Abstract
The bHLH transcription factor Olig2 is required for sequential cell fate determination of both motor neurons and oligodendrocytes and for progenitor proliferation in the central nervous system. However, the role of Olig2 in peripheral sensory neurogenesis remains unknown. We report that Olig2 is transiently expressed in the newly differentiated olfactory sensory neurons (OSNs) and is down-regulated in the mature OSNs in mice from early gestation to adulthood. Genetic fate mapping demonstrates that Olig2-expressing cells solely give rise to OSNs in the peripheral olfactory system. Olig2 depletion does not affect the proliferation of peripheral olfactory progenitors and the fate determination of OSNs, sustentacular cells, and the olfactory ensheathing cells. However, the terminal differentiation and maturation of OSNs are compromised in either Olig2 single or Olig1/Olig2 double knockout mice, associated with significantly diminished expression of multiple OSN maturation and odorant signaling genes, including Omp, Gnal, Adcy3, and Olfr15. We further demonstrate that Olig2 binds to the E-box in the Omp promoter region to regulate its expression. Taken together, our results reveal a distinctly novel function of Olig2 in the periphery nervous system to regulate the terminal differentiation and maturation of olfactory sensory neurons.
Keywords: Basic helix–loop–helix (bHLH) transcription factors, Peripheral nervous system (PNS), Tuj1, Sox2, Fabp7 (Blbp), Dcx
Introduction
Neuronal differentiation is a multi-step process involving cell fate specification, progenitor expansion, and terminal maturation. Transcription factors, particularly the basic helix–loop–helix (bHLH) transcription factors, are essential for cell fate selection at the early-phase of neuronal differentiation [1, 2]. However, it remains poorly understood whether or how bHLH transcription factors play a role in terminal differentiation or neuronal maturation processes after neuronal cell fate determination. Olfactory sensory neurogenesis is an excellent model system to address this question. Generated from the basal progenitors, immature olfactory sensory neurons (OSNs) migrate a short distance toward the apical part of the olfactory epithelium (OE) where they express the mature OSN marker proteins and receptors and respond to odorants [3]. Given the continuous turnover of OSNs in adult vertebrates and the requirement of olfaction for suckling milk by neonatal rodents, proper terminal differentiation and maturation of the newborn OSNs is an essential step for functional olfaction and neonatal survival. Like in the central nervous system (CNS), a variety of bHLH factors, such as Ascl1 (Mash1), Ngn1 and Hes1, are identified in the olfactory progenitors and demonstrated to be important in controlling the proliferation and/or the differentiation of olfactory progenitors as they do in the CNS [4–7]. However, the role of bHLH transcription factors in the later phase of OSN differentiation is poorly understood.
Olig2 is a bHLH family member originally identified as a crucial transcription factor for the sequential development of motor neurons and oligodendrocytes in the embryonic spinal cord [8–10]. Later, it has been proven to control the proliferation of neural progenitors, and the genesis of cortical GABAergic neurons and white matter astrocytes [11–13]. All support a proliferation-stimulating or gliogenic function of Olig2 in CNS. In the present study, we explored the role of Olig2 in the differentiation of OSNs and found that Olig2 is dispensable for the proliferation and fate determination of the OE progenitors, but it is specifically involved in the final maturation of OSNs, partially by regulating the expression of multiple genes, including Omp, Gnal, Adcy3, and Olfr15, which encode critical proteins for OSN maturation or odorant transduction.
Materials and methods
Genetically modified mouse lines
The Olig2CreER knock-in mice, Olig1/Olig2 double knockout, and Olig2-Cre mice were previously described [9, 10, 14]. Rosa26-LacZ mice [15] were acquired from the Jackson Laboratory. All animal experiments were carried out under protocols approved by UC Davis Animal Care and Use Committees and following NIH guidelines. Pregnant, timed mated mice were euthanized prior to cesarean section. Noon of the conception day was designated as E0.5.
Immunohistochemistry and BrdU labeling
Immunohistochemistry was conducted as described [16]. The embryos were immersion-fixed in 4% paraformaldehyde (PFA). The postnatal and adult mice were transcardially perfused and fixed by 4% PFA. 30% sucrose was used for cryoprotection and 12–14 µm frozen sections were made by a cryostat. Air-dried slides were permeabilized by 0.3% Tween-20 in PBS, blocked by 10% lamb serum, and incubated with primary antibodies at 4 °C overnight. After washing in PBS, the corresponding secondary antibodies (Alexa Fluor 488 or 594 conjugated anti-rabbit, anti-mouse, anti-guinea pig, or anti-goat, 1:500, Invitrogen) were incubated for 2 h at room temperature. Cell nuclei were stained by DAPI. The primary antibodies and dilutions are as following: rabbit anti-Olig2 (1:200, Millipore), mouse anti-Sox2 (1:50, Cell Signaling Technology), rabbit anti-Fabp7 (BLBP) (1:1000, Millipore), mouse anti-BrdU (1:50, Developmental Studies Hybridoma Bank), guinea pig anti-Dcx (1:1000, Millipore), and goat anti-OMP (1:1000, Wako). For acute BrdU labeling, pregnant mice were intraperitoneally injected with 10 mg/kg BrdU. Embryos were sampled after 1-h BrdU incorporation. Frozen sections were treated by 2 N HCl to denature DNA at 37 °C for 20 min, and by 0.1 M borate sodium buffer (pH 8.5) to neutralize the sections for subsequent immunohistochemistry with BrdU antibodies.
RNA in situ hybridization
Wholemount and section in situ hybridization were conducted as described [16]. RNA probes for Omp, Gnal (Golf), Adcy3 (AC3), Lhx2, and Ebf1 (O/E-1) were made by EST clones. Other probes were made by PCR-based in vitro transcription using primers listed on Allen Mouse Brain Atlas website, and also shown below. Olig2: forward 5′-ATATGGGAACCGAAGCAATG-3′, reverse 5′-GCTCCTGTGCTCTGAAAAGG-3′; Ncam1: CAGGTAGATATTGTTCCCAG, GTCCTTGAAGTTGATTTCCC; Gap43: GGCTCTGCTACTACCGATGC, GCAGGAGAGACAGGGTTCAG; Olfr15: GGCCTCTTACTTGTTGACGC, ATGACGCTTACTGGGACCAC; Mecp2: AGACAAGCCACTGAAGTTTAAGAAG, TTGACAACAAGTTTCCCAGG; Arfgef2: CGAGCAAGGAACACTCAACA, TGTTTGGACCATGCAGACAT; Kirrel2: GCTTGGTTTCCACTCAGCTC, CAGCAAAGGAAAACGAGGAG. Sections were counterstained by nuclear fast red.
