Abstract
Understanding when and why new species are recruited into microbial communities is a formidable problem with implications for managing microbial systems, for instance by helping us better understand whether a probiotic or pathogen would be expected to colonize a human microbiome. Much theory in microbial temporal dynamics is focused on how phylogenetic relationships between microbes impact the order in which those microbes are recruited; for example, species that are closely related may competitively exclude each other. However, several recent human microbiome studies have observed closely related bacteria being recruited into microbial communities in short succession, suggesting that microbial community assembly is historically contingent, but competitive exclusion of close relatives may not be important. To address this, we developed a mathematical model that describes the order in which new species are detected in microbial communities over time within a phylogenetic framework. We use our model to test three hypothetical assembly modes: underdispersion (species recruitment is more likely if a close relative was previously detected), overdispersion (recruitment is more likely if a close relative has not been previously detected), and the neutral model (recruitment likelihood is not related to phylogenetic relationships among species). We applied our model to longitudinal human microbiome data, and found that for the individuals we analyzed, the human microbiome generally follows the underdispersion (i.e., nepotism) hypothesis. Exceptions were oral communities and the fecal communities of two infants that had undergone heavy antibiotic treatment. None of the datasets we analyzed showed statistically significant phylogenetic overdispersion.
Subject terms: Microbial ecology, Molecular ecology, Microbiome
Introduction
A central question in microbial community assembly, especially in the human microbiome, is when and why microbes are recruited into communities. If there are patterns or general rules for which taxa have higher probabilities of recruitment, these rules can guide habitat restoration projects, help us better understand whether probiotics or pathogens will colonize, and better exploit disturbance as a tool for managing microbial systems related to human health and disease. Recruitment of new species can be studied by evaluating the order in which species are empirically detected in time-series experiments, given data such as which species have already been detected or what changes occur in an environment over time [1, 2]. Although a changing environment clearly selects for new species, it has also been shown that microbial community structure is often historically contingent on previous states of that community [1–5]. This reflects not only that microbial communities are temporally autocorrelated (gradual change over time), but also that the recruitment of a given species is a function of which species in the community are already present or have modified the local environment. Such historically contingent patterns have mainly been observed and tested within a phylogenetic context, because amplicon data naturally lend themselves to the creation of phylogenies, and because phylogenies have been shown to be predictive of genomic (and perhaps niche) overlap in human-associated microbiota [6, 7] and in general [8, 9].
Within this phylogenetic framework, a predominant hypothesis has been that closely related microbes inhibit each other’s successful recruitment [1, 3, 4]. The proposed mechanism for this hypothesis is that closely related microbes likely have similar niches (phylogenetic niche conservatism [8–10]), and species already established within a community will occupy their niches to the exclusion of ecologically similar organisms. This is also the basis of Darwin’s naturalization hypothesis [11], which proposed that new species are less likely to be recruited if a close relative is present [12]. Indeed, this assembly mode has been found to be the case in artificial nectar microcosms, where phylogenetically similar yeast species had similar nutrient requirements, and inhibited each others’ colonization [13]. If competitive exclusion of closely related species is the predominant mode of microbial community assembly in the human microbiome, close relatives would be less likely to be recruited into the same community, as compared with more distant relatives. In this paper, we refer to assembly where distant relatives are more likely to be recruited into a community than close relatives as the overdispersion hypothesis, since it predicts the preferential addition of novel phylogenetic diversity to a community (i.e., phylogenetic overdispersion).
Overdispersion is far from universal, and multiple studies have shown that extremely close relatives can coexist within the human microbiome [14–16], and may even be preferentially recruited [17]. This is consistent with simulations showing that clusters of closely related species can persist despite strong within-cluster competition, when immigration rate is high [18]. Indeed, Darwin’s preadaptation hypothesis predicts that species with a close relative present in a community will be preferentially recruited, because they are likely to already be adapted to the new environment [11]. Also, more recent ecological theory posits competition between distantly related species may result in phylogenetically clustered communities, although it may be more appropriate to include this type of competition within selection [19]. Per the empirical observations and reasoning above, we might expect that new close relatives are more likely to be detected than new distant relatives, so the amount of new phylogenetic diversity added to a community is minimized (phylogenetic underdispersion). Thus, for empirical data our underdispersion hypothesis is that species are more likely to be detected when they have a close relative that was previously detected, predicting preferential addition of minimal novel phylogenetic diversity (phylogenetic underdispersion). The over- and underdispersion hypotheses are alternatives to the null hypothesis that recruitment is independent of phylogenetic relatedness among species. Since the null hypothesis is species neutral (and phylogenetically neutral), we refer to it as the neutral hypothesis.
It should be noted that our use of the terms “overdispersion” and “underdispersion” are slightly different in this paper compared with use of the same terms elsewhere. In many cases, these words refer to the state of a community at a single timepoint or sample, with overdispersion indicating more diversity in that sample than expected by chance, and underdispersion indicating less [20]. Instead, our use of over- and underdispersion refers to the amount of newly added diversity over time. In our overdispersion hypothesis, phylogenetically novel species are preferentially added to communities, meaning more new diversity is added than expected by chance. Under our underdispersion hypothesis, the reverse is true. Following this, our question concerns the order in which new species are recruited in a time series, rather than community composition of any given sample. Furthermore, while we are interested in the biological phenomenon of species recruitment, the empirical result of recruitment is detection. Evidence of recruitment is a lack of detection, and then subsequent detection of a species via high-throughput DNA sequencing data. It is possible for a species to have been recruited into a community but not be detected, although this source of experimental error diminishes as sequencing depth increases. Also, the extent to which a species has actually been recruited into a community is questionable, if it is sufficiently rare that it is not detected in an Illumina sequencing run with tens of thousands of reads per sample (e.g., [21]). Still, we have taken care to use the term “recruitment” when discussing the biological phenomenon under investigation, and “detection” when describing empirical data and results.
