Abstract
Hardware and software advancements along with the accumulation of large amounts of data in recent years have together spurred a remarkable growth in the application of neural networks to various scientific fields. Machine learning based on neural networks with multiple (hidden) layers is becoming an extremely powerful approach for analyzing data. With the accumulation of large amounts of protein data such as structural and functional assay data, the effects of such approaches within the field of protein informatics are increasing. Here, we introduce our recent studies based on applications of neural networks for protein structure and function prediction and dynamic analysis involving: (i) inter-residue contact prediction based on a multiple sequence alignment (MSA) of amino acid sequences, (ii) prediction of protein–compound interaction using assay data, and (iii) detection of protein allostery from trajectories of molecular dynamic (MD) simulation.
Keywords: Contact prediction, Deep learning, Neural networks, Protein allostery, Protein-compound interaction
Introduction
Artificial neural networks, inspired by mechanisms of the brain, were proposed more than 60 years ago (e.g., Rosenblatt 1958). Recent years have seen the widespread use of powerful computers, the development of machine learning frameworks, and an increasing availability of data (Shi et al. 2019). That environment has elicited, in various fields, remarkable applications of deep learning using neural networks with multiple hidden layers able to learn data representations with multiple levels of abstraction (LeCun et al. 2015). Protein science is no exception. Here, we introduce three applications of neural networks for protein structure, protein function prediction, and protein dynamic analysis.
Prediction of residue contacts in proteins is an extremely active field to which deep learning has been applied (Kandathil et al. 2019). In the first section, we describe our method for predicting residue contacts in proteins (Fukuda and Tomii 2020). Similar to the other methods developed in the field, our approach relies on deep learning using multiple sequence alignments (MSAs) as inputs (Kandathil et al. 2019). Regarding protein function prediction especially protein–ligand interaction prediction, a new era has arrived that is based on the availability of large amounts of functional assay and structural data that relates to protein/small compound complex formation (Chen et al. 2018). Several groups, including our own described in the second section, have developed prediction methods using neural networks trained with such large amounts of data (Chen et al. 2018). A feature of our approach in this area is the development of novel methods based on end-to-end learning which involves assigning weights to portions of inputs shown to be important for prediction (Sutskever et al. 2014).
In the third section, we discuss our work on autoencoders (Tsuchiya et al. 2019). An autoencoder is an unsupervised neural network that is widely used for analyzing specific features of proteins (Lemke and Peter 2019). It comprises two parts: an encoder and decoder. The encoder part learns hidden relational information related to the original input data and extracts its representative features by compressing the data into a low-dimensional code. The decoder part reconstructs the original data from the low-dimensional encoded data. The autoencoder is used widely for dimensional reduction, feature extraction, representation learning, and pattern classification. For example, Lemke and Peter extracted features of generation paths in peptide conformations by dimensional reduction (Lemke and Peter 2019).
An autoencoder is also used for anomaly detection. In the third section, we introduce our autoencoder-based method, where the autoencoder was used to detect changes in protein dynamics by ligand binding and the allosteric behavior. Along with that detection, we considered the dynamics observed only in ligand-bound (holo) form during the simulations as “anomalies” from the ligand-unbound (apo) form (Tsuchiya et al. 2019).
Contact prediction
Contact prediction is inferring interacting residue pairs in protein three-dimensional structures. In recent years, vast numbers of sequences have been deposited into databases due to progress of genome or transcriptome sequencing (Karsch-Mizrachi et al. 2018). Methods for predicting residue contacts have been developed based on use of an MSA of related amino acid sequences (Kandathil et al. 2019). An MSA for a target protein can include sequences belonging to the same protein family/superfamily as the target but possessing structural differences because of their diversity according to insertions, deletions, and substitutions, especially for larger superfamilies. For instance, clustering distinct subfamilies included in an MSA of Pfam (El-Gebali et al. 2019) can be achieved using the Hopfield–Potts models (Shimagaki and Weigt 2019). Therefore, we proposed a contact prediction model based on neural networks that both calculates and uses a weight for each sequence included in the MSA to improve prediction accuracy, more specifically, to improve robustness to noisy inputs. We demonstrated that the model is effective for contact prediction when numerous amino acid sequences are included in the MSA (Fukuda and Tomii 2020).