Chromatin immunoprecipitation (ChIP) assay
ChIP assay was conducted according to the manual of the ChIP assay kit (Upstate) and a slightly modified protocol [17, 18]. Briefly, OEs of E18.5 wild-type mice were dissected and fixed by 4% PFA for 15 min on ice. Sonication was performed to break chromatin. Rabbit anti-Olig2 (1:100, Millipore) was incubated with DNA–protein complex overnight at 4 °C. Rabbit IgG at the same concentration was used as the negative control. The primer pairs targeting the E-boxes in the Omp promoter were: (1) GTGGTTCAGTTACAGAGCCC, AGAAACCCTCCTGCTTGAGC; (2) TATGTGGTTGGATCGATCAAAC, TTATCACCATCAGGACCCAG; (3) AACAAACAAATAGAACAGAGCAGGC, ATTGCCAGATGGAGGTCAAC; (4) TGTGTGTGTGTGTGATGTTC, TGTATGTGGACAGATGGCAG; (5) CCGTCTGTCTGGCAGATGATTTG, TCATAGCCCCTGTCAGGTCC.
Luciferase reporter assay
The promoter region of Omp was PCR amplified from mouse genomic DNA. Based on the ChIP data, three fragments of Omp promoter were cloned into pGL2-basic vector (Promega, Madison, WI) to construct Luci-Omp1 (containing E-box 1, or E1), Luci-Omp1–3 (containing E1–3 boxes), Luci-Omp1–6 (containing E1–6 boxes), Luci-Omp4 (containing E4 box), and Luci-Omp6 (containing E6 box) (Fig. 8a). Luciferase reporter assay was conducted as described [19, 20]. Briefly, human embryonic kidney HEK293 cells were cultured to around 80% confluence. Luci-Omp1, Luci-Omp1–3, Luci-Omp1–6, Luci-Omp4, and Luci-Omp6 were co-transfected with pcDNA3.1-Olig2 using lipofectamine 2000 for 24 h. PCR-based mutation was performed to construct Luci-Omp1-mutant. Luciferase activity of total cell lysates was measured using Dual-luciferase reporter assay system (Promega). The Renilla-luciferase reporter pRL vectors (5 ng/well) were used as an internal control.
Fig. 8.
Unchanged expression of transcription factors associated with the terminal differentiation of OSNs in Olig2 single and Olig1/Olig2 double knockout OEs at E18.5. In situ hybridization of Zfp423, Ebf1 (OE-1), Lhx2, and Mecp2 were examined on sections from three embryos per genotype
Real-time PCR
OEs of E18.5 embryos were dissected under microscope. RNA was extracted using Trizol reagent. cDNA was made by iScript™ cDNA Synthesis Kit (BIO-RAD). PCR was performed using SYBR GREEN PCR master mix (Thermofisher). The mRNA levels of Fabp7 and Omp were normalized to the mRNA level of the house-keeping gene Gapdh to allow comparisons among different experimental groups using the ΔCt method. The primer sequences are as following: Omp, TCCGTCTACCGCCTCGATTT, CGTCTGCCTCATTCCAATCCA; and Fabp7 (Blbp), TTGATGAGTACATGAAAGCTCTGG, CTTGAATGTGCATTGTGTCC.
Cell counting and statistical analyses
Cells positive for immunolabeling of Olig2, Sox2, Dcx, Omp, BrdU, and Fabp7; and in situ hybridization signals of Omp, Gnal, Olfr15, Arfgef2, Kirrel2, Adcy3, Ncam1, and Gap43 were counted under microscope along OE lining the nasal septum from dorsal to ventral in three sections for each animal. The in situ hybridization “positive” cells were identified if the positive signal is localized in cytoplasm surrounding a nucleus which was counterstained by fast red. The littermate control and mutant OE sections were mounted on the same slide for subsequent in situ hybridization and immunohistochemistry under the same condition. OE length was determined by tracing the outline of the epithelium basal lamina using Slidebook software. For OE thickness, the vertical distance from the basal cell layer to apical surface were measured in six points with 100-μm intervals along the nasal septum on each of three sections per embryo. At least three mutants and three littermate control embryos were included for each quantitative analysis. The morphological analysis was performed by a researcher blind to genotypes. The quantitative data were represented as mean ± SE (standard error). Two-tailed Student’s t test was used for comparisons of the wild-type control vs. knockout samples. One-way ANOVA was used for three-group comparisons. P ≤ 0.05 is considered significant. Related statistical details are included in the figure legends or results.
Results
Olig2 is expressed in newly differentiated Sox2(−);Tuj1(+) OSNs of the peripheral olfactory system
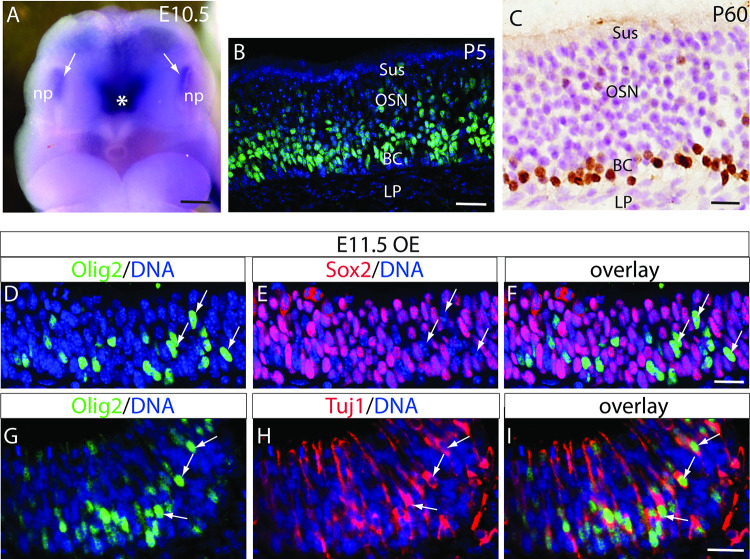
As soon as the nasal pit forms, Olig2 is expressed in the olfactory epithelium of E10.5 and E11.5 mouse embryos (Fig. 1). Considering the function of Olig2 in the CNS neural progenitors [21] and the basal localization of olfactory neural progenitors [22], we asked whether Olig2 is expressed by olfactory progenitors. Double fluorescent immunohistochemistry demonstrates that Olig2-positive cells are scattered in the early developing OE at E11.5, but they do not express Sox2, a marker for neural stem cells and progenitors [23], which are widely and uniformly expressed throughout the E11.5 OE on the same tissue section (Fig. 1d–f). In contrast, all Olig2-positive OE cells express the neuron-specific class III β-tubulin Tuj1 (Fig. 1g–i). These unexpected results suggest that Olig2 is not expressed in the OE progenitors but in newly differentiated OSNs during early gestation.
Fig. 1.