Here, we use the phylogenetic relationships among species within a time series to test the extent to which our over- or underdispersion hypotheses hold true. Instead of analyzing broad patterns of community change via beta-diversity statistics (e.g., UniFrac [22]) or analyzing patterns of select clades within the community (e.g., PhyloFactor [23], Edge PCA [24]), we model the probability of new species’ recruitment into a community for the first time as a monotonic function of their phylogenetic distances to members of the community that have already been recruited.
The model we present here can be used to estimate the degree to which the recruitment of new species is more or less likely when a close relative has been previously recruited. We fit our model to several time-series human microbiome datasets [21, 25, 26] to compare the strength of under- or overdispersion between subjects, sample sites, or time periods. We found that for the datasets we analyzed (36 individuals across three studies), the human microbiome generally follows the underdispersion hypothesis. There were exceptions where this pattern was not significantly different than the neutral model, but none of the longitudinal datasets we analyzed showed statistically significant overdispersion.
Materials and methods
Overview
With our model, our goal is to estimate the extent to which recruitment of new species over time is related to the new species’ phylogenetic similarity to (or distance from) species that were already recruited at previous timepoints. Our Statistical model describes the probabilities of detecting new species over time. We use our model with empirical data via Simulations, where we resample the empirically detected species using our model with known parameter values, to produce surrogate datasets. Specifically, we fix and record the model’s dispersion parameter (D), which determines the extent to which species with a close relative are preferentially added to the surrogate community (or, conversely, if species without a close relative are preferred). Our Parameter estimation compares the empirical pattern of species detection to that of the surrogate datasets (which have known D values), in order to determine which value of D best describes the empirical data. Hypothesis testing is done by comparing empirical data to repeated simulations under the neutral model, which is D = 0. We describe the bioinformatic and technical details of this process in our Analysis section. Code and a tutorial for our model is available at https://github.com/darcyj/pd_model.
Statistical model
At any point in time, a community is composed of many species, and other species are not present but are available to be added (“species pool”). Our model parameterizes the probability of detecting species in a local community for the first time, based on their phylogenetic distances from species that have already been detected. In a species-neutral model of community assembly, each species i in the species pool has the same probability of detection at time t, irrespective of how different it is from species that have already been detected. Thus, the neutral model for first-time species detections is a random draw without replacement of species from the species pool. We extend the species-neutral model by modeling the probability pit of species i being detected for the first time at time t as,
| 1 |
where dit is the phylogenetic distance from species i to its closest relative that has already been detected prior to timepoint t, and D is a dispersion parameter.
When D = 0, our model functions as a neutral model; all species have the same probability of being detected for the first time, since pit is the same for every species. When D < 0, pit decreases with dit meaning that species from the species pool have higher probabilities of detection when they are more closely related to species that have already been detected in the local community (underdispersion; phylogenetically constrained). When D > 0, the opposite is true (overdispersion; phylogenetically divergent). Our hypothesis testing and parameter estimation focus on the dispersion parameter, D.
Simulations
Our analysis of a dataset relies on reconstructing that dataset via simulation of our statistical model using known values of , allowing for hypothesis testing and parameter estimation (we refer to the empirical dispersion parameter as D, and use to refer to surrogate values used in simulations). Using the empirical data as a starting point, we simulate many surrogate datasets with values ranging from (underdispersed) to (neutral) to (overdispersed). This is done so that the empirical data can later be compared with the surrogate datasets, to estimate the empirical value of D.
We start each surrogate dataset with the same species present in the first sample in the time series of its corresponding empirical dataset. Then, surrogate datasets are constructed forward in time by randomly drawing rt new species from the species pool, where the probabilities of detecting those species are given by Eq. (1), and rt is the number of new species detected in the empirical dataset from times t − 1 to t. The number of new species detected from the empirical dataset is used so that species richness is kept constant between the empirical dataset and all surrogate datasets. The species pool is updated to exclude those species drawn at previous timepoints, and the newly sampled species are recorded. Surrogate datasets are produced for many different values, ranging from underdispersed to overdispersed models. We performed 500 simulations (as described above) for each dataset analyzed.
Parameter estimation
Our main goal is to estimate the empirical dispersion parameter D (Eq. (1)), which quantifies the degree to which first-time species detections are phylogenetically underdispersed (D < 0), neutral (D = 0), or overdispersed (D > 0), corresponding to our hypotheses. To this end, we use Faith’s phylodiversity [27] to compare each of the 500 surrogate datasets (described above) to the empirical dataset. Phylodiversity is the sum of branch lengths on a phylogenetic tree for a set of species, so phylodiversity of a set of highly related species is low (phylogenetically constrained) because there are no long branch lengths in the tree, but phylodiversity is higher (phylogenetically divergent) for a set of more distantly related species [27]. If D ≠ 0, then species are preferentially added if they have relatively low (D < 0) or relatively high (D > 0) phylogenetic distance to the resident community (dit, Eq. (1)), yielding accumulations of total phylodiversity that are relatively slow (D < 0) or relatively fast (D > 0) compared with the neutral model (Fig. 1a). In other words, at any timepoint t, the phylogenetic diversity of species that have already been observed is PDt, and the extent to which PDt accelerates or decelerates over a sampling effort depends on D. Because of this, we can estimate D by comparing the empirical phylodiversity curve to our surrogate phylodiversity curves, which have known values.
