Summary
With the emergence of severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2), which causes coronavirus disease 2019 (COVID-19), a high-speed and convenient detection technology should be at the forefront of medical care worldwide. This study evaluated the usefulness of GeneSoC, a compact, high-speed reciprocal flow quantitative reverse transcription polymerase chain reaction system, for the detection of SARS-CoV-2. The results support the use of this system for the rapid identification of SARS-CoV-2. This approach can contribute to the strategic selection of initial management strategies for patients with COVID-19.
Keywords: SARS-CoV-2, COVID-19, GeneSoC, RT-qPCR
Introduction
Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2), which causes coronavirus disease 2019 (COVID-19), first detected in Wuhan, China, has become a global pandemic [1,2]. With the rapid increase in the number of patients, the detection of SARS-CoV-2 is a key component of comprehensive medical care to reduce nosocomial infections, including transmission to healthcare workers. Accordingly, there is urgent demand for high-speed and practical diagnostic technologies [3].
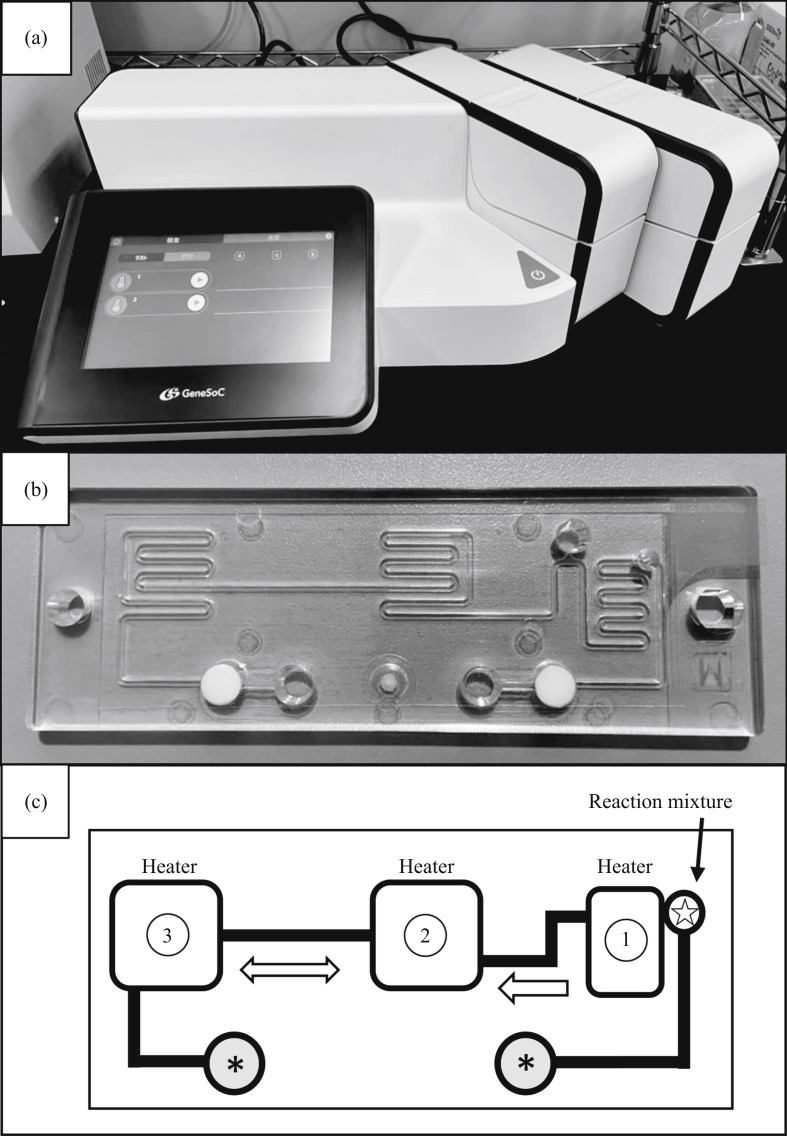
Recently, a compact, reciprocal flow polymerase chain reaction (PCR) system, GeneSoC (https://genesoc.jp/en/), has become available for specific gene amplification in a very short time (within 15 min) [4,5]. The system has one heater for the reverse transcription (RT) reaction and two heaters for thermal cycling, with two microblowers at both flow ends for the high-speed shuttle of the PCR solution. It is also possible to monitor the fluorescence intensity of the cycling solution in real time (Figure 1 ).