Expression of Olig2 in embryonic, postnatal, and adult olfactory epithelia (OE). a Wholemount in situ hybridization demonstrates Olig2 mRNA expression (white arrows) in the nasal pit (np) as early as E10.5. Asterisk indicates the high expression of Olig2 in the ventral forebrain as expected. b Immunolabeling of Olig2-positive cells (green, counterstained with DAPI in blue) in P5 OE. c Olig2-positive cells (brown, counterstained with hematoxylin and eosin in purple) in the adult OE. d–f Double immunofluorescence of Olig2 and Sox2 in E11.5 OE. Arrows indicate representative Olig2-positive cells that are Sox2 negative. g–i Double immunofluorescence of Olig2 and Tuj1 in E11.5 OE. Arrows indicate representative Olig2-positive cells with co-immunolabeling of Tuj1. BC basal cell layer, LP laminar propria, np nasal pit, OE olfactory epithelium, OSN olfactory sensory neuron layer, Sus sustentacular cell layer. Scale bars = 200 µm (a), 20 µm (b), 50 µm (c), 20 µm (d–i)
From mid-late gestation, Olig2-positive cells distribute mainly in the basal part of OE, through postnatal ages to adult (Fig. 1b, c). The seeming localization changes of the Olig2-positive cells at different ages may reflect the cellular reorganization following OE development and maturation. In the late embryonic stage, the cellular organization of OE becomes similar to the adult OE. Therefore, we further examined the details of Olig2 expression in E18.5 OE. Double immunolabeling of Olig2 with Sox2, which is expressed in olfactory stem cells of the basal OE and in sustentacular glial cells of the apical OE during late gestation and adulthood [16, 24], shows that the basal Olig2-positive cells are localized immediately atop the basal Sox2-positive cells and away from the Sox2-positive sustentacular apical OE (Fig. 2a). As with E11.5, no Olig2-positive cells express Sox2 in E18.5 OE (Fig. 2a). Double immunolabeling of Olig2 with the immature and migrating OSN marker Dcx shows that approximately 60.91 ± 0.81% of Olig2-positive cells (counted 489 from total 3 embryos) are Dcx positive (Fig. 2b). Double immunolabeling with the mature OSN marker Omp shows that approximately 11.17 ± 1.39% of Olig2-positive cells (counted 403 from total 3 embryos) are Omp positive (Fig. 2c). These results suggest that Olig2 is transiently expressed in the newly differentiated OSNs and it is down-regulated following OSN maturation.
Fig. 2.
Double immunofluorescence of Olig2 with representative OE lineage markers at E18.5. a–a′′′ At the late embryonic age, Olig2-positive cells (green) are closely located atop the Sox2 (red)-positive basal cell layer and away from the Sox2-positive sustentacular cell layer. They do not overlap each other. Dashed rectangle in a is enlarged in a′–a′′′ for better resolution. b–b′′′ More than one half of the Olig2-positive cells (green) are Dcx positive (red). Six strongly co-immunolabeled cells are marked in b′–b′′′. c–c′′′ A small portion of Olig2-positive cells are Omp positive (red). Two co-immunolabeled cells are marked in c′–c′′′. Nuclei are counterstained by DAPI (blue). Scale bars = 50 µm (a, b,c) and 20 µm (a′–c′′′)
Dispensable role of Olig2 in the proliferation of olfactory progenitor and gliogenesis in OE
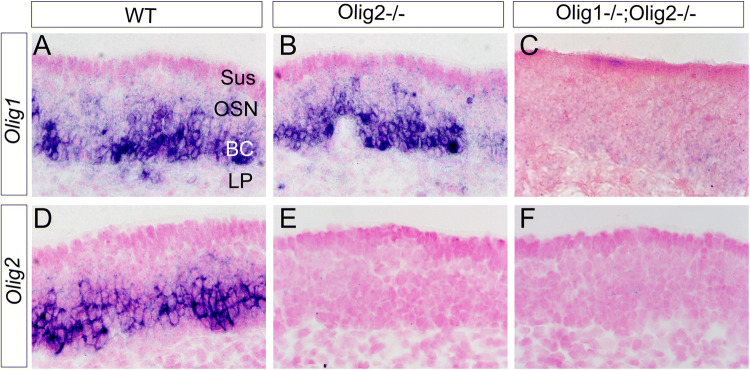
Because a key role of Olig2 in CNS is to instruct the motor neuron and oligodendrocyte lineage cell fates, and approximately 30% of the Olig2-positive cells in the basal part of OE are neither Dcx nor OMP positive (Fig. 2), we then asked whether Olig2 plays a role in gliogenesis or the proliferation of peripheral olfactory progenitors, and whether Olig1 plays a synergetic role with Olig2 in olfactory sensory neurogenesis. In situ hybridization demonstrates that Olig1 is also expressed in the embryonic OE, with a similar expression pattern as Olig2 (Fig. 3a, d). We further addressed aforementioned questions on Olig2 single- and Olig1/Olig2 double-deficient OE at E18.5 due to the neonatal lethality of these mutant mice. Olig1 expression is not affected in the Olig2-deficient OE (Fig. 3b). Morphologically, the mutant OE looks normal in either single Olig2−/−or double Olig1−/−;Olig2−/− mutants at E18.5 (Fig. 3b, c, e, f). There is no significant difference of OE thickness between Olig2−/− (159.07 ± 11.35 μm) and wild-type (171.16 ± 5.72 μm) embryos at E18.5 (n = 3 embryos for each genotype, Student’s t test, T value − 0.046, P = 0.966). Acute BrdU incorporation and TUNEL staining are adopted to evaluate proliferation and apoptosis, respectively (Fig. 4a, b). Apical Sox2 and Fabp7 (Blbp) expression are used as glial-like cell markers for sustentacular and olfactory ensheathing cells (OECs), respectively [16, 24] (Fig. 4c, d). Unexpectedly, the results show no differences in all of these parameters between the wild-type and Olig2−/− OE at E18.5 (Fig. 4), indicating that Olig2 is neither involved in progenitor proliferation nor gliogenesis in the OE, at least at the embryonic stage.
Fig. 3.
Section in situ hybridization for Olig1/Olig2 mRNAs in the normal and mutant OE at E18.5. a–c Olig1 is also expressed in OE of the wild-type (WT) and the single Olig2-KO embryos, which is ablated in the Olig1;Olig2 double KO OE. d–f Olig2 expression shows a similar pattern with Olig1 in the WT, which is ablated in either single Olig2-KO or double Olig1;Olig2 double KO OE. OE sections are counterstained with fast red. BC basal cell layer, LP laminar propria, OSN olfactory sensory neuron layer, Sus sustentacular cell layer
Fig. 4.