In our model, a weight is calculated for each sequence included in an MSA based on seven features: the number of sequences in an MSA, the sequence identity with both a target sequence and a consensus sequence of an MSA, the gap ratio for each sequence, and average values of the last three features (Fukuda and Tomii 2020). Weights are calculated using a multilayer perceptron composed of two hidden layers, each of which has seven nodes, with seven features as inputs. By using the weights and the covariance matrix calculated from the MSA as inputs, contact probabilities for every residue (−position) pair are calculated using a residual neural network (RNN). Our model is trained in an end-to-end manner: the method for calculating the weight for each sequence included in a given MSA is also trained automatically. Effects of using weights on prediction accuracy were confirmed by performing cross-validation using CASP11 (Monastyrskyy et al. 2016) and CASP12 (Schaarschmidt et al. 2018) datasets. Results show that when sufficiently numerous (200 or more) sequences were included in an MSA, a significant improvement in prediction accuracy was observed at any (i.e., short, medium, and long) range and any prediction number (i.e., L/10, L/5, L/2, and L, where L represents the target protein length) of contacts (Fukuda and Tomii 2020).
However, when the number of sequences included in an MSA was insufficient (less than 200), cases were found in which the prediction accuracy was markedly worse than the case in which no weight was used. To alleviate this shortcoming, we added five other features including a target sequence and position-specific score matrix (PSSM) in addition to the “weighted” covariance matrix as inputs for RNN in our model (Fukuda and Tomii 2020). Then, based on this model, we expanded our model to a multitask model, which simultaneously predicts contacts, secondary structures, and accessible surface areas (Fukuda and Tomii 2020). Furthermore, using ensemble averaging, we were able to develop a more accurate prediction model, which we designate as DeepECA, compared with existing contact prediction methods (Fukuda and Tomii 2020). We also confirmed the ability of our model to obtain accurate three-dimensional models based on the predicted contacts and secondary structures by the multitask model. To do so, we use CONFOLD (Adhikari et al. 2015), a method for constructing models with a set of restraints. Very recently, in CASP13, AlphaFold was able to produce accurate three-dimensional models based on predicted distances for residue pairs (Senior et al. 2020), suggesting that predicting distances in addition to contacts for residue pairs would be more helpful to protein structure prediction. Our model is available from https://github.com/tomiilab/DeepECA (Fukuda and Tomii 2020).
Protein–compound interaction prediction
In addition to protein sequences, assay and structural data of protein–compound interactions have also seen a massive increase over the last 10 years. Such data can be accessed from public database such as DrugBank (Wishart et al. 2008) and Matador (Günther et al. 2008). To use these data effectively, we developed a protein–compound interaction prediction model based on neural networks of two types, graph neural network (GNN), and convolutional neural network (CNN) for both small compound and protein sequence data, respectively (Tsubaki et al. 2019).
In our model, molecular structures of a small compound are treated as a graph with atoms as vertices and bonds between the atoms as edges. They are divided into r-radius subgraphs (Costa and De Grave 2010). Then, a “compound” vector, which is a low-dimensional real-valued representation of the compound, is calculated using a GNN. Similarly, an amino acid sequence of the target protein is projected onto a “protein” vector using a CNN. These vector representations, obtained for a compound and a protein by end-to-end learning and which have the same dimensions, are concatenated and fed into a classifier, which predicts protein–compound interaction (Tsubaki et al. 2019). We demonstrated that our model based on end-to-end learning of GNN and CNN can attain higher prediction accuracy than existing methods, irrespective of the ratio of the numbers of positive and negative samples, using positive samples from DrugBank and Matador and highly credible negative samples (Liu et al. 2015). Results indicate that our model is robust even when learning with an unbalanced dataset, typically consisting of a small number of positive samples and a huge number of negative samples, which is an all-too-common situation (Tsubaki et al. 2019).
