stimulate apoplastic alkalinization to suppress plant immunity and facilitate pathogenicity (Masachis et al., 2016). FER can associate with FLS2 and its co-receptor BAK1 (Stegmann et al., 2017), which can form complexes to detect flg22 and interfere with the assembly of FLS2–BAK1 complexes to modulate plant PTI signaling. Furthermore, ANX1 can associate with the leucine-rich repeat (NLR) proteins RPS2 and RPM1, which have a nucleotide-binding domain, to accelerate RPS2 degradation and inhibit RPS2-regulated cell death. Thus, ANX1 and ANX2 negatively control both plant PTI and effector-triggered immunity (ETI) (Mang et al., 2017). THE1 can interact with GUANINE EXCHANGE FACTOR 4 (GEF4) to positively mediate defense responses against the necrotrophic fungus Botrytis cinerea (Qu et al., 2017). However, another study verified that THE1 acts upstream of the mechanosensitive Ca2+ channel MATING PHEROMONE INDUCED DEATH 1 (MID1)-COMPLEMENTING ACTIVITY 1 (MCA1) and the RLK EFI2 to regulate cell wall integrity signaling, but is not required for PTI (Engelsdorf et al., 2018).
Our previous study showed that FLR1 and FLR2 control reproductive development in rice (Li et al., 2016), but the function of FLRs in vegetative growth and stress responses in rice is largely unknown. Here, we addressed this unknown by functionally characterizing the FLR gene family in rice.
Materials and methods
Identification of FLR genes in rice
The genome, transcript, DNA coding sequence (CDS), and peptide sequence data of wild rice (Oryza rufipogon) and Oryza sativa indica were downloaded from the Ensembl Plants database (http://plants.ensembl.org/index.html). The genomic data of O. sativa japonica Nipponbare (Nip) were downloaded from the Rice Genome Annotation Project database (RGAP; http://rice.plantbiology.msu.edu/). Seventeen Arabidopsis CrRLK1L (including FER and its relatives) protein sequences (Lindner et al., 2012) were downloaded from TAIR (ftp://ftp.arabidopsis.org). We used two methods to identify potential CrRLK1L genes in rice: (i) the sequences of the MLD PF12819 and the Pkinase domain PF07714 were downloaded from the Pfam website (https://pfam.xfam.org) and used as queries to scan rice protein databases with HMMER version 3.0; and (ii) 17 Arabidopsis CrRLK1L protein sequences were used as queries for the Basic Local Alignment Search Tool (BLAST) for searching against rice protein databases for potential CrRLK1L genes. The candidate sequences were subsequently confirmed by the Conserved Domain Database (CDD) tool within the NCBI (https://www.ncbi.nlm.nih.gov/cdd) database. Finally, each putative CrRLK1L protein sequence was manually examined via the Simple Modular Architecture Research Tool (SMART; http://smart.embl-heidelberg.de/) to ensure that it contained both the conserved MLD and Pkinase domain.
Phylogenetic tree construction and motif composition analysis
Five Physcomitrella patens CrRLK1L protein sequences (Rensing et al., 2008; Lehti-Shiu and Shiu, 2012) and two Selaginella moellendorffii CrRLK1L protein sequences (Banks et al., 2011; Lehti-Shiu and Shiu, 2012) were downloaded from Phytozome version 7.0 (https://phytozome.jgi.doe.gov/). CrRLK1L protein sequences were aligned via ClustalW software, with the default parameters (http://www.clustal.org/clustal2/), and a phylogenetic tree based on the Neighbor–Joining (NJ) method was constructed using MEGA 7.0 in accordance with the following parameters: 1000 bootstrap replications, the p-distance model, uniform rates among sites, and partial deletion (95% site coverage) for gaps and missing data. The phylogenetic tree was then optimized via iTOL (https://itol.embl.de/). Motifs were subsequently constructed via MEME (http://meme-suite.org/) by the use of the complete amino acid sequences of CrRLK1L in accordance with the following parameters: number of repetitions, any; maximum number of motifs, 20; and optimum width of each motif, 6–50 residues (Bailey et al., 2009).
Gene sequence analysis of FLR genes in rice
The chromosome positions of the FLR genes were confirmed by the rice gene annotation gff3 file downloaded from the RGAP database (http://rice.plantbiology.msu.edu/), and the FLR genes were mapped to O. sativa japonica chromosomes via Map Gene 2 Chromosome V2 (MG2C; http://mg2c.iask.in/mg2c_v2.0/). The exon–intron structures of the FLR genes were analyzed by the Gene Structure Display Server (GSDS; http://gsds.cbi.pku.edu.cn/), and the Multiple Collinearity Scan toolkit (MCScanX) package (Wang et al., 2012) was used to analyze the gene duplication events, with the default parameters. The syntenic relationships within the FLR family were displayed using Circos (Krzywinski et al., 2009).
Subcellular localization assays
FLR3, -8, -11, and -14 were inserted into pCAMBIA1300-GFP vectors. The constructs were then introduced into Arabidopsis protoplasts via polyethyleneglycol (PEG)-mediated transformation as previously described (Li et al., 2016). The protoplasts were observed via a Nikon confocal laser scanning microscope at 12–24 h after transformation.
RNA isolation and gene expression analysis
For gene expression analysis, the roots, leaf sheaths, and leaves of Nip seedlings were harvested at the three-leaf stage. Mature roots, stems, flag leaves, panicles, and developing seeds were collected after the plants were grown in a greenhouse for 3–4 months. For temperature stress analysis, seedlings at the three-leaf stage were incubated at 4 °C for cold treatment and at 42 °C for heat treatment for 12 h; 25 °C was used as a control. To induce salt stress, the roots of seedlings at the three-leaf stage were immersed in Yoshida nutrient solution supplemented with NaCl (200 mM) for 3 d, and roots immersed in unaltered Yoshida nutrient solution were used as the control. After treatment, the shoots from the control and stressed seedlings were harvested for gene expression analysis. For biotic stress, Nip seedlings were sprayed with 70-15 spores as described previously (Xing et al., 2017), and seedlings sprayed with 0.025% Tween-20 were used as the control. The inoculated and control plants were kept in a moist chamber for 24 h in the dark at 28 °C. Considering that all the aerial parts of seedlings could be infected, the shoots from the control and stressed seedlings were harvested, instantly frozen in liquid nitrogen, and then stored at –80 °C for gene expression analysis.
Total RNA extraction, reverse transcription, and quantitative real-time PCR (qRT-PCR) were performed as previously described (Li et al., 2016). PCR was performed in triplicate for expression analysis. All the qRT-PCR data were quantified by the 2–ΔΔCT method, with three technical replicates for each of the three biological replicates. The housekeeping gene OsActin (LOC_Os03g50885) was used as an internal control. The heatmaps were created by R version 3.5.1 with the pheatmap package on the basis of the log2-fold-transformed data. The primers used for qRT-PCR are listed in Table S1 at JXB online.
Plant materials and transformation
T-DNA insertion mutants of flr1 (Dongjin, DJ), flr2 (Hwayoung, HY) (Li et al., 2016), and flr4 (DJ, PFG_1B-08401. R) were obtained from the Salk Institute (http://signal.salk.edu/cgi-bin/RiceGE). For clustered regularly interspaced short palindromic repeat (CRISPR)/CRISPR-associated protein 9 (Cas9) experiments, the guide RNA (gRNAs) targets in the FLR family gene and primers were selected via the E-CRISP Design Tool (http://www.e-crisp.org) (Heigwer et al., 2014) and the CRISPR-GE tool (http://skl.scau.edu.cn) (Xie et al., 2017). The Cas9 expression vectors were constructed as described previously (Ma et al., 2015). The 20 bp target sequences were inserted into a pYLgRNA-OsU3 vector between the OsU3 promoter and the gRNA scaffolds, followed by ligation to the Cas9 expression backbone pYLCRISPR/Cas9Pubi-H vector. The sequence of the resulting vector, pYLCRISPR/Cas9Pubi-H, was verified and it was introduced into Agrobacterium tumefaciens EHA105 to infect embryogenic calli of wild-type Nip rice as previously described (Nishimura et al., 2006). The transgenic rice lines were subsequently confirmed by sequencing PCR products that were amplified with primer pairs that flanked the designated target sites. Off-target sites were predicted by the CRISPR-GE tool (Xie et al., 2017). The highest scoring off-target site was amplified from the genomic DNA of the mutated transgenic rice lines, and the resulting PCR products were analyzed by sequencing. At least two independent Cas9 knockout mutant lines were obtained for each gene, which were used for subsequent phenotypic assays.
To generate FLR2-overexpression (OE) lines, a primer pair was designed on the basis of the full-length CDS without a stop codon. The product of the CDS fragment was cloned into a pCAMBIA1300-GFP vector between the BamHI and XbaI sites. After they were validated by sequencing, the constructs were transferred into rice embryogenic calli via the Agrobacterium strain EHA105 to generate the FLR2-OE lines. The gene expression levels of the FLR2-OE transgenic lines were determined by qRT-PCR at the RNA level and by western blot at the protein level. The transgenic and wild-type rice plants were grown in a greenhouse for 2 weeks and then transplanted to a field in Taohua village, Changsha, China (28°11'N, 112°58'E).