Unchanged proliferation, apoptosis, and gliogenesis in the Olig2-deficient OE. a, a′ BrdU (red) immunolabeled OE sections and positive cell numbers in the basal and apical OE of the E18.5 wild-type (WT) and Olig2−/− embryos show no significant changes. n = 3 embryos for each genotype; Student’s t test. T value: 1.128 (basal) and − 0.873 (apical). P = 0.463 (basal) and 0.432 (apical). b, b′ No significant changes of TUNEL-labeled apoptotic cells in Olig2-deficient OE compared to the WT (n = 3 each genotype). Student’s t test, T value: 0.194, P = 0.856. c, c′ Sox2 (green) immunolabeled OE sections and positive cell numbers have no significant changes (n = 3 each). Student’s t test, T value: − 0.745 (basal) and 0.955 (apical), P = 0.497 (basal) and 0.394 (apical). d, d′ Fabp7 (Blbp) (green) immunolabeled OE and qPCR results show no significant changes (n = 3 each). Student’s t test, T value: 0.1178, P = 0.304. Scale bar = 50 µm
OSN-restricted cell lineage of Olig2-expressing OE cells
In CNS, Olig2-expressing cells generate not only neurons and oligodendrocytes, but also white matter astrocytes and even reactive astrocytes after injury [25]. We next examined whether Olig2 expressing cells in OE generate olfactory glial cells by genetic fate mapping. Olig2-Cre mice [14] were crossed with the reporter Rosa26-LacZ mice [15]. X-gal and Fabp7 (Blbp) staining were performed at E14.5 and P7 (Fig. 5). The results show that, at both time points, only neuronal progeny are detected. X-gal staining is neither seen in the sustentacular cell layer nor the Fabp7-positive OECs (Fig. 5a–d). These results are in agreement with the restricted expression of Olig2 in the newly differentiated and Tuj1-positive OSNs (Figs. 1, 2).
Fig. 5.
Genetic fate mapping of Olig2 progeny in the peripheral olfactory system. X-gal staining (green) for Olig2-Cre;Rosa26lacZ cell lineages and immunolabeling for Fabp7 (Blbp)-positive ensheathing cells (brown) are shown on the OE sections of E14.5 (a, b) and P7 (c, d) mice. b, d Enlarged from the dashed squares in a and c, respectively. Arrowhead in d indicates a Fabp7-positive ensheathing cell that is adjacent to the X-gal-positive axonal fibers (arrow). BC basal cell layer, LP laminar propria, OE olfactory epithelium, OB olfactory bulb, OSN olfactory sensory neuron layer, S nasal septum
Requirement of Olig2 for the terminal differentiation and maturation of OSNs
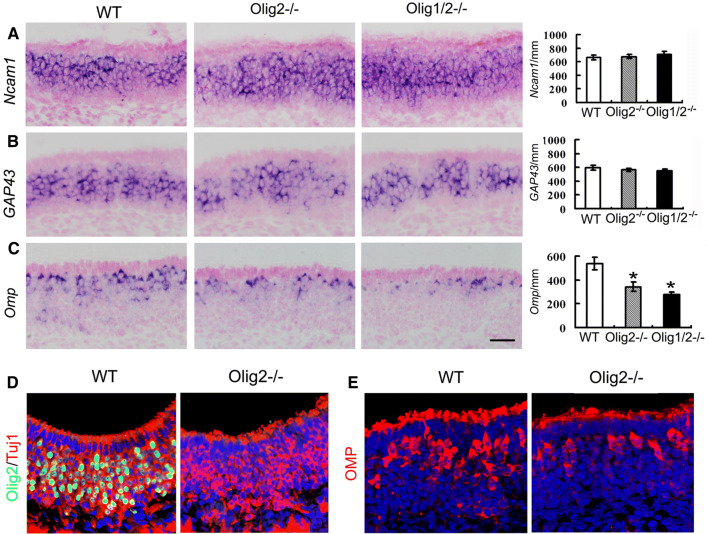
To address the role of Olig2 in peripheral sensory neurogenesis, we analyzed OSN differentiation in Olig2−/− OE by examining the expression of several critical marker genes for OSN maturation and function. In situ hybridization shows that Olig2 depletion does not affect the expression of a pan OSN marker Ncam1 and an immature OSN marker Gap43 (Fig. 6a, b). However, expression of the mature OSN marker Omp is significantly decreased in the Olig2−/− OE (Fig. 6c). Immunohistochemistry shows that regardless of Olig2 depletion, Tuj1-positive OSNs are conserved in the E18.5 Olig2−/− OE (Fig. 6d). Consistent with the in situ hybridization result, Omp immunolabeling shows a notable reduction at the protein level (Fig. 6e).
Fig. 6.
Expression of representative OSN lineage markers in Olig2 single- and Olig1;Olig2 double-deficient OE at E18.5. a–c In situ hybridization and quantification of Ncam1, Gap43, and Omp. Only Omp-expressing cells are significantly reduced in the single or double mutants. n = 3 embryos each genotype. One-way ANOVA. For Ncam1: F value, 0.206; POlig2KO vs. WT = 0.891, POlig1/2KO vs. WT = 0.562. For Gap43: F value, 0.038; POlig2KO vs. WT= 0.822, POlig1/2KO vs. WT = 0.815. For Omp: F value, 11.241; *POlig2KO vs. WT = 0.007, *POlig1/2KO vs. WT = 0.006. Double immunofluorescence of Olig2 with Tuj1 (d) and Omp (e) demonstrates ablation of Olig2, conserved Tuj1, and consistently reduced Omp-positive cells in the Olig2-deficient OE
We then examined critical genes associated with OSN odorant signaling functions. Adcy3 (AC3), encoding the type III adenylyl cyclase, is required for olfaction [26]. Gnal (Golf), encoding a stimulatory G protein alpha subunit, is required for odorant signaling and postnatal survival [27]. Both Adcy3 and Gnal mRNAs are dramatically diminished in the Olig2−/− OE (Fig. 7a, b). Moreover, the number of olfactory sensory neurons which express olfactory receptor Olfr15 is significantly decreased in Olig2−/− OE (Fig. 7c). These data indicate that the maturation and function of OSNs are impeded by Olig2 deficiency.
Fig. 7.
In situ hybridization and quantification of representative OSN differentiation marker genes Acdy3 (AC3) (a), Gnal (b), Olfr15 (c), Arfgef (d), and Kirrel2 (e) demonstrate declined expression of these differentiation marker genes in Olig2 single and Olig1/Olig2 double knockout OEs at E18.5. Sections from three embryos per genotype were used for each marker. One-way ANOVA. For Acdy3 (AC3): F value, 421.256; **POlig2KO vs. WT < 0.0001, **POlig1/2KO vs. WT < 0.0001. For Gnal: F value, 16.811; *POlig2KO vs. WT = 0.003, *POlig1/2KO vs. WT = 0.002. For Olfr15: F value, 24.873; *P Olig2KO vs. WT = 0.001, *POlig1/2KO vs. WT = 0.001. For Arfgef: F value, 25.067; *POlig2KO vs. WT = 0.002, *POlig1/2KO vs. WT = 0.001. For Kirrel2: F value, 9.865; *POlig2KO vs. WT = 0.005, *POlig1/2KO vs. WT = 0.029
We next asked whether Olig2 selectively affects the maturation of certain subgroup of OSNs. According to the expression of axonal guidance factors, OSNs can be roughly grouped as Arfgef2 (Big2)-positive and Kirrel2-positive [28]. In situ hybridization shows a similar reduction of both Arfgef2- and Kirrel2-positive cells, indicating a general role of Olig2 in the terminal differentiation of OSNs (Fig. 7d, e).
