Abstract
Microbially induced calcification is an ancient, community-driven mineralisation process that produces different types of microbialites. Symbiolites are photosynthesis-induced microbialites, formed by calcifying co-cultures of dinoflagellates from the family Symbiodiniaceae and bacteria. Symbiolites encase the calcifying community as endolithic cells, pointing at an autoendolithic niche of symbiotic dinoflagellates, and provide a rare opportunity to study the role of bacteria in bacterial–algal calcification, as symbiodiniacean cultures display either distinct symbiolite-producing (SP) or non-symbiolite-producing (NP) phenotypes. Using Illumina sequencing, we found that the bacterial communities of SP and NP cultures differed significantly in the relative abundance of 23 genera, 14 families, and 2 phyla. SP cultures were rich in biofilm digesters from the phylum Planctomycetes and their predicted metagenomes were enriched in orthologs related to biofilm formation. In contrast, NP cultures were dominated by biofilm digesters from the Bacteroidetes, and were inferred as enriched in proteases and nucleases. Functional assays confirmed the potential of co-cultures and bacterial isolates to produce biofilms and point at acidic polysaccharides as key stimulators for mineral precipitation. Hence, bacteria appear to influence symbiolite formation primarily through their biofilm-producing and modifying activity and we anticipate that symbiolite formation, as a low-complexity in vitro model, will significantly advance our understanding of photosynthesis-induced microbial calcification processes.
Subject terms: Microbial ecology, Biogeochemistry, Microbial ecology, Next-generation sequencing, Biofilms
Introduction
Interactions between bacteria and microalgae range from common trophic relationships, over pathogenesis, to specialised commensalistic, and mutualistic symbioses [1–3]. When, as in the current study, bacteria and microalgae coexist in the form of microbial biofilms, the whole range of these biological interactions can ensue [4, 5], which can create emergent features at the community level [6]. One such emergent feature that has accompanied life on Earth for hundreds of millions of years is microbially induced calcification [7–9], a mineralisation process that arises if a microbial community produces extracellular polymeric substances (EPS) that promote mineral precipitation, parallel to metabolically raising the CaCO3 saturation state (Ω) above a critical point [10–13]. For example, photosynthesis induces microbial calcification through the assimilation of CO2 and HCO3−, and the release of OH− from carbon concentrating mechanisms, as these activities raise the extracellular pH and increase Ω [9, 14]. This well-known form of photosynthesis-induced microbial calcification is involved, e.g., in the formation of stromatolites [8] and calcifying reef sediments [15].
Reef sediments are home to a diverse range of microalgae [16, 17]. Dinoflagellates from the family Symbiodiniaceae, hereafter also referred to as symbiodiniaceans, are best known for their endosymbiotic life histories with corals and other marine organisms [18] but they also exist as free-living cells in the environment [19], especially in reef sediments [17]. The recent discovery that cultures of free-living symbiodiniaceans commonly engage in photosynthesis-induced microbial calcification and produce microbialites, so-called symbiolites, into which they embed themselves as endolithic cells, led to the realisation that these ecologically important microalgae could be involved in the microbial calcification of reef sediments and that their life history could involve an autoendolithic phase [20, 21]. The concept of an autoendolithic symbiodiniacean life history still awaits verification in the environment but experimental evidence suggests that the underlying mechanisms operate under natural conditions [22] and the existence of an autoendolithic niche would provide a conceptual framework for the frequently observed association of free-living symbiodiniaceans with reef sediments [17, 23].
Evidence for an essential role of bacteria in symbiodiniacean-induced calcification stems from experiments in which symbiolite formation could be inhibited by antibiotics and reinstated at an increased rate by introducing bacterial isolates from calcifying cultures [20]. The antibiotics treatments also visibly reduced EPS, suggesting that the symbiolite-producing (SP) and non-symbiolite-producing (NP) phenotypes could be tied to the production of bacterial EPS, which are known to strongly influence the calcification potential of microbial biofilms [7, 10, 13, 24, 25]. Of particular importance in this context could be acidic polysaccharides (APs), as symbiolites co-localised with this reactive group of EPS that is known to form ionic bonds with cations such as Ca2+ and Mg2+ and that is thought to provide nucleation points for mineral precipitation [12, 26, 27].
The current study aimed at characterising symbiodiniacean-associated bacterial communities and the properties that could underpin their role in symbiolite formation. We conducted a comparative Illumina-based microbiome study to test if the SP and NP phenotypes of symbiodiniacean cultures are associated to differences in their bacterial communities. This diversity study was complemented with (1) composite metagenome inference to predict differences in community traits and (2) functional assays to relate algal growth and the production of APs with calcification dynamics. Further, we characterised the microbiomes of three strongly calcifying strains before, during, and after calcification to determine how relative community composition and inferred community traits change in relation to culture growth and calcification phase. Finally, to reach a better understanding of the potential of different microbiome constituents to produce biofilms and APs, we assessed these traits in twelve bacterial isolates from SP cultures.
Materials and methods
Symbiodiniacean strains, culturing conditions, and sampling
Experiments were conducted with 14 symbiodiniacean strains, 4 of which were represented by pairs of sister cultures, resulting in a total of 18 distinct cultures. Of these, 11 cultures had a SP phenotype and 7 a NP phenotype (Table 1). Cultures of 100 mL were grown in f/2 medium [28] at 26 °C and 130–150 µmol photons m−2 s−1 on a 12 h:12 h light/dark cycle and were monitored daily on an inverted microscope to identify the first appearance of symbiolites and other mineral deposits that did not produce endolithic cells [22]. All cultures, except 362, Mf, and 185, were sampled after 11–18 days, when SP cultures were in an actively calcifying phase. Cultures 362, Mf, and 185 were sampled in a pre-calcifying (day 10), actively calcifying (day 13), and post-calcifying (day 17–18) state (Supplementary Fig. S1). For this, whole cultures were harvested and pelleted at 15,557 g for 10 min at 4 °C. Pellets were transferred to pre-weighed 1.5-mL reaction tubes, centrifuged at 13,800 g for 10 min at 4 °C, weighed and stored at −20 °C.
Table 1.
Symbiolite-producing (SP) and non-symbiolite-producing (NP) Symbiodinium sp. and Breviolum sp. cultures.
| Culture | ITS2 | Species | Host | Location | [Reference] or GenBank accession no. |
|---|---|---|---|---|---|
| SP cultures | |||||
| 362 | A1 | Symbiodinium microadriaticum | Cassiopea andromeda | Gulf of Aquaba | [93] |
| 185 | A2 | Symbiodinium pilosum | Zoanthus sociatus | Jamaica | [93] |
| Mf | A2 | Symbiodinium sp. | Orbicella faveolata | Florida | [94] |
| 89 | A2 | Symbiodinium sp. | Gorgonia ventallina | Bermuda | [95] |
| 107 | A2 | Symbiodinium sp. | Heliopora sp. | Enewetak | [22] |
| 108 | A2 | Symbiodinium sp. | Heliopora sp. | Enewetak | [22] |
| 13 | B1 | Breviolum pseudominutum | Aiptasia tagetes | Bermuda | [96] |
| RT13 | B1 | Breviolum pseudominutum | Aiptasia tagetes | Bermuda | [96] |
| 64 | B1 | Breviolum minutum | Cassiopea xamachana | Jamaica | [96] |
| RT64 | B1 | Breviolum minutum | Cassiopea xamachana | Jamaica | [96] |
| RT146 | B1 | Breviolum pseudominutum | Oculina diffusa | Bermuda | [96] |
| NP cultures | |||||
| 379 | A4 | Symbiodinium linucheae | Plexaura homamala | Bahamas | [95] |
| Culture X | A12 | Symbiodinium sp. | Unknown | Aquarium tank | [97] |
| 146 | B1 | Breviolum pseudominutum | Oculina diffusa | Bermuda | [96] |
| 351 | B1 | Breviolum minutum | Pocillopora damicornis | Hawaii | [96] |
| RT351 | B1 | Breviolum minutum | Pocillopora damicornis | Hawaii | [96] |
| Aip24B | B1 | Breviolum sp. | Exaiptasia pallida | Aquarium tank | MN394860 |
| MACPC | B1 | Breviolum sp. | Unknown | Aquarium tank | MN394859 |
Culture names, Internal transcribed spacer 2 (ITS2) type designations, species names (if formally described), hosts of origin, geographic location of isolation, and reference for the molecular genetic identification of the strains. Underlined strains represent those used in time series experiments. Italicised strains had sister cultures included in this study (e.g., RT64 and 64).
DNA extraction, sequencing, and bioinformatic analysis
DNA was extracted using a PowerBiofilm DNA Isolation Kit (MoBio Laboratories, Carlsbad, CA, USA). Bacterial 16S rDNA hypervariable regions V5–V6–V7 were amplified using the 799F2 [29] and 1193R [30] primers. A single-indexed sequencing design with barcode on the forward primer was applied and libraries were sequenced on an Illumina MiSeq platform (MrDNA, Shallowater, TX, USA). Data were demultiplexed and quality filtered using default settings in QIIME [31]. Dereplication, sorting, and de novo operational taxonomic unit (OTU) clustering at 97% similarity was performed using VSEARCH [32]. Chimeras were filtered using the reference-based implementation of UCHIME [33] within VSEARCH against the SILVA [34] rDNA database (v132). QIIME was used to assign taxonomies to each OTU, also against the SILVA database (v132).