Figure 1.
Overview of the high-speed quantitative reverse transcription polymerase chain reaction (RT-qPCR) system. (a) GeneSoC equipment connected to two sets of PCR units on the right side. Up to four PCR units can be connected and one microfluidic chip can be loaded on each PCR unit. (b) Disposable microfluidic chip for high-speed RT-qPCR using the GeneSoC system. (c) Schematic of liquid flow on a disposable microfluidic chip. White circle with star, inlet for master mix for RT-qPCR; grey circle with asterisk, injection port for each microblower for the high-speed shuttle of the PCR solution; Heater 1, incubation for the RT reaction; Heaters 2 and 3, heaters for thermal cycling. The PCR solution cycles at high speed between Heaters 1 and 2.
The aim of this study was to clarify the utility of GeneSoC for the rapid diagnosis of COVID-19, and to consider its relative advantages and disadvantages as a point-of-care test.
Methods
Purified and quantified SARS-CoV-2 RNA isolated from an infected Japanese patient was provided by the National Institute of Infectious Diseases, Japan as a standard specimen for the molecular diagnosis of COVID-19. Analytical sensitivity was determined using 10-fold serially diluted standard RNA ranging from 1.0 × 106 to 1.0 × 100 copies/μL, stored at −30°C until required. High-speed RT-qPCR amplification using GeneSoC (Kyorin Pharmaceutical Co., Ltd., Tokyo, Japan) was performed using SpeedSTAR HS DNA Polymerase (Takara Bio Inc., Shiga, Japan), and real-time detection of the amplification product was monitored simultaneously by the measurement of emission. The detailed composition of reaction reagents, and the primers and probe targeting the specific sequence of the N gene of SARS-CoV-2 are listed in Table I [6]. Finally, 5 μL of the RNA template was added to the amplification mixture, and distilled water was added to a final volume of 20 μL. Amplification conditions were as follows: 42°C for 60 s, followed by 50 cycles of 96°C for 4 s and 58°C for 8 s. Amplification results were considered positive if an exponential amplification curve was generated that could be distinguished from that of the negative control. All reactions were performed in duplicate.
Table I.
Components of the reaction mixtures for high-speed quantitative reverse transcription polymerase chain reaction (RT-qPCR)
| Reagents | Final concentration |
|---|---|
| 2× OneStep RT-PCR Buffer IIIa | 1× |
| Primer (forward) | 2.4 μM, 5′-AAATTTTGGGGACCAGGAAC-3′ |
| Primer (reverse) | 3.2 μM, 5′-TGGCAGCTGTGTAGGTCAAC-3′ |
| Probe | 0.4 μM, FAM-ATGTCGCGCATTGGCATGGA-TAMRA |
| 10 x ROX | 0.2× |
| PrimeScript RT enzyme Mix IIa | 2 U/μL |
| SpeedSTAR HS DNA Polymerase | 0.25 U/μL |
Each reagent was included in the One Step PrimeScript RT-PCR Kit (Qiagen, Hilden, Germany).
Clinical samples were evaluated for a preliminary evaluation of the reactivity of GeneSoc in clinical practice. In total, 66 nasopharyngeal swab samples were collected from patients with suspected COVID-19 based on clinical findings. In addition, 12 nasopharyngeal swab samples positive for influenza A, as detected by RT-PCR, collected from Japanese patients in December 2019 were included as a negative control group. Viral RNA was extracted using the QIAamp Viral RNA Mini Kit (Qiagen, Hilden, Germany) following the manufacturer's instructions. Briefly, each swab was suspended in 700 μL of AVL buffer with 5.6 μL of carrier RNA, and RNA was finally eluted in 60 μL of AVE buffer. Extracts were amplified by conventional RT-qPCR with at least two technical repeats. Clinical validation of high-speed RT-qPCR using the GeneSoC system was performed simultaneously.
