Abstract
The increasing prevalence of antimicrobial resistance is a serious threat to global public health. One of the most concerning trends is the rapid spread of Carbapenemase-Producing Organisms (CPO), where colistin has become the last-resort antibiotic treatment. The emergence of colistin resistance, including the spread of mobilized colistin resistance (mcr) genes, raises the possibility of untreatable bacterial infections and motivates the development of improved diagnostics for the detection of colistin-resistant organisms. This work demonstrates a rapid response for detecting the most recently reported mcr gene, mcr−9, using a portable and affordable lab-on-a-chip (LoC) platform, offering a promising alternative to conventional laboratory-based instruments such as real-time PCR (qPCR). The platform combines semiconductor technology, for non-optical real-time DNA sensing, with a smartphone application for data acquisition, visualization and cloud connectivity. This technology is enabled by using loop-mediated isothermal amplification (LAMP) as the chemistry for targeted DNA detection, by virtue of its high sensitivity, specificity, yield, and manageable temperature requirements. Here, we have developed the first LAMP assay for mcr−9 - showing high sensitivity (down to 100 genomic copies/reaction) and high specificity (no cross-reactivity with other mcr variants). This assay is demonstrated through supporting a hospital investigation where we analyzed nucleic acids extracted from 128 carbapenemase-producing bacteria isolated from clinical and screening samples and found that 41 carried mcr−9 (validated using whole genome sequencing). Average positive detection times were 6.58 ± 0.42 min when performing the experiments on a conventional qPCR instrument (n = 41). For validating the translation of the LAMP assay onto a LoC platform, a subset of the samples were tested (n = 20), showing average detection times of 6.83 ± 0.92 min for positive isolates (n = 14). All experiments detected mcr−9 in under 10 min, and both platforms showed no statistically significant difference (p-value > 0.05). When sample preparation and throughput capabilities are integrated within this LoC platform, the adoption of this technology for the rapid detection and surveillance of antimicrobial resistance genes will decrease the turnaround time for DNA detection and resistotyping, improving diagnostic capabilities, patient outcomes, and the management of infectious diseases.
Subject terms: Lab-on-a-chip, Microbiology techniques, Infectious diseases, Diagnosis
Introduction
Antimicrobial resistance is a serious threat to global healthcare systems which poses a challenge for modern medicine and compromises effective infectious disease management. Of particular concern is the rapid spread of Carbapenemase-Producing Organisms (CPO)1–3 which are associated with high morbidity and mortality rates4. Carbapenemases are β-lactamases (bla) that are resistant to the carbapenems, a class of highly effective antibiotic agents. Therefore, therapeutic options are severely restricted with clinical management often relying upon “last line” antibiotics, primarily colistin. The emergence of colistin resistance through acquired mobilized colistin resistance (mcr) genes or chromosomal mutations is therefore a significant clinical and public health concern, raising the possibility of untreatable bacterial infections. In 2016, the first mcr variant, mcr−1, was reported in a clinical bacterial isolate5. Since then, several other mcr variants (mcr−1 to mcr−8) have been identified in over 40 countries across five different continents6. Very recently, the new variant mcr−9 has been reported and detected in a multidrug-resistant Salmonella enterica from a patient in Washington State7. Antimicrobial susceptibility testing for colistin, such as the broth microdilution methods recommended by CLSI and EUCAST guidelines, is challenging in diagnostic laboratories as current methodologies are limited by specificity/sensitivity issues and turnaround time8. The prevalence of colistin resistance is therefore likely to be under-reported, poorly characterized and outbreaks may go undetected.
Several diagnostic tests have been developed for the detection of colistin resistance9. Phenotypic assays, mostly culture-based methods, are generally recommended for under-resourced laboratories, whereas molecular tests, such as microarrays and polymerase chain reaction (PCR), are suggested for well-resourced ones9. Unfortunately, none of these methods combine all the ideal features for simple, efficient, economical and fast turnaround time10, limiting their widespread deployment. Among molecular assays, loop-mediated isothermal amplification (LAMP) is emerging as a popular nucleic acid amplification method due to its simplified thermal management, high sensitivity and high specificity11,12. Detection of mcr−1 to mcr−5 genes by LAMP has been demonstrated previously13. Moreover, other isothermal chemistries such as recombinase polymerase amplification have also been applied to mcr−114. However, these chemistries conventionally rely on fluorescent detection of the amplicon produced15, requiring complex and costly hardware, often restricting it to specialized laboratories. Alternative solutions based on colorimetric detection, enabled by microfluidics and cell-phone technology, provide promising point-of-care approaches, however, compromise dynamic range of quantification16–18.
Electrochemical-based DNA sensors offer high sensitivity, selectivity, large scale integration, detection on miniaturized hardware, fast response and low cost19. Ion-sensitive field-effect transistors (ISFETs) are emerging potentiometric sensors for nucleic-acid based applications. Implementation in unmodified complementary metal-oxide-semiconductor (CMOS) technology provides ISFETs with inherent sensitivity to pH, which has enabled DNA detection by employing an adapted version of LAMP, called pH-LAMP, that allows changes in pH to occur during nucleic acid amplification20–22. Recently, in Malpartida-Cardenas et al.23, it was shown that this approach holds significant potential for the development of a cheap, portable and quantitative diagnostic tool. This breakthrough was demonstrated in a lab-based environment using an external thermal controller in conjunction with a desktop computer. Moreover, recent work from our lab has demonstrated a fully portable Lab-on-a-Chip (LoC) platform which has integrated thermal management within the diagnostic platform and uses a smartphone application (AndroidOS) for data acquisition, visualization and cloud connectivity24–26.