Composite metagenome predictions
The functional potential of microbiomes was predicted using the R [35] package Tax4Fun [36], and Piphillin [37] (Supplementary Fig. S2), and for the latter a 90% similarity threshold was used to match OTU sequences against the Kyoto Encyclopedia of Genes and Genomes (KEGG) organism database (May 2017 release).
Assessing culture growth dynamics and AP production in bacterial–algal biofilms
Cultures were grown in f/2 medium on 96-well plates for suspension cultures (Sarstedt, Nürnbrecht, Germany) and were monitored daily on an inverted microscope to identify the first appearance of symbiolites and other mineral deposits that did not produce endolithic cells (Supplementary information). Algal growth was assessed using optical density at 675 nm (A675) as a relative proxy for cell concentration [38]. Every 3 days, triplicate wells were sacrificed to quantify AP content, using an alcian blue staining [39] and measuring [40] protocol. A xanthan gum standard curve was produced (Supplementary Fig. S3) as in Claquin et al. [40] to express AP measurements as xanthan gum equivalents (µg Xeq).
Quantification of biofilm and AP formation by bacterial isolates
Eleven previously isolated strains of bacteria (nine belonging to the phylum Proteobacteria and two from Bacteroidetes) and one newly isolated strain of Labrenzia (Table 2) were assayed for their potential to produce APs and biofilms under static conditions. Bacterial isolates were pre-cultured in Marine Broth (Difco Laboratories, Detroit, MI, USA; 48 h at 25 °C and 160 RPM), inoculated on three 96-well plates (Sarstedt) at A600 of 0.05 and grown at 25 °C for 72 h. One plate (n = 7 wells per strain) was used to produce A600 growth curves to confirm that all strains reached stationary phase within 72 h (Supplementary Fig. S4). The second plate was used to assay biofilm formation (n = 6 wells per strain) using crystal violet [41], and the third to measure AP production (n = 6 wells per strain) using the protocols described above.
Table 2.
Bacterial isolates from symbiodiniacean cultures.
| Strain | Phylum | Order | Family | Species | Accession | Strain of origin (ITS2 type) |
|---|---|---|---|---|---|---|
| 362B | Proteobacteria | Alteromonadales | Alteromonadaceae | Marinobacter salsuginis | KP645213 | 362 (A1) |
| 203A | Proteobacteria | Alteromonadales | Alteromonadaceae | Marinobacter salsuginis | KP645210 | 203 (C2) |
| 97B | Proteobacteria | Alteromonadales | Alteromonadaceae | Marinobacter salsuginis | KP645206 | 97 (A2) |
| 370 | Proteobacteria | Alteromonadales | Alteromonadaceae | Marinobacter adhaerens | KP645214 | 370 (A1) |
| 154 | Proteobacteria | Alteromonadales | Alteromonadaceae | Marinobacter adhaerens | KP645209 | 154 (A2) |
| 97A | Bacteroidetes | Flavobacteriales | Flavobacteriaceae | Muricauda aquimarina | KP645205 | 97 (A2) |
| 362A | Bacteroidetes | Flavobacteriales | Flavobacteriaceae | Muricauda aquimarina | KP645212 | 362 (A1) |
| Sym1 | Proteobacteria | Oceanospirillales | Oceanospirillaceae | Neptumonas phycophila | KM591221 | 12 (B1) |
| Sym2 | Proteobacteria | Alteromonadales | Pseudoalteromonadaceae | Pseudoalteromonas atlantica | KP645203 | 12 (B1) |
| 74 | Proteobacteria | Alteromonadales | Alteromonadaceae | Alteromonas macleodii | KP645204 | 74 (B1) |
| 203B | Proteobacteria | Rhodospirillales | Rhodospirillaceae | Thalassospira permensis | KP645211 | 203 (C2) |
| RT13P1 | Proteobacteria | Rhodobacterales | Rhodobacteraceae | Labrenzia alexandrii | MN395029 | RT13 (B1) |
List of bacterial strains used in this study, including their taxonomic affiliation, GenBank accession numbers, and the Symbiodiniaceae strain of origin (ITS2 type). The Labrenzia alexandrii strain RT13P1 was isolated during the present study. All other strains were isolated previously from cultures in Frommlet et al. [20].
Statistical analyses
OTU tables were analysed using the R packages Phyloseq [42] and Vegan [43]. Alpha diversity indices were computed with and without rarefying to determine the effect of differing library sizes, and the Kruskal–Wallis rank sum test used to contrast SP vs. NP cultures. Dunn’s Kruskal–Wallis multiple comparisons tests (with Holms correction) were used to contrast pre, active, and post-calcifying phases in each of the three SP cultures sampled in series. As rarefying did not change the alpha diversity trends, the following analyses were performed on non-rarefied data [44].
Unconstrained ordination was used to explore differences in microbial communities and inferred metagenomes. Counts were converted to proportions and subsequently to a Bray–Curtis dissimilarity matrix (function distance in Phyloseq). Permutational multivariate analysis of variance (PERMANOVA; adonis function in Vegan) was performed to test for significant differences. Homogeneity of dispersions around group centroids produced from principle coordinates was assessed using the betadisper function (Vegan) and was visualised using principal coordinate analysis. For the comparison of SP vs. NP cultures, we tested for marginal effects of state (SP and NP) after controlling for the variability attributable to symbiodiniaceae genera (Symbiodinium vs. Breviolum, see Lawson et al. [45] for proposed genus-level differences in symbiodiniacean microbiomes) using the adonis2 function. For the series data, phase was first tested as a factor (across the whole data set), and second, permutations of a second test were constrained to phase as a nested factor (three levels) within individual cultures. Differential abundance of OTUs was tested using the package DESeq2 [46] with the Wald test and a parametric fit to correct for multiple comparisons and calculate log2 fold differences. DEseq2 was also applied to Piphillin outputs and differential abundance tests of the Tax4Fun output were analysed with multtest [47].
Relationships between AP content and chlorophyll a (in vivo A675) content of the bacterial–algal biofilms were examined with a linear model and goodness of fit parameters were assessed.
Raw data and scripts to reproduce these analyses are available in the open access repositories Figshare (10.6084/m9.figshare.9782399; 10.6084/m9.figshare.9782402; 10.6084/m9.figshare.9782405) and GitHub (https://github.com/nitschkematthew/Symbiodinium-bacteria-interactions), respectively.
Results
Taxonomic and functional diversity of SP vs. NP cultures
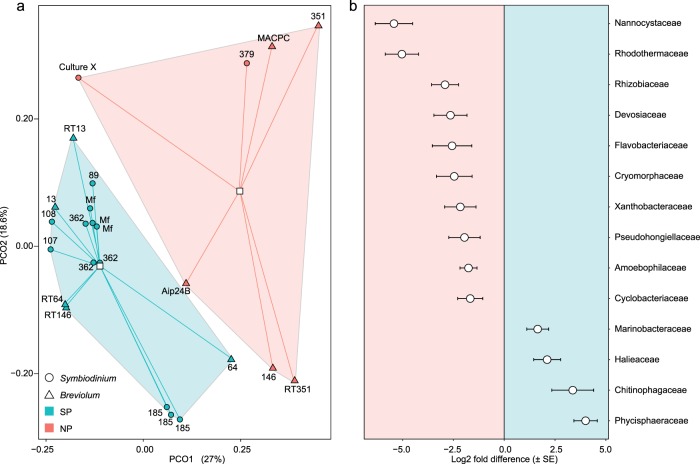
Illumina sequencing yielded 4.07 × 106 high quality 16S rRNA sequences, which clustered into 221 OTUs at 97% similarity. The microbiomes of SP and NP cultures were significantly dissimilar, with differences in dispersion around SP and NP centroids, and did not vary significantly between the genera Symbiodinium and Breviolum (Fig. 1a and Supplementary Table S1). Both OTU richness and the Inverse Simpson index were lower in SP than in NP cultures but only richness (138 ± 0.67 vs. 149 ± 3.91 OTUs, respectively) differed significantly (Supplementary Fig. S5 and Supplementary Table S2). This difference in richness was reflected in the SP-specific vs. the NP-specific core microbiomes, where only five OTUs were specific to the SP-core, while 35 OTUs were specific to the NP-core (Supplementary Table S3).
Fig. 1. Unconstrained ordination of OTUs and differential abundance of bacterial families in SP and NP cultures.
Principal coordinate analysis (a) of a Bray–Curtis dissimilarity matrix of all OTUs for SP (green symbols) and NP (red symbols) symbiodiniacean cultures belonging to the genera Symbiodinium (circles) and Breviolum (triangles). Centroids (white squares) are connected to points via lines. Significant Log2 fold differences (b) indicating which OTUs (summarised at the family taxonomic level) are of greater abundance in NP (negative Log2 values) and SP (positive Log2 values) cultures.
OTUs were assigned to four bacterial phyla (Supplementary Fig. S6). Proteobacteria were dominant (>50%) in all SP and most NP cultures. Bacteroidetes were significantly more abundant in NP cultures (30.67% vs. 11.94%), while Planctomycetes were significantly more abundant in SP cultures (14.05% vs. 4.37%), a trend driven by a single clade (SM1A02) in the family Phycisphaeraceae (order Phycisphaerales) (Fig. 1 and Supplementary Table S4). The only NP culture to contain Planctomycetes at abundances comparable to SP cultures was MACPC, but OTUs were mainly assigned to the order Planctomycetales. Actinobacteria occurred mostly at background-levels (0.01–0.47%), with notable abundances only in SP cultures RT146 (4.34%) and RT64 (6.87%).
