Abstract
Background: Endometriosis is a frequently occurring disease in women, which seriously affects their quality of life. However, its etiology and pathogenesis are still unclear. Methods: To identify key genes/pathways involved in the pathogenesis of endometriosis, we recruited 3 raw microarray datasets (GSE11691, GSE7305, and GSE12768) from Gene Expression Omnibus database (GEO), which contain endometriosis tissues and normal endometrial tissues. We then performed in-depth bioinformatic analysis to determine differentially expressed genes (DEGs), followed by gene ontology (GO), Hallmark pathway enrichment and protein-protein interaction (PPI) network analysis. The findings were further validated by immunohistochemistry (IHC) staining in endometrial tissues from endometriosis or control patients. Results: We identified 186 DEGs, of which 118 were up-regulated and 68 were down-regulated. The most enriched DEGs in GO functional analysis were mainly associated with cell adhesion, inflammatory response, and extracellular exosome. We found that epithelial-mesenchymal transition (EMT) ranked first in the Hallmark pathway enrichment. EMT may potentially be induced by inflammatory cytokines such as CXCL12. IHC confirmed the down-regulation of E-cadherin (CDH1) and up-regulation of CXCL12 in endometriosis tissues. Conclusions: Utilizing bioinformatics and patient samples, we provide evidence of EMT in endometriosis. Elucidating the role of EMT will improve the understanding of the molecular mechanisms involved in the development of endometriosis.
Subject terms: Data mining, Databases, Endocrine reproductive disorders
Introduction
Endometriosis is a frequently occurring gynaecological disease characterised by chronic pelvic pain, dysmenorrhea and infertility1. Its prevalence is estimated to be 10–15% of reproductive age females2 and around to 20–48% in infertile women3. Despite a number of theories being suggested to describe the molecular mechanisms underlying the development of endometriosis such as: Sampson’s theory of retrograde menstruation4, ectopic implantation, epigenetic factors5, immune and inflammatory factors6,7, eutopic endometrial determinism8, and stem cell factors9; disease pathogenesis is still not fully understood.
At present, there have been several studies on the gene expression profiles of endometriosis10–13, which have identified various differentially expressed genes (DEGs) involved in the development of endometriosis. However, due to heterogeneity between each independent experiment as a result of variations in tissue or specimens and/or different data processing methods, the identification of these DEGs is inconsistent. In this study, we integrated different studies using a non-biased approach, which may resolve these problems and enable the discovery of effective and reliable molecular markers.
We downloaded 3 microarray datasets GSE1169111, GSE730512, GSE1276813, from Gene Expression Omnibus database (GEO), which contain gene expression data from endometriosis tissues and normal endometrial tissues. We then performed deep bioinformatic analysis, including identifying common DEGs, gene ontology (GO), Hallmark pathway enrichment and protein-protein interaction (PPI) network analysis. The findings were further validated by immunohistochemistry (IHC) staining in endometrial tissues from endometriosis or control patients. The aim of this study was to identify common DEGs and important pathways, and to explore potential candidate biomarkers for the diagnosis and therapeutic targets in endometriosis.
Methods
Original data collection
We used “endometriosis” as a keyword on the Gene Expression Omnibus (GEO) database, and 3 datasets (GSE11691, GSE7305 and GSE12768) were collected. GSE11691 was in GPL96 platform, [HG-U133A] Affymetrix Human Genome U133A Array, which included 9 endometriosis and 9 normal endometrial samples (Control samples). GSE7305 was in GPL570 platform, [HG-U133_Plus_2] Affymetrix Human Genome U133 Plus 2.0 Array, which included 10 endometriosis and 10 normal endometrial samples (Control samples). GSE12768 was in GPL7304 platform, institute Cochin HG18 60mer expression array 47Kl, which included 2 endometriosis and 2 normal endometrial samples (Control samples). The platform and series matrix files were downloaded.
Analysis for Differentially Expressed Genes (DEGs)
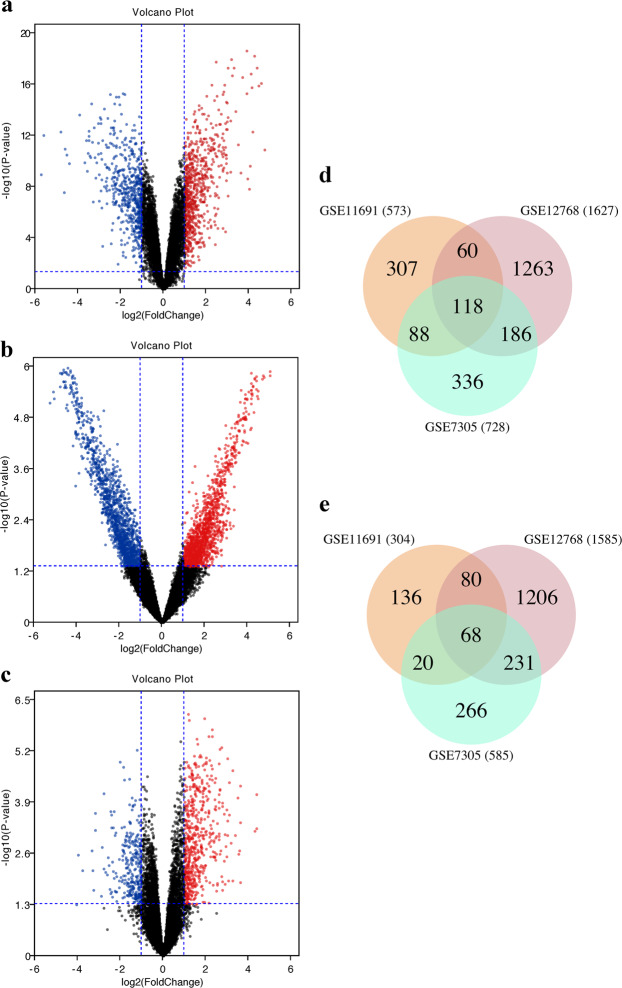
RStudio software (version 3.6) was used to process and standardise the files. The CEL files of three datasets were downloaded from GEO. Raw data of the Affymetrix platform were normalised by Robust Multi-array Average (RMA) function in the affy package (version 1.64.0). Multiple probes relating to the same gene were deleted and summarised as the median value for further analysis. These 3 datasets were analyzed using the limma package (version 3.40.6) in the RStudio14, and genes with P value <0.05 and Log[FoldChange] (Log[FC]) > 1 were considered as DEGs. Overlapping DEGs from three databases were screened for subsequent GO, Hallmark pathway enrichment and PPI analysis, and were displayed with Venn diagrams.
Analysis for GO and pathway enrichment
GO Biological Processes of DEGs were analyzed through online DAVID software15 (version 6.8), P value <0.05 as the cutoff criterion was considered statistically significant. The Hallmark pathway enrichment analysis was performed in Metascape16. P value <0.05 as the cutoff criterion was considered statistically significant.