M. oryzae isolates
The following M. oryzae isolates were used for inoculation: 70-15, which is compatible with Nip (Kim et al., 2001) and is popular at many institutions; green fluorescent protein (GFP)-tagged 70-15 (70-15GFP); Guy11, which was generously provided by Dr Xuewei Chen (Sichuan Agricultural University); and HNB7, HNB16, HNB52, HNB62, HNB119, HNB145, and HNB154, which were collected from Hunan Province (Xing et al., 2017). All strains were cultured on agar plates that contained complete medium (CM) under a 12 h light (by white fluorescent bulbs)/12 h dark photoperiod at 25 °C.
Protein extraction and immunoblot analysis
For total protein extraction, fresh leaf tissues (0.1 g) were ground in liquid nitrogen and then incubated in 200 μl of protein extraction buffer [100 mM Tris–HCl, pH 7.5, 300 mM NaCl, 1 mM EDTA, 5% glycerol, 1% Triton X-100, 1 mM phenylmethylsulfonyl fluoride (PMSF), 1:100 protease inhibitor cocktail (78430, Thermo Fisher Scientific)] at 4 °C for 1 h. Proteins were separated in a 10% SDS–PAGE gel and subsequently transferred to polyvinylidene difluoride (PVDF) membranes. Anti-GFP (AT0028, CMC) and anti-actin (AT0004, CMC) antibodies were used for immunoblotting analysis.
Pathogenicity assays
Conidia were harvested from 7- to 10-day-old cultures on complete agar medium supplemented with 0.025% Tween-20. The conidial suspension was adjusted to 1–1.5×105 conidia ml–1 before wound inoculation or spray inoculation. Wound inoculation was performed as previously described (Zhang et al., 2014), and inoculated plants were kept in a moist chamber for 24 h in the dark at 28 °C. The photoperiod was then adjusted to a 12 h light/12 h dark cycle for 5–7 d. The percentage of lesion areas (disease index) per leaf was scored via image analysis with ImageJ software. The relative fungal biomass was measured as previously described (Li et al., 2017), and DNA-based quantitative PCR (qPCR) was performed with reference to the cycle threshold value of the M. oryzae MoPot2 gene and the rice genomic ubiquitin (OsUbi) gene with the formula 2[Ct(OsUbi)–Ct(MoPot2)]. The primers used for qRT-PCR are listed in Supplementary Table S1.
To observe fungal blast infection, detached leaf sheaths from the fourth leaf of five-leaf-stage seedlings were inoculated with the 70-15GFP isolate in a spore suspension (1×105 conidia ml–1). The detached rice sheath inner epidermal sections were previously sliced to make temporary slides. The fluorescence signals of the conidia, conidia germination, appressorium development, and invasive hyphal growth in epidermal cells were observed and counted under a confocal microscope as previously described (Li et al., 2017). For the percentage of M. oryzae hyphae in each type/stage, 50 hyphae were evaluated.
For field tests, the seedlings of the tested mutants and transgenic lines were cultivated in a greenhouse for 2 weeks before being transplanted into the field at the Daweishan blast nursery (Hunan Province, 28°45'N, 114°01'E) for resistance identification. When the seedlings were transplanted into the field, the planting area of each mutant and transgenic line was 10 m2, comprising a total of four rows and 25 cm×25 cm planting space. A row of Lijiangxintuanheigu (LTH), a highly susceptible variety, was planted between and around each variety used as an inducer to ensure uniform blast infection. Normal water and fertilizer management were applied, and fungicides were not applied throughout the whole growth period. The whole identification test was completely induced under natural conditions, with no artificial inoculation. Three months later, flag leaves and the second leaf above them were harvested to analyze relative fungal biomass.
H2O2 accumulation
To visualize hydrogen peroxide (H2O2), 3,3-diaminobenzidine (DAB) staining was performed as described previously (Thordal-Christensen et al., 1997), with slight modifications. Seedlings at the five-leaf stage were sprayed with the M. oryzae isolate in a spore suspension (1×105 conidia ml–1). At 3 days post-inoculation (dpi), leaf sections were vacuum infiltrated with DAB solution [1 mg ml–1 DAB, 50 mM Tris–HCl, 0.01% Triton X-100, pH 6.5] for 10 min, after which the sections were incubated at 25 °C for 12 h in the dark. The DAB-stained leaves were cleared by boiling in 90% ethanol for 20 min and then observed under a microscope. The relative amount of H2O2 was calculated on the basis of the pixels of images via Photoshop with the following formula: H2O2 area per rectangle=pixels of H2O2 area per mycelial invasion site/pixels of the rectangle.
Results
Identification and phylogenetic analysis of FLR family CrRLK1L homologs in rice
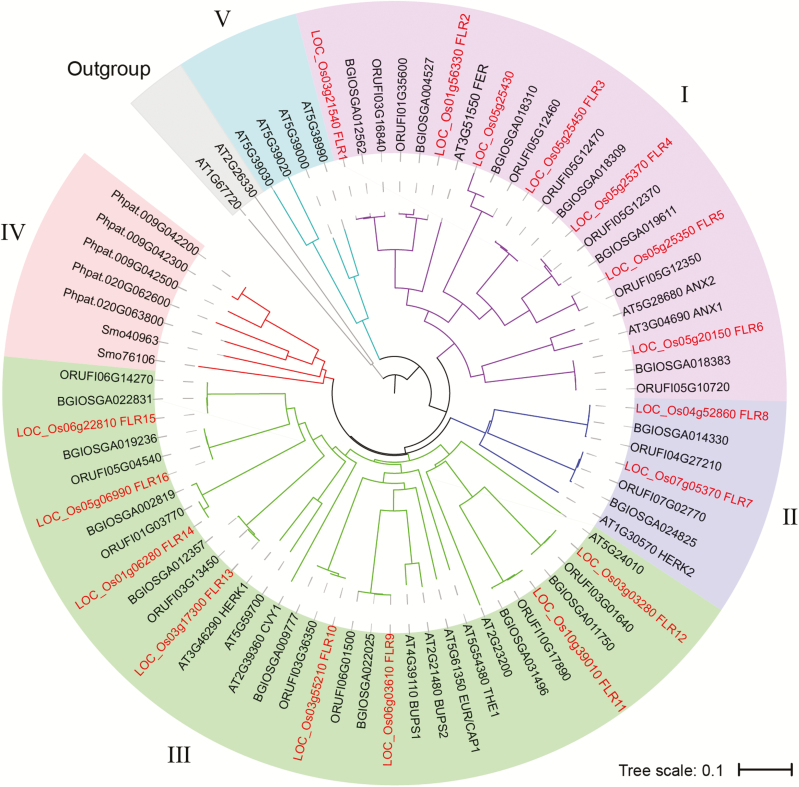
The two main rice varietal groups, O. sativa indica and O. sativa japonica, were independently domesticated from O. rufipogon wild rice populations (Londo et al., 2006; Huang et al., 2012). To investigate CrRLK1L/FLR (named FLR in rice hereafter) homologous genes in rice, we performed a homology search via BLAST (see the Materials and methods), and we identified 17, 16, and 17 potential FLR genes within the genome of O. rufipogon, O. sativa indica, and O. sativa japonica, respectively. To discover the evolutionary origin of the putative members of the CrRLK1L family, in which FLR genes in rice are included, 74 CrRLK1L genes [five P. patens, two S. moellendorffii, 17 Arabidopsis, 17 O. rufipogon, 16 O. sativa indica, and 17 O. sativa japonica CrRLK1L protein sequences, and an additional two outgroup protein sequences (AT1G67720 and AT2G26330) from Arabidopsis] were utilized to construct a phylogenetic tree via the NJ method. The results (Fig. 1) indicated that these 74 CrRLK1L genes could be divided into five subgroups (subgroups I–V), of which there were 23 members in group I, seven members in group II, 33 members in group III, seven members in group IV, and four members in group V. Subgroups I, II, and III included at least one rice CrRLK1L member and one Arabidopsis CrRLK1L member, suggesting that the divergence and emergence of the CrRLK1L family occurred early in the separation of rice and Arabidopsis. There was a one-to-one relationship between the CrRLK1L genes of wild rice and those of O. sativa japonica, while O. sativa indica rice lacked a member corresponding to wild rice ORUFI05G12350 and O. sativa japonica LOC_Os05g25350. No homologs had a similar full-length sequence to that of ORUFI05G12350/LOC_Os05g25350 while containing both the MLD PF12819 and Pkinase domain PF07714 (see the Materials and methods). These results demonstrated that rice CrRLK1L genes were conserved but were slightly divergent in terms of their evolution. Seven O. sativa japonica members in subgroup I were homologous to FER, ANX1, and ANX2, which implies that these CrRLK1Ls may have a role in mediating pollen tube growth and fertilization. Subgroup IV contained CrRLK1L members of the seedless land plants P. patens and S. moellendorffii; these members were homologous to THE1, HERK1, and HERK2, which implies that these CrRLK1Ls may have a similar role in maintaining cell wall integrity and cell growth. Subgroup V contained only Arabidopsis proteins (Fig. 1). The phylogenetic tree based only on the 74 CrRLK1L gene kinase domains (Supplementary Fig. S1) showed a similar division to that in Fig. 1, except for subgroup II, which contained 24 members (five P. patens, two S. moellendorffii, five Arabidopsis, four O. rufipogon, four O. sativa indica, and four O. sativa japonica CrRLK1L genes).