SP and NP cultures differed significantly in the relative abundances of 14 families and 23 genera (Fig. 1b and Supplementary Table S4). The genus Labrenzia was highly abundant (Supplementary Fig. S7, 68% in SP culture 89) and constituted a study-wide core microbiome member (Supplementary Table S3). The genus Marinobacter, another core member of all cultures, was significantly more abundant in SP strains, whereas the NS3a marine group, Aquibacter, and Winogradskyella (Flavobacteriaceae), were significantly more abundant in NP strains. Of particular note are the sister cultures RT146 and 146 because despite being the same clonal Breviolum pseudominutum isolate, Winogradskyella was abundant in NP 146 (22.18%) but accounted for <0.01% in SP RT146. The only other culture containing Winogradskyella at a relatively high abundance was NP culture Aip24B (3.22%).
Composite metagenomes inferred using Tax4Fun and Piphillin resulted in 5644 and 4809 predicted KEGG orthologs (KOs) across 265 and 295 metabolic pathways, respectively, with Tax4Fun predicting a significant separation of SP vs. NP compared with Piphillin (Supplementary Fig. S8 and Supplementary Tables S5 and S6). Despite differences in prediction depth, both methods indicate that SP metagenomes were significantly enriched in KOs related to (1) biofilm formation, (2) bacterial chemotaxis, and (3) the phosphotransferase system (PTS), responsible for the uptake of carbohydrates such as glucose and mannose (Supplementary Fig. S8). SP metagenomes were also inferred as significantly enriched by Tax4Fun in siderophore biosynthesis and by Piphillin in flagellar assembly. Of 37 peptidase and 19 DNase orthologs inferred as differentially abundant, 30 and 14, respectively, were of greater abundance in NP cultures (Supplementary Fig. S9). Full Tax4Fun and Piphillin outputs are available as Supplementary Tables S7 and S8.
Microbiome succession in SP cultures
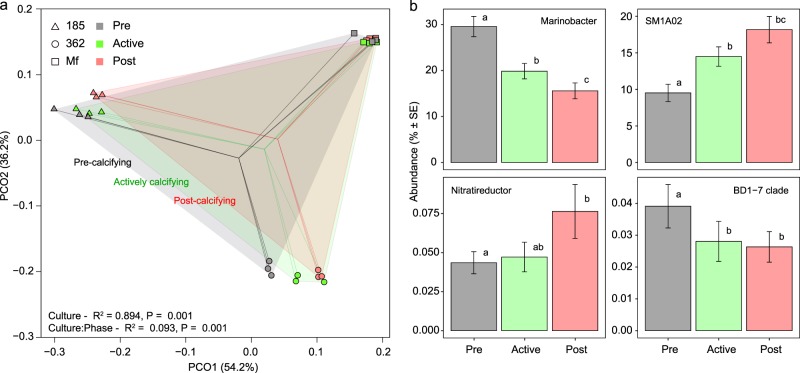
SP cultures 185, 362, and Mf showed progressive community shifts as they transitioned through pre, active, and post-calcifying phases (Fig. 2a and Supplementary Tables S9, S10). The test for study-wide differences in phase was not significant, likely due to the larger distances between culture microbiomes than within-culture phases (Supplementary Table S9). However, after constraining to within-culture permutations, the effect of phase was significant (Fig. 2a, Supplementary Table S9, F = 21.31, R2 = 0.09, P = 0.001). Succession was driven by changes in community evenness rather than richness (Supplementary Fig. S10) and four core taxa, Marinobacter, SM1A02, Nitratireductor, and BD1-7 clade, exhibited temporal changes independent of culture identity (Fig. 2b). Other changes were more strain-specific and involved less abundant OTUs (Supplementary Fig. S11). Composite metagenomes also reorganised significantly from phase to phase (Supplementary Fig. S12 and Supplementary Tables S11 and S12). A major trend, independent of inference method, was that actively calcifying and post-calcifying metagenomes were more similar to each other than to the pre-calcifying metagenomes.
Fig. 2. Succession of microbial communities as SP cultures age and calcify.
Principal coordinate analysis (a) of a Bray–Curtis dissimilarity matrix of all prokaryotic OTUs for SP strains 185 (triangles), 362 (circles), and Mf (squares) at the pre-calcifying stage (grey symbols), actively calcifying stage (green symbols), and post-calcifying stage (red symbols). The centroids of each of these stages, pooling each phase across strains (n = 9), are indicated by the convergence of matching coloured lines. Statistical outputs of the nested adonis (Supplementary Table S9) are indicated in the bottom left. OTUs summarised at the genus level (b) that exhibit temporal dynamics in all three SP strains (n = 9 at each time point).
AP accumulation in SP vs. NP cultures
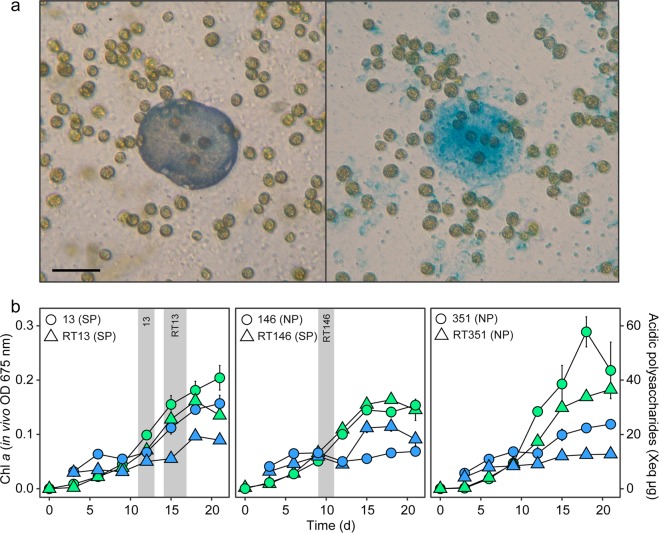
Most SP and NP cultures exhibited typical sigmoidal A675 batch culture growth curves, reaching their carrying capacities within 14–17 days (Fig. 3, Supplementary Fig. S13). Cultures 13, 108, 185, Mf, and Culture X exhibited strong positive linear relationships (R2 > 0.9) between A675 and AP. The remaining cultures had either weak linear (R2 < 0.5), or clear non-linear relationships due to late accumulation of APs (Supplementary Fig. S14 and linear model estimates in Supplementary Tables S13, S14). Furthermore, cultures 13, 64, RT64, 89, 108, RT146, 185, 146, and 351 showed bi-phasic AP accumulation, where after a first increase until day 7–9, AP-concentrations dropped slightly around day 9–13, before entering a second period of AP production (Fig. 3 and Supplementary Fig. S13). Typically, SP cultures became calcifying when A675 reached OD = 0.06–0.11 and AP 10.69–14.05 µg Xeq. The fast AP producer Mf calcified earliest, with all replicates producing symbiolites on day 9. All other SP cultures became calcifying between day 9–13, except RT13, which was delayed in both AP production and calcification compared with its sister strain (Fig. 3 and Supplementary Fig. S13). Symbiolites and APs co-localised in all tested cultures (Fig. 3 and Supplementary Fig. S15). Culture 146, despite near-identical A675 and AP values to its sister culture during the first phase, subsequently failed to accumulate comparable AP content and failed to calcify (Fig. 3). Besides, qualitative observations during microscopic monitoring of RT146 and 146 also revealed contrasting biofilm phenotypes, with thin-layered, surface-bound biofilms in the SP culture compared with loosely attached, three-dimensional aggregates in the NP culture (Supplementary Fig. S16).
Fig. 3. Co-localisation of APs and symbiolites and AP production during culture growth.
a A symbiolite (left) in SP culture 185 with endolithic cells, and the same view following alcian blue staining of acidic polysaccharides (right), showing clear co-localisation of AP patches with symbiolites. Symbiolite mineral phase is digested during staining. Scale bar = 40 µm. b Chlorophyll a content (left vertical axis, green symbols) of sister strains, measured as optical density (OD) at 675 nm, and acidic polysaccharide content (right vertical axis, blue symbols) of culture biofilms, presented as μg xanthan gum equivalents. Grey areas represent the time window of calcification onset, where at least two of three replicates had begun producing symbiolites. Values represent means (n = 3). Error bars represent SE.
Biofilm and AP production in bacterial isolates
Bacterial isolates exhibited clear differences in biofilm and AP formation (Fig. 4). Thalassospira permensis formed the least amount of biofilm of all strains (A588 = 0.46 ± 0.04). Marinobacter salsuginis and Pseudoalteromonas atlantica formed large quantities of biofilm (up to A588 = 1.83 ± 0.06) with high AP content (up to 58.49 ± 5.21 µg Xeq), whereas Labrenzia alexandrii produced large quantities of biofilm (A588 = 1.80 ± 0.05) but the lowest AP content of all strains at 4.34 ± 0.12 µg Xeq. Strains of the same species exhibited variability in biofilm formation and AP content. For example, M. salsuginis strains varied 2-fold in biofilm and 2.5-fold in AP production, and biofilms of Muricauda aquamarina strains 97A and 362A differed in AP content (36.389 ± 3.68 and 7.06 ± 1.24 µg Xeq, respectively).