Conventional RT-qPCR for specific amplification of the N gene of SARS-CoV-2 was performed using TaqMan-based QuantStudio 5 Real-Time PCR (Thermo Fisher Scientific, Waltham, MA, USA) with the same primers and probe used for the GeneSoC assay. The final reaction volume was 50 μL, including 5 μL of RNA template, 500 nM forward primer, 700 nM reverse primer and 200 nM probe. Each reaction was performed using QuantiTect Probe RT-PCR Kits (Qiagen) according to the manufacturer's instructions. The cycling conditions consisted of RT at 50°C for 30 min, initial denaturation at 95°C for 15 min, and 40 cycles of denaturation at 94°C for 15 s and annealing/extension at 60°C for 60 s. This study was approved by the Saitama Medical University Hospital Research Ethics Committee (Reference 19136).
Results
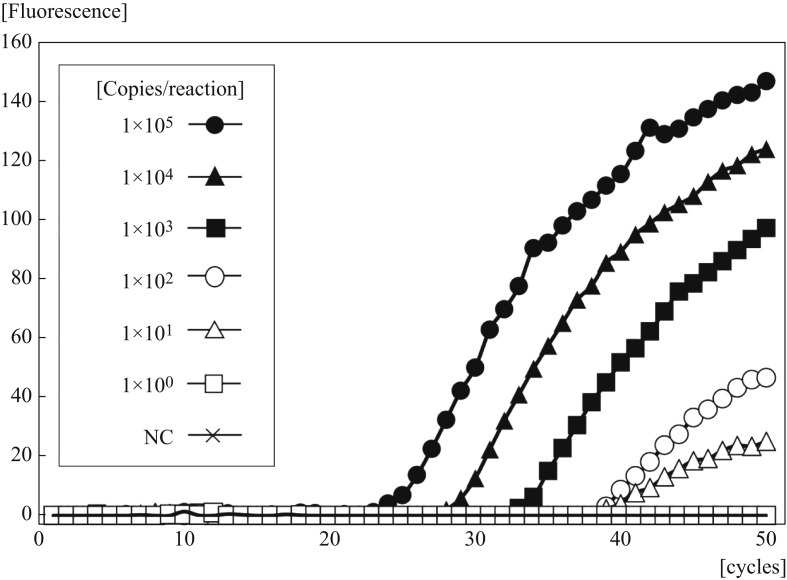
Figure 2 shows the real-time amplification plots obtained using GeneSoC. The minimum detectable amount of RNA was 1.0 × 101 copies/reaction. With this protocol, real-time amplification could be performed in less than 15 min.
Figure 2.
Limit of detection of GeneSoC for the N gene of severe acute respiratory syndrome coronavirus-2 with 10-fold serial dilutions of standard RNA. NC, negative control.
Of the 25 samples that were positive by conventional RT-qPCR, 23 (92.0%) were also identified as positive by GeneSoC, and the results for two samples were difficult to interpret as positive or negative due to weak signals. The median cycle threshold (Ct) value obtained by conventional RT-qPCR for 25 positive samples was 30.4 (range 17.2–37.0), and specific amplification was not observed for samples with a low Ct value (36.7, 36.8). In all 41 samples that were negative by conventional RT-qPCR and 12 RNA specimens derived from patients with influenza A, no amplification was observed using GeneSoC. With respect to the turnaround time, results were obtained within 1 h of RNA purification.
Discussion
Recent viral outbreaks that have spread rapidly across health infrastructures have presented major challenges in hospital settings [7]. RT-qPCR from nasopharyngeal swabs or sputum samples is still a core technology for determining the need for the hospitalization and isolation of patients with COVID-19, but this method generates false-negative results and is time-consuming [8]. A history of exposure to patients with COVID-19 and the characteristic computed tomographic (CT) findings of viral pneumonia strongly suggest COVID-19 and can provide a basis for patient management, even if a sample is negative by RT-qPCR [9,10]. However, CT images are not specific to COVID-19, emphasizing the importance of RT-qPCR with extremely high diagnostic specificity for comprehensive diagnosis. The GeneSoC assay, which requires a short test period and is not labour intensive, can contribute to the diagnosis and management of patients, and therefore to the prevention of hospital-acquired infections.