Fig. 1. Phylodiversity accumulation and model fitting in the female feces dataset [25].
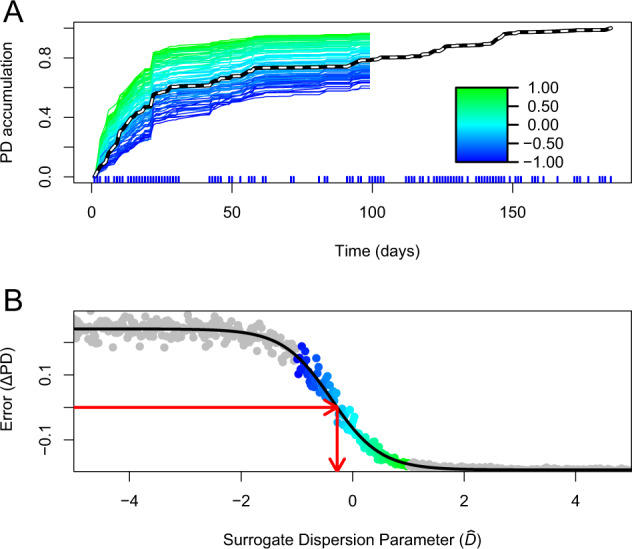
a Empirical (dashed) and surrogate phylodiversity accumulation curves. Surrogate curves are colored according to value (Eq. (1)). New species that have a previously detected close relative contribute little phylodiversity and cause slow phylodiversity accumulation (blue). New species that do not have a close relative contribute more phylodiversity and cause faster accumulation (green). The empirical model (dashed) is below the neutral model (teal), signifying underdispersion in the order of first-time species detections. The times of sampling points are shown as vertical blue lines below the X-axis. Curves are rescaled from 0 to 1 in this figure. b How empirical and surrogate data are compared with generate an estimate for D. Differences between empirical and surrogate data at time m are shown on the Y-axis, and the values used to generate surrogate datasets are shown on the X-axis. Color-coded points correspond to surrogate datasets are shown in a. Values shown in gray result from using extreme values of , which help the logistic error model (black line) fit to the data, and are not shown in a. The red arrows show the process of error minimization, yielding a D estimate. A figure showing significance testing for these data is available as Fig. S1.
For the comparison of an empirical phylodiversity accumulation curve to curves for corresponding surrogate datasets, we evaluate the amount of phylodiversity PDm accumulated at time index m, midpoint between the first and final samples. Time m is used because this leaves many species yet to be observed in the species pool, so that there can be variability in surrogate datasets. Multiple time indices are not used to compare surrogate and empirical datasets because each value is a function of all values . PDm values are calculated for all surrogate datasets, and a PDm value is calculated for the empirical dataset. The difference between the empirical PDm and PDm simulated with is , which is the error between surrogate and empirical data. We then estimate the empirical value of D by minimizing (Fig. 1b). This minimization is performed using a logistic error model,
| 2 |
where a and b are the upper and lower horizontal asymptotes, and r and i are rate and inflection parameters for the logistic model. is modeled with a logistic function because there is a maximum and minimum observable value as a function of the phylogeny; this is because there are strict minimum and maximum limits to the amount of phylodiversity obtainable by observing n species, where n is the total species richness accumulated up to time m. The two horizontal asymptotes of the logistic model are easily fit to these extremes (Fig. 1b). Once fit, the error model is solved for ΔPD = 0, giving an estimate for the empirical D. Confidence intervals for this estimate are obtained via bootstrapping our error model.
Hypothesis testing
For this test, our null hypothesis is the neutral model, where D = 0, since this model represents the absence of the effect we are testing. We test this null hypothesis competitively by simulating 1000 surrogate datasets at D = 0 (Fig. S1a) to generate a null PDm distribution. The empirical PDm is compared with this distribution (Fig. S1b), and if the empirical PDm is below the 2.5% quantile or above the 97.5% quantile, we reject the null (i.e., neutral) hypothesis. Evidence of either overdispersion (D > 0) or underdispersion (D < 0) allows us to reject. A second method of simulating communities under D = 0 was used as well, where simulated communities were drawn without replacement from the pool of all individual observations within a rarefied dataset. In this context, an “observation” is a datum within a species-by-sample matrix of count data; a given column contains d observations, where d is the sequencing depth. This “individual null” accounts for differences in total species relative abundance across a time series, while the simulation above only considers presence–absence of species. Results of this model were computed the same as above.
Analysis
This section is a summary of our data analysis. Detailed methods for this section are available as Supplementary information.
We ran our model on data from 36 individuals from three data sources. Two individuals were from Caporaso et al. [25], 33 were from Yassour et al. [21], and one was from Koenig et al. [26]. In all cases, data were downloaded and processed using the unoise3 pipeline [28], which clusters sequence data into exact sequence variants called zOTUs. The Koenig et al. infant gut dataset was split into two datasets, one for samples collected before the subject began consuming baby formula, and one after. Our model was run on these data as described above, resulting in D estimates for the before and after formula datasets.
The “moving pictures” [25] data were split into eight datasets, one for each combination of subject (n = 2) and body site (feces, right and left palms, tongue), and our model was run on each of these datasets. Analyses of these data were also done using two approaches that allowed us to test the importance of the set of species that are included in the species pool. One alternate approach analyzed communities in a “meta” context, where the species pool for a given palm was composed of all four palms in the whole dataset. If we were to estimate similar D values for both the “meta” and “self” analyses, the inclusion of extra species in the species pool would be of little importance to the model. The other alternate approach analyzed data using a sliding-window approach, wherein our model was run separately on multiple overlapping windows of five consecutive days within the same dataset, in order to see how D varied over time.