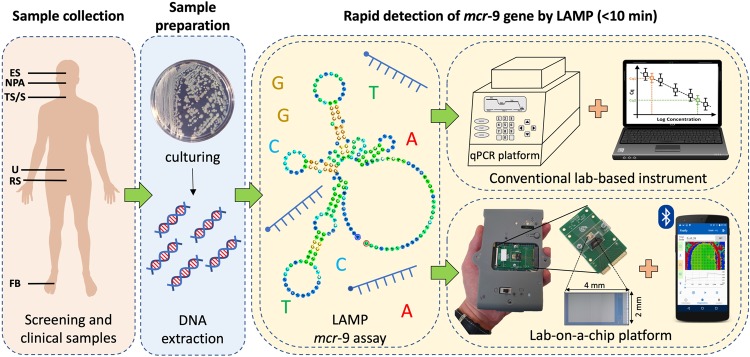
In this work, we explore the first application of LAMP for the rapid detection of mcr−9 in a conventional quantitative PCR instrument and the aforementioned LoC platform, referred to as qLAMP and eLAMP, respectively. The experimental workflow is depicted in Fig. 1 and summarized as follows: (1) through supporting a hospital investigation, 128 CPO-positive clinical and screening samples have been collected; (2) pure bacteria were cultured and nucleic acids extracted; (3) developed and characterized a new LAMP assay for detecting mcr−9 in under 10 minutes using a qPCR instrument; and (4) translated the new LAMP assay onto an ISFET-based LoC platform. We envision that the approach described in this work, when integrated with a sample preparation module and multiplexing capabilities, can be extended to other drug resistances, and its implementation in health care facilities will greatly expand diagnostic capabilities, improving patient outcomes and infection prevention & control.
Figure 1.
Diagnostic workflow for detection of mcr−9 using LAMP. Clinical and screening samples are collected from: ES, eye swab; NPA, nasopharyngeal aspirate; TS, throat swab; S, sputum; U, urine; RS, rectal swab; and FB, foot biopsy. Subsequently, samples are cultured and nucleic acids are extracted in a microbiology lab. Following this, rapid detection of mcr−9 is performed using our new LAMP assay on a conventional lab-based instrument connected to a desktop computer and a state-of-the-art lab-on-a-chip platform linked to a smartphone.
Results
LAMP assay for mcr−9 gene detection
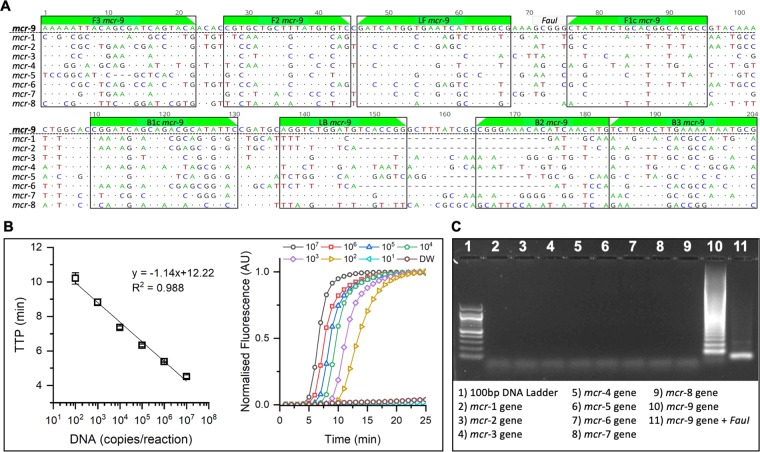
Based on the nucleotide sequence alignment of mcr gene variants, specific LAMP primers for the mcr−9 gene detection were designed. The LAMP assay has an amplicon size of 204 bp and consists of six primers targeting eight different regions: two outer primers (F3/B3), two inner primers (FIP/BIP) and two loop primers (LF/LB). Primer sequences are provided in Table S1. A sequence alignment for mcr−1 to mcr−9 indicating the location of the mcr−9 LAMP primers is shown in Fig. 2A. It can be observed that many single nucleotide polymorphisms exist across the selected region and BlastN analysis of primer sequences against the NCBI database showed 100% homology and 100% coverage for the mcr−9 variant genes only. Using synthetic DNA containing the mcr−9 gene, the LAMP assay showed an excellent lower limit of detection of 100 copies/ reaction with a good correlation curve between DNA copies per reaction and time-to-positive (TTP) values, and standard curve was linear from 107 to 102 (R2 = 0.988) corresponding to a TTP ranging from 4 to 10 min, respectively (Fig. 2B). To evaluate the specific amplification of the LAMP assay, synthetic DNA containing mcr−1 to mcr−9 sequences were tested as templates at a concentration of 105 copies/reaction. After the LAMP reaction, the amplification products were subjected to agarose gel electrophoresis. The mcr−9 synthetic DNA showed a positive LAMP pattern and the rest of DNA samples were negative (Fig. 2C). In addition, specificity of the LAMP-positive product was confirmed by digestion of the amplicon by FauI restriction enzyme digestion (sequence shown in Fig. 2A). Results presented in Fig. 2 indicate that the mcr−9 LAMP assay is highly sensitive and specific.
