Abstract
In yam (Dioscorea spp) species, bulbils at leaf axils are the most striking species-specific axillary structure and exhibit important ecological niches. Genetic regulation underlying bulbil growth remains largely unclear so far. Here, we characterize yam (Dioscorea alata L.) bulbil development using histological analysis, and perform full transcriptional profiling on key developmental stages together with phytohormone analyses. Using the stage-specific scoring algorithm, we have identified 3451 stage-specifically expressed genes that exhibit a tight link between major transcriptional changes and stages. Co-expressed gene clusters revealed an obvious over-representation of genes associated with cell division and expansion at the initiation stage of bulbils (T1). Transcriptional changes of hormone-related genes highly coincided with hormone levels, indicating that bulbil initiation and growth are coordinately controlled by multiple phytohormones. In particular, localized auxin is transiently required to trigger bulbil initiation, and be further depleted or exported from bulbils to promote growth by up-regulation of genes involved in auxinconjugation and efflux. The sharp increase in supply of sucrose and an enhanced trehalose-6-phophate pathway at T1 were observed, suggesting that sucrose probably functions as a key signal and promotes bulbil initiation. Analysis of the expression of transcription factors (TFs) predicated 149 TFs as stage-specifically expressed; several T1-specific TFs (from Aux/IAA, E2F, MYB, and bHLH families) have been shown to play key roles in triggering bulbil formation. Together, our work provides a crucial angle for in-depth understanding of the molecular programs underlying yam’s unique bulbil development processes. Stage-specific gene sets can be queried to obtain key candidates regulating bulbil growth, serving as valuable resources for further functional research.
Keywords: Bulbil, genetic regulation, growth, transcriptome, phytohormone, yam (Dioscorea alata L)
Phytohormones and sucrose coordinately control bulbil initiation, and provide forward signaling to genes that maintain the vitality of cell proliferation and expansion during growth.
Introduction
Plants exhibit a high degree of plasticity in axillary forms including dormant axillary buds, tillers, vegetative branches, or other specific axillary structures (Li et al., 2003; Evers et al., 2011). In some circumstances, plants evolve unique aerial bulbils derived from the growth of meristems in the axils of leaves or bracts, in response to the ecological or evolutionary niches in which the plant grows (Law et al., 1983; Szarek et al., 1996; Ronsheim and Bever, 2000; Deng et al., 2013). In particular, the bulbils released from the mother plants can grow into new individuals in the next growth season, and offer a failsafe strategy of asexual reproduction when seeds are absent under harsh environmental conditions (Arizaga and Ezcurra, 2002; Walck et al., 2010).
Bulbils are rarely observed in nature, but are generally seen in several plant populations from Agave tequilana, Titanotrichum oldhamii, Polygonum viviparum, Lilium lancifolium, Poa alpina, and numerous members of Dioscorea (Law et al., 1983; Murty and Purnima, 1983; Szarek et al., 1996; Wang and Cronk, 2003; Steiner et al., 2012; Yang et al., 2017). The production of these species-specific structures and their developmental patterns are distinct. In Agave and Titanotrichum plants, the floral meristems develop into numerous bulbils by repeated subdivision when the flowers fade away; this type of bulbil occurs on the sides of the inflorescence or pedicel (Wang and Cronk, 2003; Sandoval et al., 2012). In some cases, bulbils are also derived from the adjacent proximal meristems (Moody et al., 1999). For most bulbil-producing plants, such as Dioscorea and Lilium species, bulbils similar to buds are generated from axillary meristems (AMs) at the axil when apical dominance ceases (Murty and Purnima, 1983; Yang et al., 2017).
Little available information has been documented so far to explain the regulation processes of bulbil initiation and growth. Consistent with buds originated from AMs, bulbil growth involves two developmental stages, namely (i) the formation of new AMs induced by bulbil primordia; and (ii) continuous cell division of AMs for establishing bulbil form (Li et al., 2012; Yang et al., 2017; Wang and Jiao, 2018). Limited evidence has demonstrated that the fate of early bulbil formation, whether to grow or remain dormant, is strongly dependent on environmental factors, phytohormone signals, and genetic factors (Fan and Yang, 2009; Steiner et al., 2012; Abraham-Juárez et al., 2015). Bulbil-producing plants of P. alpina often occur in high altitude populations, and remain strongly vigorous (Steiner et al., 2012). Titanotrichum plants easily induce the transition from the flower to bulbil meristems when exposed to short-day conditions (autumn); and individuals under dense shade can produce more bulbils even under full light (Wang and Cronk, 2003). More importantly, plant hormones play crucial roles in regulation of bulbil formation at the early stage. In Agave plants, the reduction of auxin by direct removal of flower buds induces bulbil initiation, whereas exogenous application of auxin on cut pedicle tissue represses bulbil growth (Abraham-Juárez et al., 2015). Similarly, the short-term accumulation of auxin in upper axils activates bulbil initiation in Lilium species (Yang et al., 2017). Elevated gibberellin (GA) significantly increases bulbil yield of Chinese yam by exogenous application (Kim et al., 2003). Several genetic studies have revealed that bulbil initiation is regulated by multiple auxin-mediated transcription factors (TFs), such as KNOX, MADS, and LEAFY (Wang et al., 2004; Abraham-Juárez et al., 2010; Sandoval et al., 2012). Alternatively, the PIN proteins localized in the pedicel function by modulating auxin gradients in A. tequilana, and play a central role in the initial stage of bulbil formation (Abraham-Juárez et al., 2015). A gesneriaceae-FLORI-GAULA (GFLO) gene in T. oldhamii that switches the process of flowering to bulbil production, has been identified as a key candidate regulating bulbil formation (Wang et al., 2004). Whereas little is known in terms of understanding the regulatory mechanisms of bulbil growth, there has been a massive increase in understanding of axillary bud growth over the past 30 years (Chatfield et al., 2000; Wang and Jiao, 2018; Chabikwa et al., 2019). Two widely accepted theories, auxin transport canalization (ATC) and the second messenger (SM) models, have been proposed to describe the mechanisms regulating bud growth (Domagalska and Leyser, 2011). Central to the ATC model is that auxin from the shoot apex inhibits the outgrowth of buds, and must be depleted to activate expansion of new AMs, in which the polar auxin transport (PAT) stream is essential and driven by PIN efflux proteins (Balla et al., 2011). The SM model contends that auxin does not enter the bud, and regulates the production of axillary buds by second messengers, which directly move into the bud to control its activity (Domagalska and Leyser, 2011). These messengers include the hormones cytokinin (CK) and abscisic acid (ABA) (Yao and Finlayson, 2015; Roman et al., 2016).
Yams (Dioscorea spp) represent one of the most primitive angiosperms and differentiated species (Burkill, 1960). In >600 members recorded in this genus, the most striking axillary plasticity is the occurrence of bulbils that are termed aerial tubers or minor storage organs (Wickham et al., 1982; Murty and Purnima, 1983). Ecologically, this structure, initiated at leaf axils, enables the plants to spread rapidly and engulf the native vegetation (Mizuki and Takahash, 2009). Yam bulbils also serve as an effective means for vegetative reproduction, and are often dormant and will germinate when they drop to the ground in the following season (Main et al., 2006). In many yam species, aerial bulbils are very common and have been produced widely as food or for pharmaceutical uses (Asiedu and Sartie, 2010). Despite their importance in terms of commercial values and ecological plasticity, the gene regulatory programs throughout bulbil growth still remain largely unclear. Here, we selected a typical yam species (D. alata Linn.) and combined morphological analysis, and phytohormone- and stage-specific expression profiles, to uncover the regulatory events underlying bulbil growth. We established a regulatory model, and reported the key roles of auxin, CK, and sucrose as signals during bulbil initiation. We also identified stage-specifically expressed gene sets associated with regulation of cell division, proliferation, and expansion underlying early bulbil formation, which provide valuable genomic resources for further functional analysis.
Materials and methods
Plant materials
Homogeneous seedlings of yam (D. alata) were asexually propagated using severed tubers (100–150 g per plant) from the same mother plant. The seedlings were grown in Wenzhou botanical garden (Wenzhou city, Zhejiang province, China) under standard conditions. The bulbils were seen in the axil of leaf primordia after 130 d growth. Four staged samples of bulbil were harvested at 1 day after bulbil emergence (1 DAE) termed the initiation stage (labeled T1), early stage (7 DAE, T2), middle stage (15 DAE, T3), and mature stage (35 DAE, T4). Five plants were collected for each bulbil type.
Histological analyses of bulbil development
Bulbils across the four investigated stages were fixed in FAA solution (70% ethanol:formalin:acetic acid, 90:5:5) for 24 h at 4 °C. The fixed bulbils were dehydrated through a graded ethanol series and embedded in paraffin, as described previously (Xing et al., 2010). Longitudinal sections of 10 μm were prepared with a rotary microtome and stained in safranin–alcian green according to standard histological procedures (Gutmann,1995). Stained sections were observed and documented under an OLYMPUS BX60 light microscope (Olympus, Japan).
Determination of sucrose content
A 20 mg aliquot of bulbils was ground in liquid nitrogen and extracted with 80% ethanol for 40 min at 60 °C in cap-sealed tubes using 1 ml per 200 mg sample. The extraction was carried out twice with the same conditions. The combined suspension was centrifuged at 10 000 g for 10 min at 4 °C. The supernatant was analyzed and sucrose contents were determined with HPLC-evaporative light scattering detector (ELSD) analysis (Agilent 1200) as described previously (Ma et al., 2014). Five independent experiments were carried out for each bulbil type.
Phytohormone metabolic profiles
CK metabolites (zeatin and dihydrozeatin), free indoleacetic acid (IAA), and ABA were detected by using an LC-ESI-MS/MS system (HPLC, Shim-pack UFLC SHIMADZUCBM30A system; MS, Applied Biosystems 6500 Triple Quadrupole) (Šimura et al., 2018). A 50 mg (FW) portion of bulbils was homogenized in liquid nitrogen and extracted with methanol:water:formic acid (15:4:1, v/v/v). The extracts were further dried under a nitrogen gas stream and reconstituted in 80% methanol (v/v). The treated extracts were separated using an ACQUITY UPLC HSS T3 C18 (1.8 μm, 2.1 mm×100 mm) (Waters). Analytes were eluted by a binary gradient phase constituted of ice-cold acetonitrile (with 0.01% v/v formic acid) (A) and water (with 0.01% v/v formic acid) (B), with a flow rate of 0.35 ml min–1. The gradient programs were as follows: 0–1 min, 5% (v/v) A; 1–8 min, 5–95% (v/v) A; 8–9 min, 95% (v/v) A; 9–12 min, 5% (v/v) A. Levels of IAA, zeatin, dihydrozeatin, and ABA were determined by Metware (http://www.metware.cn/) based on the AB Sciex QTRAP 6500 LC-MS/MS platform. Three replicates of each assay were performed.
RNA sequencing and bioinformatics analyses
Total RNA of each sample was isolated using TRIzol reagent (Invitrogen) and purified using DNase I (TURBO DNase, Ambion, USA). The integrity of RNA (RIN >8.5) was detected using a Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA, USA). RNA sequencing (RNA-seq) libraries were prepared using the cDNA Synthesis kit (Illumina Inc., San Diego, CA, USA) following the standard Illumina preparation protocol. Paired-end sequencing (2×150 bp) was conducted with Illumina HiSeq 2500 (Illumina Inc.) by the Biomarker Biotechnology Corporation (Beijing, China). Three independent biological replicates were analyzed for each bulbil type. The raw reads were refined by removing reads with only adaptor and unknown nucleotides >10%, and low-quality reads with the average phred quality score <30. The clean reads were used for robust de novo assembly of a set of transcriptomes using the Trinity software package (version R2013-02-25) (Haas et al., 2013).