Finnish infant sequence data from Yassour et al. [21] were split into datasets for each of 33 individuals, and our model was run for each. Estimated D values were compared between subjects that had been treated with oral antibiotics (n = 18) and subjects that had not (n = 15) using a Mann–Whitney test. Because this data source had so many subjects, we used these data to test whether the number of zOTUs, total phylodiversity, or number of timepoints had an effect on D estimates via correlation analysis.
Results
By varying , we successfully changed the rate at which phylodiversity is added to surrogate (i.e., resampled) microbial communities over time (Fig. 1a). Compared with the neutral model where , higher values result in phylodiversity accumulating quickly, since in the overdispersed model, species that contribute more phylodiversity are preferentially sampled. Conversely, lower values result in phylodiversity accumulating slowly, since in the underdispersed model, species that contribute less phylodiversity (since they are very similar to species that are already present) are preferentially sampled. These results show that the D parameter in our model successfully corresponds to over- and underdispersion relative to the neutral model. Our error model also fits well to the differences between empirical and surrogate datasets (, Fig. 1b). Each error model fit was visually inspected for goodness of fit, to be sure that D estimates were not spurious. All datasets passed this inspection.
Results from “moving pictures” data
All time series from adult feces and palm microbiomes [25] showed significant phylogenetic underdispersion of first-time zOTU detections (Fig. 2). This means that when a zOTU was detected for the first time in one of these communities, it was more likely to be phylogenetically similar to a zOTU that had previously been detected in community. For both the male and female subject, D estimates were lower (more underdispersed) in the feces than in the palms, left and right palm D estimates were similar to each other, and tongue D estimates were higher. All sites except the tongue showed statistically significant underdispersion in both subjects, while tongue data were not significantly different than the neutral model. The results for the “individual null” were the same. In the comparison between “meta” and “self” models, “meta” models needed to be much more underdispersed than “self” in order to approximate empirical phylogenetic diversity accumulation (Fig. S2). We also observed a general upward trend in D in our sliding-window analysis of the male right palm dataset (Fig. S3), although this trend was only observed over 19 days.
Fig. 2. Dispersion parameter (D) estimates for “moving pictures” [25] datasets.
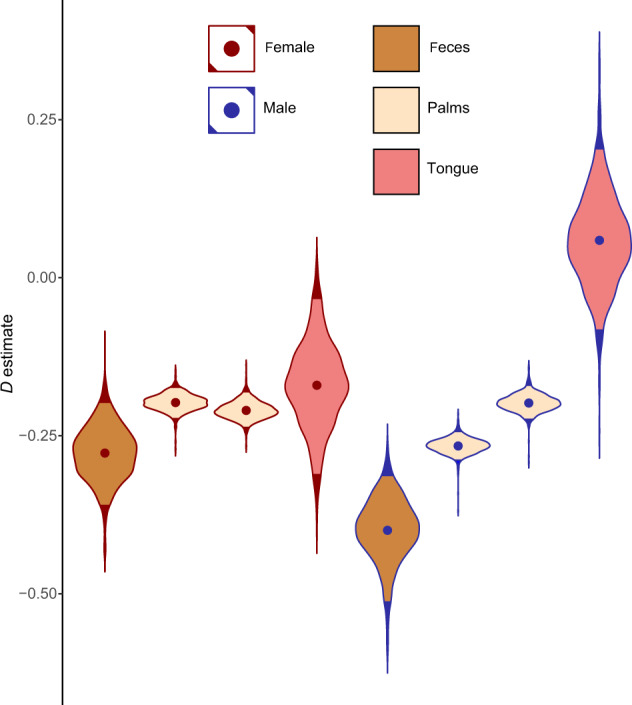
The subject’s sex is shown as the outline color of each violin, and the body site is shown as fill color. The four body sites for the female subject are shown at left, and the four body sites for the male subject are shown at right. Each violin shows the distribution of D estimates given by logistic error model bootstraps, and the dots within violins are means. Light-colored portions of violins represent 95% of bootstraps. The two subjects analyzed show parallel D estimates, with feces being the lowest, followed by palms which are all similar, followed by tongue communities. For both subjects, tongue patterns were not significantly different than the neutral model.
Results from infant gut data
Empirical phylodiversity accumulation in the infant gut microbiome [26] showed a sharp increase in phylodiversity after day 161 (Fig. 3), the same date that the subject began consuming baby formula. This suggests that baby formula changed the phylogenetic colonization patterns of the developing infant gut. We analyzed this dataset as two separate time series, one before formula use and one during, and both had negative D estimates, with the preformula D estimate being lower (Fig. 4). While the preformula dataset was significantly underdispersed (P = 0.007), the formula dataset was not significantly different from the neutral model, although this result is marginal (P = 0.107). When the “individual null” was used instead, both were significantly underdispersed. Infant gut data from Finnish infants [21] were sampled at a much lower temporal resolution, and as such were not split between formula use. Out of 33 individuals, 31 analyzed exhibited significant underdispersion, and the other two were not significantly different from the neutral model. Both nonsignificant individuals were from the group treated with heavy antibiotics, but even so, no significant difference in D values was detected between antibiotics and control groups (Fig. S4). However, when the “individual null” was used, all subjects exhibited significant underdispersion. Estimates of D did not significantly correlate with the number of zOTUs in a dataset, the total phylodiversity of the dataset, the initial phylodiversity of the dataset, or the number of samples in a dataset (Fig. S5).
Fig. 3. Empirical phylodiversity accumulation in the infant gut microbiome [26].