Figure 2.
Sequence alignment, primers location and mcr−9 LAMP assay performance. (A) Sequence alignment for mcr−1 to mcr−9 showing priming region and the location of the FauI restriction site. Compared to the reference sequence, mcr−9, mismatches in the alignment are displayed as AGTC. Matched nucleotides are represented as dots and gaps are represented as dashes. (B) Standard curve and real-time amplification profiles obtained with the mcr−9 LAMP assay and synthetic DNA at concentrations ranging from 102 to 107 copies per reaction (including non-template control). Experiments were carried-out in a real-time qPCR platform in triplicates. (C) Gel electrophoresis confirming specificity of mcr−9 LAMP assay. Line 1 shows a 100 bp DNA ladder. Lines 2 to 10 show the amplification product obtained from synthetic DNA carrying mcr−1 to mcr−9 sequences against the mcr−9 LAMP assay. Line 11 shows the specific mcr−9 LAMP product after digestion with FauI.
Detecting mcr−9 in clinical bacterial isolates using qLAMP
To look for putative clinical isolates harboring this colistin resistance mcr−9 gene, the LAMP assay was applied to extracted genomic DNA samples from 128 CPO isolates (see Table 1). All isolates were collected between 2012–2019 from clinical or screening samples routinely processed by the Microbiology Department at Charing Cross Hospital, Imperial College Healthcare NHS Trust. The pure bacterial strains were identified using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) as Acinetobacter sp. (n = 5), Citrobacter sp. (n = 7), Enterobacter spp. (n = 33), Escherichia sp. (n = 14), Klebsiella spp. (n = 61), Proteus sp. (n = 1), Pseudomonas sp. (n = 4), Raoultella sp. (n = 1) and Serratia sp. (n = 2). The carbapenemase-producing mechanisms were confirmed using commercially available diagnostic tests based on qPCR and immunochromatography. Carbapenemases were determined as a single enzyme in 121 strains (blaGES−5 = 2; blaIMP = 53; blaKPC = 8; blaNDM = 39; blaOXA−23 = 1; blaOXA−48 = 13; blaOXA−58 = 1; blaVIM = 4) and as a combination in seven isolates (blaIMP + blaOXA−48 = 1; blaIMP + blaOXA−58 = 1; blaIMP + blaNDM + blaOXA−51−like = 1; blaNDM + blaOXA−48 = 3; blaNDM + blaOXA−51 = 1).
Table 1.
Clinical carbapenemase-producing organisms analysed by the mcr−9 LAMP assay.
| Species (MALDI-TOF MS) | Carbapenemase | Positive mcr-9 Isolates/Analysed Samples |
|---|---|---|
| Acinetobacter sp. | blaIMP and blaOXA−58 | 0/1 |
| blaIMP, blaNDM and blaOXA−51-like | 0/1 | |
| blaOXA−23 | 0/1 | |
| blaOXA−51 and blaNDM | 0/1 | |
| blaOXA−58 | 0/1 | |
| Citrobacter sp. | blaIMP | 1/1 |
| blaKPC | 0/2 | |
| blaOXA−48 | 1/3 | |
| blaVIM | 0/1 | |
| Enterobacter spp. | blaIMP | 23/31 |
| blaIMP and blaOXA−48 | 1/1 | |
| blaVIM | 0/1 | |
| Escherichia coli | blaIMP | 3/3 |
| blaNDM | 0/5 | |
| blaNDM and blaOXA−48 | 0/1 | |
| blaOXA−48 | 0/5 | |
| Klebsiella spp. | blaGES−5 | 0/2 |
| blaIMP | 10/15 | |
| blaKPC | 0/5 | |
| blaNDM | 0/33 | |
| blaNDM and blaOXA−48 | 0/2 | |
| blaOXA−48 | 0/4 | |
| Proteus mirabilis | blaNDM | 0/1 |
| Pseudomonas aeruginosa | blaIMP | 1/2 |
| blaVIM | 0/2 | |
| Raoultella planticola | blaIMP | 1/1 |
| Serratia marcescens | blaKPC | 0/1 |
| blaOXA−48 | 0/1 |
All isolates were negative for mcr−1 to 8 by qPCR and whole genome sequencing. A detailed description of each isolate, including bacterial species, date of sampling, specimen type, its origin, antibiotic resistance mechanisms, TTP obtained by the mcr−9 LAMP, and WGS results can be found in Tables S2 and S3.