The expression abundance of each transcript was estimated by calculating the FPKM (fragments per kilobase of exon per million fragments mapped) value using TopHat (version 2.0.8) (Trapnell et al., 2012). Based on overall expression profiles, correlation of three biological replicates was calculated to assess the reliability of sample collection using the cor R package. Differentially expressed genes (DEGs) between differential bulbil types were identified using the Edge statistical test in terms of the following criteria: false discovery rate (FDR) <0.01, and an absolute expression foldchange (FC) ≥2 for a given gene between any comparisons (Robinson et al., 2010). We annotated biological function for DEGs using NR public databases according to BLASTX analysis with a cut-off E-value of 10−5. GO slim was conducted to obtain Gene Ontology (GO) annotations using Blast2GO (Conesa et al., 2005). To obtain enriched slims, we further performed GO term enrichment analysis using the algorithm and the Kolmogorov–Smirnov (KS) test (P-value≤0.001) in the R package topGO (Alexa et al., 2006). Based on all expressed genes, principal component analysis (PCA) was carried out to explain the relatedness among all samples. For all DEGs, we performed hierarchical cluster analysis (HCA) to present gene expression patterns using the pheatmap library in R software. In addition, TFs were identified in terms of the Arabidopsis transcription factor database (Perez-Rodriguez et al., 2010).
Identification of stage-specific (SS) gene sets
The genes specifically expressed in each stage type were identified using an SS scoring algorithm based on Zhan et al. (2015). For a given gene i, its expression values in four stages are described as Ei=+++), and the SS score of this gene in stage j is defined as follows, SS(i, j)=1-, where 1≤k≤4, k≠j. Therefore, SS scores range from 0 to 1, and genes with higher SS scores at a particular stage are likely to be more specifically expressed uding that stage. We here defined genes with an SS score >0.3 as being specifically expressed in corresponding stages.
Validation of genes using quantitative RT–PCR
Quantitative reverse transcription–PCR (qRT–PCR) was performed to verify transcriptome profiles with 20 selected genes. Three biological replicates were carried out for each selected gene. The PCR amplification primers for selected genes were designed with Primer 3 software, and sequences are listed in Supplementary Table S1 at JXB online. RNA was extracted as described above. The cDNA was prepared with SuperScript III Reverse transcriptase (Invitrogen) following the manufacturer’s protocol on 1μg of RNA. The qRT–PCR amplification was run in an ABI 7500 HT (Life Technology) with SYBR Green I Master Mix (TaKaRa), and the reaction mixture and program were carried out as described in our previous work (Wu et al., 2015). Quantification of gene expression was normalized using the EF-1a gene (accession no. JF825419) as an internal control, and counted according to the 2−ΔΔCt method (Livak and Schmittgen, 2001).
RNA in situ hybridization
RNA in situ hybridization was performed using the method described by Siciliano et al (2007). Bulbils were fixed in FAA and embedded in paraffin as described above. Gene-specific fragments for probe synthesis were amplified by PCR using designed primers: 5'-GAAGAGCACCATGCTGTGAG-3' and 5'-TAATACGACTCACTATAGGGCCACATCTCAGCAATCCAG-3' for MYB (Gene ID: c126446.graph-c0), 5'-TGGAGAGCCTTTGATCGGTT-3' and 5'-TAATACGACTCACTATAGGGCCACTGCTCTAAACGAAGG-3' for WRKY (c125026.graph_c1), and 5'-AGTGCATTACCTCTGCCGGA-3' and 5'-TAATACGACTCACTATAGGTACCTGGCAATTCCCAAGGA-3' for NAC (c116834.graph_c0). The resulting PCR fragments were used as templates for synthesis of digoxigenin (DIG)-labeled antisense and sense riboprobes with the T7/SP6 riboprobe and a DIG-RNA labeling mix (Roche). Sections (8–10 μm) were treated with 1 μg ml−1 proteinase K for 30 min at 37°C, and then washed under stringent conditions as described previously (Hsu et al., 2015).
Results
Bulbil morphology during its developmental stages
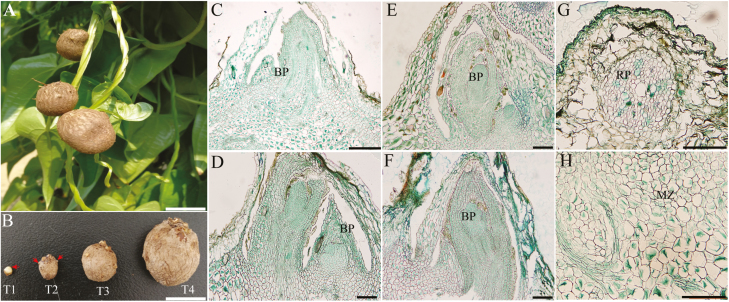
Yam bulbils occur naturally on the leaf axil when the main apex stops growing (Fig. 1A). To characterize the developmental process of bulbils in detail, we designed four growing sequences based on our observations and important literature (Murty and Purnima, 1983), namely the initiation (T1), early (T2), middle (T3), and mature (T4) stages of bulbil formation (Fig. 1B). At the T1 stage, the cells of 2–3 layers below the leaf axil have undergone periclinal and anticlinal divisions, and developed into a hump-like meristematic tissue that is termed the bulbil primordium (BP) (Fig.1C). The BP at this stage was pivotal for subsequent bulbil outgrowth and still remained partly differentiated At the same time, a dome-shaped structure was visible in the leaf axil. At the T2 stage, the cells from the BP meristematic zone (Fig. 1H) became highly meristematic and showed successive cell division and enlargement (Fig. 1D), and ultimately formed the young bulbil. The young bulbil was shaped like a spinning top, and the root primordium (RP) was seen in the cortical region at this stage (Fig. 1G). At the next stage (T3), the size of the bulbil had enlarged rapidly, as the meristem in the central region of the bulbil continuously widened (Fig. 1E). The increased cell numbers and enlarged volume by filling starch grains resulted in quasi-round bulbils. The bulbil at this stage had a distinct peripheral region covered by several rough epidermal layers (Fig. 1B). At the T4 stage, the activity of meristematic tissue was mostly depleted (Fig. 1F) and mature bulbils reached 1.2–3.0 cm in diameter. Multiple RPs had emerged on the bulbil surface, which enable bulbils to spread rapidly in the next season. Taken these findings together, bulbil morphology was distinct at the four developmental stages.
Fig. 1.
Morphology of the bulbil during key developmental stages. (A) Bulbil phenotype. (B) Photographs of the bulbil at the initiation (T1), early (T2), middle (T3), and mature stages (T4). (C–F) Paraffin sections of bulbils at T1 (C), T2 (D), T3 (E), and T4 (F) stages. The images show the zone of the junction region between the bulbil and axil. (G) Showing root primordia (RP). (H) Showing the meristematic zone (MZ). The arrows in T1 and T2 show the BP and root primordia, respectively. Scale bars (A and B), 1 cm; (C), 500 µm; (D–H), 200 µm.
Global RNA-seq analysis of yam bulbils during development
Given the histological differences between bulbil stage types, we collected three biological replicates of each type for the preparation of RNA-seq libraries. All raw reads obtained by RNA-seq have been submitted to the SRA database under accession number SRP152752. High quality RNA-Seq data were obtained for each sample, with 49–66 million paired-end reads (Supplementary Table S2). The de novo assembly generated 199 270 transcripts; ~36% of transcripts were in the size range 200–500 bp (Supplementary Table S3). The homologous transcripts were further clustered with >95% similarity, and generated 97 956 unigenes with a total length of 79 527036 bp and an average length of 812 bp. Read counts per gene were calculated and normalized as FPKM (Trapnell et al., 2012). An expressed gene was required to have an FPKM of >1.0 in at least one sequenced sample.
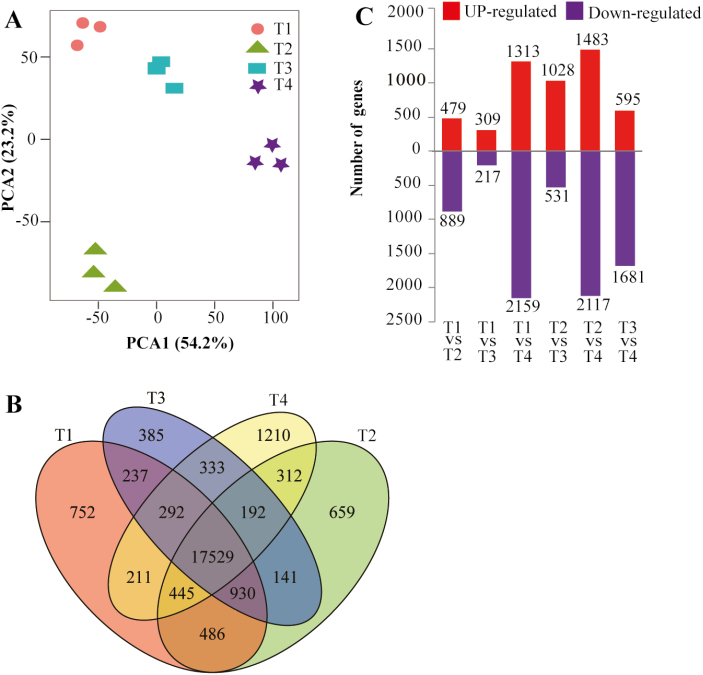
PCA visualized four stage-specific clusters, with the first two components explaining 77.4% of the total variance, and revealed distinct mRNA populations between different bulbil types (Fig. 2A). Next, we observed that 752 (in T1), 659 (T2), 385 (T3), and 1210 (T4) genes had highly stage-specific expression patterns, but most of the genes (17 529) were shared in bulbils at all stages (Fig. 2B). Meanwhile, we assessed gene expression profiles between biological triplicates and they were highly correlated (R2 >0.83), (Supplementary Fig. S1) indicating that bulbils from the same stage were well collected, avoiding the variation of samples from the same stage.
Fig. 2.
Gene expression profiles during bulbil growth. (A) Principal component analysis for 12 bulbil types showing the four stage-specific groups based on all gene expression profiles. (B) Venn diagram showing the numbers of unique and overlapping expressed genes in bulbils among different developmental stages (T1–T4). (C) Comparison of the number of up- and down-regulated genes between different stages.
To confirm the reliability of RNA-seq, we also performed a more rigorous expression measure for 20 selected genes using qRT–PCR analysis. We revealed a good agreement with a high linear correlation (R2 >0.80; see Supplementary Fig. S2) between RNA-seq and qRT–PCR technologies, suggesting the reliability of RNA-seq analyses.
Using a threshold value of FC >2 in expression difference and FDR <0.01 for filtering DEGs, we identified a total of 6112 DEGs in a least one comparison (Fig. 2C; Supplementary Table S4). These results represented substantial differences in gene expression profiles across bulbil growth. HCA using all DEGs revealed four discrete clusters corresponding to four time points (Supplementary Fig. S3), which constitute distinct gene sets, highlighting the specialized nature of different bulbil types, suggesting a tight linkage of DEGs with bulbil growth responses. Furthermore, GO enrichment analysis for DEGs indicated that GO terms including regulation of cell cycle, regulation of DNA replication, cell proliferation, and protein kinase activity, are strikingly enriched (corrected P-value <0.001) (Supplementary Table S5; Supplementary Fig. S4). Several significantly enriched KEGG (Kyoto Encyclopedia of Genes and Genomes) pathways were suggested to be linked to starch and sucrose metabolism, and plant hormone signal transduction (Supplementary Table S6; Supplementary Fig. S5).