Protein-protein interaction (PPI) network analysis
The PPI of DEGs-encoded proteins was demonstrated by STRING (version 11.0)17, with search limited to “Homo sapiens” and a score> 0.700 corresponding to high confidence interaction as significant. Network construction and analyses were performed by Cytoscape (version 3.7.1). In addition, the function and pathway enrichment analysis were performed for DEGs in the modules by ClueGo (version 2.5.4), P value <0.05 was considered to be significant.
Clinical sample collection
From June to October 2019, laparoscopic surgeries were performed in Jiangxi Maternal and Child Health Hospital (Nanchang, China), and 6 cases were pathologically diagnosed as ovarian endometriosis. On the staging criteria of endometriosis as stipulated by American Fertility Society revised (AFS-r), all patients with endometriosis were stage IV. Eutopic endometrial tissues were collected. The average age of the patients was (32.71 ± 1.12) years. Meanwhile, 6 cases of endometrial tissue were selected from patients with benign ovarian teratoma as the control group. The average age of patients was (32.18 ± 1.22) years.
All the collected endometrial tissues were diagnosed as proliferative endometrium after pathological histological diagnosis. There was no significant difference in the age of patients in each group (P value> 0.05). All menstrual cycles were normal, non-pregnant or non-lactation, and no hormonal medication was taken 6 months before the operation, and no obvious medical and surgical diseases and complications were found.
This study was approved by the Ethics Committee of Jiangxi Maternal and Child Health Hospital, China (No. EC-KT-201904). All patients had signed the informed consent for the study protocol. The experimental scheme was approved by the academic committee of Jiangxi Maternal and Child Health Hospital, and the experimental methods were carried out in accordance with the guidelines of the academic committee.
Immunohistochemistry (IHC) and image analysis
Fresh tissue specimens were taken during the operation, rinsed with physiological saline to remove blood and other impurities, fixed with 10% formaldehyde, dehydrated with conventional gradient ethanol and embedded in paraffin, continuously sliced with a paraffin microtome, and baked at 65 °C for 1 h to dewax, and removed the glass. Tablets, soak in xylene for 40 min, and soak in absolute ethanol for 20 min. Rinse once in PBS, add the configured sodium citrate solution (pure water: sodium citrate = 1000:1), and heat to boiling. Discard the sodium citrate solution after cooling, wash with PBS, and anti-CXCL12 antibody (1:200; Proteintech, Wuhan, China, 17402-1-AP) or anti-E-cadherin (CDH1) antibody (1:200; Proteintech, Wuhan, China, 20874-1-AP) was incubated, followed by incubation with goat anti-mouse/rabbit IgG polymer antibody. After rinsing with PBS three times, staining was visualised using the peroxide substrate solution diaminobenzidine. Counterstained by haematoxylin, the slides were dehydrated in graded alcohol and mounted.
Image-pro Plus software was used to convert the image format and the grayscale units into optical density (IOD) units. Then area, density and IOD were selected for measure according to the manufactor’s protocol.
Statistical analysis
Student’s t-test was used for statistical analysis between two different groups when variables were normally distributed, which was confirmed by Q-Q plots and the Shapiro-Wilk test (SPSS 18.0, Armonk, NY, USA). P value <0.05 was considered statistically significant.
Ethics approval and consent to participate
This study was approved by the Ethics Committee of Jiangxi Provincial Maternal and Child Health Hospital, China (No. EC-KT-201904). All patients have signed the informed consent for the study protocol and reserve the right to withdraw at any time.
Results
Identification of Differentially Expressed Genes (DEGs) using integrated bioinformatics
All datasets (GSE7305, GSE11691 and GSE12768) were first normalised by Robust Multi-array Average (RMA) (Supplementary Figs. 1–3). Differential expression analysis was performed on these datasets in limma, and those genes with P value <0.05 and Log[FoldChange] (Log[FC]) > 1 were considered as DEGs. In GSE7305, 1,313 DEGs were identified, of which 728 genes were up-regulated and 585 down-regulated. In GSE11691, 877 DEGs were identified, with 573 up-regulated and 304 down-regulated. In GSE12768, 3,212 DEGs were identified, with 1,627 up-regulated and 1,585 down-regulated. The expression of the top 50 DEGs for all three datasets were visualised on heat maps (Fig. 1a–c). All DEGs were highlighted in Volcano plots (Fig. 2a–c). By comparing DEGs, which appeared in all 3 datasets, 186 DEGs were identified (Table 1), including 118 up-regulated (Fig. 2d) and 68 down-regulated (Fig. 2e).
Figure 1.
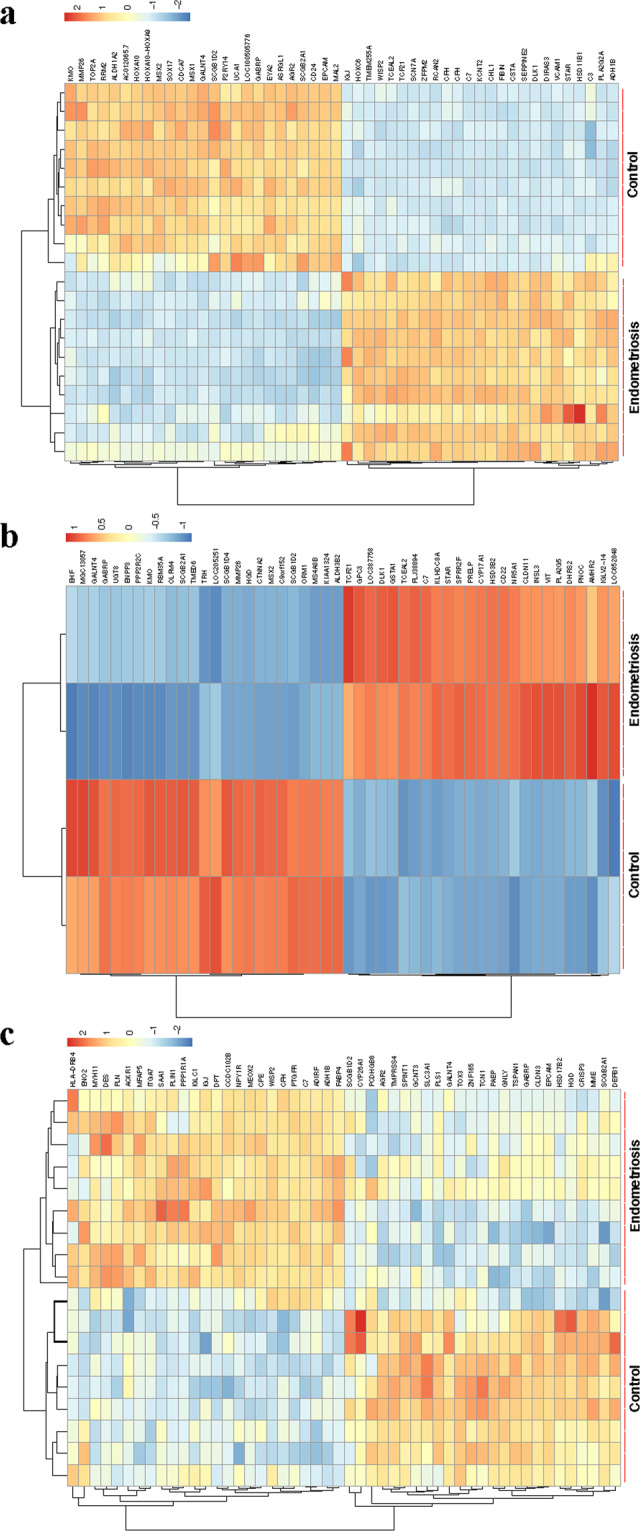
Heat maps and hierarchical clustering of the top 50 DEGs in endometriosis microarray datasets. Heat maps and hierarchal clustering analysis of top 50 DEGs in microarray datasets GSE7305 (a), GSE12768 (b), and GSE11691 (c). DEGs are those genes with P value <0.05 and Log[FC] > 1. Red indicates up-regulation and blue down-regulation.