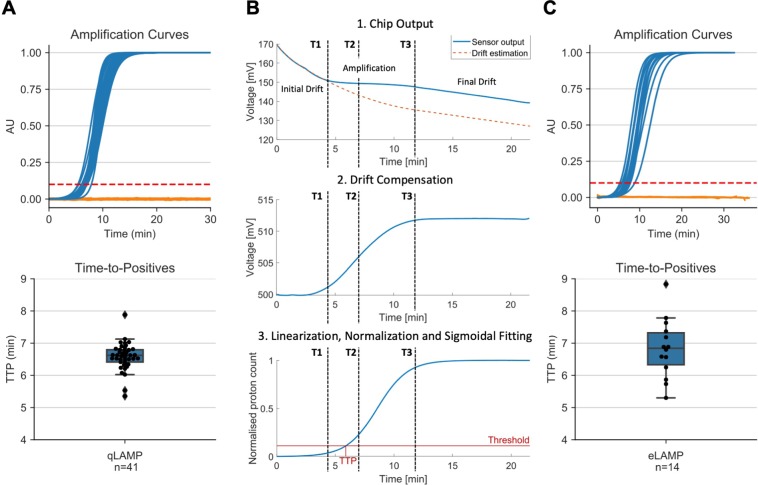
The performed LAMP assay was positive for 41 samples. Interestingly, from these mcr−9 positive samples, 39 were carrying blaIMP, one was blaIMP + blaOXA−48 and another one was carrying blaOXA−48. This suggests the horizontal transfer of a plasmid encoding both mcr−9 and blaIMP, which could result in resistance to the carbapenems and reduced susceptibility to colistin, limiting therapeutic options. However, this analysis is outside the scope of this study. The mcr gene was present in Citrobacter sp., Enterobacter sp., Escherichia sp., Klebsiella sp. and Pseudomonas sp., and all positive isolates were collected between 2016–2019. To validate these findings, we performed whole genome sequencing (WGS) analysis on 56 bacterial isolates, including all mcr−9 encoding isolates and 15 negative controls. The WGS data showed 100% concordance with the LAMP results. All samples were negative for mcr−1 to mcr−8 by qPCR and WGS. As shown in Fig. 3A, the TTP for all positive samples were under 10 minutes with an average value of 6.58 ± 0.42 minutes. The results presented in this section demonstrate that the developed mcr−9 LAMP assay allows for rapid, sensitive and specific detection of this emerging colistin resistant gene, making this assay a promising starting point for the development of a near-patient screening test.
Figure 3.
Performance of mcr−9 LAMP assay using qLAMP and eLAMP. (A) Output from qLAMP. Top panel shows the normalized negative (n = 87) and positive (n = 41) amplification curves in orange and blue, respectively. Bottom panel shows the distribution of qLAMP TTP for positive isolates. (B) Algorithm to extract amplification curve and TTP from LoC ISFET array output. (C) Output from eLAMP. Top panel shows the normalized negative (n = 6) and positive (n = 14) amplification curves in orange and blue, respectively. Bottom panel shows the distribution of eLAMP TTP for positive isolates. TTP was extracted using a threshold at 10% of the maximum. See Figures S1–S4 and Table S4 for detailed information.
Translating the assay into eLAMP
In order to translate the LAMP assay onto the pH-based Lab-on-a-Chip platform, we use an adapted version of LAMP, called pH-LAMP, that allows changes in pH to occur during nucleic acid amplification (see Materials and Methods). The output of the on-chip amplification (measured in mV) consists of 4368 time-series (associated to each individual sensor) which are proportional to the pH change during the reaction (plus inherent sensor drift). The algorithm for processing the data from the ISFET array is as follows, and illustrated in Fig. 3B. Firstly, the outputs from all exposed sensors are averaged to yield the raw output curve. To extract the signal, the algorithm finds the inflection point of the curve (T2), i.e. when the rate of pH change is at its maximum. Subsequently, three reaction regions are then identified using the following method:
0–1: no amplification; the output is only drift.
T1–T3: amplification is occurring; the output is the sum of drift and pH signal.
T3–end: amplification products have saturated; the output is only drift.
The drift is compensated by using an exponential approximation with the data points in regions 1 and 3. Finally, the output is fitted using a 4-parameter sigmoid and normalized such that a threshold can be applied to extract a time-to-positive, similar to that of qLAMP.
To validate our in-house platform, twenty of the isolates were analyzed, including 14 out of the 41 mcr−9 LAMP positive isolates. As shown in Fig. 3C, the TTP for all positive samples were also under 10 minutes with an average value of 6.83 ± 0.92 minutes. The mean between the TTP distributions obtained from qLAMP and eLAMP was found not to be statistically significant (p = 0.34) - indicating both instruments provide comparable results.
Discussion
To effectively address the threats of emerging multi-drug resistant pathogens, efforts are needed to use state-of-the-art technologies in order to provide a rapid response to prevent the misuse of antibiotics. In this paper, we investigate the use of loop-mediated isothermal amplification to detect the most recently identified variant of mobilized colistin resistance in carbapenemase-producing organisms (i.e. multi-drug resistances). With current available antibiotics, bacteria carrying carbapenem- and colistin- resistance, become potentially untreatable and represents a global threat.
To date, there have been no reports on the detection of mcr−9 using any isothermal amplification chemistry. In this study, we first develop a novel LAMP assay for mcr−9 using a recently reported sequence7. The proposed assay was evaluated in a conventional lab-based instrument through supporting a hospital investigation where 128 carbapenemase-producing organisms were tested. We found 41 positive for mcr−9 across multiple bacterial species - demonstrating the ubiquitous presence of this colistin resistant gene. Furthermore, we used a subset of the positive samples, to investigate the use of a cutting-edge Lab-on-a-Chip platform which integrates semiconductor technology, avoiding bulky and complex hardware, coupled with our LAMP assay, removing the need for sophisticated thermal management. Results show that both instruments are capable of detecting mcr−9 in under 10 minutes.