Stage-specific gene clusters contributing to bulbil initiation and growth
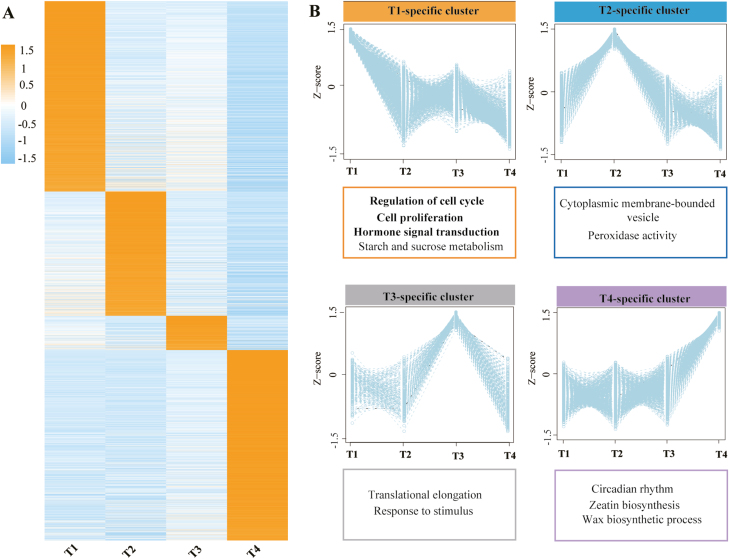
Our aim in this study was to identify genes associated with yam bulbil growth from the different stages, especially for genes promoting bulbil initiation at the early stage. Therefore, we further narrowed the DEG set and identified sets of tissue-specifically expressed genes through the SS scoring algorithm (see the Materials and methods). In this analysis, we identified a total of 3451 SS genes (Fig. 3A; Supplementary TableS7). We found that the numbers of detected SS genes are obviouslydifferent among the four stage types. T1 and T4 types had the most SS genes, with 1218 genes being identified as stage specific at the two stages. In contrast, the T3 type had the lowest number of SS genes (220). These SS genes exhibited consistent expression patterns and peak abundances in the corresponding specific stages (Fig. 3B). We also found that most SS genes are over-represented in these enriched GO terms and KEGG pathways for all DEGs (Fig. 3B), suggesting that these SS genes may be important candidates for regulating bulbil growth. Accordingly, we classified SS genes into these enriched GO terms and pathways to elucidate their functions.
Fig. 3.
Stage-specific gene clusters. (A) Heat map of scaled FPKM values of 3451 stage-specific genes (with SS score >0.3) during bulbil growth. (B) Average expression profiles of each gene cluster and its corresponding enriched GO functional category. The sets of stage-specific genes are available in Supplementary Table S7.
The T1-specific cluster highly expressed a large number of genes associated with cell cycle and proliferation
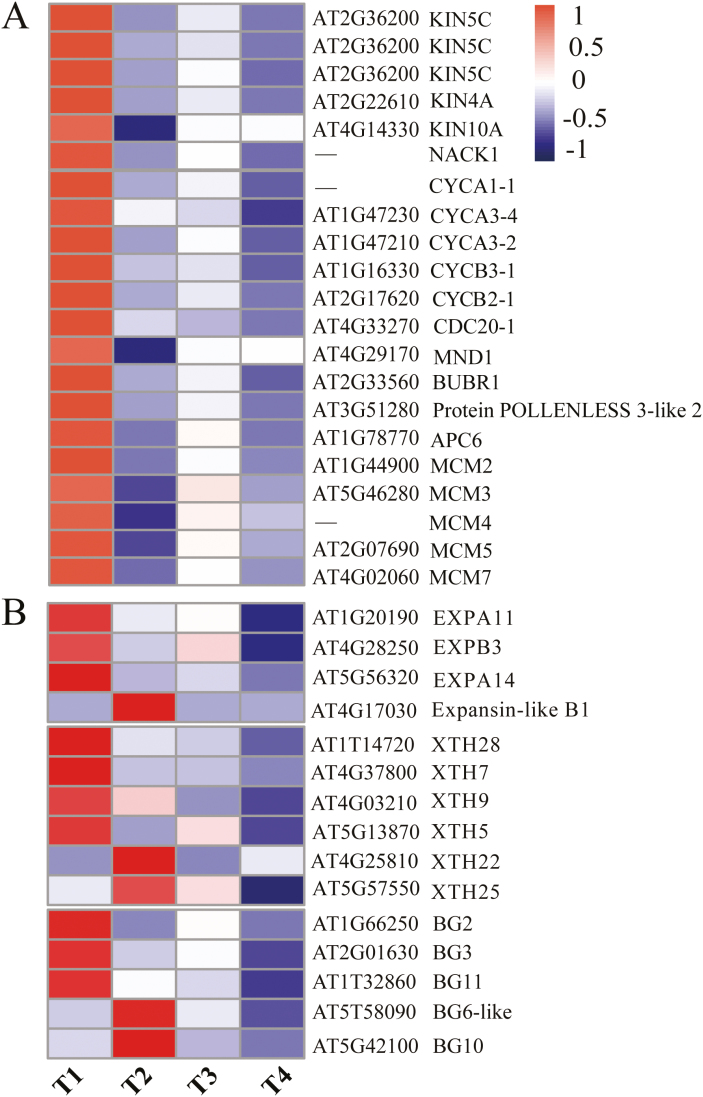
Interestingly, GO terms including regulation of the cell cycle, cytokinesis by cell plate formation, and cell proliferation were highly co-enriched in the T1-specific gene cluster (Fig. 3B), which hints at the co-regulation of genes coupled with these terms during transcriptomic responses to bulbil initiation. To better understand the control, we screened for SS genes involved in these enriched terms and detected 22 marker genes (Fig. 4A; Supplementary Dataset S1). These genes included seven genes encoding multiple kinesin-related proteins (three homologs of KIN5C, KIN4A, KIN10A, and NACK1) and a TIO kinase (AT1G50240), which are required to support cell plate growth in meristem tissue (Oh et al., 2012; Tan et al., 2019). Ten genes involved in the regulation of cell cycle or division were also observed, including genes encoding A/B-type cyclin proteins (CYCA1-1, CYCA3-2, CYCA3-4, CYCB2-1, and CYCB3-1), and multiple proteins (MND1, BUBR1, APC6, and POLLENLESS 3-like 2 protein) that control progression through mitosis in the G1/2 phase of the cell cycle (Devitt and Stafstrom, 1995; Berckmans and Veylder, 2009).
Fig. 4.
Heat maps of gene sets involved in the cell cycle and proliferation (A), and cell wall expansion (B). The gene expression levels are standardized into Z-scores and colored in red and blue for high and low expression, respectively. Gene names (shown on the right) are described in detail in Supplementary Dataset S1.
In addition, we found five members of DNA replication licensing factors (MCM2, MCM3, MCM4, MCM5, and MCM7), which play crucial roles in the control of cell proliferation processes required for lateral meristem formation (Ni et al., 2009). Unexceptionally, these genes were consistently induced and exhibited the highest expression at the initial stage of bulbil growth (T1) (Fig. 4A). For instance, as compared with the T2 stage, the transcript accumulation levels for the representative genes KIN5C (c132756.graph_c0), CYCA3-4 (c122639.graph_c0), and MCM4 (c105199.graph_c0) were up-regulated 4.8-, 2-, and 8-fold, respectively. Taken together, the trend in accumulation of cell cycle- and proliferation-related genes performed a quality control function to produce the large number of new cells required on the flanks of the stem and to ultimately produce the obvious visible bulbil meristem (Devitt and Stafstrom, 1995).
Genes related to cell wall modification were induced at the early stage of bulbil growth
Cell wall extension, as a secondary event followed by cell division, plays a crucial regulatory role in inducing early primordium initiation in meristems and determining the size and shape of cell- and plant-specific organs (Chebli and Geitmann, 2017). We here searched known genes involved in modification of the cell wall network through expansion and loosening, and identified 15 SS genes in this process (Supplementary Dataset S1). These genes included four EXPANSINS (EXPA11, EXPA14, EXPB3, and EXPB1-like), six genes encoding xyloglucan endotransglucosylase/hydrolase (XTH5, XTH7, XTH9, XTH22, XTH25, and XTH28), and five members of the glycosyl hydrolase family 17 (BG2, BG3, BG6, BG10, and BG11), which can loosen wall-like networks and control multiple types of growth and development in plants (Cosgrove, 2005). Most of them showed higher expression levels at the early stage of bulbil growth (T1 and T2) (Fig. 4B). Importantly, EXPANSINS can control the formation of the AM (Fleming et al., 1997; Zenoni et al., 2011). We found that three EXPANSINS (EXPA11, EXPA14, and EXPB3) showed peak abundance at T1 and subsequently were reduced 2.3-, 2.2-, and 4.8-fold at T2, respectively, suggesting a localized drive for initiating the hump-like BP (Fig. 1C).
Transcriptional profiles of hormone candidates: auxin, CK, and ABA
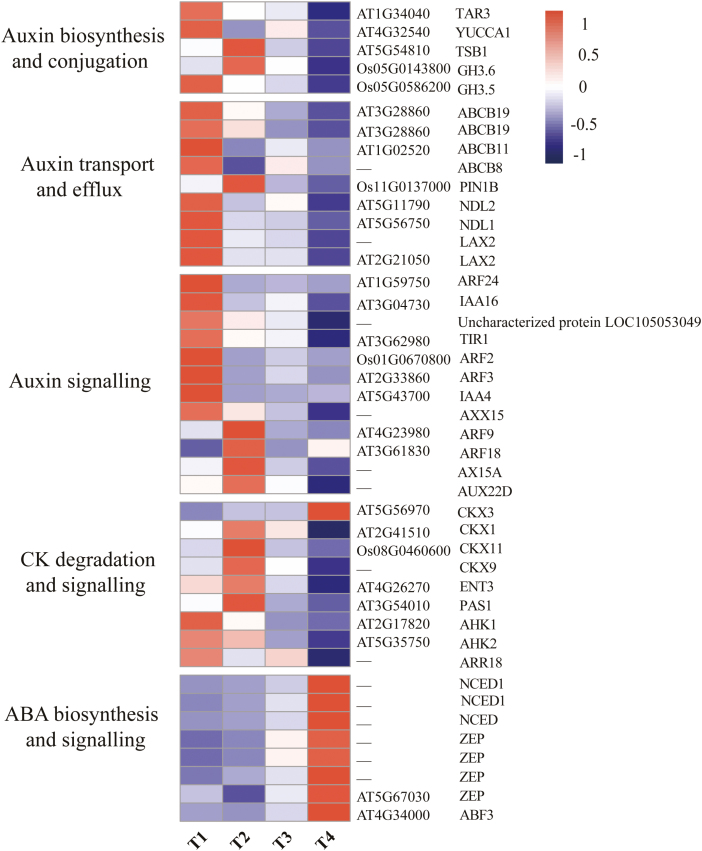
To investigate hormonal regulation during bulbil growth, we analyzed expression changes of genes involved in auxin, CK, and ABA biosynthesis, transport, and signaling (Supplementary Dataset S1). For auxin, three genes, TRYPTOPHAN AMINOTRANSFERASE RELATED3 (TAR3), YUCCA1, and tryptophan synthase (TSB1), encoding intermediate enzymes involved in auxin biosynthesis, were detected. Among them, TAR3 and YUCCA1were T1-specifically expressed and showed peak abundance at the T1 stage; TSB1 was identified as a T2-specifically expressed gene, showing the highest expression at this stage. In addition, two INDOLE-3-ACETIC ACID-AMIDO SYNTHETASES (GH3.5 and GH3.6) involved in auxin conjugation (Chen et al., 2010), were identified as T1- and T2-specifically expressed, and exhibited peak expression at the T1 and T2 stages, respectively (Fig. 5).