Figure 2.
Volcano plots and Venn diagrams of DEGs in endometriosis microarray datasets. Volcano plots showing DEGs in GSE7305 (a), GSE12768 (b) and GSE11691 (c). DEGs are those genes with P value <0.05 and [logFC]> 1. Red indicates relative up-regulated genes and blue indicates down-regulated genes. Venn diagrams of up-regulated (d) or down-regulated (e) DEGs from these three datasets, as indicated.
Table 1.
DEGs in endometriosis are identified by integrated bioinformatics.
| DEGs | Gene Names |
|---|---|
| Up-regulated |
FMOD | BGN | CXCL12 | MEIS2 | ELMO1 | AEBP1 | MCAM | GPR116 | LYZ | MMRN2 | WISP | DPYSL3 | ITM2A | NUAK1 | TPSB2 | COL8A2 | CPVL | FMO2 | KCTD12 | TSPAN7 | AQP1 | MEOX2 | AGTR1 | HLA-DPB1 | GPNMB | FRZB | FZD7 | LY96 | FMO1 | PLSCR4 | NRN1 | CPA3 | GAS1 | AOC3 | COLEC12 | TPSAB1 | KIAA1462 | CPE | SH3BP5 | SULF1 | PDGFRL | IGJ | IGFBP6 | C3 | OLFML1 | GLT8D2 | CFH | THBS2 | FXYD1 | C7 | PLP1 | LHFP | ENO2 | ITGA7 | ACACB | PDLIM3 | PRELP | MN1 | FABP4 | ROBO3 | CSTA | RNASE1 | IFI44L | PROS1 | CHL1 | VCAM1 | VWF | ACTA2 | MS4A4A | ARHGAP6 | SUSD5 | CCL | SELE | LTBP2 | TAGLN | RGS2 | SGCE | PTX3 | TCF21 | ADH1B | TNFSF14 | MYH11 | GPM6A | KLF2 | GATA6 | CNN1 | PTPRZ1 | CCDC69 | CLDN5 | TCEAL2 | PDE2A | SLC16A4 | FHL5 | MYL9 | GIMAP4 | EPHA4 | CYBRD1 | CD163 | FCGR2B | NID2 | CFB | NFASC | HSD17B6 | COL11A1 | PLN | NTRK2 | IGHM | IFIT1 | ZFPM2 | DES | ACTG2 | ITPR1 | CCL21 | SCN7A | PLA2G2A | CHI3L1 | HOXC6 | HP |
| Down-regulated |
SPINT2 | HPN | GRHL2 | ELF3 | SH3YL1 | TCN1 | PPM1H | TSPAN1 | ACSL5 | PRSS16 | BTBD3 | TOM1L1 | AP1M2 | PAPSS1 | HMGCR | HOXB6 | IL20RA | SFN | EDN3 | IRF6 | ARG2 | ITGB8 | PRSS8 | HOOK1 | PLS1 | PTPN3 | PAEP | DEFB1 | CLDN10 | KIF18A | HSD17B2 | SLC34A2 | KIAA1324 | MME | TPD52L1 | GABRP | SLC1A1 | ASRGL1 | DSP | CDH1 | PDZK1 | SLC44A4 | STX18 | KRT19 | DUSP4 | DLX5 | RAB25 | PPAP2C | SALL1 | HGD | PSAT1 | PAX2 | RORB | SORD | AGR2 | ST14 | TPD52 | HOMER2 | WFDC2 | SLC15A2 | CLDN3 | GRAMD1C | EHF | CRISP3 | PROM1 | SLC26A2 | CD24 | ELP3 |
Gene Ontology (GO) functional enrichments in DEGs
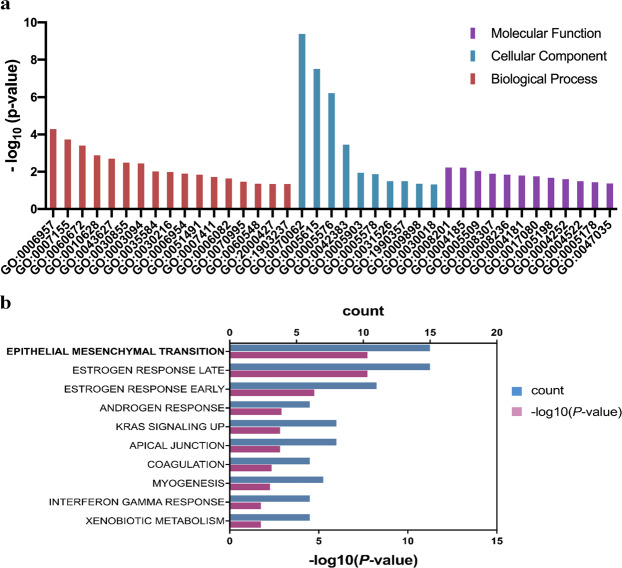
We then performed gene ontology (GO) enrichment analysis of DEGs in endometriosis using DAVID. The results were grouped into three categories: including molecular functions (MF), cellular component (CC) and biological process (BP) (Tables 2–4). The molecular functions of DEGs were mainly involved in calcium ion binding, heparin binding and structural molecule activity (Fig. 3a; Table 2). In the cellular component, DEGs were mainly involved in extracellular exosome, extracellular space and extracellular region (Fig. 3a; Table 3). In the biological process, DEGs were mainly involved in cell adhesion, epithelial cell differentiation, inflammatory response and extracellular exosome (Fig. 3a; Table 4).
Table 2.
Molecular Function (MF) analysis of DEGs in endometriosis.
| Term | Description | counts | P-value (<0.05) |
|---|---|---|---|
| GO:0008201 | heparin binding | 6 | 0.0058 |
| GO:0004185 | serine-type carboxypeptidase activity | 3 | 0.0060 |
| GO:0005509 | calciumion binding | 14 | 0.0091 |
| GO:0008307 | structural constituent of muscle | 3 | 0.0126 |
| GO:0008236 | serine-type peptidase activity | 3 | 0.0143 |
| GO:0004181 | metallocarboxypeptidase activity | 3 | 0.0159 |
| GO:0017080 | sodium channel regulator activity | 3 | 0.0176 |
| GO:0005198 | structural molecule activity | 6 | 0.0210 |
| GO:0004252 | serine-type endopeptidase activity | 6 | 0.0248 |
| GO:0004522 | ribonuclease A activity | 2 | 0.0322 |
| GO:0005178 | integrin binding | 3 | 0.0367 |
| GO:0047035 | testosterone dehydrogenase (NAD+) activity | 2 | 0.0427 |
Table 4.