Although the results in this manuscript present a milestone towards providing a rapid diagnostic solution at the bedside, there are several aspects that need to be further addressed. Firstly, due to a wide range of sample types in this study, the sample preparation (including culturing and nucleic acid extraction) is not integrated within the LoC platform, thereby presenting a bottleneck on the turnaround time (currently 8–24 hours). Several rapid sample preparation methods are commercially available to extract nucleic acids directly from samples, such as QuickExtract DNA Extraction Solution from Epicenter (3–8 min), however coupling them with the LoC platform is outside the scope of this manuscript. Secondly, samples were analyzed individually as only a single chamber was mounted on top of the chip, limiting the throughput of experiments. More importantly, the single chamber limits the number of targets per sample, such as detecting other mcr or carbapenem-resistant genes. There are multiple avenues for increasing the number of targets per experiment, such as: (i) Microfluidic solutions (e.g. SlipChip) which offer an affordable mechanism for generating multiple chambers27–30; or (ii) data-driven approaches which have recently been shown to perform single-channel multiplexing in a single chamber for carbapenemase-producing organisms using only real-time amplification data31–33. Finally, it is important to further investigate the relationship between carrying the mcr−9 gene and its phenotypic implications, i.e. being resistant to colistin.
The results presented in this work suggest that this approach could provide a versatile solution for the rapid detection of colistin and other antimicrobial resistances, supporting clinical management, surveillance and antimicrobial stewardship. Future work will consider combining this work with a sample preparation module and high-throughput mechanisms, providing a fully integrated platform to rapidly respond to emerging global threats outside the laboratory.
Materials and Methods
Samples
In this study, the mcr−9 LAMP assay was designed using synthetic DNA, and bacterial strains were used to validate its performance.
-
(i)
Synthetic DNA. Double-stranded synthetic DNA (gBlock Gene fragments) containing mcr−1 to mcr−9 gene sequences (ranging from 1,617 to 1,898 bp) were purchased from Life Technologies (ThermoFisher Scientific) and resuspended in Tris-EDTA buffer to 10 ng/μL stock solutions (stored at −20 °C until further use). The concentrations of all DNA stock solutions were determined using a Qubit 3.0 fluorimeter (Life Technologies). Sequences are provided in Table S5.
-
(ii)
Bacterial strains and culture conditions. A total of 128 non-duplicated Enterobacteriaceae isolates were collected between 2012–2019 from clinical or screening samples routinely processed by the hospital Microbiology Department at Imperial College Healthcare NHS Trust (Ethics protocol 06/Q0406/20). All isolates were identified as carbapenemase producers. Species identification was performed using MALDI-TOF MS and carbapenemase mechanisms were determined using the Xpert Carba-R (Cepheid) or Resist-3 O.K.N assay (Corisbio). The blaGES−5 and Acinetobacter species were further characterized at the national reference laboratory, Public Health England. The isolates were subcultured on appropriate growth media and incubated at 37 °C overnight, and the genomic DNA was extracted using GenElute Bacterial Genomic DNA kit (Sigma-Aldrich) following the manufacturer’s instructions.
LAMP primer design
A set of LAMP primers specific to the mcr−9 gene was designed based on the sequences reported by Carroll et al.7 (GenBank: MK791138.1 and NG_064792.1) using the Primer Explorer software, version 5.0 (http://primerexplorer.jp/lampv5e/index.html). Thirty-nine other sequences including variants of mcr−1 to mcr−8 strains were retrieved from NCBI’s GenBank database (https://www.ncbi.nlm.nih.gov/genbank/) to analyze primer specificity in silico. GenBank accession numbers can be found in Table S6. All sequences were aligned using the MUSCLE algorithm34 in Geneious 10.0.5 software35. All primers used in this study were purchased from Life Technologies (ThermoFisher Scientific).
LAMP reaction conditions
The amplification reaction conditions are as follows:
-
(i)
qLAMP. Each mix contained: 1.5 μL of 10× isothermal buffer (New England BioLabs), 0.9 μL of MgSO4 (100mM stock), 2.1 μL of dNTPs (10 mM stock), 0.375 μL of BSA (20 mg/μL stock), 2.4 μL of Betaine (5 M stock), 0.375 μL of SYTO 9 Green (20 μM stock), 0.6 μL of Bst 2.0 DNA polymerase (8000 U/μL stock), 3 μL of different concentrations of synthetic DNA or gDNA, 1.5 μL of 10× LAMP primer mixture (20 μL of BIP/FIP, 10 μM of LF/LB, and 2.5 μM B3/F3) and enough nuclease-free water (ThermoFisher Scientific) to bring the volume to 15 μL. Reactions were performed at 63 °C for 20 min. One melting cycle was performed at 0.1 °C/s from 65 °C up to 97 °C for validation of the specificity of the products. Experiments were carried out in triplicates (using 5 μL for each reaction) loading the reactions into 96-well plates and using a LightCycler 96 Real-Time PCR System (LC96) (Roche Diagnostics).