Fig. 5.
Heat maps of gene sets associated with hormone (auxin, CK, and ABA) biosynthsis and catabolism, transport, and signaling. The expression levels are standardized per gene into Z-scores and colored in red and blue for high and low expression, respectively. Gene names (shown on the right) are described in detail in Supplementary Dataset S1.
We also detected nine genes encoding proteins involved in auxin transport and efflux, namely transmembrane-targeted efflux carrier PIN1B, members of the ABC transporter B family (ABCB8/11 and two homologs of ABCB19), two homologs of auxin transporter-like protein 2 (LAX2), and NDL proteins (NDL1and NDL2). As expected, PIN1B displayed peak abundance at T2 as a T2-specific gene, and the remaining genes were specifically expressed at T1 and had the highest expression levels at this stage (Fig. 5). In particular, NDL proteins (NDL1 and NDL2) acting as positive regulators in the signaling pathway that modulate auxin transport (Mudgil et al., 2009), were highly expressed at the bulbil initiation stage (T1) and reduced ~2-fold at T2. Interestingly, key components of auxin signaling include AUX/IAA co-receptors (Aux/IAAs) and (ARFs) (Guilfoyle and Hagen, 2007), which had similar expression trends with obvious evidence of stage specificity (Fig. 5). Among them, multiple IAA and ARF genes (IAA4, IAA16, ARF2, ARF3, Axx15, and AX15A-like) were grouped into T1-spcific genes, which have also been implicated in other vegetative developmental growth (Faivre-Rampant et al., 2004; Deng et al., 2012; Jung et al., 2015), and another four genes (ARF9, ARF18, AX15A, and AUX22D) were grouped as T2-specific genes with peak expression levels at T2.
CKs can promote axillary bud outgrowth (Ferguson and Beveridge, 2009). We found that four CK dehydrogenases (CKXs) that decrease CK accumulation (Frébortová et al., 2010) had lower expression levels at T1 relative to other stages (Supplementary Dataset S1). The four genes appeared to be obviously stage specific (namely T2, CKX1, CKX9, and CKX11; and T4, CKX3), showing the highest expression at T2 or T4 (Fig. 5). Remarkably, no CK biosynthetic genes were detected. Although not an SS gene, ENT3 that participates in CK transport (Hirose et al., 2005) was highly expressed (339 FPKM) at T2 but then was strongly reduced 5.6-fold at T4. PAS1 (AT3G54010) that controls the cell division and differentiation mediated by CK (Harrar et al., 2003) was highly induced at T1 and T2, with peak expression at T2. Two histidine kinases (AHK1/2) as CK receptors and a CK-inducible response regulator (ARR18) were observed, and had the highest expression levels at T1.
Interestingly, all SS genes involved in ABA synthesis and signaling were identified as T4-specifically expressed and had the highest expression levels at T4 (Fig. 5; Supplementary Dataset S1). For instance, three genes encoding the rate-limiting ABA biosynthetic enzyme 9-cis-epoxycarotenoid dioxygenase (NECD) were expressed with an average 17-fold increase from T1 to T4. Similarly, the average expression level of four zeaxanthin epoxidases (ZEPs) responsible for regulating intermediates in ABA biosynthesis was elevated 2.5-fold at T4 relative to T1. In addition, an ABA-INSENSITIVE protein (ABF3) binding to the ABA-responsive element (Kim et al., 2004) was up-regulated 3-fold at T4 relative to T1. The expression changes for ABA-related genes provided obvious evidence that enhanced ABA biosynthesis and signaling exist at the later stage of bulbil growth (T4).
Further, to validate the involvement of auxin, CKs, and ABA during bulbil growth, we tested their contents and analyzed their correlation with the expression of hormone-related genes (Table 1; Supplementary Fig. S6). The highest amount (37.54 ng g−1) of IAA at T1 was observed and showed a high correlation with increased expression of auxin biosynthetic genes (TAR3 and YUCCA1) (R2 >0.83), demonstrating the localized production of auxin in bulbils. GH3.5 and several of the transport proteins mentioned above (ABCB19, LAX2, and NDL1) were also tightly linked to auxin levels (Supplementary Fig. S6), which provides a mechanism for coping with excess auxin by conjugation and transportation. The high contents of CKs (zeatin and dihydrozeatin) existing at T1 and T2 (Table 1) were coincident with the repression of CKX1 and CKX9, and the activation of ENT3. The ABA content was lowest at T1 and T2, but rose sharply at subsequent stages, which coincides with expression profiles of NCEDs and ZEPs (R2 >0.89) (Supplementary Fig. S6).
Table 1.
Levels of phytohormones (IAA, CK, and ABA) in 1g of bulbil tissue
| Time point | IAA | CK | Total CKs | ABA | |
|---|---|---|---|---|---|
| Zeatin | Dihydrozeatin | ||||
| T1 | 37.54±2.25*** | 101.92±8.49 | 26.05±1.58 | 127.97±8.66 | 1.47±0.21 |
| T2 | 10.90±1.20 | 122.90±3.44** | 29.71±1.77* | 152.61±4.34** | 4.54±0.74 |
| T3 | 1.70±0.58 | 56.35±3.72 | 19.03±0.99 | 75.38±4.43 | 20.72±2.45 |
| T4 | 0.49±0.45 | 46.14±3.07 | 6.00±1.75 | 52.14±2.10 | 32.82±3.69** |
Values are shown in ng g–1 (means ±SD). Asterisks represent statistical differences between stages (*P<0.05; **P<0.01;***P<0.001).
Starch and sucrose metabolic processes
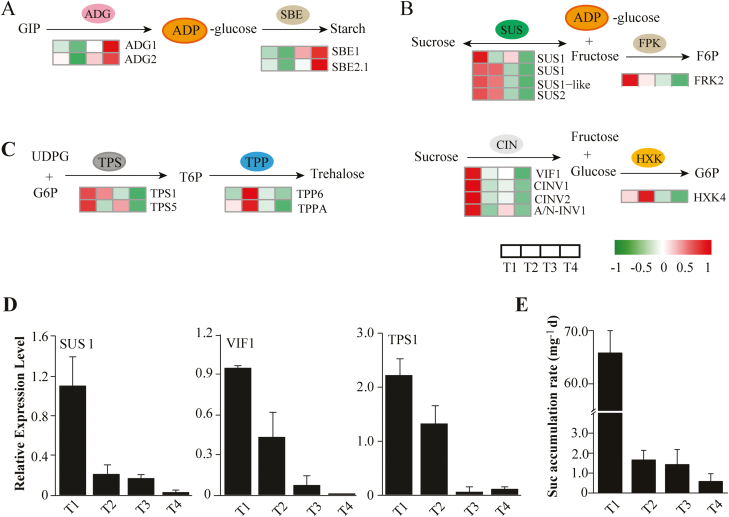
We predicted several marker genes involved in starch synthesis (Supplementary Dataset S1), such as genes encoding the small subunit of Glu-1-P adenylyltransferase (ADG1/2), and glucan-branching enzymes (SBE1 and SBE2.1), which consistently presented the highest expression levels at T4 (Fig. 6A). Two genes (ADG2 and SBE2.1) were T4-specifically expressed according to the SS method. This result was consistent with that of GO-enriched analysis, demonstrating an enriched wax biosynthetic process at T4 (Fig. 3B). In addition, there were multiple genes encoding sucrose synthases (SUSs) and invertases, representing key genes that participate in sucrose metabolism (Fig. 6B; Supplementary Dataset S1). Four genes encoding SUSs (three homologs of SUS1 plus SUS2-like) that can reversibly catalyze sucrose biosynthesis were significantly up-regulated at T1 and gradually decreased at the next stages. In particular, SUS1 (c130514.graph_c0) showed a highly expressed level with an FPKM value >2000. Meanwhile, we found that four T1-specific SS genes encoding invertases (VIF1, CINV1, CINV2, and A/N-INV1) peaked at T1, which can degrade sucrose and produce free hexoses. More importantly, we observed several members of the trehalose-phosphate synthase (TPS) and trehalose-phosphate phosphatase (TPP) families which have been implicated in the regulation of axillary bud development through sugar signaling (Fichtner et al., 2017). Among them, two TPS genes (TPS1 and TPS6) peaked at T1 (Fig. 6C); these can produce trehalose-6-phophate (T6P) as an important sucrose signal affecting plant development (O’Hara et al., 2013; Figueroa and Lunn, 2016). In contrast, two TPP genes (TPP6 and TPPA) with the role of dephosphorylating T6P showed lower expression levels at T1 and peaked at T2. The result of this appeared to be an enhanced T6P pathway in bulbils at the early stage (T1).
Fig. 6.
Gene and metabolite regulation involved in starch and sucrose metabolic processes, and sucrose signaling. (A) Starch biosynthesis process. (B) Sucrose metabolism. (C) Sucrose signaling involved in the trehalose metabolism pathway. Heat maps next to the arrows represent changes in expression of genes encoding corresponding enzymes for reactions. The expression levels are standardized per gene into Z-scores and colored in red and green for high and low expression, respectively. (D) Verification of expression levels of genes encoding SUS1, VIF1, and TPS1 determined by qRT–PCR. (E) Sucrose accumulation in bulbils, demonstrating that sucrose is sharply increased at the early stage of bulbil formation (T1). Data show the mean ±SD (n=5). ADG, glucose-1-phosphate adenylyltransferase; CIN, cell wall invertase; FRK, fructokinase; HXK, hexokinase; SBE, glucan-branching enzyme; SUS, sucrose synthase; TPP, trehalose-phosphate phosphatase; TPS, trehalose-phosphate synthase; VIF1, vacuolar invertase1.
Given the crucial role in triggering axillary bud outgrowth by sucrose (Barbier et al., 2015), we further detected the expression levels of some key genes involved in sucrose metabolism by qRT–PCR technology, and detected sucrose accumulation in bulbils at different stages. Three representative genes SUS1, VIF1, and TPS1 involved in this process showed peak expression profiles at T1 (Fig. 6D), which was consistent with the results of transcriptome analyses. Importantly, we observed that the level of sucrose was sharply elevated with an accumulation rate of 65.8 mg d−1 during bulbil initiation and increased slowly in the subsequent stages (Fig. 6E).
Transcription regulators (TFs) involved in yam bulbil growth
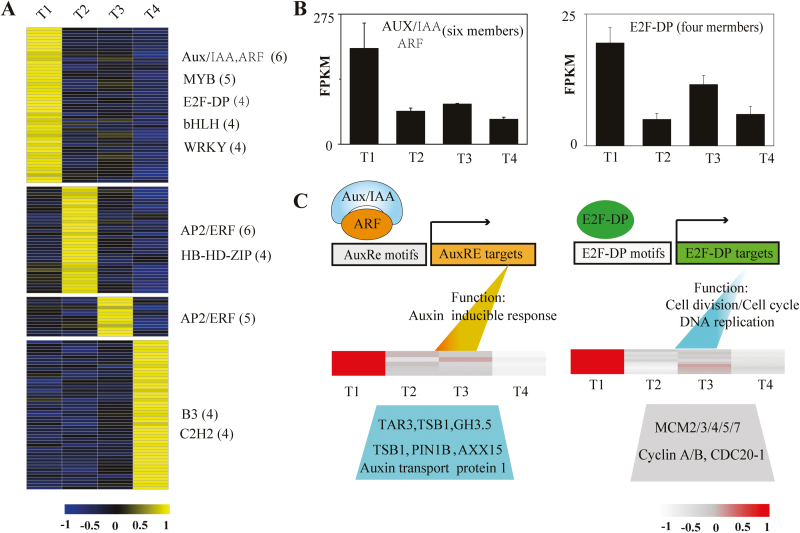
We searched the SS gene set for the over-representation of families of TFs from The Arabidopsis Information Resource (TAIR) (Perez-Rodriguez et al., 2010), and predicted 149 TFs as stage-specifically expressed. Most of them are members of the AUX/IAA, AP2/ERF, WRKY, bHLH, MYB, C2H2, and C3H families (Supplementary Dataset S1). Consistent with the proportion of all SS genes captured during bulbil developmental stages (Fig. 3A), T1 and T4 type bulbils had greater numbers of stage-specific TFs relative to the other two types, with 52 and 49 TFs, respectively. In contrast, the T3 type had the lowest number of TFs (13), and 35 TFs were identified as T2-specifically expressed. The expression profiles of these stage-specific TFs are shown in Fig. 7A.