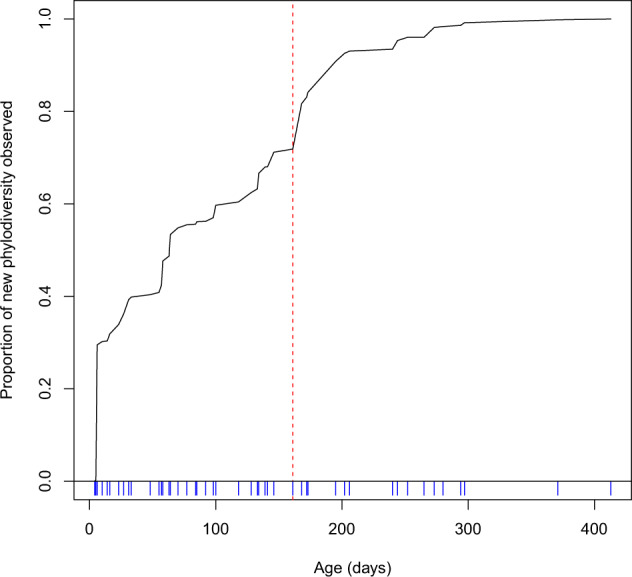
Phylodiversity increases sharply after day 161 of the infant’s life, then plateaus. This timing coincides with the day the subject began consuming baby formula. The times of sampling points are shown as vertical lines below the X-axis.
Fig. 4. Dispersion parameter (D) estimates in the infant gut, preformula, and during formula use.
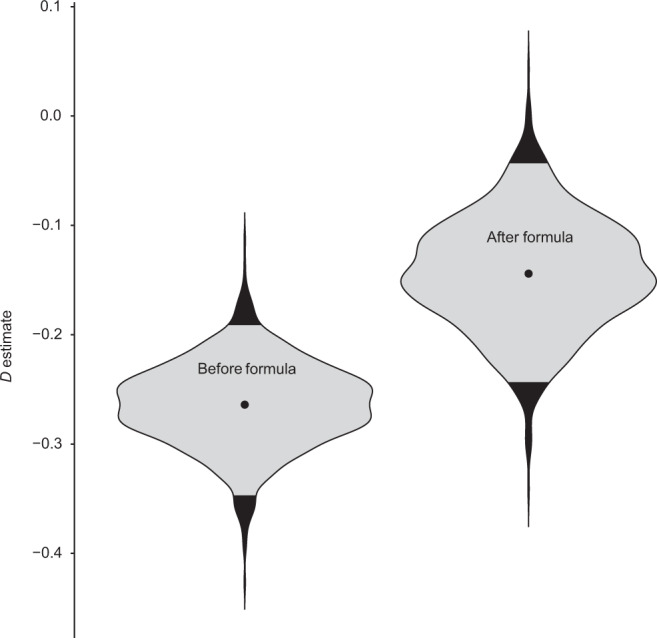
Formula use began on day 161, thus the first 160 days of the subject’s life were analyzed separately. Community assembly was significantly underdispersed in the preformula dataset, but was not significantly different from the neutral model during formula use (P = 0.107).
Discussion
Any organism of interest in a human microbiome dataset, from the pathogenic to the probiotic, will at some point be recruited for the first time, and the order in which these organisms are detected in the community is determined by community assembly processes [1]. Predicting which lineages of organisms can be recruited into a given environment has far-reaching implications for ecosystem remediation and management, especially in microbial communities, where the medical and ecological importances of many microbes are still largely unknown [29, 30]. Identifying conditions under which assembly mechanisms change, or under which nonneutral assembly is particular strong, may facilitate microbial community rehabilitation by understanding when and how microbial communities can be colonized by close/distant relatives. We found that assembly during primary succession of the infant gut (Figs. 4, S4) and during turnover of the microbial communities on the adult palms and gut (Fig. 2) follows a predictable pattern: new species are more likely to be recruited if a close relative has been recruited previously.
The generally “nepotistic” pattern we observed supports our underdispersion hypothesis, which follows Darwin’s preadaptation hypothesis [11] and more recent ecological theory as well [18, 19]. Much work in phylogenetic community ecology posits that competition tends to be strongest among closely related species due to phylogenetic niche conservatism [8], so many closely related species are able to coexist in a community, competition must not be an important factor structuring that community [20]. However, strong competition between distantly related species may actually cause groups of phylogenetically similar species to coexist, especially when immigration is high [18, 19, 31]. This type of competition is perhaps better conceptualized as selection (i.e., environmental filtering) instead [19], especially since studies showing evidence for competitive exclusion in microbial communities focus on competition between closely related species [2, 13].
Our model investigates the extent to which newly recruited species are likely to be similar to previously recruited close relatives, but “previously recruited” may include a significant time span. Thus, the observation of underdispersion may not reflect a lack of importance of competition between close relatives per se. However, asking whether new species recruitment is likely after recruitment of a close relative has relevance; for instance in human microbiome systems it may be beneficial to understand if a pathogen’s probability of recruitment may be higher if a conspecific strain was previously observed [14, 16]. Approaches that consider only recent community membership may more directly inform hypotheses regarding direct competition between close relatives, or regarding more recent recruitment of close relatives. For this reason, we included a sliding-window analysis of 5-day intervals for a subset of intensively sampled data, and showed significant underdispersion in a majority of windows analyzed (Fig. S3). This type of analysis can satisfy the issue of recency when using our model, but only when data collection is sufficiently frequent.