-
(ii)
eLAMP. Each mix contained: 3.0 μL of 10× isothermal pH-based buffer (pH 8.5–9), 1.8 μL of MgSO4 (100mM stock), 1.68 μL of dNTPs (25 mM stock), 1.8 μL of BSA (20mg/μL stock), 4.8 μL of Betaine (5M stock), 0.15 μL of Bst 2.0 DNA polymerase (120000 U/μL stock), 3 μL of different concentrations of synthetic DNA or gDNA, 0.75 μL of NaOH (0.2M stock), 3.0 μL of 10× LAMP primer mixture and enough nuclease- free water (ThermoFisher Scientific) to bring the volume to 30 μL. Reactions were performed at 63 °C for 20 min. Reactions of 10 μL each were loaded into a disposable cartridge and experiments were carried-out using our in-house platform.
Limit of detection for mcr−9 LAMP assay
Analytical sensitivity was evaluated with 10-fold dilutions of synthetic DNA containing the mcr−9 sequence, ranging from 102 to 107 copies per reaction. Each experimental condition was run in triplicate. The LAMP assays were performed in a LightCycler 96 and the data was analyzed using LC96 System software version SW1.1.
Cross-reactivity of mcr−9 LAMP assay
The specificity of the assay was validated through multiple methods as follows:
-
(i)
Restriction digestion. The specificity of mcr−9 amplification was determined by the restriction digestion of the LAMP products using the FauI enzyme (New England BioLabs). The restriction was performed for 1 h at 55 °C, using the CutSmart buffer provided by the manufacturer. The digested LAMP products were subjected to electrophoresis on a 1.5% agarose gel at 120 V for 1 h and then visualized under UV light, using the BioSpectrum Imaging System (Ultra-Violet Products Ltd.), after staining with SYBR Safe DNA Gel Stain (InvitroGen). Experiments were performed using mcr−1 to mcr−9 synthetic DNA.
-
(ii)
PCR analysis. The presence of mcr−1 to mcr−8 was tested by using published primer sets and reaction conditions36.
-
(iii)
Whole genome sequencing. Bacterial isolates positive for the mcr−9 LAMP assay were further analyzed by whole genome sequencing. Multiplexed Illumina Nextera XT-generated libraries were sequenced on the NextSeq. 500 platform (Illumina) using 2× 150-bp paired-end mode with a target average coverage of 100-fold. Read quality was assessed using FastQC v0.11.637. Raw reads were assembled using SPAdes v3.1138 and annotated with Prokka v.1.1239. Antimicrobial resistance genes, including mcr−9 were identified using the Basic Local Alignment Search Tool (BLAST) searching against the databases of ResFinder v3.1.040 and the Comprehensive Antibiotic Research Database41 with a threshold of 98% identity and 60% query match length.
Lab-on-a-chip Platform
The LoC system (depicted in Fig. S5) is comprised of23–26: (1) Diagnostic platform powered by a rechargeable battery containing: a Peltier heating module with an embedded PID controller to enable on-board isothermal nucleic acid amplification; digital algorithms for data acquisition; and a Bluetooth module to transmit information to a smartphone. (2) Single-use cartridge consisting of a CMOS ISFET microchip (4,368 sensors, 2 × 4 mm) for DNA sensing and a microfluidic chamber made of acrylic to contain the amplification reaction. To guarantee proper sealing, biocompatible double-sided tape is used to attach the manifold to the chip and PCR tape is used to cover the chamber during the amplification reaction. A reference electrode (0.03 mm Ag/AgCl) is placed between the chip and the double-sided tape such that it is in contact with the chemical solution, and then soldered on the PCB track to allow electrical contact with the motherboard. The diagnostic platform and single-use cartridge is shown in Fig. 1. (3) Smartphone application (developed for AndroidOS) to interface with the chip via Bluetooth in order to gather the diagnostic data and perform geotagging of the diagnostic location through the phone GPS. The application supports optional storage of the data on a cloud server (Amazon web services).
Statistical analysis
Time-to-positive data is presented as mean TTP ± standard deviation; p-values were calculated by Welch’s unequal variance t-test in Python (v3.6) SciPy Package (v1.4.1). A p-value equal to 0.05 was considered as a threshold for statistical significance. To estimate the number of required samples for translating the LAMP assay onto the LoC platform we used the following formula42:
| 1 |
where p is the suspected sensitivity and x is the desired margin of error. We define the true-positive rate (sensitivity) as the proportion of mcr−9 positive which are correctly identified by the lab-on-a-chip platform compared to the gold standard qPCR instrument. We suspected the sensitivity and specificity to be 95% with a desired margin of error of 10%. Under these conditions, the number of required samples is 18.2 (rounded up to 19) therefore we tested a total 20 samples (14 positive and 6 negative).
Supplementary information
Acknowledgements
This work was supported by the National Institute for Health Research Health Protection Research Unit (NIHR HPRU) [HPRU-2012–10047] in Healthcare Associated Infection and Antimicrobial Resistance at Imperial College London in partnership with Public Health England; the NIHR Imperial Biomedical Research Centre (BRC) [P80763]; and the Centre for Antimicrobial Optimization (CAMO). We would also like to thank the Imperial College Healthcare NHS Trust microbiology lab for providing the samples. Moreover, the Imperial BRC Genomics Facility has provided resources and support that have contributed to the research results reported within this paper. The Imperial BRC Genomics Facility is supported by NIHR funding to the Imperial Biomedical Research Centre. We would also like to acknowledge EPSRC HiPEDS CDT (EP/L016796/1 to K.M.C) and EPSRC DTP (EP/N509486/1 to A.M.) for supporting this work.