Fig. 7.
Expression profiles of transcription factors (TFs) and hypothetical regulatory modules. (A) Heat map of scaled FPKM values of 149 stage-specific TFs (with SS score >0.3) identified during bulbil growth. The TF families listed on the right show predominantly expression in corresponding stage-specific clusters. The numbers in parentheses represent the number of members from TF families in this cluster. (B) Cumulative expression profiles of AUX/IAA (six members) and E2F-DP (four members) TFs in the T1-specific cluster. (C) Hypothetical models of AUX/IAA- and E2F-DP- mediated gene regulation in the T1-specific cluster. The heat map shows the expression of genes involved in GO functional categories by AUX/IAA and E2F-DP induction, suggesting that the two TFs and their targets are co-over-represented in the T1-specific cluster.
Notably, we here focused on T1-specifically expressed TFs due to their possible roles in regulating the initiation of the bulbil. Strikingly, six members of the Aux/IAA and ABF protein families showed 3-fold accumulation induction at T1 relative to the next stage (T2) (Fig. 7B), and are required for regulating auxin response genes in plant developmental processes including AM initiation (Deng et al., 2012; Jung et al., 2015; Kato et al., 2017). Generally, TFs and their targets are co-expressed in the same gene cluster. We found that some genes involved in auxin metabolism, signaling, and transport, including GH3.5, TAR3, PIN1B, AXX15, and LAX2, were grouped into the same cluster and over-represented at T1 (Fig. 7C). Most of them are potentially targeted by Aux/IAAs and ARFs (Guilfoyle and Hagen, 2007). In addition, the E2F-DP family are important regulators of the cell cycle and directly target various genes related to cell division and DNA replication (De Veylde et al., 2002). Four E2F-DP TFs exhibited a 6.8-fold induction of accumulation at T1 compared with T2 (Fig. 7B). Genes including multiple CYCA/B genes and DNA replication licensing factors (MCM2/3/4/5/7) associated with the cell cycle and DNA replication were also enriched at T1 and peaked in expression levels, which probably resulted from the over-representation of E2F-DP TFs at T1 (Fig. 7C).
In addition to AUX/IAA and E2F-DP families, MYB, WRKY, and bHLH were other TF families over-represented in the T1 cluster and had the highest expression at T1. For instance, WRKY TFs have been implicated in the regulation of AMs (Guo et al., 2015); the accumulated expression of four WRKY TFs at T1 was 1.6-fold higher relative to other stages. Some MYB TFs affect the pattern of lateral meristem initiation (Hsu et al., 2015), five MYB TFs represented a functional category at T1, all of which had 2-fold higher expression at T1 relative to other stages. Similarly, four bHLH TFs were highly induced, with 2- to 11-fold higher expression at T1. To further confirm the regulatory role of TFs, we selected three TFs, MYB (C126446.graph-c0), WRKY (C125026.graph_c1), and NAC (C116834.graph_c0), due to their higher expression levels and key roles in promoting AM initiation in other plants (Yang and Jiao, 2016), and examined their expression at the early initiation of bulbil formation by in situ hybridization (Supplementary Fig. S8). Consistent with the RNA-seq results, we found that all of them were specifically expressed in the AM initiation zone (a dome-shaped tissue) at the T1 stage, and strongly decreased with the thin chromogenic signals at the next developmental stages, indicating the key roles that they play during bulbil initiation.
Discussion
Identification of gene sets associated with yam bulbil growth
In this study, we performed comprehensive transcriptome profiling of yam bulbil, demonstrating a dramatic shift in gene expression for different bulbil types. In particular, we used the SS scoring algorithm to obtain stage-specifically expressed gene sets that highlight the nature of different bulbil types. These gene sets were enriched in some known biological processes and KEGG pathways associated with gene regulation of axillary bud growth, providing a good resource for identifying key candidates controlling bulbil growth. As shown in Fig. 4A, the T1-specific cluster identified a set of genes encoding cell cyclin proteins (CYCA1-1, CYCA3-2, CYCA3-4, CYCB2-1, and CYCB3-1), kinesins (KIN5C, KIN4A, KIN10A, and NACK1), and DNA replication licensing factors (MCM2/3/4/5/7) involved in the cell cycle and proliferation (Supplementary Dataset S1). In the control of axillary buds, cell cycle-related genes perform a quality control function in promoting the resumption of meristem organogenic activity. This fact is directly supported by a variety of experiments showing that increased expression of cell cycle genes in dormant buds from pea and Arabidopsis by decapitation stimulates bud outgrowth (Devitt and Stafstrom, 1995; Shimizu and Mori, 1998). Similar evidence has shown that decreased expression of cell cycle genes (CYCB, CYCD2, and CDKB) represses bud growth in Sorghum bicolor (Kebrom et al., 2010). In apple plant, the expression of cell proliferation genes (kinesins) is up-regulated at the early stage of axillary bud formation (Tan et al., 2019). Consistent with these findings, a large set of cell cycle/division genes were T1-specifically expressed and showed peak abundance at this stage (Fig. 4A), which were probably regulated by E2F-DP TFs over-represented at this stage (Fig. 7C). This gene expression profile increases the vitality of cell division in leaf primordia meristems and allows the resumption of organogenesis of young bulbils.
Several lines of evidence suggest that the expression of genes associated with cell wall expansion, such as EXPANSINS, determines the rate of the initiation and elongation of premature AMs, and exerts a profound influence on plant development and morphology (Fleming et al., 1997). In a reported experiment from Petunia hybrida, overexpression of the PHEXPA1 gene promotes AM release, whereas silencing PHEXPA1 produces opposite phenotypes (Zenoni et al., 2011). The enhanced expression of RhEXP by a CK signal activates the shoot apical meristem (SAM) organogenic activity in rose axillary buds and further promotes bud elongation (Roman et al., 2016). In addition to EXPANSINS, glycosyl hydrolase family 16/17 (XTHs/BGs) involved in cell wall modification have been implicated in the regulation of cell elongation during development of various plant organs (Cosgrove, 2005). Consistent with these observations, we detected 15 gene sets of EXPANSINS from EXPA/B and GH 16/17 families, and found a distinctly up-regulated expression at the early stage of bulbil formation (T1) (Fig. 4B; Supplementary Dataset S1). Most of them were defined as T1-specifically expressed. Such results may be explained by the fact that the set of EXPANSINS, XTH, and BG genes contributes a unique role in maintaining cell enlargement during bulbil initiation and building the special architectural form.
Starch constitutes most of the biomass of the mature bulbil, accounting for 50–80% of its dry matter, and is a main trait being improved by breeding (Tamiru et al., 2008). As expected, we observed multiple marker genes (ADG1/2 and SBE1/2.1) involved in the starch biosynthesis pathway (Fig. 6A). Of particular interest are the SBE genes that control amylopectin production, which showed relatively high expression levels at the later stages of bulbil growth (T4). These results are consistent with previous observations in maize embryo and endosperm development (Chen et al., 2014). The highly expressed SBE genes may be used to improve the quality of the bulbil by further genetic manipulation.
Bulbil growth requires the coordinated control of hormone-related genes
Plant hormones and their interactions have long been considered to play central regulatory roles in controlling axillary bud growth (Domagalska and Leyser, 2011; Yang and Jiao, 2016). Here, T1-specifically expressed TAR3 and YUCCA1 genes involved in auxin biosynthesis were highly up-regulated at T1 (Fig. 5); these genes were directly responsible for the localized production of auxin in bulbils at the early stage (Table1). Consistent with our observation, localized auxin biosynthesis is required for AM initiation in maize and Arabidopsis (Gallavotti et al., 2008; Zhao, 2010). Knockout of YUCCA genes leads to fewer branches due to the absence of AMs (Cheng et al., 2006). A similar result had been found in that auxin promotes the initiation of upper bulbils in L. lancifolium, but inhibits their further growth (Yang et al., 2017). On the other hand, the ATC model supports that auxin needs to be exported from axillary buds for its outgrowth by establishing the localized PAT stream in the bud stem (not in the main stem) (Blilou et al., 2005). The PAT stream is driven by PIN proteins, auxin transporter-like protein 2 (LAX2), and members of the ABC transporter B family belonging to auxin efflux carriers that can facilitate auxin export out of cells (Petrášek and Friml, 2009). Arabidopsis mutants with more axillary growth increase PIN protein levels and the amount of auxin moving by the PAT stream (Bennett et al., 2006). In pea plants, increased auxin export from buds is accompanied by PIN1 polarization after decapitation, and further activates bud outgrowth (Balla et al., 2011). In accordance with this evidence, we observed the over-representation of auxin efflux proteins, including ABCB-type transporters (ABCB8/11/19), LAX2, and NDL1/2 that were closely correlated with the auxin level (Supplementary Fig. S6), whereby these transport proteins facilitate transport of excess auxin and trigger bulbil growth (Table1).
Outgrowth of axillary buds is positively correlated with CK levels that are inhibited by the mobile auxin in the main stem (Ferguson and Beveridge, 2009; Domagalska and Leyser, 2011). The CK levels in chickpea buds increase 25-fold after decapitation, suggesting that CKs are necessary to initiate bud growth (Turnbull et al., 1997). In some cases, CKs can stimulate bud outgrowth by direct application to buds, even in the presence of apical auxin (Dun et al., 2012). In phyB sorghum mutants, reduced expression of genes involved in CK biosynthesis and signaling leads to resistance to bud outgrowth (Kebrom and Mullet, 2016). Mutations in rice CKX that degrade CK give rise to increased panicle branches and spikelet numbers in the inflorescence (Ashikari et al., 2005). Here, we did not find any CK synthetic genes, but observed the repression of degradation genes (CKX1, CKX3, CKX9, and CKX11) in bulbils at the T1 stage (Supplementary Dataset S1), indicating that the presence of CK (Table 1) in the bulbil probably stemmed from the adjacent stem by CK flow. This speculation was strongly supported by the fact that the highly expressed CK transporter (ENT3) is tightly linked to CK levels (R2 >0.95) (Supplementary Fig. S6). In addition, a CK-inducible response regulator (ARR18) and two CK receptors (AHK1/2) were identified as T1-specifically expressed, suggesting an enhanced CK signaling at the T1 stage. Taken as a whole, the expression of the CK-related genes above could drive the promotion of bulbil growth at the initiation stage.