Biological process analysis of DEGs in endometriosis.
| Term | Description | counts | P-value (<0.05) |
|---|---|---|---|
| GO:0006957 | complement activation, alternative pathway | 4 | 5.15E-05 |
| GO:0007155 | cell adhesion | 10 | 1.84E-04 |
| GO:0060672 | epithelial cell morphogenesis involved in placenta | 3 | 3.96E-04 |
| GO:0010628 | positive regulation of gene expression | 8 | 0.0013 |
| GO:0043627 | response to estrogen | 4 | 0.0019 |
| GO:0030855 | epithelial cell differentiation | 5 | 0.0032 |
| GO:0003094 | glomerular filtration | 3 | 0.0035 |
| GO:0035584 | calcium-mediated signaling using intracellular cal | 3 | 0.0095 |
| GO:0030216 | keratinocyte differentiation | 4 | 0.0102 |
| GO:0006954 | inflammatory response | 8 | 0.0125 |
| GO:0051491 | positive regulation of filopodium assembly | 3 | 0.0143 |
| GO:0007411 | axon guidance | 5 | 0.0190 |
| GO:0006082 | organic acid metabolic process | 2 | 0.0230 |
| GO:0070995 | NADPH oxidation | 2 | 0.0343 |
| GO:0060548 | negative regulation of cell death | 3 | 0.0441 |
| GO:2000427 | positive regulation of apoptotic cell clearance | 2 | 0.0455 |
| GO:1903237 | negative regulation of leukocyte tethering or roll | 2 | 0.0455 |
Figure 3.
GO analysis and Hallmark pathway enrichment of DEGs in endometriosis. (a) GO analysis of DEGs in endometritis visualised on a bar chart clustered by molecular functions, cellular component and biological process. (b) Hallmark pathway enrichment of DEGs in endometriosis visualised on a bar chart, showing number of shared genes (count) and -Log10 (P value).
Table 3.
Cellular component analysis of DEGs in endometriosis.
| Term | Description | counts | P-value (<0.05) |
|---|---|---|---|
| GO:0070062 | extracellular exosome | 56 | 4.18E-10 |
| GO:0005615 | extracellular space | 32 | 3.13E-08 |
| GO:0005576 | extracellular region | 21 | 6.03E-07 |
| GO:0042383 | sarcolemma | 6 | 3.48E-04 |
| GO:0005903 | brush border | 4 | 0.0113 |
| GO:0005578 | proteinaceous extracellular matrix | 7 | 0.0135 |
| GO:0031526 | brush border membrane | 3 | 0.0321 |
| GO:1990357 | terminal web | 2 | 0.0322 |
| GO:0009898 | cytoplasmic side of plasma membrane | 3 | 0.0444 |
| GO:0030018 | Z disc | 4 | 0.0482 |
Signaling pathway enrichment in DEGs
Signaling pathway enrichment of DEGs in endometriosis was performed using Metascape. The most significantly enriched pathways were submitted to Hallmark genes hit analysis. Hallmark pathway enrichment analysis identified epithelial mesenchymal transition (EMT), estrogen response late and estrogen response early as top pathways (Fig. 3b; Table 5).
Table 5.
Hallmark pathway enrichment analysis of DEGs in endometriosis.
| ID | Description | Counts | P-value | Gene |
|---|---|---|---|---|
| M5930 | Epithelial mesenchymal transition | 15 | 4.75E-11 |
CXCL12 | TAGLN | ACTA2 | MYL9 | VCAM1 | DPYSL3 | FMOD | GAS1 | PTX3 | ENO2 | BGN | COL8A2 | COL11A1 | THBS2 | NID |
| M5907 | Estrogen response late | 15 | 4.75E-11 |
CDH1 | CPE | SLC26A2 | SFN | CXCL12 | KRT19 | PDZK1 | SORD | ST14 | TPD52L1 | TPSAB1 | CCN5 | HOMER2 | AGR2 | PDLIM3 |
| M5906 | Estrogen response early | 11 | 9.58E-09 |
SLC26A2 | ELF3 | SFN | KRT19 | PDZK1 | CXCL12 | SLC1A1 | TPD52L1 | CCN5 | SH3BP5 | PDLIM3 |
| M5908 | Androgen response | 6 | 0.0001 |
SLC26A2 | HMGCR | KRT19 | SORD | TPD52 | HOMER2 |
| M5953 | Kras signaling up | 8 | 0.0002 |
CFB | CPE | CFH | TSPAN7 | TSPAN1 | GPNMB | BTBD3 | LY96 |
| M5915 | Apical junction | 8 | 0.0002 |
VWF | SGCE | MYL9 | NFASC | ACTG2 | CDH1 | CLDN5 | VCAM1 |
| M5946 | Coagulation | 6 | 0.0007 | CFB | C3 | CFH | HPN | PROS1 | VWF |
| M5909 | Myogenesis | 7 | 0.0009 |
AEBP1 | DES | ITGA7 | MYH11 | FXYD1 | TAGLN | TPD52L1 |
| M5913 | Interferon gamma Response | 6 | 0.0046 |
CFB | CFH | IFIT1 | CCL2 | VCAM1 | IFI44 |
| M5934 | Xenobitoic metabolism | 6 | 0.0046 |
ARG2 | CFB | FMO1 | HSD17B2 | PROS1 | SPINT2 |
Protein-protein interaction (PPI) network analysis in DEGs
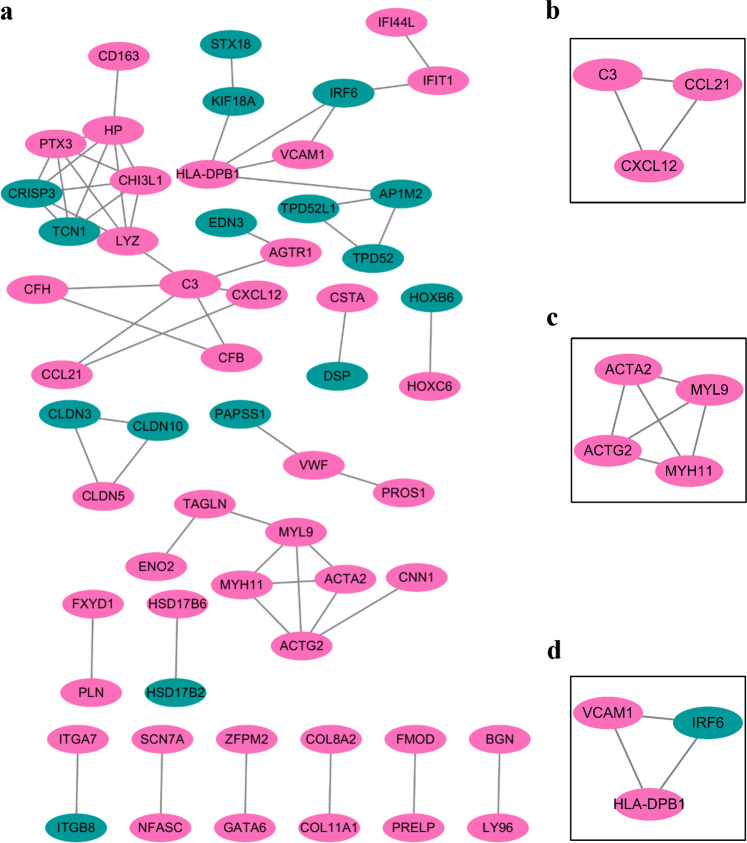
PPI analysis was performed using the online STRING database and Cytoscape software. After removing the isolated nodes and the partially connected nodes, a grid network was constructed using the Cytoscape software (Fig. 4). Pathway enrichment analysis revealed that the genes were mainly involved in vascular smooth muscle contraction, cell adhesion molecules, NF-κB pathway, complement and coagulation cascade.