Author contributions
J.R.-M., A.H. and P.G. conceived the idea. J.R.-M., A.H., P.G., F.B. and F.D. planned the experiments. J.R.-M., N.M., A.M., K.M.-C., E.J. and A.B. curated the data. J.R.-M., K.M.-C., L.F., A.A., I.P., E.J. and A.B. conducted the experiment(s). J.R.-M., A.M., K.M.-C., L.F., X.D., J.O., E.J. and A.B. analyzed the result(s). J.R.-M., A.M. and F.B. wrote the initial manuscript. J.R-M., A.H. and P.G. acquired the funding. All authors reviewed and edited the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-64612-1.
References
- 1.European Centre for Disease Prevention and Control. Carbapenem-resistant enterobacteriaceae, second update. https://www.ecdc.europa.eu/sites/default/files/documents/carbapenem-resistant-enterobacteriaceae-risk-assessment-rev-2.pdf (2019).
- 2.Nellums LB, et al. Antimicrobial resistance among migrants in europe: a systematic review and meta-analysis. The Lancet Infectious Diseases. 2018;18:796–811. doi: 10.1016/S1473-3099(18)30219-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Osei Sekyere J, Govinden U, Essack S. Review of established and innovative detection methods for carbapenemase-producing gram-negative bacteria. Journal of applied microbiology. 2015;119:1219–1233. doi: 10.1111/jam.12918. [DOI] [PubMed] [Google Scholar]
- 4.Neuner EA, et al. Treatment and outcomes in carbapenem-resistant klebsiella pneumoniae bloodstream infections. Diagnostic microbiology and infectious disease. 2011;69:357–362. doi: 10.1016/j.diagmicrobio.2010.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu Y-Y, et al. Emergence of plasmid-mediated colistin resistance mechanism mcr-1 in animals and human beings in china: a microbiological and molecular biological study. The Lancet infectious diseases. 2016;16:161–168. doi: 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- 6.Lima T, Domingues S, Da Silva GJ. Plasmid-mediated colistin resistance in salmonella enterica: a review. Microorganisms. 2019;7:55. doi: 10.3390/microorganisms7020055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carroll LM, et al. Identification of novel mobilized colistin resistance gene mcr-9 in a multidrug-resistant, colistin-susceptible salmonella enterica serotype typhimurium isolate. mBio. 2019;10:e00853–19. doi: 10.1128/mBio.00853-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.European Committee on Antimicrobial Susceptibility Testing and Clinical and Laboratory Standards Institute. Recommendations for MIC determination of colistin (polymyxin E) as recommended by the joint CLSI-EUCAST Polymyxin Breakpoints Working Group. http://www.eucast.org/guidance_documents (2016).
- 9.Osei Sekyere J. Mcr colistin resistance gene: a systematic review of current diagnostics and detection methods. MicrobiologyOpen. 2019;8:e00682. doi: 10.1002/mbo3.682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ming D, et al. Connectivity of rapid-testing diagnostics and surveillance of infectious diseases. Bulletin of the World Health Organization. 2019;97:242. doi: 10.2471/BLT.18.219691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tomita N, Mori Y, Kanda H, Notomi T. Loop-mediated isothermal amplification (lamp) of gene sequences and simple visual detection of products. Nature protocols. 2008;3:877–882. doi: 10.1038/nprot.2008.57. [DOI] [PubMed] [Google Scholar]
- 12.Mori Y, Notomi T. Loop-mediated isothermal amplification (lamp): a rapid, accurate, and cost-effective diagnostic method for infectious diseases. Journal of infection and chemotherapy. 2009;15:62–69. doi: 10.1007/s10156-009-0669-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhong L-L, et al. Multiplex loop-mediated isothermal amplification (multi-lamp) assay for rapid detection of mcr-1 to mcr-5 in colistin-resistant bacteria. Infection and drug resistance. 2019;12:1877. doi: 10.2147/IDR.S210226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu J, Wang X, Yang L, Kan B, Lu X. Rapid detection of mcr-1 by recombinase polymerase amplification. Journal of medical microbiology. 2018;67:1682–1688. doi: 10.1099/jmm.0.000865. [DOI] [PubMed] [Google Scholar]
- 15.Craw P, Balachandran W. Isothermal nucleic acid amplification technologies for point-of-care diagnostics: a critical review. Lab on a Chip. 2012;12:2469–2486. doi: 10.1039/c2lc40100b. [DOI] [PubMed] [Google Scholar]
- 16.Yetisen AK, Martinez-Hurtado J, Garcia-Melendrez A, da Cruz Vasconcellos F, Lowe CR. A smartphone algorithm with inter-phone repeatability for the analysis of colorimetric tests. Sensors and Actuators B: Chemical. 2014;196:156–160. doi: 10.1016/j.snb.2014.01.077. [DOI] [Google Scholar]
- 17.Goto M, Honda E, Ogura A, Nomoto A, Hanaki K-I. Colorimetric detection of loop-mediated isothermal amplification reaction by using hydroxy naphthol blue. Biotechniques. 2009;46:167–172. doi: 10.2144/000113072. [DOI] [PubMed] [Google Scholar]