Fig. 4. Biofilm formation and AP production by bacterial isolates.
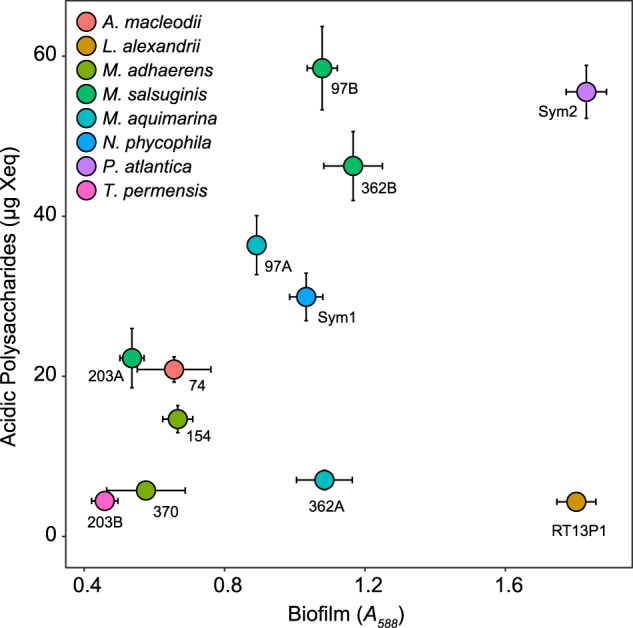
Biofilm formation, presented as absorbance of solubilized crystal violet at 588 nm (horizontal axis), and acidic polysaccharide content of biofilms, presented as μg xanthan gum equivalents (vertical axis), in 12 monoclonal bacterial isolates (symbols). Species are indicated by symbol colour and annotated according to strain. Values represent means (n = 6). Error bars represent SE.
Discussion
Symbiodiniacean microbiomes in culture
The studied microbiomes were composed of four common marine bacterial phyla [48, 49]. Proteobacteria and Bacteroidetes, the two most abundant phyla, are ubiquitous members of marine bacterioplankton that were found to dominate symbiodiniacean microbiomes before [45]. This earlier study identified Marinobacter, the purple sulfur bacteria family Chromatiaceae, and Labrenzia as core members of the symbiodiniacean microbiome; a combination of taxa known as biocathode MCL community [50]. We did not detect Chromatiaceae but Marinobacter and Labrenzia were study-wide core members, as were an additional 25 bacterial genera/clades, 8 of which had not been found previously in association with symbiodiniaceans [45] and 7 uncultured taxa, from 7 different families (Supplementary Table S3). In part, these differences can be due to the different genetic markers that were targeted. However, the fact that microbiomes of sister cultures of the same algal strain also differed significantly within our study indicates that symbiodiniaceans associate with a wide range of bacteria and that communities are likely to diverge from their ancestral state over time. Hence, the full range of bacteria that can associate with symbiodiniacean cultures and influence emergent properties such as symbiolite formation is probably not yet characterised.
Compositional and functional differences of SP vs. NP microbiomes
Our study revealed that SP and NP microbiomes differed significantly in the abundance of 23 bacterial genera, 14 families, and 2 phyla, providing first comprehensive evidence for the importance of distinct microbiome constituents in the formation of symbiolites.
At the phylum level, SP cultures contained an abundance of Planctomycetes, which are known for their association with particles and surfaces in aquatic environments and for their affiliation with micro- and macro-algae, which provide them with sulphated polysaccharides as carbon sources [51]. Planctomycetes have been shown to increase in abundance during blooms of diatoms, cyanobacteria, and dinoflagellates [52, 53] and they are commonly enriched in microbial mats, biofilms and calcifying microbial systems, including stromatolites [54, 55], ooids [56], and other forms of microbialites [57]. However, as discussed by Diaz et al. [56], natural calcifying communities are often too complex and metabolically redundant to allow for a clear deduction of functional contributions to mineral precipitation by specific taxonomic groups. Symbiodiniacean cultures provide a rare opportunity to overcome this limitation because their SP and NP phenotypes are linked to distinct differences in bacterial microbiome structure. Hence, the prevalence of Planctomycetes in SP cultures, represented by a single clade (SM1A02) within the family Phycisphaeraceae, suggests that the SM1A02 clade could have an actual function in producing symbiolites.
The SM1A02 clade is thought to comprise important degraders of particulate and high-molecular weight organic matter in permeable surface sediments [58, 59], which means that this clade could support symbiolite formation through its impact on the bacterial–algal biofilm structure. Furthermore, the fact that high permeability and high rates of remineralization are characteristic features of reef sediments [60]; a habitat that constitutes a hotspot for free-living symbiodiniaceans [61], combined with the knowledge that the resulting high primary productivity induces a calcification process in reef sediments that is analogous to symbiolite formation in vitro [15, 22], provide further clues for a connection between the SM1A02 clade and microbially induced calcification.
At the family level, the high-relative abundance of Chitinophagaceae (Bacteroidetes) and of Halieaceae and Marinobacteraceae (Gammaproteobacteria) in SP cultures suggests that these bacteria could also have a functional role in symbiolite formation. Functional features that could be relevant in this context and which are further discussed below include: (1) the capacity of chitinophagaceans to digest cellulose [62], a constituent of dinoflagellate cell walls [63], which points at a function similar to that of the Planctomycetes; (2) the capacity of halieaceans, such as the core microbiome clade OM60(NOR5), of aerobic photoheterotrophy [64, 65], similar to Labrenzia [66]; and (3) the propensity of marinobacteraceans to produce biofilms [67].
Members of the phylum Bacteroidetes, which were significantly more abundant in NP cultures, also commonly associate with phytoplankton cells and particulates of algal origin [68] and metabolise complex polymers [69]. Their genomes reflect this by featuring a general enrichment of proteases, glycosidases, and polysaccharide binding domains [49], which could explain the enrichment of peptidase and nuclease orthologs in NP metagenomes. The Bacteroidetes family with the highest differential abundance in NP cultures, the Rhodothermaceae, is not well enough studied to infer whether it represents these phylum-level features [70] and the Cryomorphaceae are not known to degrade complex organic matter [71]. However, the Flavobacteriaceae and the Cytophagales families Amoebophiliaceae and Cyclobacteriaceae are typical representatives of the Bacteroidetes and they share their potential to utilise macromolecules with the most differentially abundant family in NP cultures, the Nannocystaceae (deltaproteobacteria) [72].
The fact that the most differentially abundant taxa all degrade high-molecular weight polymers suggests that SP and NP phenotypes could result directly from the outcome of resource competition between these taxa and consequently, how the EPS matrix is modified and how it influences mineralisation [10, 73]. The surface-attached, thin-layered biofilms of RT146 and its SP phenotype vs. the loosely structured biofilms of its NP sister culture 146 could be a manifestation of such differences in EPS modification (Supplementary Fig. S16), in this case likely caused by the high-relative abundance of Winogradskyella (Flavobacteriaceae) in culture 146, as Winogradskyella sp. extracts are known to inhibit biofilm production [74].
Evidence for a strong potential of symbiodiniacean-associated bacterial communities to produce substrate for EPS digesters stems from (1) the presence of known biofilm producers such as Marinobacter [67], (2) the significant enrichment of orthologs related to biofilm formation in SP metagenome predictions, and (3) the high levels of biofilm formation by bacterial isolates from these communities. Considering the crucial role of EPS in mineralising microbial systems [10], this propensity to produce EPS must lay an important foundation for the formation of symbiolites [22].
Another feature of SP metagenomes was an enrichment in orthologs of the PTS, containing fructose, glucose, mannose, mannitol, and cellobiose components. This suggests that SP communities might be adapted to utilise simple sugars, the source of which could be symbiodiniaceans, as these microalgae are well-known for their potential to exude sugars and other small molecules when in hospite with cnidarian hosts [75]. Endozoicomonas, a genus commonly found within coral holobionts, have expanded PTS genes specific for cellobiose [76], a sugar component of algal cell walls, suggesting that specific sugar-transport systems may be diagnostic of a symbiodiniacean-associated niche. Beyond a direct role in carbon metabolism, the PTS system also regulates cellular functions, such as chemotaxis [77] and biofilm formation [78]. Mannitol release from algal mats, for example, is hypothesised to induce biofilm formation in Vibrio spp., where the sensing of mannitol by the PTS system induces a switch to biofilm synthesis at the transcriptional level [78]. This, combined with an abundance of biofilm synthesis orthologs, may thus predispose SP microbiomes to use the release of sugars from symbiodiniaceans as a cue for settlement and EPS production, ultimately facilitating calcification.