Fig. 1.
Phylogenetic tree of CrRLK1L family proteins. Complete amino acid sequences were aligned using ClustalW. The phylogenetic tree was constructed via the NJ method with 1000 bootstrap values in MEGA7 and was optimized with the iTOL online tool. The analyzed CrRLK1Ls were distributed in five main subgroups: I–V, marked with different background colors. Smo, Selaginella moellendorffii; Phpat, Physcomitrella patens; AT, Arabidopsis thaliana; ORUFI, Oryza rufipogon; BGIOSGA, Beijing Genomics Institute Oryza sativa Genome Annotation (indica); LOC_Os, Oryza sativa locus (japonica). The CrRLK1Ls (FLRs) of Oryza sativa japonica rice are highlighted in red.
Multiple sequence alignment and motif analysis of CrRLK1L proteins
We analyzed the domain conservation by performing a motif analysis using the MEME tool. We identified 20 conserved motif sequences from all of the CrRLK1L proteins. As shown in Supplementary Fig. S2, all CrRLK1L proteins had a similar motif composition with similar motif arrangements. Most members of the same subgroup had the same motifs; for instance, LOC_Os01g56330, LOC_Os03g21540, and AT3G51550 shared the same motifs. Motifs 13 and 19 were present within N-terminal signal peptides. Nine conserved motifs (motifs 6, 8, 10, 11, 14, 15, 16, 17, and 18) were distributed within the malectin domain region. The remaining nine motifs (motifs 1, 2, 3, 4, 5, 7, 9, 12, and 20) were distributed within the C-terminal kinase domain region. Notably, three proteins (AT5G39020, AT5G39030, and LOC_Os05g25430) had lost more than half of the conserved motifs within their C-terminal kinase domain region. All CrRLK1L proteins were similar in length except for LOC_Os05g25430 of O. sativa japonica, which had a relatively short protein length; thus, LOC_Os05g25430 was excluded as a canonical CrRLK1L member. The remaining CrRLK1L members of O. sativa japonica were denoted as FLR1–FLR16 (Fig. 1), corresponding to the distribution of their phylogenetic relationships with FER. We focused on the role of these 16 members (Supplementary Table S2) in the balance between growth and the stress response in subsequent experiments.
Gene structure, chromosomal distribution, and duplication in the FLR family
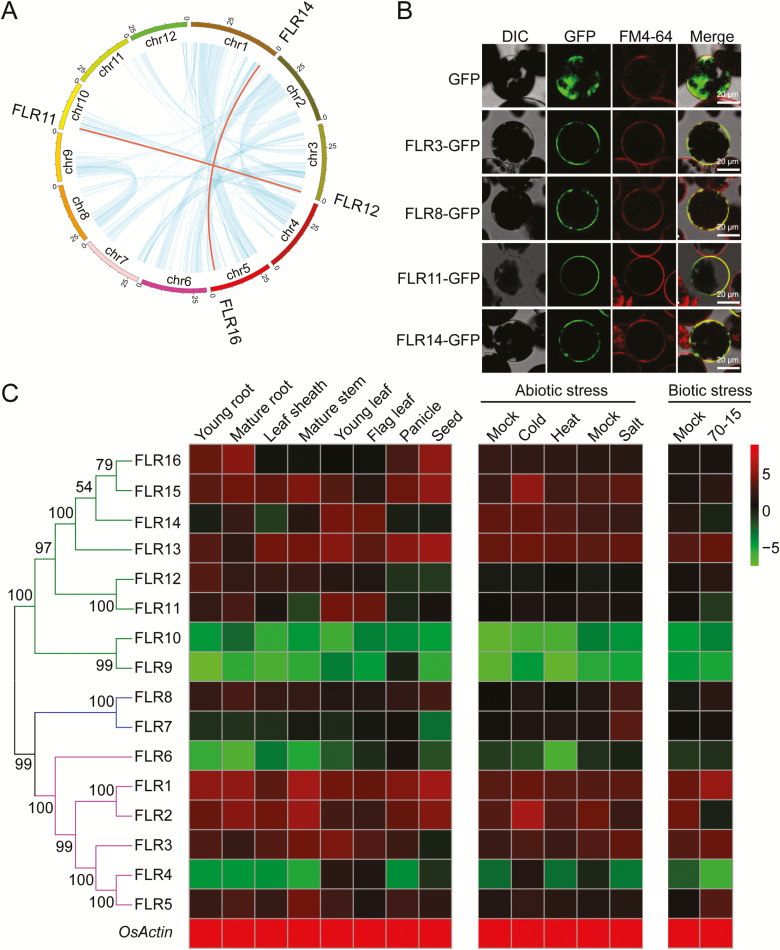
The exon–intron structures of the FLR genes were examined to understand the gene structure evolution of the FLR family (Supplementary Fig. S3). All FLR genes had similar coding sequence lengths, and the vast majority of the FLR genes had no introns, except FLR13 and FLR15, each of which contained one intron (Supplementary Fig. S3). Sixteen FLR genes were unevenly distributed among the 12 rice chromosomes (Supplementary Fig. S4), with the exception of chromosomes 2, 8, 9, 11, and 12. Chromosomes 3 and 5 contained relatively more FLR genes, with four and five genes, respectively, than did chromosomes 1 and 6, each of which contained two FLR genes. Chromosomes 4, 7, and 10 contained only one FLR gene each. Genomic duplications are considered an essential method to expand gene families and to increase the likelihood of evolutionary diversification. Ancient whole-genome duplication, chromosomal segmental duplication, and tandem duplication are the main dramatic forms of genomic duplication (Epstein, 1971; Lynch and Conery, 2000). In this study, we identified tandem and segmental duplication events in the FLR family. In accordance with the criterion that a chromosomal region of 200 kb containing two genes is defined as a tandem duplication event, three FLR genes (FLR3, -4, and -5) were clustered into such an event on chromosome 5 (Supplementary Fig. S3) (Holub, 2001). Another two segmental duplication events (FLR11 to FLR12 and FLR14 to FLR16) among the 16 FLR genes were also identified by MCScanX (Fig. 2A). These results suggest that gene duplication occurred in the FLR family. The duplicated genes may have expanded the function of the family members to adapt to a more complex environment during the evolution of rice.
Fig. 2.
Analysis of duplication, subcellular localization, and gene expression in the rice FLR family. (A) Synteny analysis of FLR genes in Oryza sativa japonica rice. Chromosome numbers are shown on the inner side, and the blue lines show all synteny blocks in the rice genome. FLR gene pairs with syntenic relationships are linked by red lines. (B) The subcellular localization of FLR3–GFP, FLR8–GFP, FLR11–GFP, and FLR14–GFP in Arabidopsis protoplasts. (C) Expression patterns of FLR genes in different organs/stages and under different abiotic and biotic stresses (n=3 for each group). These experiments were performed three times, each yielding similar results.
Subcellular localization of four FLR family members
We previously reported that FLR1 and FLR2 localized to the PM (Li et al., 2016) and are the closest homolog to FER in Arabidopsis. To determine the subcellular localization of FLR3, -8, -11, and -14, which form different subgroups (Fig. 1), we constructed a GFP fusion protein with a 35S promoter and transiently expressed the construct in Arabidopsis protoplasts. The GFP fluorescence of FLR3–GFP, FLR8–GFP, FLR11–GFP, and FLR14–GFP was detected on the PM (indicated by FM4-64 membrane staining) of Arabidopsis protoplasts (Fig. 2B). These results are consistent with the conclusion that CrRLK1L proteins act as sensors located on the cell membrane to perceive external signals and phosphorylate downstream proteins (Stegmann et al., 2017).
Expression patterns of FLR genes under normal growth and stress conditions
We analyzed the expression patterns of FLR family genes in different organs/stages and under various abiotic and biotic stresses via qRT-PCR. To compare the expression patterns, we converted the qRT-PCR gene expression values into a heatmap (Fig. 2C). Under normal growth conditions, FLR1, -2, and -3 were relatively highly expressed in all tissues, while the expression levels of FLR6, FLR9/RUPO, and FLR10 were extremely low in most tested tissues. In the vegetative organs, FLR1, -2, -15, and -16 were highly expressed in the young roots and mature roots, and FLR1, -2, -3, -5, -13, and -15 were highly expressed in the leaf sheaths and mature stems. FLR1, -3, -11, -13, and -14 were highly expressed in the leaves and flag leaves. In the reproductive organs, FLR1, -2, -13, and -15 were relatively abundant in the panicles, whereas FLR1, -2, -13, -15, and -16 were predominantly highly expressed in developing seeds. Remarkably, FLR9/RUPO tended to be expressed in the panicles and, according to previous reports, FLR9/RUPO functions in pollen tube growth and integrity (Liu et al., 2016)
Plants adapt to environmental changes and microbial attack by regulating gene expression. Under abiotic stress, compared with those in the control treatment, the expression levels of FLR2, -4, and -15 in the cold treatment were up-regulated by >2-fold. Under high-temperature conditions, the expression of FLR6 significantly decreased by 10-fold. Salinity stress caused a multifold up-regulation in the expression of FLR7 and -8, while the expression levels of FLR2 and -4 were 2-fold down-regulated. To analyze the response of FLR genes to biotic stress, Nip seedlings were sprayed with the compatible M. oryzae strain 70-15. The transcription of FLR2, -4, -11, and -14 was down-regulated by >2-fold after fungal treatment, while the expression of FLR1, -5, and -8 greatly increased in response to this fungal attack (Fig. 2C).