Figure 4.
PPI network analysis of DEGs in endometriosis. Protein-Protein Interaction Network of DEGs from all datasets generated in String.db (v. 11) and visualised in Cytoscape (v. 3.7.1). (a) PPI network analysis of DEGs. (b–d) Representative local association graphs in PPI network analysis. Nodes indicate proteins/genes and lines indicate protein-protein interaction. Pink indicates up-regulation and green indicates down-regulation.
Candidate gene expression analysis and validations
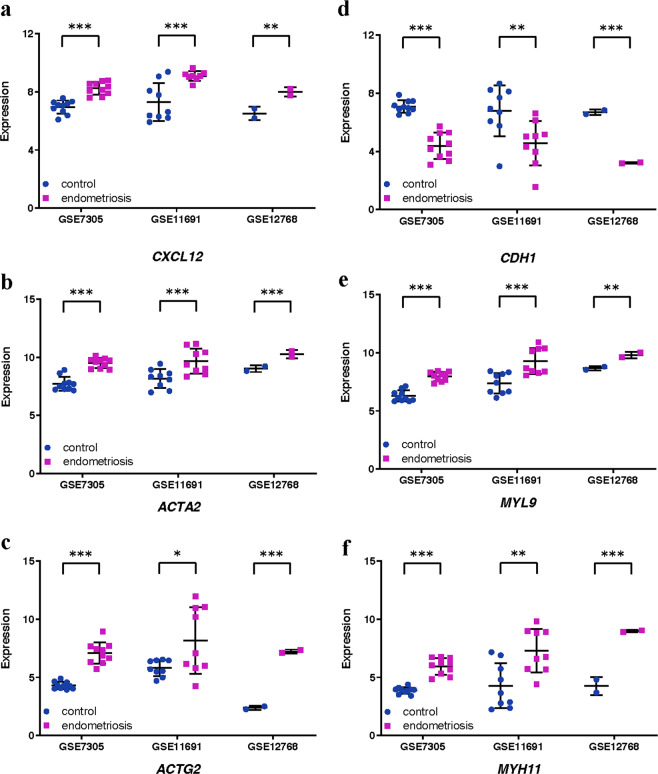
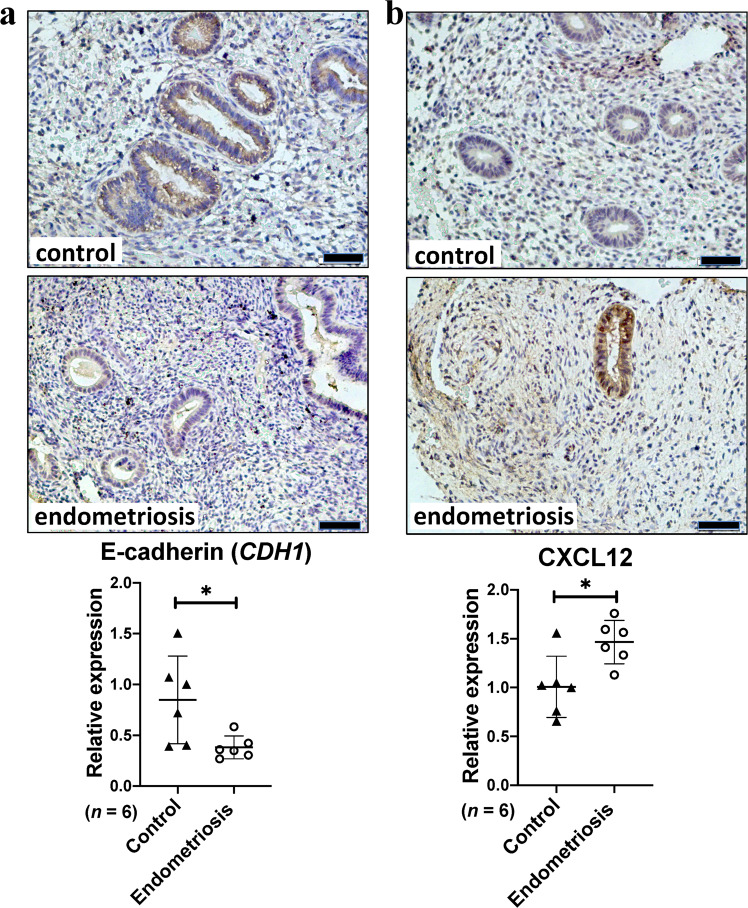
Hallmark pathway enrichment analysis of DEGs in endometriosis identified 15 EMT-associated genes (CXCL12, TAGLN, ACTA2, MYL9, VCAM1, DPYSL3, FMOD, GAS1, PTX3, ENO2, BGN, COL8A2, COL11A1, THBS2, NID) (Table 5). In PPI network analysis, CXCL12 was found to be connected to a hub gene C3, while ACTG2, ACTA2, MYL9 and MYH11 formed a connected component sub-network. In addition, a change in the expression of E-cadherin (CDH1) is the prototypical epithelial cell marker of EMT. As a result, although CDH1 is not listed in Gene Set Hallmark_EMT, it was included in further analysis. Expression levels of these 6 genes (CXCL2, ACTA2, MYL9, ACTG2, MYH11 and CDH1) were analysed in these three databases (Fig. 5). Significant increases were observed in CXCL2, ACTA2, MYL9, ACTG2 and MYH11 across all three databases. A significant decrease in CDH1 was observed in all three databases. We further investigated the expression of E-cadherin (CDH1) and CXCL12 in endometriosis or control tissues by IHC. As shown in Fig. 6, E-cadherin was significantly down-regulated in endometriosis (Fig. 6a; P value = 0.028), while CXCL12 was significantly increased in endometriosis (Fig. 6b; P value = 0.015).
Figure 5.
Expression levels of 6 genes in endometriosis microarray datasets. Graphs showing expression levels of CXCL12 (a), ACTA2 (b), ACTG2 (c), CDH1 (d), MYL9 (e) and MYH11 (f) in endometrial tissues from control (blue) or endometriosis (purple) patients in three endometriosis microarray datasets, as indicated. Data are mean ± s.d. *P value <0.05.** P value <0.01. *** P value <0.001.
Figure 6.