ABA has been thought to be a key component of regulating axillary organ growth. For a variety of plant species, the decline of ABA levels after decapitation precedes the onset of bud outgrowth (Zheng et al., 2015). In Arabidopsis, elevated ABA delays bud outgrowth and decreases elongating buds under low red:far-red (R:FR) conditions (Reddy et al., 2013; Yao and Finlayson, 2015). Several hypotheses have been postulated suggesting that ABA acts downstream of auxin and stringonolactone, possibly as a second messenger (Cline and Oh, 2006; López-Ráez et al., 2010). Independently of this presumption, it is undoubted that increased expression of genes involved in ABA biosynthesis is linked to repression of axillary bud growth. Consistent with previous reports, our data revealed that three NECD genes and four ZEP genes responsible for regulating ABA synthesis were down-regulated at T1 and then strongly up-regulated at T4 (as T4-specifically expressed genes) (Fig. 5). In particular, the ABA level was closely linked to the expression of NECDs and ZEPs (Supplementary Fig. S6). Thus, these genes could decrease ABA accumulation in bulbils at the initiation stage (Table 1), thereby promoting bulbil growth.
On the other hand, we attempted to establish a linkage of phytohormone signals and functional gene sets associated with bulbil growth. We found that levels of multiple phytohormones (auxin, CKs, and ABA) were tightly correlated with transcriptional expression of most identified genes involved in the functional categories cell division, proliferation, and expansion (Supplementary Fig. S7). This close linkage was also found in other investigations on axillary bud development (Reddy et al., 2013; Roman et al., 2016; Majda and Robert, 2018), which provides evidence that mutiple phytohormones are likely to act as integrated signals that provide a forward force to genes related to cell division, proliferation, and expansion, thus promoting bulbil growth.
The key mechanism of bulbli initiation is modulated through sucrose supply and signaling
In addition to phytohormone signals, sugar (sucrose or its analogs), as a novel player, contributes to the activation of axillary bud growth (Barbier et al., 2015). In diverse plant species after decapitation, the progressive decrease of auxin levels in the stem is too slow to dominate the early bud formation, whereas sugars are rapidly redistributed and enter buds to promote their growth (Barbier et al., 2015). From a representative experiment in pea plants, the rate of PAT is 1 cm h−1 when apical dominance is lost, yet the outgrowth of the bud reaches 40 cm at 2.5 h after decapitation (Mason et al., 2014). In Narcissus plants, direct application of sucrose greatly increases the percentage of bulbil production (Staikidou et al., 1994). In our study, the rapid rate of accumulationof sucrose was observed at T1 (Fig. 6E), which facilitates triggering the release of bulbils. Moreover, the sucrose accumulation level had positive effects on transcriptional expression of gene sets associated with cell division, and expansion (Supplementary Fig. S7). Transcriptome analysis revealed that the expression of key genes (SUSs and CINs) involved in sucrose metabolism was highly up-regulated at T1 (Fig. 6B), which can unblock the process of sucrose supply in a timely manner. More importantly, increased CIN expression can positively regulate axillary bud initiation by generating sugars for trophic uptake under the interplay of light and phytohormones (Rabot et al., 2014).
On the other hand, sucrose functions as a critical signal through regulating the pathways involving T6P and HXK1 (O’Hara et al., 2013). HXK1-overexpressing Arabidopsis lines show enhanced branching (Kelly et al., 2012). In particular, the elevated T6P level has been implicated to be the signal that increases sugar influx into buds after decapitation, whereby buds are released from dormancy and elongated (Yadav et al., 2014; Figueroa and Lunn, 2016). T6P is synthesized by TPS and dephosphorylated to TPP by TPP genes. Overexpression of Escherichia coli TPS (OtsA) in Arabidopsis results in a rise in T6P levels and triggers the proliferation of shoot branching; on the other hand, overexpression of E. coli TPP (OtsB) decreases both the T6P level and shoot branching (Schluepmann et al., 2003). Similar evidence demonstrated that Arabidopsis lines constitutively affected in synthesis and degradation of T6P increase and reduce branching phenotypes, respectively (Yadav et al., 2014). In garden pea, T6P was found to be the signal of sucrose availability to promote outgrowth of axillary buds (Fichtner et al., 2017). In our study, we observed the induction of TPS genes (TPS1 and TPS5) and the inhibition of TPP genes (TPP6 and TPPA) at the initiation stage of the bulbil (T1) (Fig. 6C), which is more likely to be a consequence of enhanced sucrose signaling. These up-regulated TPS genes could increase T6P levels, thereby contributing to the promotion of bulbil growth.
Requirement for transcription regulators for bulbil initiation
The initiation of AMs is tightly linked to the activity of bud-specific regulators that can regulate transcription responses of functional genes through binding motifs in their promoters. Most of the regulators are members of multiple TF families, such as known R2R3 MYB proteins (REGULATOR OF AXILLARY MERISTERMS, RAX) from Arabidopsis, NAC domain TFs (CUP-SHAPED COTYLEDON, CUC), WRKY domain protein (EXB1), GRAS domain protein (LATERAL SUPPRESSOR, LS) in tomato, and TCP TFs (BRANCHED1/2) in Arabidopsis and in sorghum (TB1) (Janssen et al., 2014; Yang and Jiao, 2016). Genetic studies of mutant plants have demonstrated that these transcriptional activators are specifically expressed in the boundary zone between the leaf primordium and the SAM, and control the fate of AM initiation and the production of buds (Keller et al., 2006; Yang et al., 2012; Guo et al., 2015). For instance, loss of the RAX1 gene encoding MYB37 in Arabidopsis or its orthologous gene BLIND (BL) in tomato leads to failure to generate lateral buds during vegetative development (Keller et al., 2006; Naz et al., 2013). Overexpression of EXB1 encoding WRKY71 increases excessive AM initiation and bud activities, and produces bushy and dwarf phenotypes by transcriptionally regulating RAX genes (Guo et al., 2015). In this study, we verified a large set of TF genes during bulbil developmental stages, especially the over-represented TFs from Aux/IAA, E2F-DP, MYB,WRKY, and bHLH families that are T1-specifically expressed with the highest expression at this stage (Fig. 7A). In the same cluster, these TFs directly resulted in the over-representation of their targeted genes by putative regulatory modules. In the T1 cluster, two regulatory modules of Aux/IAA and E2F-DP were speculated to exist, and caused the over-representation of genes linked to auxin-inducible responses, cell division, and DNA replication (Fig. 7C), respectively. Three T1-specifically expressed TFs from WRKY, NAC, and MYB were confirmed to be highly expressed in the meristematic cell zone at the early stage of bulbil formation (Supplementary Fig. S8). These data suggested that transcriptional regulators are required for the early step of bulbil expansion. However, regulatory modules, and the downstream genes targeted by them, still need to be explored by genetic interaction experiments in future studies.
Conclusion
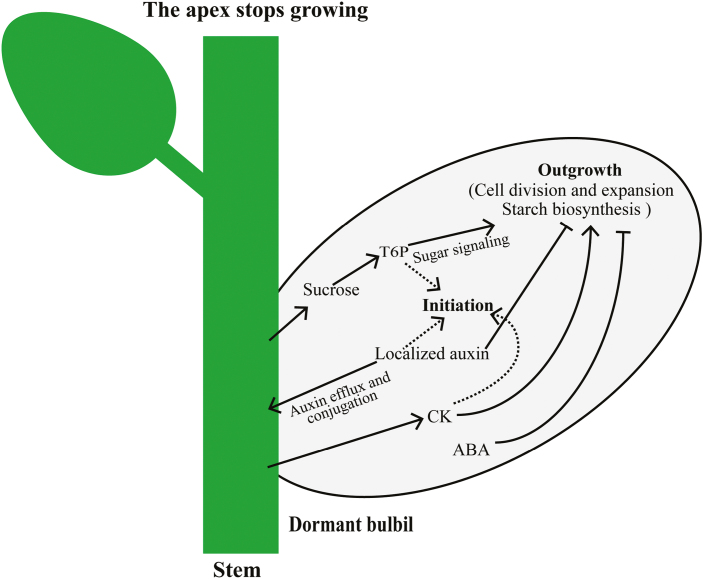
Taken together, bulbil transcriptomic data, especially for stage-specifically expressed gene sets, provide a valuable new resource that can be queried to obtain important candidates regulating yam bulbil initiation and growth. A schematic model based on our results is proposed in Fig. 8. Compared with previous reports discussed above, more direct evidence was presented here to reveal the key regulatory programs of bulbil formation. We found that metabolite levels of multiple phytohormones (auxin, CKs, and ABA) and sucrose are highly coincident with transcription changes of genes involved in cell division, proliferation, and expansion. Furthermore, the integration of auxin, CKs, ABA, and sucrose provides a forward signaling to these functional genes that contribute unique roles in maintaining cell enlargement and lead to bulbil initiation and growth. In particular, the localized production of auxin in bulbils is transiently required to trigger bulbil formation, but can be exported to maintain bulbil outgrowth by up-regulating various auxin efflux proteins (PIN1B, LAX2, and the ABC transporter B family). In addition, we have identified several T1-specifically expressed TFs from Aux/IAA, E2F, MYB, WRKY, and bHLH families, and described their key role in triggering bulbil formation by the prediction of regulatory modules or in situ hybridization experiments. Such work as presented here allows, to our knowledge for the first time, an increased understanding of the complex gene regulatory framework underlying yam bulbil growth. Further work will contribute to identification of the role of candidate genes in regulating bulbil growth by misregulation and genetic transformation.
Fig. 8.
Schematic model of gene regulation by auxin, CK, ABA, and sucrose during bulbil growth. The localized auxin in the bulbil is transiently produced by stimulation of auxin synthesis (TAR3 and YUCCA1), and activates bulbil initiation. Further, the excess auxin is depleted and transported to maintain bulbil outgrowth by up-regulation of auxin-conjugated genes (GH3.5 and GH3.6) and auxin efflux proteins (PIN1B, LAX2, and the ABC transporter B family). CKs promote its growth through rapid activation of the CK transporter (ENT3) gene, together with suppression of CK degradative genes (CKX1/3/9/11). ABA contributes to bulbil growth through repression of ABA synthesis genes (NECDs and ZEPs) at the early stage of bulbil formation. Sucrose probably acts as a signal, and promotes bulbil growth through up-regulation of genes involved in the T6P pathway (TPS1 and TPS5). The integration of these hormones and sucrose provides a forward signaling to activate genes associated with cell division, proliferation, and expansion, which contributes key roles in maintaining cell enlargement during bulbil growth.
Supplementary data
Supplementary data are available at JXB online.
Table S1. List of the primer sequences used for qRT–PCR analyses.
Table S2. Summary of RNA-seq reads in the yam bulbil transcriptome.
Table S3. Assembly statistics for the yam bulbil transcriptome.
Table S4. Lists of differentially expressed genes.
Table S5. List of the most enriched GO terms for DEGs.
Table S6. Lists of significantly enriched KEGG pathways.
Table S7. Gene set of stage-specifically expressed genes.
Dataset S1. Tables of candidate genes and regulators associated with bulbil growth.
Fig. S1. Pearson correlation relationship between biological replicates.
Fig. S2. Validations of gene expression profiles by qRT–PCR.
Fig. S3. Hierarchical clustering of all DEGs across different stages.
Fig. S4. The most enriched GO terms for all DEGs.
Fig.S5. Enriched KEGG pathways.
Fig. S6. Correlations between hormone-related gene expression and its metabolite levels.
Fig. S7. Correlations between levels of hormones, sucrose, and the expression of genes involved in cell division, proliferation, and expansion.
Fig. S8. RNA in situ hybridization for MYB, WRKY, and NAC transcription factors.
Acknowledgements
This study was supported by Zhejiang Provincial Key Laboratory for Genetic Improvement and Quality Control of Medicinal Plants (no. 2011E10015) in China, and Zhejiang Provincial Applied Research Program for Commonweal Technology (no. 2016C32SA300040).