Regardless, nonneutral patterns of phylogenetic community structure have been interpreted to mean that traits are under ecological selection [20, 32–34]. If traits are not driving community assembly [35] or if the traits driving community assembly are largely horizontally transferred between taxa independent of their relatedness (as estimated by a 16S rDNA phylogeny), we would expect no phylogenetic signature, and a D estimate that is not significantly different from 0 (the neutral model). Instead, we observed a very strong and significant phylogenetic signal in species recruitment order for almost all datasets we analyzed. However, if selection on traits is driving this pattern, selection itself may not occur within the host environment. An alternative explanation for the underdispersion we observed is that selection is external to the host environment (i.e., selection occurs within the neighboring species pool from which emigration occurs), causing change in the community entering the host to already be underdispersed. Similarly, phylogenetic dispersion of community structure has been unable to distinguish between selection and differences in migration rates [36], so a preunderdispersed community entering the host is a plausible mechanism for phylogenetic underdispersion of species recruitment. But, selection of microbial communities within the host has been shown by multiple studies [37–39], so it is our opinion that selection within the host is a more likely scenario.
A similar point is that species compositions of these datasets do not reflect the actual species pool from which they originate, which is likely not static over time. Our model draws species from the pool of all species observed over the course of a time series, and with that model we ask whether the order in which those species were recruited is consistent with under- or overdispersion hypotheses. It is for this reason that we discuss our model in the context of the order of recruitment, regardless of whether selection took place in the host or was external as described above. A null model that accounts for a dynamic species pool that is external to the host could be used with the type of analysis we present here, in order to understand if the processes underlying over-/underdispersion take place in the host or in the host’s environment. Our “meta vs. self” comparison (Fig. S2) uses the combined habitat-specific microbiomes of two co-habitating individuals as a broader species pool, and we found even stronger underdispersion when empirical “self” patterns were compared with the broader “meta” species pool, suggesting strong selection within the host environment. Still, this broader species pool is not a substitute for a thorough inventory of the real species pool, which would be required to ascertain whether over-/underdispersion originated in the host or elsewhere. As such, future studies may wish to intensively sample a subject’s environment in order to better catalog the species pool, and perhaps use qPCR to obtain environmental concentrations of species within the species pool [40].
As to why no datasets analyzed showed significant phylogenetic overdispersion (D > 0), we are not certain. At the beginning of development of this model, we expected microbial communities in the human microbiome to follow the overdispersion hypothesis, partly from microbiome studies suggesting competition among closely related bacteria is an important factor in human gut microbial community assembly [1, 41, 42], and also because of work in experimental microcosms [13]. However, the human microbiome environments analyzed here are environments that undergo constant physical disturbance, unlike aqueous microcosms. Palm communities are physically disturbed with every use of the hands, and by the sampling procedure itself. Gut (fecal) communities are also disturbed constantly by the movement of feces through the gut. It may be possible that continuous disturbance allows for underdispersion via constant reassembly of communities. In this case, niches may be filled by random “winners” after each disturbance, as in a competitive lottery scenario [4]. These “winners” would still need to be preadapted to their environment, so they would be more likely to be closely related to previous “winners”, as in our findings. Similarly, environments with fluctuating resource profiles may result in clusters of organisms occupying the same niche [43]. The datasets we used are also somewhat limited in terms of phylogenetic resolution, as short reads of the 16S marker gene are insufficient to detect strain-level variation [15, 42, 44]. Thus, competitive exclusion could occur at the extreme tips of the bacterial phylogenetic tree, and this would not be detectable using 16S rDNA data. Even so, broader patterns of underdispersion at phylogenetic depths accessible with 16S data could still result in significantly underdispersed model fits.
A strength of our model is that it estimates values of D that can be compared among datasets (Fig. 2) or potentially across time (Figs. 4, S3) in order to learn how differences between datasets impact community assembly. We found that gut and palm communities were almost universally underdispersed (Figs. 2, 4, S4), and that the D value for a community appears to be a function of body site (Fig. 2). Although this result is only shown across two subjects, the parallel patterns between the male and female subject are striking, in that fecal communities are the most strongly underdispersed (lowest D), palm communities are similar to each other, and tongue communities had the highest D estimates. Similarly, comparing D before and after an event can be used within an experimental framework to see how that event may affect community assembly. Our analysis of infant gut microbiome data [26] before and during the use of baby formula (Fig. 4) showed that while the preformula community was significantly underdispersed, community assembly during formula consumption was more neutral. While the postformula trend was not significantly different from the neutral model, this finding was marginal (P = 0.107).
In addition to showing that our model can be a useful tool for future studies, our findings also hint that phylogenetic underdispersion may be a common trend for the human gut microbiome, although demonstrating a general trend would require analysis of more than the 36 individuals we analyzed. Indeed, recent research has shown that for fecal transplants, donor strains are able to integrate into the recipient’s gut community when a conspecific strain is already present, but novel donor strains are unlikely to successfully integrate into the recipient [14]. Congeneric bacteria have also been shown to be predictors of each others’ recruitment in the mouse gut microbiome, both for pathogens and commensals [16]. Different body sites—as we saw with the skin—may have qualitatively similar patterns of underdispersion, yet quantitatively different magnitudes of this effect. Thus, the efficacy of an engineered probiotic based on similarity to organisms already present in the community for which it was engineered may largely depend on the body site for which it is intended, although again more exhaustive study is needed.
Supplementary information
Acknowledgements
The authors thank D.R. Nemergut for her help and support, and also thank P. Sommers, E. Gendron, A. Solon, E. Pruesse, A. Armstrong, C. Martin, K. Hazleton, and S. Sauce for many helpful discussions. Funding was provided by an NSF grant for studying microbial community assembly following disturbance (DEB-1258160) and by a NIH NLM Computational Biology training grant (5 T15 LM009451-12). The funding bodies had no role in study design, analysis, interpretation, or in the preparation of this paper.