Expression levels of E-cadherin (CDH1) and CXCL12 in endometriosis. Representative E-cadherin (a) or CXCL12 (b) expression in endometrial tissues from control or endometriosis patients. Scale bars: 50 μm. Graphs showing comparisons of E-cadherin (a, P = 0.028) or CXCL12 (b, P = 0.015) expression in endometrial tissues from 6 control or endometriosis patients. Data are mean ± s.d.
Discussion
Endometriosis occurs in about 10–15% of reproductive age females and the etiology is unknown1,2. At present there is no cure and the treatment options available are limited. The disease has a high recurrence rate, which adds to its large socio-economic impact18. Endometriosis is the growth of cells derived from the endometrium outside the uterus, such as the ovaries, peritoneum, intestines and vagina19. In a small number of cases (0.5–1%) endometriosis can lead to tumor formation20. The underlying mechanisms of the disease are similar to malignant tumors such as cell proliferation, differentiation, apoptosis, migration, cell adhesion, invasion, and neurovascularisation21.
Utilising data from 3 microarray datasets (GSE1169111, GSE730512, GSE1276813), we identified DEGs between endometriosis tissues and normal endometrial samples, including 118 up-regulated and 68 down-regulated genes. GO functional analysis based on these DEGs shows that DEGs are mainly enriched in cell adhesion, inflammatory response, and extracellular exosome. These findings are similar to those previously published22.
Importantly, Hallmark pathway enrichment analysis identified EMT as the most significant pathway. A number of studies have implicated EMT in the development of endometriosis23–25. EMT is a biological process where immotile epithelial cells acquire phenotypes of motile mesenchymal cells, this is accompanied by changes in cell morphology and gene expression26. It creates favourable conditions for the implantation and growth of endometriotic lesions27. During EMT the expression of a number of epithelial surface markers are lost including E-cadherin (CDH1), keratin, Desmoplakin, Mucin-1 and claudin; whilst a number of mesenchymal makers are up-regulated such as N-cadherin, vimentin, and fibronectin28,29. Numerous signaling pathways are suggested to participate in EMT induction, including transforming growth factor β (TGF-β)30, Wnt/β-catenin signaling pathway31, estrogen receptor β (ER-β)32, epidermal growth factor (EGF)33, mitogen-activated protein kinase (MAPK)/extracellular signal-regulated kinase (ERK)34, NF-κB35, estrogen receptor (ER)-α36 and hypoxia-inducible factor (HIF)-1α37. The activities of these pathways appear to be interconnected to one another, and depend on the particular epithelial or endothelial cell type affected, different signaling molecules mediate their interconnection or crosstalk. Previous studies have also found that EMT can be induced by pro-inflammatory cytokines in endometriosis, such as TGF-β38, tumor necrosis factor (TNF)-α39 and interleukin (IL)-640. The mechanisms that present or activate TGF-β in the tissue microenvironment are of importance for the EMT response41. TGF-β induced EMT mediated by inflammatory cells in the tumor microenvironment is promoted by leukotriene B4 receptor 2, which, in response to leukotriene B4, activates reactive oxygen species (ROS) and NF-κB transcriptional activity that facilitates the establishment of EMT by TGF-β42.
In this unbiased study, we found EMT in endometriosis could be potentially induced by inflammatory cytokines such as C-X-C motif chemokine ligand 12 (CXCL12), also known as stromal cell-derived factor 1 (SDF1). CXCL12 is highly expressed in endometriosis in our analysis, which is consistent with a previous report43. CXCL12 interacts with its specific receptor, C-X-C motif chemokine receptor 4 (CXCR4), which is not consistently over-expressed in these three datasets though. The CXCL12-CXCR4 axis promotes proliferation, migration, and invasion of endometriotic cells44,45. In human papillary thyroid carcinoma, the CXCL12-CXCR4 axis promotes EMT processes by activating the NF-κB signaling pathway46. In a murine model of endometriosis both C-X-C motif chemokine receptor 7 (CXCR7) and CXCL12 expression increased with grafting time47. Expression of CXCR7 is enhanced during pathological inflammation and tumor development, and CXCR7 mediates TGFβ1-induced EMT48. However, there were no probes for CXCR7 in the microarrays analysed in our studies. In endometriosis, it is still unclear whether CXCL12 promotes EMT through the CXCL12-CXCR4 axis or the CXCL12-CXCR7 axis. PPI analysis showed that CXCL12 interacts directly with complement C3 and C-C motif chemokine ligand 21 (CCL21), and a previous study showede CCL21 is up-regulated in endometriosis, which acts through inflammatory responses49. In TGF-β-induced EMT, the expression of C-C motif chemokine receptor 7 (CCR7), the CCL21 receptor, is increased and this facilitates breast cancer cell migration50. Through IHC, we confirmed that CXCL12 is significantly increased in endometriosis, accompanied by a decrease in the expression E-cadherin (CDH1), which is consistent with bioinformatics analysis. These findings, together, suggest that CXCL12 may lead to endometriosis through EMT, although further research is required.
EMT in endometriosis has been suggested to be associated with smooth muscle metaplasia and fibrogenesis51,52. We found various markers for smooth muscle cells in our analysis, including ACTA2 and MYL9, which interact with ACTG2 and MYH11 in the PPI network analysis. ACTA2 (α-SMA), is considered to be a marker of fibrosis and is up-regulated in endometriosis53, which is consistent with our findings. Previous studies54,55 have shown that platelet-derived TGF-β1 can activate the TGF-β1/Smad3 signaling pathway, subsequently promoting EMT and fibroblast-to-myofibroblast trans-differentiation (FMT) in endometriotic lesions in turn, promoting smooth muscle metaplasia and ultimately leading to fibrosis.
Conclusion
By comparing 3 microarray datasets, we have identified 186 DEGs (118 up-regulated, 68 down-regulated) which may be involved in the progression of endometriosis. GO functional analysis determined DEGs were mainly enriched in cell adhesion, inflammatory response, and extracellular exosome. EMT was the highest ranked Hallmark pathway enrichment and we proposed that it could be induced by inflammatory cytokines and associated with smooth muscle metaplasia and fibrogenesis. Further elucidating the underlying mechanisms of endometriosis is key for the development of new treatments and bio-markers.
Supplementary information
Acknowledgements
This research was funded by Medical Research Council (MR/S025480/1), an Academy of Medical Sciences/the Wellcome Trust Springboard Award (SBF002\1038) and National Natural Science Foundation of China (81860267). Y.Z. was supported by an Institute for Life Sciences PhD Studentship. C.H. was supported by Gerald Kerkut Charitable Trust and University of Southampton Central VC Scholarship Scheme. The funding sponsors had no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Author contributions
Conceptualisation: M.C., X.Z. and Y.W.; Methodology: M.C. and Y.Z.; Formal analysis: M.C. and Y.Z.; Experiment: H.X. and D.H.; Writing: M.C., C.H., Y.Z. and Y.W.; Supervision: R.E., X.Z. and Y.W.
Data availability
Data and materials from this study are available upon a written request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Meihong Chen and Yilu Zhou.
Contributor Information
Xiaoling Zhang, Email: xlzzz777@126.com.
Yihua Wang, Email: yihua.wang@soton.ac.uk.