Microbiome successions in SP cultures
Temporal changes in SP microbiomes indicate that the community composition during the actively calcifying phase formed part of a continuous development from young, pre-calcifying to older, post-calcifying communities. Actively calcifying communities were more similar to post-calcifying than to pre-calcifying communities, which is consistent with earlier results that showed that symbiodiniacean cultures enter the calcifying phase during late stages of batch culture growth [22]. The significant decrease of Marinobacter and increase of SMA1A02 in all cultures over the course of the three calcification stages indicates that communities shifted from EPS production [67] to EPS consumption [58] and secondary production processes [71]. Furthermore, the decrease of the BD1-7 clade and the increase of the Nitratireductor over time, indicate a shift from organically poor pre-calcifying to organically rich post-calcifying cultures, as (1) BD1-7 and other Cellvibrionales are adapted to grow under oligotrophic conditions and many are mixotrophic, using light as an alternative energy source [65], and (2) Nitratireductor and other Phyllobacteriaceae are indicators of nutrient rich habitats [79]. The fact that study-wide core microbiome members such as Owenweeksia and Mf105b01, which were significantly more abundant in NP cultures, also increased over time in certain SP cultures, further supports the concept that the NP and SP phenotypes are not primarily the result of fundamentally different microbiomes but are rather related to relative differences in microbiome composition.
Importance of APs in symbiolite formation
The reversible metal-binding properties of APs play an important role in microbial calcification, as they either inhibit or promote mineral nucleation, depending on the EPS microstructure [12, 26, 73]. Earlier work on SP biofilms revealed a patchy AP distribution that co-localised with symbiolites [20]. The current study corroborated this observation in three additional strains (Supplementary Fig. S15), affirming that AP must play an important role in symbiodiniacean-induced calcification. The observed non-linear AP production dynamics (Supplementary Fig. S14) and the fact that AP patches did not match the comparatively uniform distribution of algal cells in the biofilm (Fig. 3 and Supplementary Fig. S15) further suggest that bacteria are the main AP producers in the studied bacterial–algal co-cultures [40]. Therefore, the patchy AP distribution could be due to (1) a patchiness of bacterial communities, either established stochastically, or through chemotactic community assembly processes, as SP metagenomes were enriched in chemotaxis and flagellar assembly orthologs, or (2) reflects functional dependencies between bacterial groups.
Remarkably, the size and shape of AP patches precisely matched that of the associated symbiolites. This suggests that either symbiolite size or shape is controlled by AP production, but without AP production being influenced by symbiolite formation, or alternatively, that the interaction is reciprocal, in that symbiolite formation itself or the driving forces underlying the process, create a microenvironment that is conducive to, or even forces bacteria to produce more AP. The latter mechanism could be linked to the hyperalkaline state that photosynthesis-induced carbonate precipitation is based on [14], as alkalitolerant bacteria have been shown to survive pH stress by buffering their local microenvironment via the production of EPS flocs that contain acidic functional groups [80]. Finally, the congruence of AP patches and symbiolites may be further preserved when AP become entombed between growth laminae, similar to organics within ooids [81, 82].
Quantitative measurements confirmed the importance of APs, as SP cultures had produced 10.69–14.05 µg Xeq when calcification occurred, whereas NP cultures generally remained below these values. Sister cultures RT146 and 146 exemplify this difference well, with the SP culture RT146 producing more APs than its NP sister culture, which is likely related to heterotrophic breakdown of APs by the high proportion of Bacteroidetes in the latter. However, symbiolite formation cannot simply be a function of AP concentrations either, as Culture X reached amongst the highest levels of APs yet failed to produce symbiolites. As the formation of non-motile, vegetative cells are thought to play a decisive role in establishing bacterial–algal biofilms that produce symbiolites [22], this NP phenotype, despite high levels of APs, could be related to the fact that motility patterns of symbiodiniacean strains vary [83]. Interestingly, Culture X occasionally produced small mineral deposits that did not incorporate cells, i.e., did not form functional symbiolites (Supplementary Fig. S17). This phenotype could potentially emerge from an excess of AP nucleation points relative to the amount of non-motile, vegetative cells and suggests that symbiolite formation probably requires a balance between photosynthesis-induced changes in Ω and the number of AP nucleation sites.
Bacterial isolates from SP communities clearly varied in their potential to produce biofilms and APs. Particularly noteworthy are Marinobacter and Labrenzia isolates, as these genera were among the most abundant OTUs across all cultures and isolates of both genera produced high amounts of biofilm, while only Marinobacter spp. produced significant quantities of APs. These genera may thus represent different roles in symbiolite formation: both producing an EPS scaffold, but only Marinobacter producing nucleation points for mineral precipitation. Nevertheless, the role of Marinobacter spp. in symbiolite formation is probably more complex, as not all strains of this genus produced equally large quantities of APs and because M. salsuginis is also known for its algicidal compounds [84]. Another well-known producer of EPS [85] and APs [86] is P. atlantica, which could explain that our strain of this bacterium was foremost in combining these two traits and that it effectively promoted symbiolite formation in an earlier study [20].
SP in vitro communities vs. calcifying communities in nature
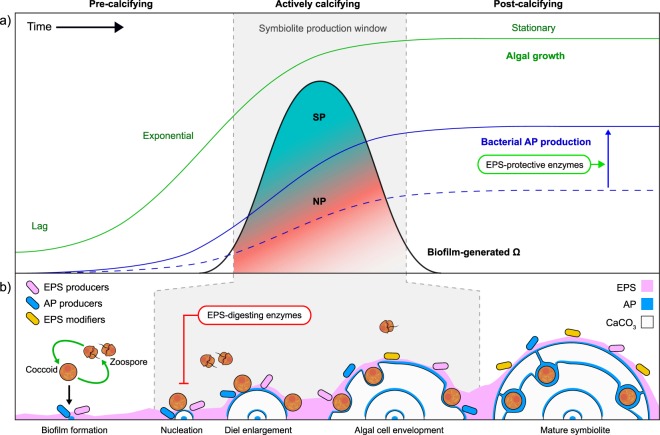
Microbially induced calcification is driven by the net-metabolic activity of a community on Ω [11]. Often, calcifying communities are organised as mats that are vertically structured into 5–7 functional groups [87, 88]. The studied SP communities harboured only three of these groups: phototrophs, aerobic heterotrophs, and anoxygenic phototrophs, the latter being represented by aerobic anoxygenic phototrophs (AAPs), not by the typical purple and green sulfur bacteria. The communities lacked obligate fermenters and obligate anaerobic heterotrophs like sulfate reducers/oxidisers. Thus, the role of fermenters in breaking down complex organics into substrates for SRBs and of the latter in inducing mineral precipitation by raising Ω [87] are evidently not required for the formation of symbiolites in vitro. Aerobic heterotrophs and AAPs are also not likely to contribute to the metabolic induction of symbiolite formation, as heterotrophy decreases Ω. Hence, our study consolidates the view that symbiolite formation is induced by symbiodiniacean photosynthesis and that bacteria support symbiolite formation through their production and modification of EPS components (Fig. 5) [20, 22]. To what extent AAPs and other bacteria that were common in both SP and NP cultures contribute to the production of symbiolite-promoting EPS components cannot be answered yet but the high level of biofilm production by isolate RT13P1 hints at a strong potential of AAPs in producing EPS under oligotrophic conditions.
Fig. 5. Symbiolite formation: a model for the formation of microbialites by calcifying bacterial–algal communities.
a Symbiodiniacean growth, biofilm-generated CaCO3 saturation state (Ω) and bacterial AP production create a window for the production of symbiolites. Symbiodiniacean batch cultures transition through lag, exponential, and stationary phase (green line). As growth slows during the mid-to-late exponential phase, photosynthetic activity shifts to the biofilm (see below), raising the biofilm-generated Ω (black line). During late exponential/early stationary phase, if Ω is sufficiently high and suitable amounts of bacterial APs (solid blue line) are present, a window of opportunity for symbiolite production (SP) arises (grey area) [22]. A lack of APs (blue dashed line), either through low AP production or AP degradation by EPS-digesting bacteria, or a mismatch in timing between biofilm-generated Ω and AP accumulation, may lead to a non-symbiolite producing phenotype (NP, indicated by the changing colour gradient). b Schematic of key steps involved in symbiolite formation. Non-motile (coccoid) algal cells diurnally produce motile daughter cells (zoospores) which again transition into non-motile cells before further division [92]. As cultures age, cell division rates decrease and the proportion of cells that remain in the coccoid stage during the day phase increases. These photosynthetically active, coccoid cells establish a bacterial–algal biofilm, together with EPS/AP-producing and EPS-digesting/modifying bacteria. CaCO3 nucleation marks the beginning of the symbiolite production window, followed by diel enlargement (seen as daily growth rings [20]) and the envelopment of algae as endolithic cells, at which point the spherulitic aragonite deposits are termed symbiolites. Mature, post-calcifying symbiolites (typically with several growth rings containing endolithic cells) signal a shift in the metabolic state of the culture, with diminishing biofilm-generated Ω and a shift towards bacterial groups indicative of a nutrient/organic compound rich media, including EPS-digesting bacteria. Through the manipulation of bacterial communities of NP and SP cultures and through the use of e.g., EPS-digesting enzymes (red box) or EPS-protective enzymes (green box), symbiolite formation provide a unique opportunity to further untangle the role of bacteria in photosynthesis-induced microbial calcification.