Author contributions
Z-GW conceived the program,designed the experiment, and supervised the writing; WJ analyzed the transcriptome data and dealt with the figures; Z-MT, W-HY, and H-LH performed the experiments and wrote the text with their contributions; X-JP helped to analyze bioinformatics. All authors read and approved the final manuscript.
References
- Abraham Juárez MJ, Hernández Cárdenas R, Santoyo Villa JN, O’Connor D, Sluis A, Hake S, Ordaz-Ortiz J, Terry L, Simpson J. 2015. Functionally different PIN proteins control auxin flux during bulbil development in Agave tequilana. Journal of Experimental Botany 66, 3893–3905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abraham-Juárez MJ, Martínez-Hernández A, Leyva-González MA, Herrera-Estrella L, Simpson J. 2010. Class I KNOX genes are associated with organogenesis during bulbil formation in Agave tequilana. Journal of Experimental Botany 61, 4055–4067. [DOI] [PubMed] [Google Scholar]
- Alexa A, Rahnenführer J, Lengauer T. 2006. Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics 22, 1600–1607. [DOI] [PubMed] [Google Scholar]
- Arizaga S, Ezcurra E. 2002. Propagation mechanisms in Agave macroacantha (Agavaceae), a tropical arid-land succulent rosette. American Journal of Botany 89, 632–641. [DOI] [PubMed] [Google Scholar]
- Ashikari M, Sakakibara H, Lin S, Yamamoto T, Takashi T, Nishimura A, Angeles ER, Qian Q, Kitano H, Matsuoka M. 2005. Cytokinin oxidase regulates rice grain production. Science 309, 741–745. [DOI] [PubMed] [Google Scholar]
- Asiedu R, Sartie A. 2010. Crops that feed the world 1. Yams. Food Security 2, 305–315. [Google Scholar]
- Balla J, Kalousek P, Reinöhl V, Friml J, Procházka S. 2011. Competitive canalization of PIN-dependent auxin flow from axillary buds controls pea bud outgrowth. The Plant Journal 65, 571–577. [DOI] [PubMed] [Google Scholar]
- Barbier F, Péron T, Lecerf M, et al. 2015. Sucrose is an early modulator of the key hormonal mechanisms controlling bud outgrowth in Rosa hybrida. Journal of Experimental Botany 66, 2569–2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett T, Sieberer T, Willett B, Booker J, Luschnig C, Leyser O. 2006. The Arabidopsis MAX pathway controls shoot branching by regulating auxin transport. Current Biology 16, 553–563. [DOI] [PubMed] [Google Scholar]
- Berckmans B, De Veylder L. 2009. Transcriptional control of the cell cycle. Current Opinion in Plant Biology 12, 599–605. [DOI] [PubMed] [Google Scholar]
- Blilou I, Xu J, Wildwater M, Willemsen V, Paponov I, Friml J, Heidstra R, Aida M, Palme K, Scheres B. 2005. The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 433, 39–44. [DOI] [PubMed] [Google Scholar]
- Burkill IH. 1960. The organography and the evolution of Dioscoreaceae, the family of the yams. Botanical Journal of the Linnean Society 56, 319–412. [Google Scholar]
- Chabikwa TG, Brewer PB, Beveridge CA. 2019. Initial bud outgrowth occurs independent of auxin flow from out of buds. Plant Physiology 179, 55–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatfield SP, Stirnberg P, Forde BG, Leyser O. 2000. The hormonal regulation of axillary bud growth in Arabidopsis. The Plant Journal 24, 159–169. [DOI] [PubMed] [Google Scholar]
- Chebli Y, Geitmann A. 2017. Cellular growth in plants requires regulation of cell wall biochemistry. Current Opinion in Cell Biology 44, 28–35. [DOI] [PubMed] [Google Scholar]
- Chen J, Zeng B, Zhang M, Xie S, Wang G, Hauck A, Lai J. 2014. Dynamic transcriptome landscape of maize embryo and endosperm development. Plant Physiology 166, 252–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Westfall CS, Hicks LM, Wang S, Jez JM. 2010. Kinetic basis for the conjugation of auxin by a GH3 family indole-acetic acid-amido synthetase. Journal of Biological Chemistry 285, 29780–29786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Dai X, Zhao Y. 2006. Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Genes & Development 20, 1790–1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline MG, Oh C. 2006. A reappraisal of the role of abscisic acid and its interaction with auxin in apical dominance. Annals of Botany 98, 891–897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, Robles M. 2005. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21, 3674–3676. [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ. 2005. Growth of the plant cell wall. Nature Reviews. Molecular Cell Biology 6, 850–861. [DOI] [PubMed] [Google Scholar]
- De Veylder L, Beeckman T, Beemster GT, et al. 2002. Control of proliferation, endoreduplication and differentiation by the Arabidopsis E2Fa-DPa transcription factor. The EMBO Journal 21, 1360–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng T, Kim C, Zhang DG, Zhang JW, Li ZM, Nie ZL, Sun H. 2013. Zhengyia shennongensis: a new bulbiliferous genus and species of the nettle family (Urticaceae) from central China exhibiting parallel evolution of the bulbil trait. Taxon 62, 89–99. [Google Scholar]
- Deng W, Yang Y, Ren Z, Audran-Delalande C, Mila I, Wang X, Song H, Hu Y, Bouzayen M, Li Z. 2012. The tomato SlIAA15 is involved in trichome formation and axillary shoot development. New Phytologist 194, 379–390. [DOI] [PubMed] [Google Scholar]
- Devitt ML, Stafstrom JP. 1995. Cell cycle regulation during growth–dormancy cycles in pea axillary buds. Plant Molecular Biology 29, 255–265. [DOI] [PubMed] [Google Scholar]
- Domagalska MA, Leyser O. 2011. Signal integration in the control of shoot branching. Nature Reviews. Molecular Cell Biology 12, 211–221. [DOI] [PubMed] [Google Scholar]
- Dun EA, de Saint Germain A, Rameau C, Beveridge CA. 2012. Antagonistic action of strigolactone and cytokinin in bud outgrowth control. Plant Physiology 158, 487–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evers JB, van der Krol AR, Vos J, Struik PC. 2011. Understanding shoot branching by modelling form and function. Trends in Plant Science 16, 464–467. [DOI] [PubMed] [Google Scholar]
- Faivre-Rampant O, Cardle L, Marshall D, Viola R, Taylor MA. 2004. Changes in gene expression during meristem activation processes in Solanum tuberosum with a focus on the regulation of an auxin response factor gene. Journal of Experimental Botany 55, 613–622. [DOI] [PubMed] [Google Scholar]
- Fan DM, Yang YP. 2009. Altitudinal variations in flower and bulbil production of an alpine perennial, Polygonum viviparum L. (Polygonaceae). Plant Biology 11, 493–497. [DOI] [PubMed] [Google Scholar]
- Ferguson BJ, Beveridge CA. 2009. Roles for auxin, cytokinin, and strigolactone in regulating shoot branching. Plant Physiology 149, 1929–1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fichtner F, Barbier FF, Feil R, Watanabe M, Annunziata MG, Chabikwa TG, Höfgen R, Stitt M, Beveridge CA, Lunn JE. 2017. Trehalose 6-phosphate is involved in triggering axillary bud outgrowth in garden pea (Pisum sativum L.). The Plant Journal 92, 611–623. [DOI] [PubMed] [Google Scholar]
- Figueroa CM, Lunn JE. 2016. A tale of two sugars: trehalose 6-phosphate and sucrose. Plant Physiology 172, 7–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming AJ, McQueen-Mason S, Mandel T, Kuhlemeier C. 1997. Induction of leaf primordia by the cell wall protein expansin. Science 276, 1415–1418. [Google Scholar]
- Frébortová J, Novák O, Frébort I, Jorda R. 2010. Degradation of cytokinins by maize cytokinin dehydrogenase is mediated by free radicals generated by enzymatic oxidation of natural benzoxazinones. The Plant Journal 61, 467–481. [DOI] [PubMed] [Google Scholar]
- Gallavotti A, Barazesh S, Malcomber S, Hall D, Jackson D, Schmidt RJ, McSteen P. 2008. sparse inflorescence1 encodes a monocot-specific YUCCA-like gene required for vegetative and reproductive development in maize. Proceedings of the National Academy of Sciences, USA 105, 15196–15201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilfoyle TJ, Hagen G. 2007. Auxin response factors. Current Opinion in Plant Biology 10, 453–460. [DOI] [PubMed] [Google Scholar]
- Guo D, Zhang J, Wang X, et al. 2015. The WRKY transcription factor WRKY71/EXB1 controls shoot branching by transcriptionally regulating RAX genes in Arabidopsis. The Plant Cell 27, 3112–3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutmann M. 1995. Improved staining procedures for photographic documentation of phenolic deposits in semithin sections of plant tissue. Journal of Microscopy 179, 277–281. [Google Scholar]
- Haas BJ, Papanicolaou A, Yassour M, et al. 2013. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nature Protocols 8, 1494–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrar Y, Bellec Y, Bellini C, Faure JD. 2003. Hormonal control of cell proliferation requires PASTICCINO genes. Plant Physiology 132, 1217–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose N, Makita N, Yamaya T, Sakakibara H. 2005. Functional characterization and expression analysis of a gene, OsENT2, encoding an equilibrative nucleoside transporter in rice suggest a function in cytokinin transport. Plant Physiology 138, 196–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu CC, Chen YY, Tsai WC, Chen WH, Chen HH. 2015. Three R2R3-MYB transcription factors regulate distinct floral pigmentation patterning in Phalaenopsis spp. Plant Physiology 168, 175–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen BJ, Drummond RS, Snowden KC. 2014. Regulation of axillary shoot development. Current Opinion in Plant Biology 17, 28–35. [DOI] [PubMed] [Google Scholar]
- Jung H, Lee DK, Choi YD, Kim JK. 2015. OsIAA6, a member of the rice Aux/IAA gene family, is involved in drought tolerance and tiller outgrowth. Plant Science 236, 304–312. [DOI] [PubMed] [Google Scholar]
- Kato H, Kouno M, Takeda M, Suzuki H, Ishizaki K, Nishihama R, Kohchi T. 2017. The roles of the sole activator-type auxin response factor in pattern formation of Marchantia polymorpha. Plant & Cell Physiology 58, 1642–1651. [DOI] [PubMed] [Google Scholar]
- Kebrom TH, Brutnell TP, Finlayson SA. 2010. Suppression of sorghum axillary bud outgrowth by shade, phyB and defoliation signalling pathways. Plant, Cell & Environment 33, 48–58. [DOI] [PubMed] [Google Scholar]
- Kebrom TH, Mullet JE. 2016. Transcriptome profiling of tiller buds provides new insights into PhyB regulation of tillering and indeterminate growth in sorghum. Plant Physiology 170, 2232–2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller T, Abbott J, Moritz T, Doerner P. 2006. Arabidopsis Regulator Of Axillary Meristems1 controls a leaf axil stem cell niche and modulates vegetative development. The Plant Cell 18, 598–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly G, David-Schwartz R, Sade N, Moshelion M, Levi A, Alchanatis V, Granot D. 2012. The pitfalls of transgenic selection and new roles of AtHXK1: a high level of AtHXK1 expression uncouples hexokinase1-dependent sugar signaling from exogenous sugar. Plant Physiology 159, 47–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Kang JY, Cho DI, Park JH, Kim SY. 2004. ABF2, an ABRE-binding bZIP factor, is an essential component of glucose signaling and its overexpression affects multiple stress tolerance. The Plant Journal 40, 75–87. [DOI] [PubMed] [Google Scholar]