Supplementary information
is available for this paper at 10.1038/s41598-020-65606-9.
References
- 1.Zannoni L, Forno SD, Paradisi R, Seracchioli R. Endometriosis in Adolescence: Practical Rules for an Earlier Diagnosis. Pediatric annals. 2016;45:e332–335. doi: 10.3928/19382359-20160727-03. [DOI] [PubMed] [Google Scholar]
- 2.Johnson NP, et al. World Endometriosis Society consensus on the classification of endometriosis. Human reproduction. 2017;32:315–324. doi: 10.1093/humrep/dew293. [DOI] [PubMed] [Google Scholar]
- 3.Liang Y, Yao S. Potential role of estrogen in maintaining the imbalanced sympathetic and sensory innervation in endometriosis. Molecular and cellular endocrinology. 2016;424:42–49. doi: 10.1016/j.mce.2016.01.012. [DOI] [PubMed] [Google Scholar]
- 4.Sourial S, Tempest N, Hapangama DK. Theories on the pathogenesis of endometriosis. International journal of reproductive medicine. 2014;2014:179515. doi: 10.1155/2014/179515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sofo V, et al. Correlation between dioxin and endometriosis: an epigenetic route to unravel the pathogenesis of the disease. Archives of gynecology and obstetrics. 2015;292:973–986. doi: 10.1007/s00404-015-3739-5. [DOI] [PubMed] [Google Scholar]
- 6.Bruner-Tran KL, Herington JL, Duleba AJ, Taylor HS, Osteen KG. Medical management of endometriosis: emerging evidence linking inflammation to disease pathophysiology. Minerva ginecologica. 2013;65:199–213. [PMC free article] [PubMed] [Google Scholar]
- 7.Zhao Y, et al. Dual suppression of estrogenic and inflammatory activities for targeting of endometriosis. Science translational medicine. 2015;7:271ra279. doi: 10.1126/scitranslmed.3010626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Evian Annual Reproduction Workshop G, et al. Contemporary genetic technologies and female reproduction. Human reproduction update. 2011;17:829–847. doi: 10.1093/humupd/dmr033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Du H, Taylor HS. Contribution of bone marrow-derived stem cells to endometrium and endometriosis. Stem cells. 2007;25:2082–2086. doi: 10.1634/stemcells.2006-0828. [DOI] [PubMed] [Google Scholar]
- 10.Wren JD, Wu Y, Guo SW. A system-wide analysis of differentially expressed genes in ectopic and eutopic endometrium. Human reproduction. 2007;22:2093–2102. doi: 10.1093/humrep/dem129. [DOI] [PubMed] [Google Scholar]
- 11.Hull ML, et al. Endometrial-peritoneal interactions during endometriotic lesion establishment. The American journal of pathology. 2008;173:700–715. doi: 10.2353/ajpath.2008.071128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hever A, et al. Human endometriosis is associated with plasma cells and overexpression of B lymphocyte stimulator. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:12451–12456. doi: 10.1073/pnas.0703451104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Borghese B, et al. Gene expression profile for ectopic versus eutopic endometrium provides new insights into endometriosis oncogenic potential. Molecular endocrinology. 2008;22:2557–2562. doi: 10.1210/me.2008-0322. [DOI] [PubMed] [Google Scholar]
- 14.Ritchie ME, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic acids research. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature protocols. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 16.Zhou Y, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nature communications. 2019;10:1523. doi: 10.1038/s41467-019-09234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Szklarczyk D, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic acids research. 2019;47:D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Flores I, et al. Molecular profiling of experimental endometriosis identified gene expression patterns in common with human disease. Fertility and sterility. 2007;87:1180–1199. doi: 10.1016/j.fertnstert.2006.07.1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baranov VS, Ivaschenko TE, Liehr T, Yarmolinskaya MI. Systems genetics view of endometriosis: a common complex disorder. European journal of obstetrics, gynecology, and reproductive biology. 2015;185:59–65. doi: 10.1016/j.ejogrb.2014.11.036. [DOI] [PubMed] [Google Scholar]
- 20.Aznaurova YB, Zhumataev MB, Roberts TK, Aliper AM, Zhavoronkov AA. Molecular aspects of development and regulation of endometriosis. Reproductive biology and endocrinology: RB&E. 2014;12:50. doi: 10.1186/1477-7827-12-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reis FM, Petraglia F, Taylor RN. Endometriosis: hormone regulation and clinical consequences of chemotaxis and apoptosis. Human reproduction update. 2013;19:406–418. doi: 10.1093/humupd/dmt010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Z, Ruan L, Lu M, Yao X. Analysis of key candidate genes and pathways of endometriosis pathophysiology by a genomics-bioinformatics approach. Gynecological endocrinology: the official journal of the International Society of Gynecological Endocrinology. 2019;35:576–581. doi: 10.1080/09513590.2019.1576609. [DOI] [PubMed] [Google Scholar]
- 23.Yang YM, Yang WX. Epithelial-to-mesenchymal transition in the development of endometriosis. Oncotarget. 2017;8:41679–41689. doi: 10.18632/oncotarget.16472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu H, et al. Autophagy contributes to hypoxia-induced epithelial to mesenchymal transition of endometrial epithelial cells in endometriosis. Biology of reproduction. 2018;99:968–981. doi: 10.1093/biolre/ioy128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu RF, et al. High expression of ZEB1 in endometriosis and its role in 17beta-estradiol-induced epithelial-mesenchymal transition. International journal of clinical and experimental pathology. 2018;11:4744–4758. [PMC free article] [PubMed] [Google Scholar]
- 26.Polyak K, Weinberg RA. Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nature reviews. Cancer. 2009;9:265–273. doi: 10.1038/nrc2620. [DOI] [PubMed] [Google Scholar]
- 27.Wu RF, et al. Lipoxin A4 Suppresses Estrogen-Induced Epithelial-Mesenchymal Transition via ALXR-Dependent Manner in Endometriosis. Reproductive sciences. 2018;25:566–578. doi: 10.1177/1933719117718271. [DOI] [PubMed] [Google Scholar]
- 28.Bilyk O, Coatham M, Jewer M, Postovit LM. Epithelial-to-Mesenchymal Transition in the Female Reproductive Tract: From Normal Functioning to Disease Pathology. Frontiers in oncology. 2017;7:145. doi: 10.3389/fonc.2017.00145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nature reviews. Molecular cell biology. 2014;15:178–196. doi: 10.1038/nrm3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Soni UK, et al. A high level of TGF-B1 promotes endometriosis development via cell migration, adhesiveness, colonization, and invasivenessdagger. Biology of reproduction. 2019;100:917–938. doi: 10.1093/biolre/ioy242. [DOI] [PubMed] [Google Scholar]