Also noteworthy is that the close spatial proximity of photoautotrophic and heterotrophic microorganisms in the thin SP biofilms, which can prevent mineral precipitation in natural systems [11], does evidently not prevent symbiolite formation. The fact that SP communities lack obligate fermenters and SOBs, two functional groups that decrease Ω, could constitute an important community feature to achieve net calcification. Whether analogues of SP communities in nature would reflect this low functional complexity awaits testing in the field. However, independent of this open question, SP and NP cultures in themselves provide a unique in vitro model to study how single clones of eukaryotic phototrophs and bacterial communities of low functional complexity achieve microbially induced calcification. To further elucidate the role of biofilm modification in symbiolite formation, SP cultures could be treated with exogenous peptidases or DNAses [89], or NP cultures could be treated with protease or DNAse inhibitors [90] to potentially invert their phenotypes (Fig. 5), and the metal-binding properties of biofilms could be quantified through the use of mass spectrometry [91]. Also, we envisage that through the generation of axenic symbiodiniacean cultures and improved bacterial culturing techniques this model system can reach a state where bacterial strains can be added in a modular fashion to create calcifying and non-calcifying communities of varying complexity.
Supplementary information
Acknowledgements
This work is a contribution to the project Symbiolite (PTDC/MAR-EST/3726/2014—POCI-01-0145-FEDER-016748), funded by FEDER, through COMPETE2020—Programa Operacional Competitividade e Internacionalização (POCI), and by national funds (OE), through FCT/MCTES. Funding was also provided by CESAM (UID/AMB/50017/2019), FCT/MCTES through national funds. MRN was supported by a postdoctoral grant (BPD/CESAM/PTDC/MAREST/3726/2014) and JS, CF by research assistant grants (BI/CESAM/PTDC/MAREST/3726/2014) of the Symbiolite project. CB was supported by a FCT-funded doctoral fellowship (SFRH/BD/119270/2016). JCF was supported by a FCT-funded postdoctoral fellowship (SFRH/BPD/111685/2015) and by national funds (OE), through FCT—Fundação para a Ciência e a Tecnologia, I.P., in the scope of the framework contract foreseen in the numbers 4, 5 and 6 of the article 23, of the Decree-Law 57/2016, of August 29, changed by Law 57/2017, of July 19.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version of this article (10.1038/s41396-020-0629-z) contains supplementary material, which is available to authorized users.
References
- 1.Cooper MB, Smith AG. Exploring mutualistic interactions between microalgae and bacteria in the omics age. Curr Opin Plant Biol. 2015;26:147–53. doi: 10.1016/j.pbi.2015.07.003. [DOI] [PubMed] [Google Scholar]
- 2.Not Fabrice, Probert Ian, Gerikas Ribeiro Catherine, Crenn Klervi, Guillou Laure, Jeanthon Christian, Vaulot Daniel. The Marine Microbiome. Cham: Springer International Publishing; 2016. Photosymbiosis in Marine Pelagic Environments; pp. 305–332. [Google Scholar]
- 3.Zhou J, Lyu Y, Richlen ML, Anderson DM, Cai Z. Quorum sensing is a language of chemical signals and plays an ecological role in algal-bacterial interactions. Crit Rev Plant Sci. 2016;35:81–105. doi: 10.1080/07352689.2016.1172461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Krohn-Molt I, Wemheuer B, Alawi M, Poehlein A, Güllert S, Schmeisser C, et al. Metagenome survey of a multispecies and alga-associated biofilm revealed key elements of bacterial-algal interactions in photobioreactors. Appl Environ Microbiol. 2013;79:6196–206. doi: 10.1128/AEM.01641-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hays SG, Patrick WG, Ziesack M, Oxman N, Silver PA. Better together: engineering and application of microbial symbioses. Curr Opin Biotechnol. 2015;36:40–49. doi: 10.1016/j.copbio.2015.08.008. [DOI] [PubMed] [Google Scholar]
- 6.Flemming H-C, Wingender J, Szewzyk U, Steinberg P, Rice SA, Kjelleberg S. Biofilms: an emergent form of bacterial life. Nat Rev Microbiol. 2016;14:563. doi: 10.1038/nrmicro.2016.94. [DOI] [PubMed] [Google Scholar]
- 7.Riding R. Microbial carbonates: the geological record of calcified bacterial–algal mats and biofilms. Sedimentology. 2000;47:179–214. [Google Scholar]
- 8.Arp G, Reimer A, Reitner J. Photosynthesis-induced biofilm calcification and calcium concentrations in Phanerozoic oceans. Science. 2001;292:1701–4. doi: 10.1126/science.1057204. [DOI] [PubMed] [Google Scholar]
- 9.Riding R. Cyanobacterial calcification, carbon dioxide concentrating mechanisms, and Proterozoic–Cambrian changes in atmospheric composition. Geobiology. 2006;4:299–316. [Google Scholar]
- 10.Dupraz C, Reid RP, Braissant O, Decho AW, Norman RS, Visscher PT. Processes of carbonate precipitation in modern microbial mats. Earth-Sci Rev. 2009;96:141–62. [Google Scholar]
- 11.Visscher PT, Stolz JF. Microbial mats as bioreactors: populations, processes, and products. Palaeogeogr Palaeocl. 2005;219:87–100. [Google Scholar]
- 12.Braissant O, Decho AW, Dupraz C, Glunk C, Przekop KM, Visscher PT. Exopolymeric substances of sulfate-reducing bacteria: interactions with calcium at alkaline pH and implication for formation of carbonate minerals. Geobiology. 2007;5:401–11. [Google Scholar]
- 13.Decho AW. Overview of biopolymer-induced mineralization: what goes on in biofilms? Ecol Eng. 2010;36:137–44. [Google Scholar]
- 14.Shiraishi F. Chemical conditions favoring photosynthesis-induced CaCO3 precipitation and implications for microbial carbonate formation in the ancient ocean. Geochim Cosmochim Acta. 2012;77:157–74. [Google Scholar]
- 15.Werner U, Blazejak A, Bird P, Eickert G, Schoon R, Abed RMM, et al. Microbial photosynthesis in coral reef sediments (Heron Reef, Australia) Estuar Coast Shelf Sci. 2008;76:876–88. [Google Scholar]
- 16.Heil CA, Chaston K, Jones A, Bird P, Longstaff B, Costanzo S, et al. Benthic microalgae in coral reef sediments of the southern Great Barrier Reef, Australia. Coral Reefs. 2004;23:336–43. [Google Scholar]
- 17.Yamashita H, Koike K. Genetic identity of free-living Symbiodinium obtained over a broad latitudinal range in the Japanese coast. Phycol Res. 2013;61:68–80. [Google Scholar]
- 18.Trench R. Microalgal–invertebrate symbioses-a review. Endocytobiosis Cell Res. 1993;9:135–75. [Google Scholar]
- 19.Coffroth MA, Lewis CF, Santos SR, Weaver JL. Environmental populations of symbiotic dinoflagellates in the genus Symbiodinium can initiate symbioses with reef cnidarians. Curr Biol. 2006;16:R985–R987. doi: 10.1016/j.cub.2006.10.049. [DOI] [PubMed] [Google Scholar]
- 20.Frommlet JC, Sousa ML, Alves A, Vieira SI, Suggett DJ, Serôdio J. Coral symbiotic algae calcify ex hospite in partnership with bacteria. Proc Natl Acad Sci USA. 2015;112:6158–63. doi: 10.1073/pnas.1420991112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marlow J, Peckmann J, Orphan V. Autoendoliths: a distinct type of rock-hosted microbial life. Geobiology. 2015;13:303–7. doi: 10.1111/gbi.12131. [DOI] [PubMed] [Google Scholar]
- 22.Frommlet JC, Wangpraseurt D, Sousa ML, Guimarães B, Medeiros da Silva M, Kühl M, et al. Symbiodinium-induced formation of microbialites: mechanistic insights from in vitro experiments and the prospect of its occurrence in nature. Front Microbiol. 2018;9:998. doi: 10.3389/fmicb.2018.00998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nitschke MR, Davy SK, Ward S. Horizontal transmission of Symbiodinium cells between adult and juvenile corals is aided by benthic sediment. Coral Reefs. 2016;35:335–44. [Google Scholar]
- 24.Kawaguchi T, Decho AW. A laboratory investigation of cyanobacterial extracellular polymeric secretions (EPS) in influencing CaCO3 polymorphism. J Cryst Growth. 2002;240:230–5. [Google Scholar]
- 25.Arp G, Helms G, Karlinska K, Schumann G, Reimer A, Reitner J, et al. Photosynthesis versus exopolymer degradation in the formation of microbialites on the atoll of Kiritimati, Republic of Kiribati, Central Pacific. Geomicrobiol J. 2012;29:29–65. [Google Scholar]
- 26.More T, Yadav J, Yan S, Tyagi R, Surampalli R. Extracellular polymeric substances of bacteria and their potential environmental applications. J Environ Manag. 2014;144:1–25. doi: 10.1016/j.jenvman.2014.05.010. [DOI] [PubMed] [Google Scholar]
- 27.Shiraishi F, Bissett A, de Beer D, Reimer A, Arp G. Photosynthesis, respiration and exopolymer calcium-binding in biofilm calcification (Westerhöfer and Deinschwanger Creek, Germany) Geomicrobiol J. 2008;25:83–94. [Google Scholar]
- 28.Guillard Robert R. L. Culture of Marine Invertebrate Animals. Boston, MA: Springer US; 1975. Culture of Phytoplankton for Feeding Marine Invertebrates; pp. 29–60. [Google Scholar]
- 29.Bulgarelli D, Rott M, Schlaeppi K, van Themaat EVL, Ahmadinejad N, Assenza F, et al. Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature. 2012;488:91. doi: 10.1038/nature11336. [DOI] [PubMed] [Google Scholar]