- 18.Rodriguez-Manzano J, et al. Reading out single-molecule digital rna and dna isothermal amplification in nanoliter volumes with unmodified camera phones. ACS nano. 2016;10:3102–3113. doi: 10.1021/acsnano.5b07338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Drummond TG, Hill MG, Barton JK. Electrochemical dna sensors. Nature biotechnology. 2003;21:1192–1199. doi: 10.1038/nbt873. [DOI] [PubMed] [Google Scholar]
- 20.Toumazou C, et al. Simultaneous dna amplification and detection using a ph-sensing semiconductor system. Nature methods. 2013;10:641. doi: 10.1038/nmeth.2520. [DOI] [PubMed] [Google Scholar]
- 21.Gurrala R, et al. Novel ph sensing semiconductor for point-of-care detection of hiv-1 viremia. Scientific reports. 2016;6:1–6. doi: 10.1038/srep36000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Duarte-Guevara C, et al. On-chip electrical detection of parallel loop-mediated isothermal amplification with dg-biofets for the detection of foodborne bacterial pathogens. RSC advances. 2016;6:103872–103887. doi: 10.1039/C6RA19685C. [DOI] [Google Scholar]
- 23.Malpartida-Cardenas K, et al. Quantitative and rapid plasmodium falciparum malaria diagnosis and artemisinin-resistance detection using a cmos lab-on-chip platform. Biosensors and Bioelectronics. 2019;145:111678. doi: 10.1016/j.bios.2019.111678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Au, A., Moser, N., Rodriguez-Manzano, J. & Georgiou, P. Live demonstration: a mobile diagnostic system for rapid detection and tracking of infectious diseases. In 2018 IEEE International Symposium on Circuits and Systems (ISCAS), 1–1 (IEEE, 2018).
- 25.Moser N, Rodriguez-Manzano J, Lande TS, Georgiou P. A scalable isfet sensing and memory array with sensor auto-calibration for on-chip real-time dna detection. IEEE transactions on biomedical circuits and systems. 2018;12:390–401. doi: 10.1109/TBCAS.2017.2789161. [DOI] [PubMed] [Google Scholar]
- 26.Miscourides N, Yu L-S, Rodriguez-Manzano J, Georgiou P. A 12.8 k current-mode velocity-saturation isfet array for on-chip real-time dna detection. IEEE transactions on biomedical circuits and systems. 2018;12:1202–1214. doi: 10.1109/TBCAS.2018.2851448. [DOI] [PubMed] [Google Scholar]
- 27.Witters D, et al. Digital biology and chemistry. Lab on a Chip. 2014;14:3225–3232. doi: 10.1039/C4LC00248B. [DOI] [PubMed] [Google Scholar]
- 28.Sun B, et al. Measuring fate and rate of single-molecule competition of amplification and restriction digestion, and its use for rapid genotyping tested with hepatitis c viral rna. Angewandte Chemie International Edition. 2014;53:8088–8092. doi: 10.1002/anie.201403035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Du W, Li L, Nichols KP, Ismagilov RF. Slipchip. Lab on a Chip. 2009;9:2286–2292. doi: 10.1039/b908978k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shen F, et al. Nanoliter multiplex pcr arrays on a slipchip. Analytical chemistry. 2010;82:4606–4612. doi: 10.1021/ac1007249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rodriguez-Manzano J, et al. Simultaneous single-channel multiplexing and quantification of carbapenem-resistant genes using multidimensional standard curves. Analytical chemistry. 2019;91:2013–2020. doi: 10.1021/acs.analchem.8b04412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moniri A, et al. Framework for dna quantification and outlier detection using multidimensional standard curves. Analytical chemistry. 2019;91:7426–7434. doi: 10.1021/acs.analchem.9b01466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Georgiou, P., Moniri, A. & Rodriguez-Manzano, J. A method for analysis of real-time amplification data. https://patents.google.com/patent/WO2019234247A1/ (2019).
- 34.Edgar RC. Muscle: multiple sequence alignment with high accuracy and high throughput. Nucleic acids research. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kearse M, et al. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang F, et al. Plasmid-mediated colistin resistance gene mcr-1 in escherichia coli and klebsiella pneumoniae isolated from market retail fruits in guangzhou, china. Infection and drug resistance. 2019;12:385. doi: 10.2147/IDR.S194635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Andrews, S. & FastQC. Quality control tool for high throughput sequence data (2015).
- 38.Bankevich A, et al. Spades: a new genome assembly algorithm and its applications to single-cell sequencing. Journal of computational biology. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 40.Zankari E, et al. Identification of acquired antimicrobial resistance genes. Journal of antimicrobial chemotherapy. 2012;67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McArthur AG, et al. The comprehensive antibiotic resistance database. Antimicrobial agents and chemotherapy. 2013;57:3348–3357. doi: 10.1128/AAC.00419-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Banoo S, et al. Evaluation of diagnostic tests for infectious diseases: general principles. Nature Reviews Microbiology. 2007;5:S21–S31. doi: 10.1038/nrmicro1523x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.