Screening the FLR gene family-mediated response to M. oryzae
To determine whether FLR family members function in rice immunity, we analyzed the function of FLR genes in response to M. oryzae. CRISPR/Cas9 technology was used to mutate 12 FLR family genes (Supplementary Fig. S5) in Nip; 11 (FLR3, -5, -7, -8, -10, -11, -12, -13, -14, -15, and -16) homozygous mutants were obtained, with at least two independent lines obtained for each gene. FLR9/RUPO mutants were heterozygous, which was in accordance with previous reports (Liu et al., 2016). Moreover, we obtained three T-DNA insertion mutants: one was FLR4 (DJ) from RiceGE (Supplementary Fig. S5), and the other two were FLR1 (DJ) and FLR2 (HY) mutants, which were identified in our previous works (Yang et al., 2015; Li et al., 2016).
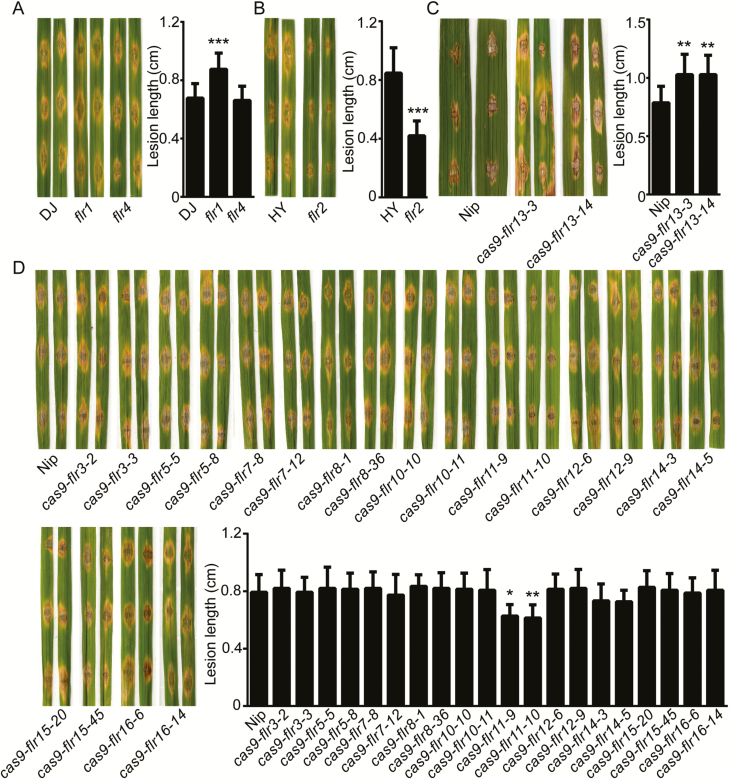
To analyze the variation in FLR family mutations in response to rice blast fungus via these FLR family mutants, the leaves of the mutants were inoculated with M. oryzae by the wound method (Zhang et al., 2014). As shown in Fig. 3, the lesion lengths of flr1 and flr13 were significantly longer than those of the wild-type control. These results suggest that FLR1 and FLR13 may be related to rice immunity. Nevertheless, the lesions of the flr2 and flr11 mutants were significantly shorter and smaller than those of the wild-type HY and Nip plants, respectively. flr14 was slightly resistant to M. oryzae, but its resistance was not significantly different from the control. The basal resistance of the transgenic plants of nine other genes (FLR3, -4, -5, -7, -8, -10, -12, -15, and -16) was similar to that of the wild-type plants, indicating that the loss of these genes did not cause a defect in rice blast fungus disease resistance. A similar conclusion was confirmed in two independent CRISPR/Cas9 lines with mutations in FLR family members. These results suggest that FLR2 and FLR11 negatively regulate rice blast fungus response. FLR2 was chosen for further analysis because it has a very high similarity to FER, which plays important roles in both growth and biotic stress responses (Stegmann et al., 2017).
Fig. 3.
Function of the FLR gene family members in rice blast resistance. (A–D) Disease symptoms and lesion lengths of FLR mutant plants and wild-type plants at 7 dpi; the plants were inoculated (by wounding) with the compatible Magnaporthe oryzae isolate 70-15. (A) Disease symptoms and lesion lengths of flr1 and flr4 mutants and wild-type DJ plants. (B) Disease symptoms and lesion lengths of flr2 and wild-type HY. (C and D) Disease symptoms and lesion lengths of 11 (FLR3, -5, -7, -8, -10, -11, -12, -13, -14, -15, and -16) mutants and wild-type Nip plants. (A–D) Statistical analysis of average lesion lengths, measured for five leaves (n=15 lesions for each group). The error bars correspond to 1 SD as determined by the duplicate analyses. The data are shown as the mean ±SD. The asterisks indicate significant differences from the control, as determined by a t-test or one-way ANOVA followed by Tukey’s test (*P<0.05, **P<0.01, ***P<0.001). These experiments were performed with three independent biological replicates, each yielding similar results, while wound inoculation of flr13 with the rice blast fungus had only technical replicates, as flr13 is sterile. (This figure is available in color at JXB online.)
FLR2 negatively regulates rice resistance to M. oryzae
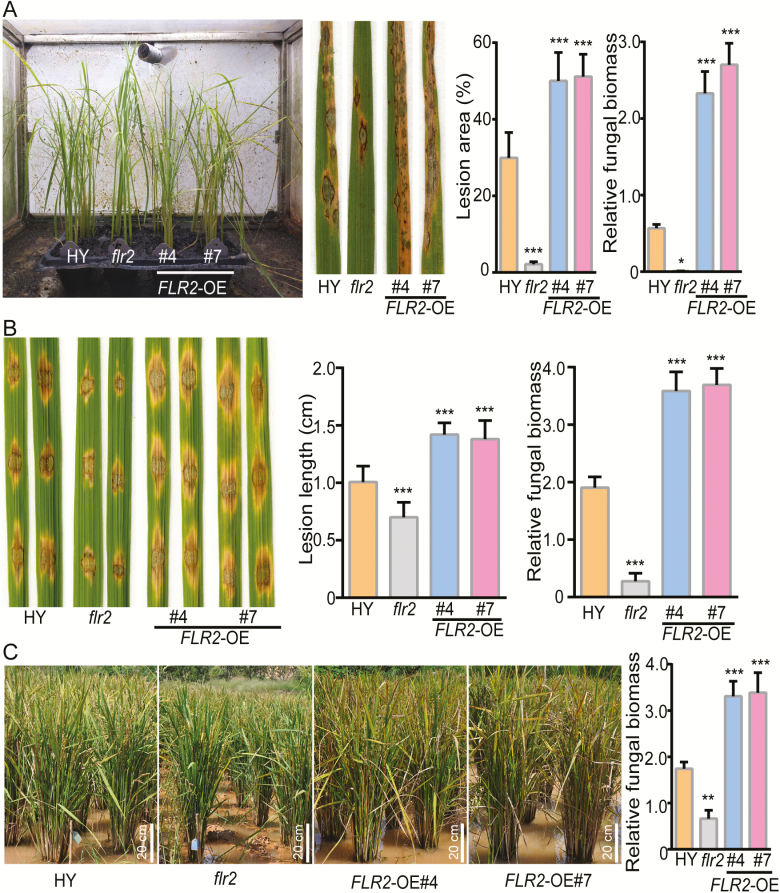
To further determine the function of FLR2 in the response to rice blast disease, we created two FLR2-OE lines (FLR2-OE#4 and #7) by using a GFP fusion gene under the control of the 35S promoter, and their up-regulated FLR2 mRNA levels and protein levels were confirmed (Supplementary Fig. S6). The flr2 (Li et al., 2016), OE lines, and wild-type plants were inoculated with the compatible M. oryzae 70-15 isolate by spraying at the seedling stage (Fig. 4A, left panel). flr2 exhibited increased blast resistance, as reflected by the significantly decreased lesion area and fungal biomass in the flr2 mutant compared with the wild-type HY plants at 7 dpi (Fig. 4A, middle and right panels). FLR2-OE#4 and #7 exhibited increased plant susceptibility to fungal infection, as reflected by the increased lesion area and fungal biomass in the OE plants compared with the wild-type plants. We then inoculated the leaves (via the wound method) with the compatible M. oryzae isolate 70-15 (Fig. 4B). flr2 developed 30–40% smaller and shorter lesions at the wound-inoculated sites than did HY (Fig. 4B, left and middle panel). The fungal biomass of flr2 was reduced to 10–20% of that of HY (Fig. 4B, right panel). In contrast, FLR2-OE#4 and #7 displayed 30–40% longer lesions and greater fungal biomass than did HY (Fig. 4B, middle and right panel). To further determine the resistance of the transgenic rice, the materials were replanted in a blast nursery. As shown in Fig. 4C, compared with the wild-type plants, the flr2 was more resistant to rice blast in the field, whereas the FLR2-overexpressing plants displayed highly susceptible phenotypes.
Fig. 4.