- 31.Matsuzaki S, Darcha C. Involvement of the Wnt/beta-catenin signaling pathway in the cellular and molecular mechanisms of fibrosis in endometriosis. PloS one. 2013;8:e76808. doi: 10.1371/journal.pone.0076808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Han SJ, et al. Estrogen Receptor beta Modulates Apoptosis Complexes and the Inflammasome to Drive the Pathogenesis of Endometriosis. Cell. 2015;163:960–974. doi: 10.1016/j.cell.2015.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chatterjee K, Jana S, DasMahapatra P, Swarnakar S. EGFR-mediated matrix metalloproteinase-7 up-regulation promotes epithelial-mesenchymal transition via ERK1-AP1 axis during ovarian endometriosis progression. FASEB journal: official publication of the Federation of American Societies for Experimental Biology. 2018;32:4560–4572. doi: 10.1096/fj.201701382RR. [DOI] [PubMed] [Google Scholar]
- 34.Huang M, et al. MAPK pathway mediates epithelial-mesenchymal transition induced by paraquat in alveolar epithelial cells. Environmental toxicology. 2016;31:1407–1414. doi: 10.1002/tox.22146. [DOI] [PubMed] [Google Scholar]
- 35.Pires BR, et al. NF-kappaB Is Involved in the Regulation of EMT Genes in Breast Cancer Cells. PloS one. 2017;12:e0169622. doi: 10.1371/journal.pone.0169622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen YJ, et al. Oestrogen-induced epithelial-mesenchymal transition of endometrial epithelial cells contributes to the development of adenomyosis. The Journal of pathology. 2010;222:261–270. doi: 10.1002/path.2761. [DOI] [PubMed] [Google Scholar]
- 37.Xiong Y, et al. Hypoxia-inducible factor 1alpha-induced epithelial-mesenchymal transition of endometrial epithelial cells may contribute to the development of endometriosis. Human reproduction. 2016;31:1327–1338. doi: 10.1093/humrep/dew081. [DOI] [PubMed] [Google Scholar]
- 38.Young VJ, Brown JK, Saunders PT, Duncan WC, Horne AW. The peritoneum is both a source and target of TGF-beta in women with endometriosis. PloS one. 2014;9:e106773. doi: 10.1371/journal.pone.0106773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Khan KN, et al. 17beta-estradiol and lipopolysaccharide additively promote pelvic inflammation and growth of endometriosis. Reproductive sciences. 2015;22:585–594. doi: 10.1177/1933719114556487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alvarado-Diaz CP, Nunez MT, Devoto L, Gonzalez-Ramos R. Iron overload-modulated nuclear factor kappa-B activation in human endometrial stromal cells as a mechanism postulated in endometriosis pathogenesis. Fertility and sterility. 2015;103:439–447. doi: 10.1016/j.fertnstert.2014.10.046. [DOI] [PubMed] [Google Scholar]
- 41.Moustakas, A. & Heldin, C. H. Mechanisms of TGFbeta-Induced Epithelial-Mesenchymal Transition. Journal of clinical medicine5, 10.3390/jcm5070063 (2016). [DOI] [PMC free article] [PubMed]
- 42.Kim H, Choi JA, Kim JH. Ras promotes transforming growth factor-beta (TGF-beta)-induced epithelial-mesenchymal transition via a leukotriene B4 receptor-2-linked cascade in mammary epithelial cells. The Journal of biological chemistry. 2014;289:22151–22160. doi: 10.1074/jbc.M114.556126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Leconte M, et al. Role of the CXCL12-CXCR4 axis in the development of deep rectal endometriosis. Journal of reproductive immunology. 2014;103:45–52. doi: 10.1016/j.jri.2013.12.121. [DOI] [PubMed] [Google Scholar]
- 44.Ruiz A, et al. Pharmacological blockage of the CXCR4-CXCL12 axis in endometriosis leads to contrasting effects in proliferation, migration, and invasion. Biology of reproduction. 2018;98:4–14. doi: 10.1093/biolre/iox152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moridi I, Mamillapalli R, Cosar E, Ersoy GS, Taylor HS. Bone Marrow Stem Cell Chemotactic Activity Is Induced by Elevated CXCl12 in Endometriosis. Reproductive sciences. 2017;24:526–533. doi: 10.1177/1933719116672587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lin Y, Ma Q, Li L, Wang H. The CXCL12-CXCR4 axis promotes migration, invasiveness, and EMT in human papillary thyroid carcinoma B-CPAP cells via NF-kappaB signaling. Biochemistry and cell biology = Biochimie et biologie cellulaire. 2018;96:619–626. doi: 10.1139/bcb-2017-0074. [DOI] [PubMed] [Google Scholar]
- 47.Pluchino N, Mamillapalli R, Moridi I, Tal R, Taylor HS. G-Protein-Coupled Receptor CXCR7 Is Overexpressed in Human and Murine Endometriosis. Reproductive sciences. 2018;25:1168–1174. doi: 10.1177/1933719118766256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu YC, Tang SJ, Sun GH, Sun KH. CXCR7 mediates TGFbeta1-promoted EMT and tumor-initiating features in lung cancer. Oncogene. 2016;35:2123–2132. doi: 10.1038/onc.2015.274. [DOI] [PubMed] [Google Scholar]
- 49.Sundqvist J, et al. Endometriosis and autoimmune disease: association of susceptibility to moderate/severe endometriosis with CCL21 and HLA-DRB1. Fertility and sterility. 2011;95:437–440. doi: 10.1016/j.fertnstert.2010.07.1060. [DOI] [PubMed] [Google Scholar]
- 50.Pang MF, et al. TGF-beta1-induced EMT promotes targeted migration of breast cancer cells through the lymphatic system by the activation of CCR7/CCL21-mediated chemotaxis. Oncogene. 2016;35:748–760. doi: 10.1038/onc.2015.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yan D, Liu X, Guo SW. The establishment of a mouse model of deep endometriosis. Human reproduction. 2019;34:235–247. doi: 10.1093/humrep/dey361. [DOI] [PubMed] [Google Scholar]
- 52.Ibrahim MG, et al. Arrangement of myofibroblastic and smooth muscle-like cells in superficial peritoneal endometriosis and a possible role of transforming growth factor beta 1 (TGFbeta1) in myofibroblastic metaplasia. Archives of gynecology and obstetrics. 2019;299:489–499. doi: 10.1007/s00404-018-4995-y. [DOI] [PubMed] [Google Scholar]
- 53.Xu Z, et al. The estrogen-regulated lncRNA H19/miR-216a-5p axis alters stromal cell invasion and migration via ACTA2 in endometriosis. Molecular human reproduction. 2019;25:550–561. doi: 10.1093/molehr/gaz040. [DOI] [PubMed] [Google Scholar]
- 54.Zhang Q, Duan J, Liu X, Guo SW. Platelets drive smooth muscle metaplasia and fibrogenesis in endometriosis through epithelial-mesenchymal transition and fibroblast-to-myofibroblast transdifferentiation. Molecular and cellular endocrinology. 2016;428:1–16. doi: 10.1016/j.mce.2016.03.015. [DOI] [PubMed] [Google Scholar]
- 55.Zhang Q, Duan J, Olson M, Fazleabas A, Guo SW. Cellular Changes Consistent With Epithelial-Mesenchymal Transition and Fibroblast-to-Myofibroblast Transdifferentiation in the Progression of Experimental Endometriosis in Baboons. Reproductive sciences. 2016;23:1409–1421. doi: 10.1177/1933719116641763. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data and materials from this study are available upon a written request.