We further examined the expression of Zfp423, Lhx2, Mecp2, and Ebf1 (OE-1), which encode transcription factors involved in the terminal differentiation of OSNs [29–32]. No differencein the expression of all these factors is found between the wild-type and Olig2−/− OE by in situ hybridization (Fig. 8).
Considering that Olig1 is expressed in Olig2−/− OE (Fig. 3), we asked whether functional redundancy between Olig1 and Olig2 may occur in the OE as in the spinal cord [9, 33]. The OSN differentiation was then investigated in the Olig1/Olig2 double knockout embryos [9] (Fig. 5). In situ hybridization of all aforementioned OSN marker genes in Olig1/Olig2 double knockout OE demonstrates almost identical phenotypes as seen in the Olig2 single knockout (Figs. 6, 7, 8), suggesting the requirement of Olig2, not Olig1 in terminal differentiation and functional maturation of OSNs.
Direct regulation of Omp expression by Olig2
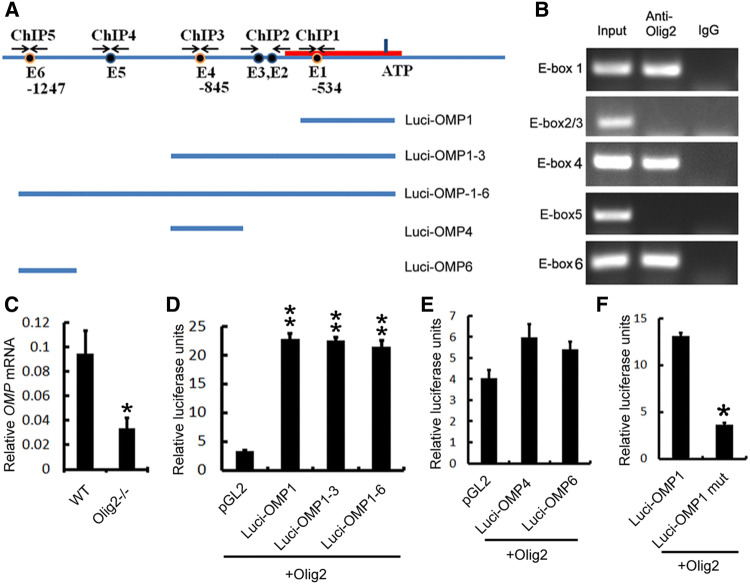
To address the mechanism of Olig2 in regulation of critical OSN maturation genes, we conducted in vitro molecular biological analyses. As a bHLH transcription factor, Olig2 regulates its downstream factors by binding to the E-box sequences on the promoter of the target genes [34]. Among the significantly down-regulated OE genes in the Olig2−/− mutants, Omp plays a crucial role in the final maturation of OSNs and also is a widely used early marker for mature OSNs [35, 36]. Therefore, we choose Omp as an exemplary candidate of downstream target genes to address whether it is directly regulated by Olig2. We searched the promoter region of Omp and found six E-boxes (E1, CACCTG, − 534 bp; E2, E3, CAGATG, − 592 bp, − 623 bp; E4, E5, CATCTG, − 845 bp, − 1043 bp; E6, CACGTG, − 1247 bp from ATG, respectively) (Fig. 9a). Chromatin immunoprecipitation assay shows that Olig2 is bound to E1, E4 and E6 among six E-boxes, indicating that Olig2 may directly regulate Omp expression (Fig. 9b). Real-time RT-PCR confirms that the Omp mRNAs are decreased significantly in the Olig2−/− OE (Fig. 9c). To identify which E-box may contribute to the Olig2-regulated Omp expression, we co-transfected Olig2 full-length cDNAs with three Omp luciferase reporter constructs (Luci-Omp1 containing E1, Luci-Omp1–3 containing E1–3, and Luci-Omp1–6 containing E1–6) (Fig. 9a). The results demonstrate that Olig2 significantly activates the transcriptional activity of all of these luciferase reporter constructs at the similar level (Fig. 9d), suggesting that E4 and E6 might be dispensable for Olig2-regulated Omp expression. Indeed, co-transfection of Olig2 with constructs containing individual E4 (Luci-Omp4) or E6 (Luci-Omp6) shows no significant effects of both constructs on Omp expression as compared to pGL2 control (Fig. 9e). On the other hand, the construct with mutated E1 box dramatically abolishes the Olig2-induced Omp transcription (Fig. 9f). Collectively, these results suggest that Olig2 directly regulates the Omp expression by binding to the E1 box, which might partially account for the compromised OSN maturation in the Olig2-deficient OE.
Fig. 9.
Olig2 binds to E-box and regulates Omp promoter activity. a Illustration of E-boxes in presumptive Omp promoter region, and ChIP and luciferase assay strategies. Red line represents the core promoter region of Omp. E1–6 stand for the E-boxes 1–6. ChIP1–5 show the PCR amplification regions containing different E-boxes in ChIP assays. Luci-Omp1–6 represent Omp luciferase constructions containing one, three, or all six E-boxes. b ChIP results demonstrate that Olig2 binds to the E-boxes 1, 4, 6, but not 2/3 and 5 of the Omp promoter. c Real-time PCR results confirm the significantly diminished expression of Omp in Olig2-deficient OE. n = 3 embryos for each genotype, Student’s t test, T value: 3.219, P = 0.032. d Luciferase assay results demonstrate that Olig2 promotes the transcription of three Omp promoter constructs that all contain E-box 1. n = 3 batches of cells. One-way ANOVA. F value, 161.268; **PLuci-Omp1 vs. pGL < 0.0001, **PLuci-Omp1–3 vs. pGL < 0.0001, *PLuci-Omp1–6 vs. pGL < 0.0001. e Constructs which contain E-box 4 or 6 have no significant effects on the expression of Omp promoter luciferase activities. n = 3 batches of cells. One-way ANOVA. F value, 2.773; PLuci-Omp4 vs. pGL = 0.099, PLuci-Omp6 vs. pGL = 0.079. f Mutation in E-box1 significantly abolished the effects of Olig2 on the Omp promoter luciferase activity. n = 3 batches of cells. Student’s t test. T value: 9.633. *P = 0.001
Discussion
The bHLH transcription factors have been well documented as cell fate instructors, but they are poorly studied in the late stages of neurogenesis. In the present study, we demonstrate a novel role of Olig2 in the terminal differentiation of sensory neurons by analyzing the cell identity of Olig2-positive cells in the OE and the phenotypes of Olig2 single and Olig1/Olig2 double knockout mutants during peripheral sensory neurogenesis.