FLR2 functions in rice blast resistance. (A) Disease symptoms of HY, flr2, and FLR2-OE plants at 7 dpi; the plants were inoculated (by spraying) with the compatible M. oryzae isolate 70-15. The percentage of lesion areas (disease index) was scored via image analysis with ImageJ software (n=5 leaves). Fungal growth was measured by the expression level of the Magnaporthe oryzae MoPot2 gene in the inoculated leaves via qRT-PCR, and the levels were normalized to the expression level of the OsUbi gene (n=3). (B) Phenotypes of HY, flr2, and FLR2-OE plants at 7 dpi after they were inoculated (by wounding) with the compatible M. oryzae isolate 70-15 (n=15 lesions). (C) Disease resistance of HY, flr2, and FLR2-OE plants in the blast nursery (n=3). Scale bar=20 cm. (A–C) The error bars correspond to 1 SD as determined by the duplicate analyses. The data are shown as the mean ±SD. The asterisks indicate significant differences from the control, as determined by one-way ANOVA followed by Tukey’s test (*P<0.05, **P<0.01, ***P<0.001). Three independent biological replicates of (A–C) were tested, each yielding similar results.
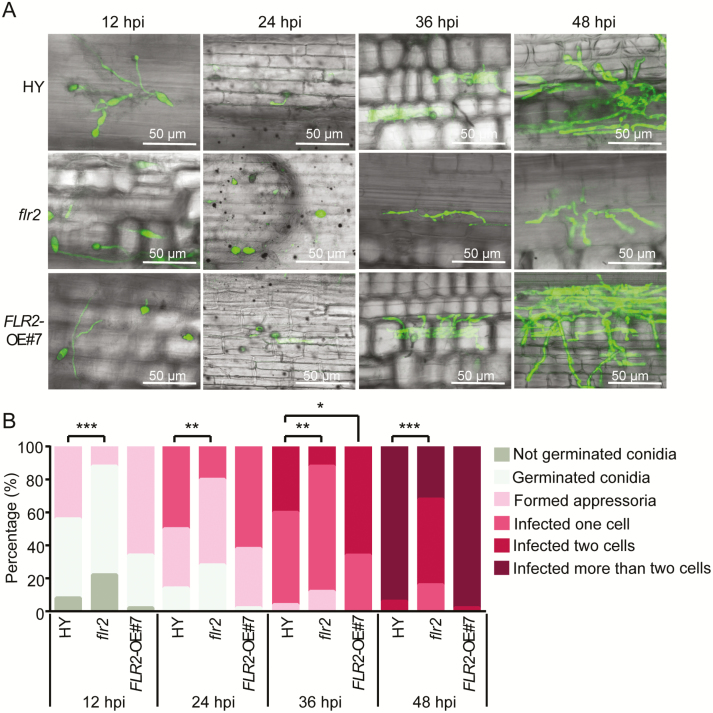
We then investigated the invasive hyphal growth of the virulent isolate 70-15GFP in the leaf sheaths. We observed that more penetration sites formed appressoria inside the epidermal cells of the wild-type HY plants and FLR2-OE#7 lines than inside those of the flr2 at 12 h post-inoculation (hpi). More invasive hyphae formed primary hyphae on FLR2-OE#7 lines than on wild-type HY plants at 24 hpi, which was opposite to the results observed for the flr2. The primary hyphae branched out into secondary invasive hyphae and extended to the neighboring cells at 36 hpi (Fig. 5A). Notably, the number of invasive hyphae in the flr2 was significantly lower than that in the HY plants and FLR2-OE lines at 36 and 48 hpi (Fig. 5B).
Fig. 5.
Fluorescence signals of GFP-tagged Magnaporthe oryzae strain 70-15GFP invasive hyphae in the leaf sheaths of flr2, FLR2-OE, and HY wild-type plants. (A) Confocal images showing the invasive M. oryzae hyphae in the epidermal cells of flr2, FLR2-OE#7, and HY leaf sheaths. Scale bar=50 μm. (B) Percentage of invasive hyphae growth at 12, 24, 36, and 48 hpi. The asterisks indicate significant differences from the control as determined by Crosstabs, followed by a χ 2 test (*P<0.05, **P<0.01, ***P<0.001). Fifty hyphae were counted per replication, and the experiment was repeated three times, each yielding similar results. (This figure is available in color at JXB online.)
The use of broad-spectrum resistant and robust resistant varieties represents an environmentally friendly and cost-effective method for controlling rice blast fungi. Dozens of broad-spectrum resistance genes have been reported in recent works (Azizi et al., 2016; Deng et al., 2017; Li et al., 2017), providing information for breeding new disease-resistant varieties. To determine whether knocking out FLR2 conferred broad-spectrum blast resistance to plants, we performed inoculation experiments with six blast isolates (HNB7, HNB52, HNB119, HNB145, HNB154, and Guy11); the HNB52 strain belongs to the ZC16 physiological race, and HNB7, HNB119, HNB145, and HNB154 belong to the ZB13 physiological race (Xing et al., 2017). We found that the flr2 was more resistant to these strains than were the HY plants, while the FLR2-OE lines were more susceptible to these strains than were the HY plants (Supplementary Fig. S7). Thus, the FLR2 mutation has a certain degree of broad-spectrum resistance.
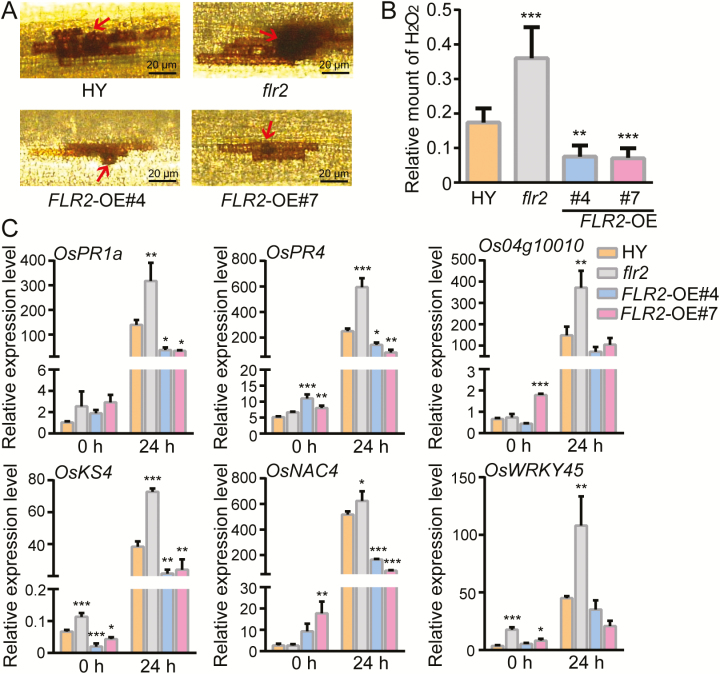
ROS signaling can modulate a broad range of biological processes involved in cell expansion, development, and responses to biotic and abiotic stimuli (Monshausen et al., 2007; Baxter et al., 2014; Duan et al., 2014; Qi et al., 2017). FER regulation has a profound role in the control of ROS in different biological processes (Wong et al., 2007; Duan et al., 2010; Li et al., 2015); thus, H2O2 accumulation was tested in response to FLR2-mediated rice blast disease. DAB staining revealed that the flr2 produced much greater amounts of H2O2 at the penetration sites of leaf cells than did the wild-type plants, while less H2O2 was produced in the leaf cells of the FLR2-OE lines in response to rice blast (Fig. 6A, B).
Fig. 6.
FLR2 negatively regulates resistance to disease caused by Magnaporthe oryzae. (A) DAB staining of the infection sites in flr2, FLR2-OE, and HY wild-type plants at 3 dpi. The arrows indicate the infection structures of appressoria. Scale bar=20 μm. (B) Quantification of H2O2. The relative amount of H2O2 was calculated on the basis of the pixels of images via Photoshop with the following formula: H2O2 area per rectangle=pixels of H2O2 area per mycelial invasion site/pixels of the rectangle. (C) M. oryzae-induced expression of defense-related genes in 2-week-old transgenic rice lines and wild-type lines. The OsActin gene was used as an internal control, and relative expression levels were normalized to the level of the internal control at 0 h in the wild-type plants (n=3). The error bars correspond to 1 SD as determined by the duplicate analyses. The data are shown as the mean ±SD. The asterisks indicate significant differences from the control, as determined by one-way ANOVA followed by Tukey’s test (*P<0.05, **P<0.01, ***P<0.001). These experiments were performed three times, each yielding similar results. (This figure is available in color at JXB online.)
Upon pathogen attack, plants increase their immunity by activating sets of defense-related genes, including the salicylic acid (SA) signaling pathway marker gene OsPR1a (Agrawal et al., 2000), the jasmonate (JA) signaling pathway marker gene OsPR4 (Wang et al., 2011), the dehydrogenase gene OsO4g10010, and the kaurene synthase4 gene OsKS4 (Agrawal et al., 2000; Miyamoto et al., 2016). Transcription factor genes such as OsNAC4 and OsWKRY45 also play roles as defense-related genes (Shimono et al., 2007; Kaneda et al., 2009). We therefore analyzed the expression of these defense-related genes in the transgenic lines and the wild-type plants at 0 h and 24 h after the leaves were inoculated with rice blast. The expression levels of the six defense-related genes were greater in flr2 than in HY at 24 hpi. In contrast, the FLR2-OE lines had much lower expression levels of OsPR1a, OsPR4, OsO4g10010, OsKS4, OsNAC4, and OsWKRY45 than did the HY plants (Fig. 6C). These results suggest that FLR2-mediated susceptibility was associated with decreased expression of defense-related genes induced by rice blast fungi.