- Kim SK, Lee SC, Lee BH, Choi HJ, Kim KU, Lee IJ. 2003. Bulbil formation and yield responses of Chinese yam to application of gibberellic acid, mepiquat chloride and trinexapac-ethyl. Journal of Agronomy and Crop Science 189, 255–260. [Google Scholar]
- Law R, Cook RED, Manlove RJ. 1983. The ecology of flower and bulbil production in Polygonum viviparum. Nordic Journal of Botany 3, 559–566. [Google Scholar]
- Li T, Li S, Luo R. 2012. Morphological observation and anatomical study on bulbil development of Lilium sulphureum. Acta Botanica Boreali-Occidentalia Sinica 32, 85–89. [Google Scholar]
- Li X, Qian Q, Fu Z, et al. 2003. Control of tillering in rice. Nature 422, 618–621. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. [DOI] [PubMed] [Google Scholar]
- López-Ráez JA, Kohlen W, Charnikhova T, et al. 2010. Does abscisic acid affect strigolactone biosynthesis? New Phytologist 187, 343–354. [DOI] [PubMed] [Google Scholar]
- Ma C, Sun Z, Chen C, Zhang L, Zhu S. 2014. Simultaneous separation and determination of fructose, sorbitol, glucose and sucrose in fruits by HPLC-ELSD. Food Chemistry 145, 784–788. [DOI] [PubMed] [Google Scholar]
- Main CL, Beeler JE, Robinson DK, Mueller TC. 2006. Growth, reproduction, and management of Chinese yam (Dioscorea oppositifolia). Weed Technology 20, 773–777. [Google Scholar]
- Majda M, Robert S. 2018. The role of auxin in cell wall expansion. International Journal of Molecular Sciences 19, 951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason MG, Ross JJ, Babst BA, Wienclaw BN, Beveridge CA. 2014. Sugar demand, not auxin, is the initial regulator of apical dominance. Proceedings of the National Academy of Sciences, USA 111, 6092–6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuki I, Takahashi A. 2009. Secondary dispersal of Dioscorea japonica (Dioscoreaceae) bulbils by rodents. Journal of Forestry Research 14, 95–100. [Google Scholar]
- Moody A, Diggle PK, Steingraeber DA. 1999. Developmental analysis of the evolutionary origin of vegetative propagules in Mimulus gemmiparus (Scrophulariaceae). American Journal of Botany 86, 1512–1522. [PubMed] [Google Scholar]
- Mudgil Y, Uhrig JF, Zhou J, Temple B, Jiang K, Jones AM. 2009. Arabidopsis N-MYC DOWNREGULATED-LIKE1, a positive regulator of auxin transport in a G protein-mediated pathway. The Plant Cell 21, 3591–3609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murty YS, Purnima. 1983. Morphology, anatomy and development of bulbil in some Dioscoreas. Proceedings: Plant Sciences 92, 443–449. [Google Scholar]
- Naz AA, Raman S, Martinez CC, Sinha NR, Schmitz G, Theres K. 2013. Trifoliate encodes an MYB transcription factor that modulates leaf and shoot architecture in tomato. Proceedings of the National Academy of Sciences, USA 110, 2401–2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni DA, Sozzani R, Blanchet S, Domenichini S, Reuzeau C, Cella R, Bergounioux C, Raynaud C. 2009. The Arabidopsis MCM2 gene is essential to embryo development and its over-expression alters root meristem function. New Phytologist 184, 311–322. [DOI] [PubMed] [Google Scholar]
- O’Hara LE, Paul MJ, Wingler A. 2013. How do sugars regulate plant growth and development? New insight into the role of trehalose-6-phosphate. Molecular Plant 6, 261–274. [DOI] [PubMed] [Google Scholar]
- Oh SA, Allen T, Kim GJ, Sidorova A, Borg M, Park SK, Twell D. 2012. Arabidopsis Fused kinase and the Kinesin-12 subfamily constitute a signalling module required for phragmoplast expansion. The Plant Journal 72, 308–319. [DOI] [PubMed] [Google Scholar]
- Pérez-Rodríguez P, Riaño-Pachón DM, Corrêa LG, Rensing SA, Kersten B, Mueller-Roeber B. 2010. PlnTFDB: updated content and new features of the plant transcription factor database. Nucleic Acids Research 38, D822–D827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrásek J, Friml J. 2009. Auxin transport routes in plant development. Development 136, 2675–2688. [DOI] [PubMed] [Google Scholar]
- Rabot A, Portemer V, Péron T, Mortreau E, Leduc N, Hamama L, Coutos-Thévenot P, Atanassova R, Sakr S, Le Gourrierec J. 2014. Interplay of sugar, light and gibberellins in expression of Rosa hybrida vacuolar invertase 1 regulation. Plant & Cell Physiology 55, 1734–1748. [DOI] [PubMed] [Google Scholar]
- Reddy SK, Holalu SV, Casal JJ, Finlayson SA. 2013. Abscisic acid regulates axillary bud outgrowth responses to the ratio of red to far-red light. Plant Physiology 163, 1047–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roman H, Girault T, Barbier F, et al. 2016. Cytokinins are initial targets of light in the control of bud outgrowth. Plant Physiology 172, 489–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronsheim ML, Bever JD. 2000. Genetic variation and evolutionary trade-offs for sexual and asexual reproductive modes in Allium vineale (Liliaceae). American Journal of Botany 87, 1769–1777. [PubMed] [Google Scholar]
- Sandoval SDCD, Juárez MJA, Simpson J. 2012. Agave tequilana MADS genes show novel expression patterns in meristems, developing bulbils and floral organs. Sexual Plant Reproduction 25, 11–26. [DOI] [PubMed] [Google Scholar]
- Schluepmann H, Pellny T, van Dijken A, Smeekens S, Paul M. 2003. Trehalose 6-phosphate is indispensable for carbohydrate utilization and growth in Arabidopsis thaliana. Proceedings of the National Academy of Sciences, USA 100, 6849–6854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu S, Mori H. 1998. Analysis of cycles of dormancy and growth in pea axillary buds based on mRNA accumulation patterns of cell cycle-related genes. Plant & Cell Physiology 39, 255–262. [DOI] [PubMed] [Google Scholar]
- Siciliano V, Genre A, Balestrini R, Cappellazzo G, deWit PJ, Bonfante P. 2007. Transcriptome analysis of arbuscular mycorrhizal roots during development of the prepenetration apparatus. Plant Physiology 144, 1455–1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Šimura J, Antoniadi I, Široká J, Tarkowská D, Strnad M, Ljung K, Novák O. 2018. Plant hormonomics: multiple phytohormone profiling by targeted metabolomics. Plant Physiology 177, 476–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staikidou I, Selby C, Arvey B. 1994. Stimulation by auxin and sucrose of bulbil formation in vitro by single leaf cultures of Narcissus. New Phytologist 127, 315–320. [DOI] [PubMed] [Google Scholar]
- Steiner BL, Armbruster GF, Scheepens JF, Stöcklin J. 2012. Distribution of bulbil- and seed-producing plants of Poa alpina (Poaceae) and their growth and reproduction in common gardens suggest adaptation to different elevations. American Journal of Botany 99, 2035–2044. [DOI] [PubMed] [Google Scholar]
- Szarek SR, Driscoll B, Shohet C, Priebe S. 1996. Bulbil production in Agave (Agavaceae) and related genera. The Southwestern Naturalist 41, 465–469. [Google Scholar]
- Tamiru M, Maass BL, Pawelzik E. 2008. Characterizing diversity in composition and pasting properties of tuber flour in yam germplasm (Dioscorea spp.) from Southern Ethiopia. Journal of the Science of Food and Agriculture 88, 1675–1685. [Google Scholar]
- Tan M, Li G, Chen X, Xing L, Ma J, Zhang D, Ge H, Han M, Sha G, An N. 2019. Role of cytokinin, strigolactone, and auxin export on outgrowth of axillary buds in apple. Frontiers in Plant Science 10, 616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. 2012. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nature Protocols 7, 562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbull CG, Raymond MA, Dodd IC, Morris SE. 1997. Rapid increases in cytokinin concentration in lateral buds of chickpea (Cicer arietinum L.) during release of apical dominance. Planta 202, 271–276. [Google Scholar]
- Walck JL, Cofer MS, Hidayati SN. 2010. Understanding the germination of bulbils from an ecological perspective: a case study on Chinese yam (Dioscorea polystachya). Annals of Botany 106, 945–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CN, Cronk QC. 2003. Meristem fate and bulbil formation in Titanotrichum (Gesneriaceae). American Journal of Botany 90, 1696–1707. [DOI] [PubMed] [Google Scholar]
- Wang CN, Möller M, Cronk QC. 2004. Altered expression of GFLO, the Gesneriaceae homologue of FLORICAULA/LEAFY, is associated with the transition to bulbil formation in Titanotrichum oldhamii. Development Genes and Evolution 214, 122–127. [DOI] [PubMed] [Google Scholar]
- Wang Y, Jiao Y. 2018. Axillary meristem initiation—a way to branch out. Current Opinion in Plant Biology 41, 61–66. [DOI] [PubMed] [Google Scholar]
- Wickham LD, Wilson LA, Passam HC. 1982. The origin, development and germination of bulbils in two Dioscorea species. Annals of Botany 50, 621–627. [Google Scholar]
- Wu ZG, Jiang W, Mantri N, Bao XQ, Chen SL, Tao ZM. 2015. Transciptome analysis reveals flavonoid biosynthesis regulation and simple sequence repeats in yam (Dioscorea alata L.) tubers. BMC Genomics 16, 346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing M, Du Y, Wang X, Niu L, Chen X. 2010. A simplified paraffin embedding method for small botanical samples. Biotechnic & Histochemistry 85, 241–246. [DOI] [PubMed] [Google Scholar]
- Yadav UP, Ivakov A, Feil R, et al. 2014. The sucrose–trehalose 6-phosphate (Tre6P) nexus: specificity and mechanisms of sucrose signalling by Tre6P. Journal of Experimental Botany 65, 1051–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F, Wang Q, Schmitz G, Müller D, Theres K. 2012. The bHLH protein ROX acts in concert with RAX1 and LAS to modulate axillary meristem formation in Arabidopsis. The Plant Journal 71, 61–70. [DOI] [PubMed] [Google Scholar]
- Yang M, Jiao Y. 2016. Regulation of axillary meristem initiation by transcription factors and plant hormones. Frontiers in Plant Science 7, 183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang P, Xu L, Xu H, Tang Y, He G, Cao Y, Feng Y, Yuan S, Ming J. 2017. Histological and transcriptomic analysis during bulbil formation in Lilium lancifolium. Frontiers in Plant Science 8, 1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao C, Finlayson SA. 2015. Abscisic acid is a general negative regulator of arabidopsis axillary bud growth. Plant Physiology 169, 611–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenoni S, Fasoli M, Tornielli GB, Dal Santo S, Sanson A, de Groot P, Sordo S, Citterio S, Monti F, Pezzotti M. 2011. Overexpression of PhEXPA1 increases cell size, modifies cell wall polymer composition and affects the timing of axillary meristem development in Petunia hybrida. New Phytologist 191, 662–677. [DOI] [PubMed] [Google Scholar]
- Zhan J, Thakare D, Ma C, et al. 2015. RNA sequencing of laser-capture microdissected compartments of the maize kernel identifies regulatory modules associated with endosperm cell differentiation. The Plant Cell 27, 513–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y. 2010. Auxin biosynthesis and its role in plant development. Annual Review of Plant Biology 61, 49–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng C, Halaly T, Acheampong AK, Takebayashi Y, Jikumaru Y, Kamiya Y, Or E. 2015. Abscisic acid (ABA) regulates grape bud dormancy, and dormancy release stimuli may act through modification of ABA metabolism. Journal of Experimental Botany 66, 1527–1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.