In CNS, Olig2 is expressed by neural progenitors from early embryonic stage. Our data show that Olig2 is expressed in the nascent olfactory placode as early as E10.5 in mice. Although distributed sparsely, Olig2 expression is restricted to the newly differentiated Tuj1-positive OSNs, without expression in the Sox2-positive OE progenitors at E11.5. The co-immunolabeling of Olig2 with Tuj1 has also been shown in rat E14.5 OE [37]. In late gestation of mouse embryos, the Olig2-positive cells become layered and locate atop Sox2-positive progenitors in the basal cell layer. Our double immunolabeling of Olig2 with Dcx and Omp further reveals that it is expressed by the newly differentiated OSNs and down-regulated with the maturation of OSNs. The neuronal lineage-restricted expression of Olig2 in newly differentiated OSNs and its absence in OE progenitors/glial lineage cells suggest a distinctly novel role of Olig2 in PNS development. Given that Olig2-positive cells give birth to both neurons and glia in CNS, the solely neuronal progeny of Olig2 fate mapping in OE is in line with its expression pattern, and further suggests a potential role of Olig2 in late stages of peripheral sensory neurogenesis.
To determine the new role of Olig2 in peripheral neurogenesis, we examined alterations of the proliferation, apoptosis, and differentiation of the OE in Olig2-deficient mutants. Because Olig2-KO mice die shortly after birth, we conducted analysis at E18.5. In line with the expression pattern and fate-tracing results, Olig2 deficiency does not affect the proliferation of progenitors and the generation of glial cells in the OE. Interestingly, the expression of the pan OSN markers (Tuj1 and Ncam1) and immature marker (Gap43) remains unchanged, but the expression of multiple critical genes (Omp, Gnal, Adcy3, and Olfr15) required for OSN maturation and odorant signaling transduction is significantly reduced in Olig2-deficient OE.
Considering that Olig2 expression in the olfactory placode starts as early as E10.5, and the requirement of olfaction for the suckling behavior of neonatal pups, it is necessary that OSN maturation must occur before birth. Our data, therefore, suggest a key role of Olig2 in the maturation process of OSNs during embryonic development, in preparation of functional readiness of olfaction for neonatal survival in rodents. The neuronal maturation role of Olig2 is indirectly supported by a previous study reporting an essential role of another bHLH transcription factor NeuroD1 in terminal differentiation of subventricular zone-derived neurons in the olfactory bulb, which act through different molecular mechanisms [38]. Among the significantly down-regulated factors in Olig2-deficient OE as demonstrated in the current study, the GTP-binding G protein Golf (encoded by Gnal) and the type III adenylyl cyclase AC3 (encoded by Adcy3) are located in the olfactory cilia of the mature OSNs to transduce odorant signaling through the Golf-AC3-cAMP cascade [26, 39, 40]. Together, these factors strongly support a novel role of Olig2 in functional maturation of OSNs during embryonic development.
The half reduction of Omp in Olig2−/− OE promoted us to assess if there is a redundant function of Olig1 with Olig2 in peripheral sensory neurogenesis. Indeed, Olig1 is also expressed in embryonic OE in a similar pattern with Olig2 expression. However, the similar defects of OSN maturation in Olig1/Olig2-double knockout OE suggest a dispensable or minimal role of Olig1 in embryonic OE development, but its function remains to be determined in the future. Olig1 and Olig2 have been shown to play distinct roles in postnatal CNS development and disease [41]. Therefore, it is highly possible that Olig1 and Olig2 also play different roles in postnatal and adult olfactory sensory neurogenesis in normal and pathological conditions and during regeneration processes.
The incomplete decline of OSN maturation in Olig2-deficient OE is quite intriguing, as both Arfgef2- and Kirrel2-positive subgroups of OSNs are diminished by about 50%. It remains to be explored whether Olig2 is responsible for maturation of all or certain subgroups of OSNs, or other bHLH transcription factors may play a synergistic role with Olig2 in OSN maturation. Previous studies have reported that transcription factors Zfp423, Lhx2, Mecp2, and Ebf1 are involved in the terminal differentiation of OSNs [6, 29, 30, 32]. However, the expression of these transcription factors remains unchanged in the Olig2-deficient OE, indicating that these transcription factors are not regulated by Olig2, but they may work with Olig2 together or in parallel to regulate OSN terminal differentiation and maturation. For instance, co-transfection of Olig2 and Nkx2.2 can induce oligodendrocytes, and co-transfection of Olig2 with HB9 but not Ngn2 can increase the motor neuron marker Isl1/2 from adult human olfactory epithelial-derived progenitors [42, 43]. Olig2 and Sox10 induce oligodendrocyte differentiation through reciprocal interactions and dosage-dependent mechanisms in embryonic chicken spinal cord [44]. The interactive and regulatory mechanisms of Olig2 with other transcription factors during sensory neurogenesis remain to be investigated.
During peripheral sensory neurogenesis, OSNs express several critical genes in the following sequence: from Gap43 to Adcy3 then Omp within 6–8 days after cell mitosis in postnatal and young adult mice [36, 45]. Adcy3 acts downstream of Gnal during odorant signaling transduction [40]. Omp plays a crucial role in the final maturation of OSNs and also is an early marker for mature OSNs [35, 36]. Based on results showing unaltered Gap43 and significantly diminished Gnal, Adcy3, and Omp expression in the Olig2-deficient OE, it is logical to propose that Olig2 directly or sequentially regulates Gnal, Adcy3, and Omp to promote OSN maturation. In the current study, chromatin immunoprecipitation and luciferase reporter assays demonstrate the direct regulation of Olig2 for one of these genes, Omp. It is highly possible that Olig2 also directly regulates additional downstream target genes, at least Gnal and Adcy3 during OSN maturation. Therefore, how the expression of whole profile of the mature OSN-associated genes is regulated by Olig2, and whether Olig2 plays conserved or pleiotropic roles in neurogenesis of PNS remain to be studied.
Acknowledgements
We are grateful to David J. Anderson (HHMI and Caltech) for providing the Olig1/Olig2 frozen embryos, Jordan Hui, Ben Palmer, Arjun Stokes, Huan Zhao, Taylor Imai, Rebecca Duncan, Yue Liu, Santosh Kumar, Saharul Islam, Sarwat Amina and the rest of Zhou lab members for technical assistance or general support. This work was partially supported by grants from the NIH (R01DE021696, R01DE026737 and R01NS102261 to C.J.Z.), the Shriners Hospitals for Children (86600 and 85105 to C.J.Z., and postdoctoral fellowships to Y.Z.W. and R.G.), and the National Science Foundation of China (31970907 to Y.Z.W.). Q.G. and T.Y. received postdoctoral fellowships from the California Institute for Regenerative Medicine (CIRM) Stem Cell Training Program.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Ya-Zhou Wang, Hong Fan, and Yu Ji contribute equally.