Growth traits related to the FLR2/11 genes involved in response to M. oryzae
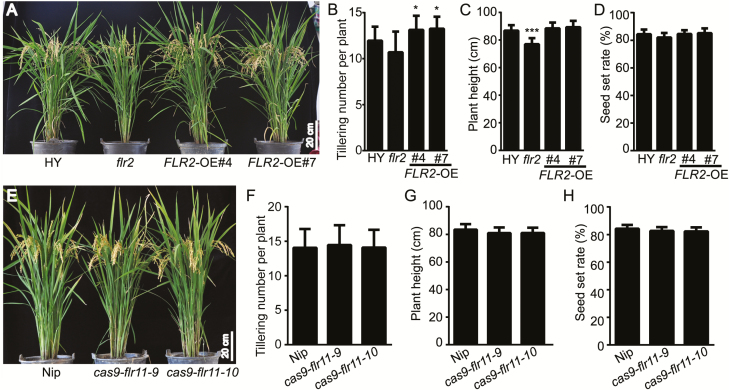
Compared with the wild-type plants, the flr2 and flr11 mutants were more resistant to rice blast (Fig. 3); therefore, we investigated whether the improved blast resistance would affect rice plant growth. At the mature stage, compared with those of the wild-type HY plants, the tiller numbers of the flr2 was not significantly different, while the FLR2-OE lines had increased tiller numbers (Fig. 7A, B). Compared with that of wild-type HY, the plant heights of flr2 were ~9% lower (Fig. 7A, C). FER functions in pollen tube reception and fertility in Arabidopsis (Escobar-Restrepo et al., 2007). However, the flr2 undergo normal pollen development (Li et al., 2016), and the percentage of seed set did not differ between the flr2 and wild-type HY plants (Fig. 7A, D). The number of tillers was similar between the FLR11 mutants and the wild-type Nip plants (Fig. 7F). The seed setting rate and height did not obviously differ between the flr11 mutants and the wild-type Nip plants (Fig. 7G, H). These results suggest that flr2 and flr11 exhibited increased disease resistance without significantly negative effects on their vegetative growth.
Fig. 7.
FLR2/11 gene function in rice growth and development. (A) Morphology of mature flr2, FLR2-OE lines, and wild-type HY plants in normal field conditions. Scale bar=20 cm. (B–D) Tiller numbers, plant heights, and seed setting rates of flr2, FLR2-OE lines, and wild-type HY plants (n=25). (E) Morphology of mature flr11 mutants and wild-type Nip plants. Scale bar=20 cm. (F–H) Tiller number, plant height, and seed setting rate of flr11 mutants and wild-type Nip plants in normal field conditions (n=25). The error bars correspond to 1 SD as determined by the duplicate analyses. The data are shown as the mean ±SD. The asterisks indicate significant differences from the control, as determined by one-way ANOVA followed by Tukey’s test (*P<0.05, **P<0.01, ***P<0.001). Three independent biological replicates of (A–G) were tested, each yielding similar results. (This figure is available in color at JXB online.)
Discussion
Members of the CrRLK1L family have multiple functions and have been highly conserved throughout the evolution of plants (Lindner et al., 2012; Liao et al., 2017; Franck et al., 2018). FER, a CrRLK1L family member, is highly involved in most aspects of the plant life cycle, such as cell growth, root hair development, female fertility, and immunity (Lindner et al., 2012; Kessler et al., 2015; Franck et al., 2018). The FER homologs FLR1 and FLR2 (Li et al., 2016) controlled rice height, tillering, branching, and fructification to varying degrees. In the present study, we found that CrRLK1L gene members were identified in indica, japonica, and O. rufipogon wild rice (Fig. 1), suggesting that CrRLK1L is highly conserved during evolution. Moreover, expression analysis revealed that members of the CrRLK1L/FLR family were ubiquitously expressed in different organs/stages in rice (Fig. 2C); this result might suggest its functional divergence. In addition, we systematically analyzed the functions of FLR family genes in rice resistance to M. oryzae (Fig. 3). The results showed that FLR2 and FLR11 are potential regulators involved in rice blast resistance without significant impact on growth. Our studies of FLRs can provide a reference for molecular breeding practices that can improve rice resistance and production through genetic manipulation.
The abundance of ROS and the pH of rice cells are two critical elements related to fungal pathogenicity. ROS bursts are a typical phenomenon in PTI and ETI pathways (Jones and Dangl, 2006; Couto and Zipfel, 2016). FER directly interacts with Rop-guanine exchange factors (RopGEFs), which then act as upstream regulators of RHO GTPases (RAC/ROPs). Activated RAC/ROPs further interact with several signal mediators, including NADPH oxidases, to regulate the production of ROS and plant immunity (Wong et al., 2007; Duan et al., 2010; Nibau and Cheung, 2011; Nagano et al., 2016). FLR family members mediate rice immunity by possibly functioning as upstream regulators of Rac/ROP, which bind to the NADPH oxidase gene OsRbohB to regulate the production of ROS during rice blast infection (Wong et al., 2007; Chen et al., 2010; Kosami et al., 2014; Nagano et al., 2016). The possible mechanism is as follows: FLR may function as a PRR to detect microbial/PAMPs and may trigger ROS bursts, and certain effectors secreted by M. oryzae may directly or indirectly associate with FLR to suppress the production of ROS and evade plant immune defenses (Engelsdorf et al., 2018).
Apoplastic pH is not only crucial for growth but is also important for immune responses in plants (Haruta et al., 2014; Barbez et al., 2017; Fernandes et al., 2017). For pathogens, the ability to sense and adapt to or alter the pH of their environment is essential for their survival and pathogenicity (Fernandes et al., 2017). During M. oryzae infection in rice and barley, once the infecting hyphae form and penetrate the plant epidermis, ammonia is rapidly released to stimulate apoplastic alkalinization through the PacC signaling pathway (Landraud et al., 2013). Environmental alkalinization requires significant amounts of infected mycelia to release ammonia, but only a small portion of infected mycelia develops during the early stages of infection (Prusky and Yakoby, 2003; Fernandes et al., 2017). To more rapidly infect the host and evade the host’s immune response, the pathogenic fungus F. oxysporum secretes a RALF1 homolog (F-RALF), which is involved in the FER-dependent signaling pathway to stimulate apoplastic alkalinization and suppress plant immunity to facilitate pathogenicity (Masachis et al., 2016; Thynne et al., 2017). Fungal RALF homologs are widely conserved among a diverse range of Ascomycota and Basidiomycota (Thynne et al., 2017). Whether M. oryzae contains RALF homologs, which may interact with FLR to increase the extracellular pH, inhibit plant growth, and promote pathogenesis, is an open question. Therefore, determining whether RALF homologs exist in M. oryzae is warranted.
Both plant growth and immunity are related to several hormone signaling pathways, such as those of SA, JA, auxin, ABA, ethylene, and brassinosteroids (Ning et al., 2017). In response to biotic stress, plants integrate multiple hormone signaling pathways to boost immunity, which can negatively affect plant development and growth, and ultimately affect grain yield. In Arabidopsis, FER is involved in the crosstalk between several hormone pathways that regulate cell growth, seed yield, and stress responses (Liao et al., 2017; Franck et al., 2018). In the present study, after plants were infected with the fungal strain 70-15, higher levels of OsPR1a (a marker gene of the SA pathway) and OsPR4 (a marker gene of the JA pathway) were detected in flr2 than in wild-type plants (Fig. 6C). Moreover, the flr2 and flr11 mutants did not exhibit a significant reduction in growth (Fig. 7). Thus, as immune receptors, FLR gene family members (e.g. FLR2 and -11) have the potential to balance disease resistance with plant growth. The detailed molecular mechanisms of FLR2- and FLR11-mediated immune regulation still require further study. The CrRLK1L/FLR family has been highly conserved throughout evolution (Franck et al., 2018); thus, our work provides a reference for studying the role of the CrRLK1L/FLR family in the function of immunity and growth in other crop species.
Supplementary data
Supplementary material is available at JXB online
Fig. S1. Phylogenetic tree of CrRLK1L family protein kinase domains from cultivated rice (Oryza sativa japonica and O. sativa indica), wild rice (Oryza rufipogon), Arabidopsis, Physcomitrella patens, and Selaginella moellendorffii.
Fig. S2. Conserved motif analyses.
Fig. S3. Exon–intron structure of FLR genes.
Fig. S4. Chromosomal localization of FLR genes.
Fig. S5. Identification of FLR mutants.
Fig. S6. Expression levels of FLR2 in FLR2-OE lines, as detected by qRT-PCR and immunoblot analysis.
Fig. S7. Analysis of the resistance of the FLR2 transgenic lines to different rice blast strains.
Table S1. List of primers used in this research.
Table S2. Members of the FLR family.