- 30.Bodenhausen N, Horton MW, Bergelson J. Bacterial communities associated with the leaves and the roots of Arabidopsis thaliana. PLoS ONE. 2013;8:e56329. doi: 10.1371/journal.pone.0056329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high–throughput community sequencing data. Nat Methods. 2010;7:335–6. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rognes T, Flouri T, Nichols B, Quince C, Mahé F. VSEARCH: a versatile open source tool for metagenomics. PeerJ. 2016;4:e2584. doi: 10.7717/peerj.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2012;41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.R Core Team . R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2019. [Google Scholar]
- 36.Aßhauer KP, Wemheuer B, Daniel R, Meinicke P. Tax4Fun: predicting functional profiles from metagenomic 16S rRNA data. Bioinformatics. 2015;31:2882–4. doi: 10.1093/bioinformatics/btv287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Iwai S, Weinmaier T, Schmidt BL, Albertson DG, Poloso NJ, Dabbagh K, et al. Piphillin: improved prediction of metagenomic content by direct inference from human microbiomes. PLoS ONE. 2016;11:e0166104. doi: 10.1371/journal.pone.0166104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roesler CS, Barnard AH. Optical proxy for phytoplankton biomass in the absence of photophysiology: Rethinking the absorption line height. Methods Oceanogr. 2013;7:79–94. [Google Scholar]
- 39.Jain R, Raghukumar S, Tharanathan R, Bhosle N. Extracellular polysaccharide production by thraustochytrid protists. Mar Biotechnol. 2005;7:184–92. doi: 10.1007/s10126-004-4025-x. [DOI] [PubMed] [Google Scholar]
- 40.Claquin P, Probert I, Lefebvre S, Veron B. Effects of temperature on photosynthetic parameters and TEP production in eight species of marine microalgae. Aquat Microb Ecol. 2008;51:1–11. [Google Scholar]
- 41.Merritt JH, Kadouri DE, O’Toole GA. Growing and analyzing static biofilms. Curr Protoc Microbiol. 2011;22:1B. 1.1–1B. 1.18. doi: 10.1002/9780471729259.mc01b01s00. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McMurdie PJ, Holmes S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE. 2013;8:e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Oksanen J, Kindt R, Legendre P, O’Hara B, Stevens MHH, Oksanen MJ, et al. The vegan package. Community Ecol Package. 2007;10:631–7. [Google Scholar]
- 44.McMurdie PJ, Holmes S. Waste not, want not: why rarefying microbiome data is inadmissible. PLoS Comput Biol. 2014;10:e1003531. doi: 10.1371/journal.pcbi.1003531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lawson CA, Raina JB, Kahlke T, Seymour JR, Suggett DJ. Defining the core microbiome of the symbiotic dinoflagellate, Symbiodinium. Environ Microbiol Rep. 2018;10:7–11. doi: 10.1111/1758-2229.12599. [DOI] [PubMed] [Google Scholar]
- 46.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pollard K. S., Dudoit S., van der Laan M. J. Bioinformatics and Computational Biology Solutions Using R and Bioconductor. New York, NY: Springer New York; 2005. Multiple Testing Procedures: the multtest Package and Applications to Genomics; pp. 249–271. [Google Scholar]
- 48.DeLong EF, Franks DG, Alldredge AL. Phylogenetic diversity of aggregate-attached vs. free-living marine bacterial assemblages. Limnol Oceanogr. 1993;38:924–34. [Google Scholar]
- 49.Yilmaz P, Yarza P, Rapp JZ, Glöckner FO. Expanding the world of marine bacterial and archaeal clades. Front Microbiol. 2016;6:1524. doi: 10.3389/fmicb.2015.01524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Malanoski AP, Lin B, Eddie BJ, Wang Z, Hervey WJ, IV, Glaven SM. Relative abundance of ‘Candidatus Tenderia electrophaga’ is linked to cathodic current in an aerobic biocathode community. Microb Biotechnol. 2018;11:98–111. doi: 10.1111/1751-7915.12757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wegner C-E, Richter-Heitmann T, Klindworth A, Klockow C, Richter M, Achstetter T, et al. Expression of sulfatases in Rhodopirellula baltica and the diversity of sulfatases in the genus Rhodopirellula. Mar Genom. 2013;9:51–61. doi: 10.1016/j.margen.2012.12.001. [DOI] [PubMed] [Google Scholar]
- 52.Cai H-Y, Yan Z-S, Wang A-J, Krumholz LR, Jiang H-L. Analysis of the attached microbial community on mucilaginous cyanobacterial aggregates in the eutrophic Lake Taihu reveals the importance of Planctomycetes. Microb Ecol. 2013;66:73–83. doi: 10.1007/s00248-013-0224-1. [DOI] [PubMed] [Google Scholar]
- 53.Pizzetti I, Fuchs BM, Gerdts G, Wichels A, Wiltshire KH, Amann R. Temporal variability of coastal Planctomycetes clades at station Kabeltonne, North Sea. Appl Environ Microbiol. 2011;77:5009–17. doi: 10.1128/AEM.02931-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Havemann SA, Foster JS. Comparative characterization of the microbial diversities of an artificial microbialite model and a natural stromatolite. Appl Environ Microbiol. 2008;74:7410–21. doi: 10.1128/AEM.01710-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Proemse BC, Eberhard RS, Sharples C, Bowman JP, Richards K, Comfort M, et al. Stromatolites on the rise in peat-bound karstic wetlands. Sci Rep. 2017;7:15384. doi: 10.1038/s41598-017-15507-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Diaz MR, Piggot AM, Eberli GP, Klaus JS. Bacterial community of oolitic carbonate sediments of the Bahamas Archipelago. Mar Ecol Prog Ser. 2013;485:9–24. [Google Scholar]
- 57.Saghaï A, Zivanovic Y, Zeyen N, Moreira D, Benzerara K, Deschamps P, et al. Metagenome-based diversity analyses suggest a significant contribution of non-cyanobacterial lineages to carbonate precipitation in modern microbialites. Front Microbiol. 2015;6:797. doi: 10.3389/fmicb.2015.00797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Probandt D, Knittel K, Tegetmeyer H, Ahmerkamp S, Holtappels M, Amann R. Permeability shapes bacterial communities in sublittoral surface sediments. Environ Microbiol. 2017;19:1584–99. doi: 10.1111/1462-2920.13676. [DOI] [PubMed] [Google Scholar]
- 59.Probandt D, Eickhorst T, Ellrott A, Amann R, Knittel K. Microbial life on a sand grain: from bulk sediment to single grains. ISME J. 2018;12:623. doi: 10.1038/ismej.2017.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rasheed M, Wild C, Franke U, Huettel M. Benthic photosynthesis and oxygen consumption in permeable carbonate sediments at Heron Island, Great Barrier Reef, Australia. Estuar Coast Shelf Sci. 2004;59:139–50. [Google Scholar]
- 61.Takabayashi M, Adams LM, Pochon X, Gates RD. Genetic diversity of free-living Symbiodinium in surface water and sediment of Hawai’i and Florida. Coral Reefs. 2012;31:157–67. [Google Scholar]
- 62.Rosenberg E. The family Chitinophagaceae. In: Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson F (eds). The Prokaryotes. Springer: Berlin, Heidelberg, 2014. p. 493–5.
- 63.Levin RA, Suggett DJ, Nitschke MR, Oppen MJ, Steinberg PD. Expanding the Symbiodinium (Dinophyceae, Suessiales) toolkit through protoplast technology. J Eukaryot Microbiol. 2017;64:588–97. doi: 10.1111/jeu.12393. [DOI] [PubMed] [Google Scholar]
- 64.Yan S, Fuchs BM, Lenk S, Harder J, Wulf J, Jiao N-Z, et al. Biogeography and phylogeny of the NOR5/OM60 clade of Gammaproteobacteria. Syst Appl Microbiol. 2009;32:124–39. doi: 10.1016/j.syapm.2008.12.001. [DOI] [PubMed] [Google Scholar]
- 65.Spring S, Scheuner C, Goeker M, Klenk H-P. A taxonomic framework for emerging groups of ecologically important marine gammaproteobacteria based on the reconstruction of evolutionary relationships using genome-scale data. Front Microbiol. 2015;6:281. doi: 10.3389/fmicb.2015.00281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Biebl H, Pukall R, Lünsdorf H, Schulz S, Allgaier M, Tindall BJ, et al. Description of Labrenzia alexandrii gen. nov., sp. nov., a novel alphaproteobacterium containing bacteriochlorophyll a, and a proposal for reclassification of Stappia aggregata as Labrenzia aggregata comb. nov., of Stappia marina as Labrenzia marina comb. nov. and of Stappia alba as Labrenzia alba comb. nov., and emended descriptions of the genera Pannonibacter, Stappia and Roseibium, and of the species Roseibium denhamense and Roseibium hamelinense. Int J Syst Evol Microbiol. 2007;57:1095–107. doi: 10.1099/ijs.0.64821-0. [DOI] [PubMed] [Google Scholar]
- 67.Klein B, Grossi V, Bouriat P, Goulas P, Grimaud R. Cytoplasmic wax ester accumulation during biofilm-driven substrate assimilation at the alkane–water interface by Marinobacter hydrocarbonoclasticus SP17. Res Microbiol. 2008;159:137–44. doi: 10.1016/j.resmic.2007.11.013. [DOI] [PubMed] [Google Scholar]
- 68.Grossart HP, Levold F, Allgaier M, Simon M, Brinkhoff T. Marine diatom species harbour distinct bacterial communities. Environ Microbiol. 2005;7:860–73. doi: 10.1111/j.1462-2920.2005.00759.x. [DOI] [PubMed] [Google Scholar]
- 69.González JM, Fernández-Gómez B, Fernàndez-Guerra A, Gómez-Consarnau L, Sánchez O, Coll-Lladó M, et al. Genome analysis of the proteorhodopsin-containing marine bacterium Polaribacter sp. MED152 (Flavobacteria) Proc Natl Acad Sci USA. 2008;105:8724–9. doi: 10.1073/pnas.0712027105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Park Sanghwa, Akira Yokota, Kogure Kazuhiro. The Prokaryotes. Berlin, Heidelberg: Springer Berlin Heidelberg; 2014. The Family Rhodothermaceae; pp. 849–856. [Google Scholar]
- 71.Bowman JP. The family Cryomorphaceae. In: Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson F (eds). The Prokaryotes. Springer: Berlin, Heidelberg, 2014. p. 539–50.