Furthermore, we sought to identify regions in a target protein that are important for predicting protein–compound interaction using a neural attention mechanism (Bahdanau et al. 2014). To this end, we used a neural attention mechanism to assign weights to the subsequences according to their importance for interaction prediction. Such weights can be assumed to represent interaction strength between a subsequence in a protein and a compound. In fact, using the DUD-E benchmark (a dataset originally constructed for evaluating structure-based virtual screening methods (Mysinger et al. 2012)), we confirmed and demonstrated that most of regions with high-value attention weights correspond to actual compound–binding sites (Tsubaki et al. 2019). Results suggest that our model is useful to identify compound-binding sites by consideration of weight values calculated using the neural attention mechanism. The model is available from https://github.com/masashitsubaki (Tsubaki et al. 2019).
Analysis of protein allostery
Dynamic allostery, triggered by ligand binding or introduction of mutations on proteins, transmits a signal from the binding or mutation site to distant regions, thereby dramatically altering protein function (Cooper and Dryden 1984). Allosteric effects are often accompanied by subtle changes in side-chain conformations of the protein (Liu and Nussinov 2016). Therefore, a precise analysis of the changes of the dynamics, rather than the static conformational changes, is of fundamental importance for elucidating the regulation of protein function. We adopted an autoencoder, unsupervised neural network, to detect dynamic changes in the PDZ2 protein domain in human PTPN13, as triggered by binding of the peptide of RAPGEF6 (Tsuchiya et al. 2019).
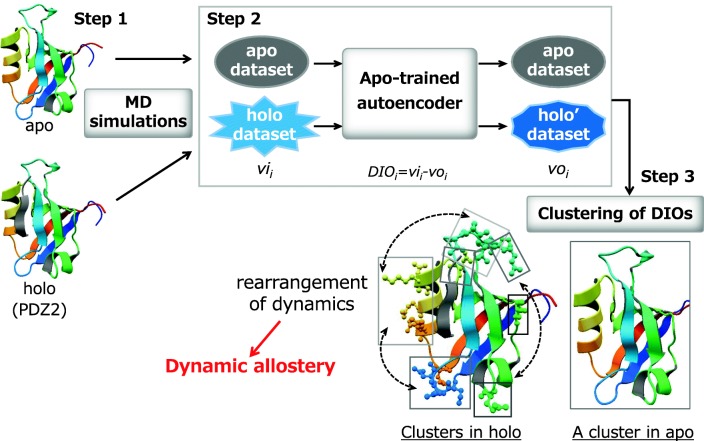
This study was composed of three steps (Fig. 1). For the first step of obtaining protein dynamic data, molecular dynamic (MD) simulations of PDZ2 were executed twice for each of ligand-unbound (apo) and ligand-bond (holo) forms. Both crystal structures of apo and holo forms (PDB IDs 3lnx and 3lny, respectively (Zhang et al. 2010)) were used as the initial structures for the simulations. All simulations were performed using the Gromacs package 2018 (Toxvaerd et al. 2012). The 1500-dimensional vector of time fluctuations of the side-chain distances in a pair of residues, from 50.1 to 200.0 ns in 0.1-ns increments obtained from the MD trajectories, was prepared as an input for the autoencoder. It is noteworthy that the previous NMR study (Fuentes et al. 2004) measured side-chain methyl dynamic parameters on a ps–ns time scale and showed long-range fluctuations in several residues only in holo forms. PDZ2 is a small protein that consists of 94 residues; therefore, we decided to use 200 ns MD trajectories of PDZ2 for the development of the autoencoder-based method.
Fig. 1.
Autoencoder-based analysis of dynamic allostery
The second step involved processing the data reflecting the time-dependent fluctuations of side-chain distances. For this step, the autoencoder was trained using the input vectors in apo form. Finally, the nine-layer autoencoder (1500–1000–500–200–100–200–500–1000–1500 nodes; hereafter designated as the apo-trained autoencoder) was chosen because it had the lowest error among several combinations of nodes and layers. Then, the apo-trained autoencoder was used to inspect the apo and holo data. For the apo data inspection, the output vectors from the autoencoder were almost identical to the apo input vectors because the autoencoder tried to reconstruct the input vectors precisely. In contrast, the output vectors for the holo data, regions for which features differed from those in apo form, were exaggerated and modified according to the features observed in apo form, as shown in the upper right of Fig. 1. Such findings suggest that analysis of the differences between the input and output (DIO) vectors can provide useful information to detect differences between apo and holo forms, i.e., the dynamic changes by ligand binding.