Change history
6/22/2021
A Correction to this paper has been published: 10.1007/s00018-021-03870-2
References
- 1.Kageyama R, Ohtsuka T, Hatakeyama J, Ohsawa R. Roles of bHLH genes in neural stem cell differentiation. Exp Cell Res. 2005;306(2):343–348. doi: 10.1016/j.yexcr.2005.03.015. [DOI] [PubMed] [Google Scholar]
- 2.Powell LM, Jarman AP. Context dependence of proneural bHLH proteins. Curr Opin Genet Dev. 2008;18(5):411–417. doi: 10.1016/j.gde.2008.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Murdoch B, Roskams AJ. Olfactory epithelium progenitors: insights from transgenic mice and in vitro biology. J Mol Histol. 2007;38(6):581–599. doi: 10.1007/s10735-007-9141-2. [DOI] [PubMed] [Google Scholar]
- 4.Sammeta N, Yu TT, Bose SC, McClintock TS. Mouse olfactory sensory neurons express 10,000 genes. J Comp Neurol. 2007;502(6):1138–1156. doi: 10.1002/cne.21365. [DOI] [PubMed] [Google Scholar]
- 5.Suzuki Y. Expression of bHLH transcription factors and IGFs in the non-sensory patches, olfactory epithelium and vomeronasal organ. Chem Senses. 2005;30(Suppl 1):i125–i126. doi: 10.1093/chemse/bjh146. [DOI] [PubMed] [Google Scholar]
- 6.Cau E, Casarosa S, Guillemot F. Mash1 and Ngn1 control distinct steps of determination and differentiation in the olfactory sensory neuron lineage. Development. 2002;129(8):1871–1880. doi: 10.1242/dev.129.8.1871. [DOI] [PubMed] [Google Scholar]
- 7.Cau E, Gradwohl G, Casarosa S, Kageyama R, Guillemot F. Hes genes regulate sequential stages of neurogenesis in the olfactory epithelium. Development. 2000;127(11):2323–2332. doi: 10.1242/dev.127.11.2323. [DOI] [PubMed] [Google Scholar]
- 8.Rowitch DH, Lu QR, Kessaris N, Richardson WD. An ‘oligarchy’ rules neural development. Trends Neurosci. 2002;25(8):417–422. doi: 10.1016/s0166-2236(02)02201-4. [DOI] [PubMed] [Google Scholar]
- 9.Zhou Q, Anderson DJ. The bHLH transcription factors OLIG2 and OLIG1 couple neuronal and glial subtype specification. Cell. 2002;109(1):61–73. doi: 10.1016/s0092-8674(02)00677-3. [DOI] [PubMed] [Google Scholar]
- 10.Takebayashi H, Nabeshima Y, Yoshida S, Chisaka O, Ikenaka K, Nabeshima Y. The basic helix–loop–helix factor olig2 is essential for the development of motoneuron and oligodendrocyte lineages. Curr Biol. 2002;12(13):1157–1163. doi: 10.1016/s0960-9822(02)00926-0. [DOI] [PubMed] [Google Scholar]
- 11.Miyoshi G, Butt SJ, Takebayashi H, Fishell G. Physiologically distinct temporal cohorts of cortical interneurons arise from telencephalic Olig2-expressing precursors. J Neurosci. 2007;27(29):7786–7798. doi: 10.1523/JNEUROSCI.1807-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cai J, Chen Y, Cai WH, Hurlock EC, Wu H, Kernie SG, et al. A crucial role for Olig2 in white matter astrocyte development. Development. 2007;134(10):1887–1899. doi: 10.1242/dev.02847. [DOI] [PubMed] [Google Scholar]
- 13.Sun Y, Meijer DH, Alberta JA, Mehta S, Kane MF, Tien AC, et al. Phosphorylation state of Olig2 regulates proliferation of neural progenitors. Neuron. 2011;69(5):906–917. doi: 10.1016/j.neuron.2011.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harrington EP, Zhao C, Fancy SP, Kaing S, Franklin RJ, Rowitch DH. Oligodendrocyte PTEN is required for myelin and axonal integrity, not remyelination. Ann Neurol. 2010;68(5):703–716. doi: 10.1002/ana.22090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21(1):70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- 16.Wang YZ, Yamagami T, Gan Q, Wang Y, Zhao T, Hamad S, et al. Canonical Wnt signaling promotes the proliferation and neurogenesis of peripheral olfactory stem cells during postnatal development and adult regeneration. J Cell Sci. 2011;124(Pt 9):1553–1563. doi: 10.1242/jcs.080580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao T, Gan Q, Stokes A, Lassiter RN, Wang Y, Chan J, et al. beta-catenin regulates Pax3 and Cdx2 for caudal neural tube closure and elongation. Development. 2014;141(1):148–157. doi: 10.1242/dev.101550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang YP, Stokes A, Duan ZJ, Hui J, Xu Y, Chen YP, et al. LDL receptor-related protein 6 modulates Ret proto-oncogene signaling in renal development and cystic dysplasia. J Am Soc Nephrol. 2016;27(2):417–427. doi: 10.1681/ASN.2014100998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Song L, Li Y, Wang K, Wang YZ, Molotkov A, Gao L, et al. Lrp6-mediated canonical Wnt signaling is required for lip formation and fusion. Development. 2009;136(18):3161–3171. doi: 10.1242/dev.037440. [DOI] [PubMed] [Google Scholar]
- 20.Gan Q, Lee A, Suzuki R, Yamagami T, Stokes A, Nguyen BC, et al. Pax6 mediates ss-catenin signaling for self-renewal and neurogenesis by neocortical radial glial stem cells. Stem Cells. 2014;32(1):45–58. doi: 10.1002/stem.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ligon KL, Fancy SP, Franklin RJ, Rowitch DH. Olig gene function in CNS development and disease. Glia. 2006;54(1):1–10. doi: 10.1002/glia.20273. [DOI] [PubMed] [Google Scholar]
- 22.Schwob JE, Jang W, Holbrook EH, Lin B, Herrick DB, Peterson JN, et al. Stem and progenitor cells of the mammalian olfactory epithelium: taking poietic license. J Comp Neurol. 2017;525(4):1034–1054. doi: 10.1002/cne.24105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ellis P, Fagan BM, Magness ST, Hutton S, Taranova O, Hayashi S, et al. SOX2, a persistent marker for multipotential neural stem cells derived from embryonic stem cells, the embryo or the adult. Dev Neurosci. 2004;26(2–4):148–165. doi: 10.1159/000082134. [DOI] [PubMed] [Google Scholar]