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (NSFC-31871396, 31400232, and 31571444), Hunan Provincial Science & Technology funds (2015JJ3048, 2017RS3053), Young Elite Scientist Sponsorship Program by CAST (YESS20160001), Science and Technology Project of Changsha (KQ1706017), and Hunan Technology Major Project (2018NK1020).
Author contributions
FY conceived the project; Z-HY, LW, YL, J-NQ, YT, X-QF, and Q-LL performed the research; H-FD contributed new reagents/analytical tools; Z-HY, J-JX, and FY designed the research, analyzed the data, and wrote the paper; all authors reviewed and approved the manuscript for publication.
References
- Agrawal GK, Jwa NS, Rakwal R. 2000. A novel rice (Oryza sativa L.) acidic PR1 gene highly responsive to cut, phytohormones, and protein phosphatase inhibitors. Biochemical and Biophysical Research Communications 274, 157–165. [DOI] [PubMed] [Google Scholar]
- Azizi P, Rafii MY, Abdullah SN, Nejat N, Maziah M, Hanafi MM, Latif MA, Sahebi M. 2016. Toward understanding of rice innate immunity against Magnaporthe oryzae. Critical Reviews in Biotechnology 36, 165–174. [DOI] [PubMed] [Google Scholar]
- Bai L, Ma X, Zhang G, Song S, Zhou Y, Gao L, Miao Y, Song CP. 2014. A receptor-like kinase mediates ammonium homeostasis and is important for the polar growth of root hairs in Arabidopsis. The Plant Cell 26, 1497–1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. 2009. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Research 37, W202–W208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banks JA, Nishiyama T, Hasebe M, et al. 2011. The Selaginella genome identifies genetic changes associated with the evolution of vascular plants. Science 332, 960–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbez E, Dünser K, Gaidora A, Lendl T, Busch W. 2017. Auxin steers root cell expansion via apoplastic pH regulation in Arabidopsis thaliana. Proceedings of the National Academy of Sciences, USA 114, E4884–E4893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter A, Mittler R, Suzuki N. 2014. ROS as key players in plant stress signalling. Journal of Experimental Botany 65, 1229–1240. [DOI] [PubMed] [Google Scholar]
- Boisson-Dernier A, Roy S, Kritsas K, Grobei MA, Jaciubek M, Schroeder JI, Grossniklaus U. 2009. Disruption of the pollen-expressed FERONIA homologs ANXUR1 and ANXUR2 triggers pollen tube discharge. Development 136, 3279–3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Shiotani K, Togashi T, Miki D, Aoyama M, Wong HL, Kawasaki T, Shimamoto K. 2010. Analysis of the Rac/Rop small GTPase family in rice: expression, subcellular localization and role in disease resistance. Plant & Cell Physiology 51, 585–595. [DOI] [PubMed] [Google Scholar]
- Couto D, Zipfel C. 2016. Regulation of pattern recognition receptor signalling in plants. Nature Reviews. Immunology 16, 537–552. [DOI] [PubMed] [Google Scholar]
- Deng Y, Zhai K, Xie Z, et al. 2017. Epigenetic regulation of antagonistic receptors confers rice blast resistance with yield balance. Science 355, 962–965. [DOI] [PubMed] [Google Scholar]
- Du C, Li X, Chen J, et al. 2016. Receptor kinase complex transmits RALF peptide signal to inhibit root growth in Arabidopsis. Proceedings of the National Academy of Sciences, USA 113, E8326–E8334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan Q, Kita D, Johnson EA, Aggarwal M, Gates L, Wu HM, Cheung AY. 2014. Reactive oxygen species mediate pollen tube rupture to release sperm for fertilization in Arabidopsis. Nature Communications 5, 3129. [DOI] [PubMed] [Google Scholar]
- Duan Q, Kita D, Li C, Cheung AY, Wu HM. 2010. FERONIA receptor-like kinase regulates RHO GTPase signaling of root hair development. Proceedings of the National Academy of Sciences, USA 107, 17821–17826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelsdorf T, Gigli-Bisceglia N, Veerabagu M, McKenna JF, Vaahtera L, Augstein F, Van der Does D, Zipfel C, Hamann T. 2018. The plant cell wall integrity maintenance and immune signaling systems cooperate to control stress responses in Arabidopsis thaliana. Science Signaling 11, eaao3070. [DOI] [PubMed] [Google Scholar]
- Epstein CJ. 1971. Evolution by gene duplication. American Journal of Human Genetics 23, 541. [Google Scholar]
- Escobar-Restrepo JM, Huck N, Kessler S, Gagliardini V, Gheyselinck J, Yang WC, Grossniklaus U. 2007. The FERONIA receptor-like kinase mediates male–female interactions during pollen tube reception. Science 317, 656–660. [DOI] [PubMed] [Google Scholar]
- Fernandes TR, Segorbe D, Prusky D, Di Pietro A. 2017. How alkalinization drives fungal pathogenicity. PLoS Pathogens 13, e1006621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franck CM, Westermann J, Boisson-Dernier A. 2018. Plant malectin-like receptor kinases: from cell wall integrity to immunity and beyond. Annual Review of Plant Biology 69, 301–328. [DOI] [PubMed] [Google Scholar]
- Gachomo EW, Jno Baptiste L, Kefela T, Saidel WM, Kotchoni SO. 2014. The Arabidopsis CURVY1 (CVY1) gene encoding a novel receptor-like protein kinase regulates cell morphogenesis, flowering time and seed production. BMC Plant Biology 14, 221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge Z, Bergonci T, Zhao Y, et al. 2017. Arabidopsis pollen tube integrity and sperm release are regulated by RALF-mediated signaling. Science 358, 1596–1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gravois KA, McNew RW. 1993. Genetic relationships among and selection for rice yield and yield components. Crop Science 33, 249–252. [Google Scholar]
- Guo H, Li L, Ye H, Yu X, Algreen A, Yin Y. 2009. Three related receptor-like kinases are required for optimal cell elongation in Arabidopsis thaliana. Proceedings of the National Academy of Sciences, USA 106, 7648–7653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haruta M, Monshausen G, Gilroy S, Sussman MR. 2008. A cytoplasmic Ca2+ functional assay for identifying and purifying endogenous cell signaling peptides in Arabidopsis seedlings: identification of AtRALF1 peptide. Biochemistry 47, 6311–6321. [DOI] [PubMed] [Google Scholar]
- Haruta M, Sabat G, Stecker K, Minkoff BB, Sussman MR. 2014. A peptide hormone and its receptor protein kinase regulate plant cell expansion. Science 343, 408–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heigwer F, Kerr G, Boutros M. 2014. E-CRISP: fast CRISPR target site identification. Nature Methods 11, 122–123. [DOI] [PubMed] [Google Scholar]
- Holub EB. 2001. The arms race is ancient history in Arabidopsis, the wildflower. Nature Reviews. Genetics 2, 516–527. [DOI] [PubMed] [Google Scholar]
- Huang X, Kurata N, Wei X, et al. 2012. A map of rice genome variation reveals the origin of cultivated rice. Nature 490, 497–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JD, Dangl JL. 2006. The plant immune system. Nature 444, 323–329. [DOI] [PubMed] [Google Scholar]
- Kaneda T, Taga Y, Takai R, Iwano M, Matsui H, Takayama S, Isogai A, Che FS. 2009. The transcription factor OsNAC4 is a key positive regulator of plant hypersensitive cell death. The EMBO Journal 28, 926–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler SA, Lindner H, Jones DS, Grossniklaus U. 2015. Functional analysis of related CrRLK1L receptor-like kinases in pollen tube reception. EMBO Reports 16, 107–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler SA, Shimosato-Asano H, Keinath NF, Wuest SE, Ingram G, Panstruga R, Grossniklaus U. 2010. Conserved molecular components for pollen tube reception and fungal invasion. Science 330, 968–971. [DOI] [PubMed] [Google Scholar]
- Kim S, Ahn IP, Lee YH. 2001. Analysis of genes expressed during rice–Magnaporthe grisea interactions. Molecular Plant-Microbe Interactions 14, 1340–1346. [DOI] [PubMed] [Google Scholar]
- Kosami K, Ohki I, Nagano M, et al. 2014. The crystal structure of the plant small GTPase OsRac1 reveals its mode of binding to NADPH oxidase. Journal of Biological Chemistry 289, 28569–28578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA. 2009. Circos: an information aesthetic for comparative genomics. Genome Research 19, 1639–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landraud P, Chuzeville S, Billon-Grande G, Poussereau N, Bruel C. 2013. Adaptation to pH and role of PacC in the rice blast fungus Magnaporthe oryzae. PLoS One 8, e69236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehti-Shiu MD, Shiu SH. 2012. Diversity, classification and function of the plant protein kinase superfamily. Philosophical Transactions of the Royal Society B: Biological Sciences 367, 2619–2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Liu X, Qiang X, et al. 2018. EBP1 nuclear accumulation negatively feeds back on FERONIA-mediated RALF1 signaling. PLoS Biology 16, e2006340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Wang L, Cui Y, et al. 2016. Two FERONIA-like receptor (FLR) genes are required to maintain architecture, fertility, and seed yield in rice. Molecular Breeding 36, 151. [Google Scholar]