Data availability
R code and data to replicate our analysis, or to perform a similar analysis on other data, are available on GitHub, at https://github.com/darcyj/pd_model.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version of this article (10.1038/s41396-020-0613-7) contains supplementary material, which is available to authorized users.
References
- 1.Nemergut DR, Schmidt SK, Fukami T, O’Neill SP, Bilinski TM, Stanish LF, et al. Patterns and processes of microbial community assembly. Microbiol Mol Biol Rev. 2013;77:342–56. doi: 10.1128/MMBR.00051-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sprockett D, Fukami T, Relman DA. Role of priority effects in the early-life assembly of the gut microbiota. Nat Rev Gastroenterol Hepatol. 2018;15:197–205. doi: 10.1038/nrgastro.2017.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fukami T. Historical contingency in community assembly: integrating niches, species pools, and priority effects. Annu Rev Ecol Evol Syst. 2015;46:1–23. [Google Scholar]
- 4.Verster AJ, Borenstein E. Competitive lottery-based assembly of selected clades in the human gut microbiome. Microbiome. 2018;6:186. doi: 10.1186/s40168-018-0571-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Litvak Y, Bäumler AJ. The founder hypothesis: a basis for microbiota resistance, diversity in taxa carriage, and colonization resistance against pathogens. PLOS Pathog. 2019;15:e1007563. doi: 10.1371/journal.ppat.1007563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zaneveld JR, Lozupone C, Gordon JI, Knight R. Ribosomal RNA diversity predicts genome diversity in gut bacteria and their relatives. Nucleic Acids Res. 2010;38:3869–79. doi: 10.1093/nar/gkq066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Langille MGI, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol. 2013;31:814–21. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wiens JJ, Ackerly DD, Allen AP, Anacker BL, Buckley LB, Cornell HV, et al. Niche conservatism as an emerging principle in ecology and conservation biology. Ecol Lett. 2010;13:1310–24. doi: 10.1111/j.1461-0248.2010.01515.x. [DOI] [PubMed] [Google Scholar]
- 9.Pyron RA, Costa GC, Patten MA, Burbrink FT. Phylogenetic niche conservatism and the evolutionary basis of ecological speciation. Biol Rev. 2015;90:1248–62.. doi: 10.1111/brv.12154. [DOI] [PubMed] [Google Scholar]
- 10.Losos JB. Phylogenetic niche conservatism, phylogenetic signal and the relationship between phylogenetic relatedness and ecological similarity among species. Ecol Lett. 2008;11:995–1003. doi: 10.1111/j.1461-0248.2008.01229.x. [DOI] [PubMed] [Google Scholar]
- 11.Darwin C. On the origin of species. London: Murray; 1859. [Google Scholar]
- 12.Ma C, Li S-p PuZ, Tan J, Liu M, Zhou J, et al. Different effects of invader–native phylogenetic relatedness on invasion success and impact: a meta-analysis of Darwin’s naturalization hypothesis. Proc R Soc B Biol Sci. 2016;283:20160663. doi: 10.1098/rspb.2016.0663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peay KG, Belisle M, Fukami T. Phylogenetic relatedness predicts priority effects in nectar yeast communities. Proc R Soc B Biol Sci. 2012;279:5066. doi: 10.1098/rspb.2011.1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li SS, Zhu A, Benes V, Costea PI, Hercog R, Hildebrand F, et al. Durable coexistence of donor and recipient strains after fecal microbiota transplantation. Science. 2016;352:586–9. doi: 10.1126/science.aad8852. [DOI] [PubMed] [Google Scholar]
- 15.Tett A, Huang KD, Asnicar F, Fehlner-Peach H, Pasolli E, Karcher N, et al. The prevotella copri complex comprises four distinct clades that are underrepresented in westernised populations. Cell Host Microbe. 2019;26:666–79. doi: 10.1016/j.chom.2019.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stecher B, Chaffron S, Käppeli R, Hapfelmeier S, Freedrich S, Weber TC, et al. Like will to like: abundances of closely related species can predict susceptibility to intestinal colonization by pathogenic and commensal bacteria. PLoS Pathog. 2010;6:e1000711. doi: 10.1371/journal.ppat.1000711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brown CT, Xiong W, Olm MR, Thomas BC, Baker R, Firek B, et al. Hospitalized premature infants are colonized by related bacterial strains with distinct proteomic profiles. mBio. 2018;9:e00441–18. doi: 10.1128/mBio.00441-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.D’Andrea R, Riolo M, Ostling AM. Generalizing clusters of similar species as a signature of coexistence under competition. PloS Comput Biol. 2019;15:e1006688. doi: 10.1371/journal.pcbi.1006688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mayfield MM, Levine JM. Opposing effects of competitive exclusion on the phylogenetic structure of communities. Ecol Lett. 2010;13:1085–93. doi: 10.1111/j.1461-0248.2010.01509.x. [DOI] [PubMed] [Google Scholar]
- 20.Webb CO, Ackerly DD, McPeek MA, Donoghue MJ. Phylogenies and community ecology. Annu Rev Ecol Evol Syst. 2002;33:475–505. [Google Scholar]
- 21.Yassour M, Vatanen T, Siljander H, Hämäläinen AM, Härkönen T, Ryhänen SJ, et al. Natural history of the infant gut microbiome and impact of antibiotic treatment on bacterial strain diversity and stability. Sci Transl Med. 2016;8:343ra81. doi: 10.1126/scitranslmed.aad0917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 2005;71:8228–35. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Washburne AD, Silverman JD, Leff JW, Bennett DJ, Darcy JL, Mukherjee S, et al. Phylogenetic factorization of compositional data yields lineage-level associations in microbiome datasets. PeerJ. 2017;5:e2969. doi: 10.7717/peerj.2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matsen FA, Evans SN, Gilks W, Ghodsi M, Kingsford C. Edge principal components and squash clustering: using the special structure of phylogenetic placement data for sample comparison. PLoS ONE. 2013;8:e56859. doi: 10.1371/journal.pone.0056859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Caporaso JG, Lauber CL, Costello EK, Berg-Lyons D, Gonzalez A, Stombaugh J, et al. Moving pictures of the human microbiome. Genome Biol. 2011;12:R50. doi: 10.1186/gb-2011-12-5-r50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koenig JE, Spor A, Scalfone N, Fricker AD, Stombaugh J, Knight R, et al. Succession of microbial consortia in the developing infant gut microbiome. Proc Natl Acad Sci USA. 2011;108:4578–85. doi: 10.1073/pnas.1000081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Faith DP. Conservation evaluation and phylogenetic diversity. Biol Conserv. 1992;61:1–10. [Google Scholar]
- 28.Edgar RC. UNOISE2: improved error-correction for Illumina 16S and ITS amplicon sequencing. bioRxiv [Preprint]. 2016. http://www.biorxiv.org/content/early/2016/10/15/081257.