- 72.Reichenbach H. Nannocystaceae fam. nov. In: Whitman WB, Rainey F, Kämpfer P, Trujillo M, Chun J, DeVos P, Hedlund B, and Dedysh S (eds). Bergey’s Manual of Systematics of Archaea and Bacteria. John Wiley & Sons: Hoboken, New Jersey, 2015. p. 1–2.
- 73.Flemming H-C, Wingender J. The biofilm matrix. Nat Rev Microbiol. 2010;8:623. doi: 10.1038/nrmicro2415. [DOI] [PubMed] [Google Scholar]
- 74.Dash S, Nogata Y, Zhou X, Zhang Y, Xu Y, Guo X, et al. Poly-ethers from Winogradskyella poriferorum: antifouling potential, time-course study of production and natural abundance. Bioresour Technol. 2011;102:7532–7. doi: 10.1016/j.biortech.2011.05.034. [DOI] [PubMed] [Google Scholar]
- 75.Yellowlees D, Rees TAV, Leggat W. Metabolic interactions between algal symbionts and invertebrate hosts. Plant Cell Environ. 2008;31:679–94. doi: 10.1111/j.1365-3040.2008.01802.x. [DOI] [PubMed] [Google Scholar]
- 76.Neave MJ, Michell CT, Apprill A, Voolstra CR. Endozoicomonas genomes reveal functional adaptation and plasticity in bacterial strains symbiotically associated with diverse marine hosts. Sci Rep. 2017;7:40579. doi: 10.1038/srep40579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lux R, Jahreis K, Bettenbrock K, Parkinson JS, Lengeler JW. Coupling the phosphotransferase system and the methyl-accepting chemotaxis protein-dependent chemotaxis signaling pathways of Escherichia coli. Proc Natl Acad Sci USA. 1995;92:11583–7. doi: 10.1073/pnas.92.25.11583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ymele-Leki P, Houot L, Watnick PI. Mannitol and the mannitol-specific enzyme IIB subunit activate Vibrio cholerae biofilm formation. Appl Environ Microbiol. 2013;79:4675–83. doi: 10.1128/AEM.01184-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Willems Anne. The Prokaryotes. Berlin, Heidelberg: Springer Berlin Heidelberg; 2014. The Family Phyllobacteriaceae; pp. 355–418. [Google Scholar]
- 80.Charles C, Rout SP, Patel K, Akbar S, Laws AP, Jackson B, et al. Floc formation reduces the pH stress experienced by microorganisms living in alkaline environments. Appl Environ Microbiol. 2017;83:e02985–16. doi: 10.1128/AEM.02985-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Edgcomb V, Bernhard J, Beaudoin D, Pruss S, Welander P, Schubotz F, et al. Molecular indicators of microbial diversity in oolitic sands of Highborne Cay, Bahamas. Geobiology. 2013;11:234–51. doi: 10.1111/gbi.12029. [DOI] [PubMed] [Google Scholar]
- 82.O’Reilly S, Mariotti G, Winter A, Newman S, Matys E, McDermott F, et al. Molecular biosignatures reveal common benthic microbial sources of organic matter in ooids and grapestones from Pigeon Cay, The Bahamas. Geobiology. 2017;15:112–30. doi: 10.1111/gbi.12196. [DOI] [PubMed] [Google Scholar]
- 83.Jeong HJ, Lee SY, Kang NS, Yoo YD, Lim AS, Lee MJ, et al. Genetics and morphology characterize the Dinoflagellate Symbiodinium voratum, n. sp., (Dinophyceae) as the sole representative of Symbiodinium Clade E. J Eukaryot Microbiol. 2014;61:75–94. doi: 10.1111/jeu.12088. [DOI] [PubMed] [Google Scholar]
- 84.Keawtawee T, Fukami K, Songsangjinda P, Muangyao P. Isolation and characterization of Noctiluca-killing bacteria from a shrimp aquaculture pond in Thailand. Fish Sci. 2011;77:657–64. [Google Scholar]
- 85.Holmström C, Kjelleberg S. Marine Pseudoalteromonas species are associated with higher organisms and produce biologically active extracellular agents. FEMS Microbiol Ecol. 1999;30:285–93. doi: 10.1111/j.1574-6941.1999.tb00656.x. [DOI] [PubMed] [Google Scholar]
- 86.Uhlinger DJ, White DC. Relationship between physiological status and formation of extracellular polysaccharide glycocalyx in Pseudomonas atlantica. Appl Environ Microbiol. 1983;45:64–70. doi: 10.1128/aem.45.1.64-70.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Dupraz C, Visscher PT. Microbial lithification in marine stromatolites and hypersaline mats. Trends Microbiol. 2005;13:429–38. doi: 10.1016/j.tim.2005.07.008. [DOI] [PubMed] [Google Scholar]
- 88.Spadafora A, Perri E, McKenzie JA, Vasconcelos C. Microbial biomineralization processes forming modern Ca:Mg carbonate stromatolites. Sedimentology. 2010;57:27–40. [Google Scholar]
- 89.Beenken KE, Spencer H, Griffin LM, Smeltzer MS. Impact of extracellular nuclease production on the biofilm phenotype of Staphylococcus aureus under in vitro and in vivo conditions. Infect Immun. 2012;80:1634–8. doi: 10.1128/IAI.06134-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tseng BS, Reichhardt C, Merrihew GE, Araujo-Hernandez SA, Harrison JJ, MacCoss MJ, et al. A biofilm matrix-associated protease inhibitor protects Pseudomonas aeruginosa from proteolytic attack. mBio. 2018;9:e00543–00518. doi: 10.1128/mBio.00543-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Aguilera A, Souza-Egipsy V, San Martín-Úriz P, Amils R. Extracellular matrix assembly in extreme acidic eukaryotic biofilms and their possible implications in heavy metal adsorption. Aquat Toxicol. 2008;88:257–66. doi: 10.1016/j.aquatox.2008.04.014. [DOI] [PubMed] [Google Scholar]
- 92.Fitt W, Trench R. The relation of diel patterns of cell division to diel patterns of motility in the symbiotic dinoflagellate Symbiodinium microadriaticum Freudenthal in culture. New Phytol. 1983;94:421–32. [Google Scholar]
- 93.Lee SY, Jeong HJ, Kang NS, Jang TY, Jang SH, Lajeunesse TC. Symbiodinium tridacnidorum sp. nov., a dinoflagellate common to Indo-Pacific giant clams, and a revised morphological description of Symbiodinium microadriaticum Freudenthal, emended Trench & Blank. Eur J Phycol. 2015;50:155–72. [Google Scholar]
- 94.Steinke M, Brading P, Kerrison P, Warner ME, Suggett DJ. Concentrations of dimethylsulfoniopropionate and dimethyl sulfide are strain-specific in symbiotic dinoflagellates (Symbiodinium sp., Dinophyceae) J Phycol. 2011;47:775–83. doi: 10.1111/j.1529-8817.2011.01011.x. [DOI] [PubMed] [Google Scholar]
- 95.LaJeunesse TC. Investigating the biodiversity, ecology, and phylogeny of endosymbiotic dinoflagellates in the genus Symbiodinium using the its region: In search of a “species” level marker. J Phycol. 2001;37:866–80. [Google Scholar]
- 96.Parkinson JE, Coffroth MA, LaJeunesse TC. New species of Clade B Symbiodinium (Dinophyceae) from the greater Caribbean belong to different functional guilds: S. aenigmaticum sp. nov., S. antillogorgium sp. nov., S. endomadracis sp. nov., and S. pseudominutum sp. nov. J Phycol. 2015;51:850–8. doi: 10.1111/jpy.12340. [DOI] [PubMed] [Google Scholar]
- 97.LaJeunesse TC, Lambert G, Andersen RA, Coffroth MA, Galbraith DW. Symbiodinium (Pyrrhophyta) genome sizes (DNA content) are smallest among dinoflagellates. J Phycol. 2005;41:880–6. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.