- Li C, Yeh FL, Cheung AY, et al. 2015. Glycosylphosphatidylinositol-anchored proteins as chaperones and co-receptors for FERONIA receptor kinase signaling in Arabidopsis. eLife 4, e06587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Zhu Z, Chern M, et al. 2017. A natural allele of a transcription factor in rice confers broad-spectrum blast resistance. Cell 170, 114–126.e15. [DOI] [PubMed] [Google Scholar]
- Liao H, Tang R, Zhang X, Luan S, Yu F. 2017. FERONIA receptor kinase at the crossroads of hormone signaling and stress responses. Plant & Cell Physiology 58, 1143–1150. [DOI] [PubMed] [Google Scholar]
- Lindner H, Müller LM, Boisson-Dernier A, Grossniklaus U. 2012. CrRLK1L receptor-like kinases: not just another brick in the wall. Current Opinion in Plant Biology 15, 659–669. [DOI] [PubMed] [Google Scholar]
- Liu L, Zheng C, Kuang B, Wei L, Yan L, Wang T. 2016. Receptor-like kinase RUPO interacts with potassium transporters to regulate pollen tube growth and integrity in rice. PLoS Genetics 12, e1006085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Londo JP, Chiang YC, Hung KH, Chiang TY, Schaal BA. 2006. Phylogeography of Asian wild rice, Oryza rufipogon, reveals multiple independent domestications of cultivated rice, Oryza sativa. Proceedings of the National Academy of Sciences, USA 103, 9578–9583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M, Conery JS. 2000. The evolutionary fate and consequences of duplicate genes. Science 290, 1151–1155. [DOI] [PubMed] [Google Scholar]
- Ma X, Zhang Q, Zhu Q, et al. 2015. A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Molecular Plant 8, 1274–1284. [DOI] [PubMed] [Google Scholar]
- Mang H, Feng B, Hu Z, et al. 2017. Differential regulation of two-tiered plant immunity and sexual reproduction by ANXUR receptor-like kinases. The Plant Cell 29, 3140–3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao D, Yu F, Li J, et al. 2015. FERONIA receptor kinase interacts with S-adenosylmethionine synthetase and suppresses S-adenosylmethionine production and ethylene biosynthesis in Arabidopsis. Plant, Cell & Environment 38, 2566–2574. [DOI] [PubMed] [Google Scholar]
- Masachis S, Segorbe D, Turrà D, et al. 2016. A fungal pathogen secretes plant alkalinizing peptides to increase infection. Nature Microbiology 1, 16043. [DOI] [PubMed] [Google Scholar]
- Mecchia MA, Santos-Fernandez G, Duss NN, et al. 2017. RALF4/19 peptides interact with LRX proteins to control pollen tube growth in Arabidopsis. Science 358, 1600–1603. [DOI] [PubMed] [Google Scholar]
- Miyamoto K, Fujita M, Shenton MR, et al. 2016. Evolutionary trajectory of phytoalexin biosynthetic gene clusters in rice. The Plant Journal 87, 293–304. [DOI] [PubMed] [Google Scholar]
- Miyazaki S, Murata T, Sakurai-Ozato N, Kubo M, Demura T, Fukuda H, Hasebe M. 2009. ANXUR1 and 2, sister genes to FERONIA/SIRENE, are male factors for coordinated fertilization. Current Biology 19, 1327–1331. [DOI] [PubMed] [Google Scholar]
- Monshausen GB, Bibikova TN, Messerli MA, Shi C, Gilroy S. 2007. Oscillations in extracellular pH and reactive oxygen species modulate tip growth of Arabidopsis root hairs. Proceedings of the National Academy of Sciences, USA 104, 20996–21001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagano M, Ishikawa T, Fujiwara M, Fukao Y, Kawano Y, Kawai-Yamada M, Shimamoto K. 2016. Plasma membrane microdomains are essential for Rac1–RbohB/H-mediated immunity in rice. The Plant Cell 28, 1966–1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nibau C, Cheung AY. 2011. New insights into the functional roles of CrRLKs in the control of plant cell growth and development. Plant Signaling & Behavior 6, 655–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ning Y, Liu W, Wang GL. 2017. Balancing immunity and yield in crop plants. Trends in Plant Science 22, 1069–1079. [DOI] [PubMed] [Google Scholar]
- Nishimura A, Aichi I, Matsuoka M. 2006. A protocol for Agrobacterium-mediated transformation in rice. Nature Protocols 1, 2796–2802. [DOI] [PubMed] [Google Scholar]
- Pearce G, Moura DS, Stratmann J, Ryan CA Jr. 2001. RALF, a 5-kDa ubiquitous polypeptide in plants, arrests root growth and development. Proceedings of the National Academy of Sciences, USA 98, 12843–12847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prusky D, Yakoby N. 2003. Pathogenic fungi: leading or led by ambient pH? Molecular Plant Pathology 4, 509–516. [DOI] [PubMed] [Google Scholar]
- Pu CX, Han YF, Zhu S, et al. 2017. The rice receptor-like kinases DWARF AND RUNTISH SPIKELET1 and 2 repress cell death and affect sugar utilization during reproductive development. The Plant Cell 29, 70–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi J, Wang J, Gong Z, Zhou JM. 2017. Apoplastic ROS signaling in plant immunity. Current Opinion in Plant Biology 38, 92–100. [DOI] [PubMed] [Google Scholar]
- Qu S, Zhang X, Song Y, Lin J, Shan X. 2017. THESEUS1 positively modulates plant defense responses against Botrytis cinerea through GUANINE EXCHANGE FACTOR4 signaling. Journal of Integrative Plant Biology 59, 797–804. [DOI] [PubMed] [Google Scholar]
- Rensing SA, Lang D, Zimmer AD, et al. 2008. The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science 319, 64–69. [DOI] [PubMed] [Google Scholar]
- Schallus T, Jaeckh C, Fehér K, et al. 2008. Malectin: a novel carbohydrate-binding protein of the endoplasmic reticulum and a candidate player in the early steps of protein N-glycosylation. Molecular Biology of the Cell 19, 3404–3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze-Muth P, Irmler S, Schröder G, Schröder J. 1996. Novel type of receptor-like protein kinase from a higher plant (Catharanthus roseus). cDNA, gene, intramolecular autophosphorylation, and identification of a threonine important for auto- and substrate phosphorylation. Journal of Biological Chemistry 271, 26684–26689. [DOI] [PubMed] [Google Scholar]
- Shimono M, Sugano S, Nakayama A, Jiang CJ, Ono K, Toki S, Takatsuji H. 2007. Rice WRKY45 plays a crucial role in benzothiadiazole-inducible blast resistance. The Plant Cell 19, 2064–2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegmann M, Monaghan J, Smakowska-Luzan E, Rovenich H, Lehner A, Holton N, Belkhadir Y, Zipfel C. 2017. The receptor kinase FER is a RALF-regulated scaffold controlling plant immune signaling. Science 355, 287–289. [DOI] [PubMed] [Google Scholar]
- Thordal-Christensen H, Zhang Z, Wei Y, Collinge DB. 1997. Subcellular localization of H2O2 in plants. H2O2 accumulation in papillae and hypersensitive response during the barley–powdery mildew interaction. The Plant Journal 11, 1187–1194. [Google Scholar]
- Thynne E, Saur IML, Simbaqueba J, et al. 2017. Fungal phytopathogens encode functional homologues of plant rapid alkalinization factor (RALF) peptides. Molecular Plant Pathology 18, 811–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N, Xiao B, Xiong L. 2011. Identification of a cluster of PR4-like genes involved in stress responses in rice. Journal of Plant Physiology 168, 2212–2224. [DOI] [PubMed] [Google Scholar]
- Wang Y, Tang H, Debarry JD, et al. 2012. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Research 40, e49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong HL, Pinontoan R, Hayashi K, et al. 2007. Regulation of rice NADPH oxidase by binding of Rac GTPase to its N-terminal extension. The Plant Cell 19, 4022–4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X, Ma X, Zhu Q, Zeng D, Li G, Liu YG. 2017. CRISPR-GE: a convenient software toolkit for CRISPR-based genome editing. Molecular Plant 10, 1246–1249. [DOI] [PubMed] [Google Scholar]
- Xing J, Jia Y, Peng Z, Shi Y, He Q, Shu F, Zhang W, Zhang Z, Deng H. 2017. Characterization of molecular identity and pathogenicity of rice blast fungus in Hunan province of China. Plant Disease 101, 557–561. [DOI] [PubMed] [Google Scholar]
- Yang T, Wang L, Li C, Liu Y, Zhu S, Qi Y, Liu X, Lin Q, Luan S, Yu F. 2015. Receptor protein kinase FERONIA controls leaf starch accumulation by interacting with glyceraldehyde-3-phosphate dehydrogenase. Biochemical and Biophysical Research Communications 465, 77–82. [DOI] [PubMed] [Google Scholar]
- Yu F, Qian L, Nibau C, et al. 2012. FERONIA receptor kinase pathway suppresses abscisic acid signaling in Arabidopsis by activating ABI2 phosphatase. Proceedings of the National Academy of Sciences, USA 109, 14693–14698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Fang W, Liu C, Zhao W, Peng Y. 2014. Identification of Magnaporthe oryzae pathotypes by wounding inoculation of detached rice leaves. Plant Protection. 40, 121–125 (in Chinese). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.