- 29.Martiny JBH, Jones SE, Lennon JT, Martiny AC. Microbiomes in light of traits: a phylogenetic perspective. Science. 2015;350:649–57. doi: 10.1126/science.aac9323. [DOI] [PubMed] [Google Scholar]
- 30.Vázquez-Baeza Y, Callewaert C, Debelius J, Hyde E, Marotz C, Morton JT, et al. Impacts of the human gut microbiome on therapeutics. Ann Rev Pharm Toxicol. 2018;58:253–70. doi: 10.1146/annurev-pharmtox-042017-031849. [DOI] [PubMed] [Google Scholar]
- 31.Scheffer M, van Nes EH. Self-organized similarity, the evolutionary emergence of groups of similar species. Proc Natl Acad Sci USA. 2006;103:6230–5. doi: 10.1073/pnas.0508024103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Webb CO. Exploring the phylogenetic structure of ecological communities: an example for rain forest trees. Am Nat. 2000;156:145–55. doi: 10.1086/303378. [DOI] [PubMed] [Google Scholar]
- 33.Cavender-Bares J, Ackerly DD, Baum DA, Bazzaz FA. Phylogenetic overdispersion in Floridian oak communities. Am Nat. 2004;163:823–43. doi: 10.1086/386375. [DOI] [PubMed] [Google Scholar]
- 34.Gerhold P, Cahill JF, Winter M, Bartish IV, Prinzing A. Phylogenetic patterns are not proxies of community assembly mechanisms (they are far better) Funct Ecol. 2015;29:600–14. [Google Scholar]
- 35.Hubbell SP. The unified neutral theory of biodiversity and biogeography. Princeton, New Jersey: Princeton University Press; 2001.
- 36.Emerson BC, Gillespie RG. Phylogenetic analysis of community assembly and structure over space and time. Trends Ecol Evol. 2008;23:619–30. doi: 10.1016/j.tree.2008.07.005. [DOI] [PubMed] [Google Scholar]
- 37.Peterfreund GL, Vandivier LE, Sinha R, Marozsan AJ, Olson WC, Zhu J, et al. Succession in the gut microbiome following antibiotic and antibody therapies for Clostridium difficile. PloS ONE. 2012;7:e46966. doi: 10.1371/journal.pone.0046966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.David LA, Materna AC, Friedman J, Campos-Baptista MI, Blackburn MC, Perrotta A, et al. Host lifestyle affects human microbiota on daily timescales. Genome Biol. 2014;15:R89. doi: 10.1186/gb-2014-15-7-r89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kennedy RC, Fling RR, Robeson MS, Saxton AM, Donnell RL, Darcy JL, et al. Temporal development of gut microbiota in triclocarban exposed pregnant and neonatal rats. Sci Rep. 2016;6:33430. doi: 10.1038/srep33430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Williamson BD, Hughes JP, Willis AD. A multi-view model for relative and absolute microbial abundances. bioRxiv [Preprint]. 2019. https://www.biorxiv.org/content/early/2019/09/08/761486. [DOI] [PMC free article] [PubMed]
- 41.Chatzidaki-Livanis M, Geva-Zatorsky N, Comstock LE. Bacteroides fragilis type vi secretion systems use novel effector and immunity proteins to antagonize human gut bacteroidales species. Proc Natl Acad Sci USA. 2016;113:3627–32. doi: 10.1073/pnas.1522510113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hecht AL, Casterline BW, Earley ZM, Goo YA, Goodlett DR, Bubeck Wardenburg J. Strain competition restricts colonization of an enteric pathogen and prevents colitis. EMBO Rep. 2016;17:1281–91. doi: 10.15252/embr.201642282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sakavara A, Tsirtsis G, Roelke DL, Mancy R, Spatharis S. Lumpy species coexistence arises robustly in fluctuating resource environments. Proc Natl Acad Sci USA. 2018;115:738–43. doi: 10.1073/pnas.1705944115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Morowitz MJ, Denef VJ, Costello EK, Thomas BC, Poroyko V, Relman DA, et al. Strain-resolved community genomic analysis of gut microbial colonization in a premature infant. Proc Natl Acad Sci USA. 2011;108:1128–33. doi: 10.1073/pnas.1010992108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
R code and data to replicate our analysis, or to perform a similar analysis on other data, are available on GitHub, at https://github.com/darcyj/pd_model.