- 24.Guo Z, Packard A, Krolewski RC, Harris MT, Manglapus GL, Schwob JE. Expression of pax6 and sox2 in adult olfactory epithelium. J Comp Neurol. 2010;518(21):4395–4418. doi: 10.1002/cne.22463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen Y, Miles DK, Hoang T, Shi J, Hurlock E, Kernie SG, et al. The basic helix–loop–helix transcription factor olig2 is critical for reactive astrocyte proliferation after cortical injury. J Neurosci. 2008;28(43):10983–10989. doi: 10.1523/JNEUROSCI.3545-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wong ST, Trinh K, Hacker B, Chan GC, Lowe G, Gaggar A, et al. Disruption of the type III adenylyl cyclase gene leads to peripheral and behavioral anosmia in transgenic mice. Neuron. 2000;27(3):487–497. doi: 10.1016/s0896-6273(00)00060-x. [DOI] [PubMed] [Google Scholar]
- 27.Belluscio L, Gold GH, Nemes A, Axel R. Mice deficient in G(olf) are anosmic. Neuron. 1998;20(1):69–81. doi: 10.1016/s0896-6273(00)80435-3. [DOI] [PubMed] [Google Scholar]
- 28.Kaneko-Goto T, Yoshihara S, Miyazaki H, Yoshihara Y. BIG-2 mediates olfactory axon convergence to target glomeruli. Neuron. 2008;57(6):834–846. doi: 10.1016/j.neuron.2008.01.023. [DOI] [PubMed] [Google Scholar]
- 29.Cheng LE, Reed RR. Zfp423/OAZ participates in a developmental switch during olfactory neurogenesis. Neuron. 2007;54(4):547–557. doi: 10.1016/j.neuron.2007.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kolterud A, Alenius M, Carlsson L, Bohm S. The Lim homeobox gene Lhx2 is required for olfactory sensory neuron identity. Development. 2004;131(21):5319–5326. doi: 10.1242/dev.01416. [DOI] [PubMed] [Google Scholar]
- 31.Macdonald JL, Verster A, Berndt A, Roskams AJ. MBD2 and MeCP2 regulate distinct transitions in the stage-specific differentiation of olfactory receptor neurons. Mol Cell Neurosci. 2010;44(1):55–67. doi: 10.1016/j.mcn.2010.02.003. [DOI] [PubMed] [Google Scholar]
- 32.Tsai RY, Reed RR. Cloning and functional characterization of Roaz, a zinc finger protein that interacts with O/E-1 to regulate gene expression: implications for olfactory neuronal development. J Neurosci. 1997;17(11):4159–4169. doi: 10.1523/JNEUROSCI.17-11-04159.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu QR, Sun T, Zhu Z, Ma N, Garcia M, Stiles CD, et al. Common developmental requirement for Olig function indicates a motor neuron/oligodendrocyte connection. Cell. 2002;109(1):75–86. doi: 10.1016/s0092-8674(02)00678-5. [DOI] [PubMed] [Google Scholar]
- 34.Ligon KL, Huillard E, Mehta S, Kesari S, Liu H, Alberta JA, et al. Olig2-regulated lineage-restricted pathway controls replication competence in neural stem cells and malignant glioma. Neuron. 2007;53(4):503–517. doi: 10.1016/j.neuron.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee AC, He J, Ma M. Olfactory marker protein is critical for functional maturation of olfactory sensory neurons and development of mother preference. J Neurosci. 2011;31(8):2974–2982. doi: 10.1523/JNEUROSCI.5067-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rodriguez-Gil DJ, Bartel DL, Jaspers AW, Mobley AS, Imamura F, Greer CA. Odorant receptors regulate the final glomerular coalescence of olfactory sensory neuron axons. Proc Natl Acad Sci USA. 2015;112(18):5821–5826. doi: 10.1073/pnas.1417955112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Takebayashi H, Yoshida S, Sugimori M, Kosako H, Kominami R, Nakafuku M, et al. Dynamic expression of basic helix–loop–helix Olig family members: implication of Olig2 in neuron and oligodendrocyte differentiation and identification of a new member, Olig3. Mech Dev. 2000;99(1–2):143–148. doi: 10.1016/s0925-4773(00)00466-4. [DOI] [PubMed] [Google Scholar]
- 38.Boutin C, Hardt O, de Chevigny A, Core N, Goebbels S, Seidenfaden R, et al. NeuroD1 induces terminal neuronal differentiation in olfactory neurogenesis. Proc Natl Acad Sci USA. 2010;107(3):1201–1206. doi: 10.1073/pnas.0909015107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jones DT, Reed RR. Golf: an olfactory neuron specific-G protein involved in odorant signal transduction. Science. 1989;244(4906):790–795. doi: 10.1126/science.2499043. [DOI] [PubMed] [Google Scholar]
- 40.Qiu L, LeBel RP, Storm DR, Chen X. Type 3 adenylyl cyclase: a key enzyme mediating the cAMP signaling in neuronal cilia. Int J Physiol Pathophysiol Pharmacol. 2016;8(3):95–108. [PMC free article] [PubMed] [Google Scholar]
- 41.Meijer DH, Kane MF, Mehta S, Liu H, Harrington E, Taylor CM, et al. Separated at birth? The functional and molecular divergence of OLIG1 and OLIG2. Nat Rev Neurosci. 2012;13(12):819–831. doi: 10.1038/nrn3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang X, Cai J, Klueber KM, Guo Z, Lu C, Qiu M, et al. Induction of oligodendrocytes from adult human olfactory epithelial-derived progenitors by transcription factors. Stem Cells. 2005;23(3):442–453. doi: 10.1634/stemcells.2004-0274. [DOI] [PubMed] [Google Scholar]
- 43.Zhang X, Cai J, Klueber KM, Guo Z, Lu C, Winstead WI, et al. Role of transcription factors in motoneuron differentiation of adult human olfactory neuroepithelial-derived progenitors. Stem Cells. 2006;24(2):434–442. doi: 10.1634/stemcells.2005-0171. [DOI] [PubMed] [Google Scholar]
- 44.Liu Z, Hu X, Cai J, Liu B, Peng X, Wegner M, et al. Induction of oligodendrocyte differentiation by Olig2 and Sox10: evidence for reciprocal interactions and dosage-dependent mechanisms. Dev Biol. 2007;302(2):683–693. doi: 10.1016/j.ydbio.2006.10.007. [DOI] [PubMed] [Google Scholar]
- 45.Liberia T, Martin-Lopez E, Meller SJ, Greer CA. Sequential maturation of olfactory sensory neurons in the mature olfactory epithelium. eNeuro. 2019 doi: 10.1523/ENEURO.0266-19.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]