With respect to sensitivity, the limit of detection of the GeneSoC assay was 1.0 × 101 copies/reaction within 15 min. The main advantages of the platform are its simplicity and portability, its capacity for high-speed and real-time monitoring, and the use of a single disposable tip per analysis. GeneSoC can be easily transported and does not require highly equipped laboratories. However, it has some disadvantages. First, the analytical sensitivity was lower than that of the conventional RT-qPCR assay used as a reference. Second, the results could be influenced by the sampling procedure and timing, similar to conventional RT-qPCR. Third, simplification and refinement of the RNA extraction procedure is required.
These findings support the use of GeneSoC for rapid, low-throughput detection for strategic selection of initial management strategies for patients with COVID-19 and for prevention of hospital-acquired infections.
Conflict of interest statement
None declared.
Funding sources
This work was supported by AMED under Grant Number JP19he2202007.
Acknowledgements
The authors wish to thank the Advanced Photonics and Biosensing Open Innovation Laboratory, National Institute of Advanced Industrial Science and Technology for providing technical and scientific guidance on nucleonic amplification with GeneSoC.
References
- 1.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li Q., Guan X., Wu P., Wang X., Zhou L., Tong Y. Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N Engl J Med. 2020;382:1199–1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang J., Zhou M., Liu F. Exploring the reasons for healthcare workers infected with novel coronavirus disease 2019 (COVID-19) in China. J Hosp Infect. 2020 doi: 10.1016/j.jhin.2020.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Furutani S., Naruishi N., Hagihara Y., Nagai H. Development of an on-site rapid real-time polymerase chain reaction system and the characterization of suitable DNA polymerases for TaqMan probe technology. Anal Bioanal Chem. 2016;408:5641–5649. doi: 10.1007/s00216-016-9668-8. [DOI] [PubMed] [Google Scholar]
- 5.Furutani S., Hagihara Y., Nagai H. On-site identification of meat species in processed foods by a rapid real-time polymerase chain reaction system. Meat Sci. 2017;131:56–59. doi: 10.1016/j.meatsci.2017.04.009. [DOI] [PubMed] [Google Scholar]
- 6.National Institute of Infectious Diseases . NIID; Tokyo: 2020. Manual for the detection of pathogen 2019-nCoV Ver.2.6.https://www.niid.go.jp/niid/images/epi/corona/2019-nCoVmanual20200217-en.pdf Available at: [last accessed March 2020] [Google Scholar]
- 7.Wong G., Liu W., Liu Y., Zhou B., Bi Y., Gao G.F. MERS, SARS, and Ebola: the role of super-spreaders in infectious disease. Cell Host Microbe. 2015;18:398–401. doi: 10.1016/j.chom.2015.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Centers for Disease Control and Prevention . CDC; Atlanta, GA: 2020. Research use only 2019-novel coronavirus (2019-nCoV) real-time RT-PCR primer and probe information.https://www.cdc.gov/coronavirus/2019-ncov/downloads/rt-pcr-panel-primer-probes.pdf Available at: [last accessed March 2020] [Google Scholar]
- 9.Li Y., Xia L. Coronavirus disease 2019 (COVID-19): role of chest CT in diagnosis and management. Am J Roentgenol. 2020 doi: 10.2214/AJR.20.22954. [DOI] [PubMed] [Google Scholar]
- 10.Ai T., Yang Z., Hou H., Zhan C., Chen C., Lv W. Correlation of chest CT and RT-PCR testing in coronavirus disease 2019 (COVID-19) in China: a report of 1014 cases. Radiology. 2020 doi: 10.1148/radiol.2020200642. [DOI] [PMC free article] [PubMed] [Google Scholar]