The third step of our developed approach involves cluster analysis of the DIO vectors. Clustering of DIO vectors in both apo and holo forms was performed simultaneously using methods within the R program (R Core Team 2018) to detect the residues involved in changes to the dynamics by ligand binding. Residue pairs in apo and holo forms were clustered based on the cosine similarity of DIO vectors. In each cluster, a residue that formed pairs with more than 80% of other residues was defined as a “leading residue,” which led to cluster-specific fluctuation. Also, a residue that formed pairs with 60–80% of other residues was defined as an “accompanying residue,” which assisted cluster-specific fluctuations led by leading residues (and which played a key role in propagation of the specific fluctuations to distant regions). For this study, MD simulations were performed twice for apo and holo forms, as described above. Therefore, the clustering analysis involved four combinations of apo and holo DIO data. The DIO vectors in apo1 and holo1 and those in apo1 and holo2 were obtained from an inspection by the apo1-trained autoencoder. The DIO vectors in apo2 and holo1 and those in apo2 and holo2 were obtained using the apo2-trained autoencoder. In all of the four clustering routines, almost all residue pairs in apo form were separated from those in holo form. In the next several clustering steps, residue pairs in holo forms were divided according to the similarity in their fluctuation pattern. It is particularly interesting that, in the clustering of all four combinations of the DIO data, the same residues in holo forms were detected as “leading residues,” shown as ball and stick models at the lower right side of Fig. 1. Several leading residues were located close to the N-terminus or C-terminus of the ligand peptide, which suggests that the correlative fluctuations led by these leading residues were involved in communication of the signal from binding of the ligand. In addition, various leading and accompanying residues were shown to be able to lead to specific fluctuations in other clusters under different conditions, which led to rearrangement of the correlative fluctuations. These residues belonged not only to clusters around the N-terminus and/or C-terminus of the ligand but also to those distant from the ligand. Rearrangement of the correlative fluctuations by these residues led to propagation of the signals by ligand binding to the distant regions. These findings suggest that the leading and accompanying residues, as detected by the clustering of the DIO vectors, were involved in dynamic allostery (Tsuchiya et al. 2019).
Finally, we compared our results with those reported from an earlier nuclear magnetic resonance (NMR) study (Fuentes et al. 2004). To elucidate the allosteric behavior of PDZ2, the authors measured parameters related to side-chain methyl dynamics on a ps–ns time scale using NMR techniques for apo and holo forms. The 20 residues detected showed long-range fluctuations only in the holo form. Some of these residues were located distant from the ligand, suggesting that these residues might be involved in dynamic allostery. Our autoencoder-based method detected all 20 resides as (8) leading and (12) accompanying residues. Some of these accompanying residues were distant from the ligand, which suggests that these accompanying residues were involved not only in assisting the correlative fluctuations led by the leading residues; they were also involved in propagating the fluctuations to regions that are distant from the ligand (Tsuchiya et al. 2019).
To summarize this section, the autoencoder-based method can elicit important clues to elucidate the dynamic allostery of PDZ2 occurring as a result of the ligand binding. The success might rely on distance matrix information reflecting time fluctuations of protein motions. Success might also rely on the use of the autoencoder based–regression method for the detection of subtle conformational and dynamic changes in comparison with the fluctuation data in the apo and holo forms. This autoencoder-based method can be applied to detection of important signals in the signal transduction and altered signals in mutated proteins involved in both normal protein function and altered disease states.
Conclusion
Due to increases in the amount of protein data, available computational resources, and frameworks developed for deep learning, the application of neural networks to protein informatics is becoming increasingly popular. Our studies and the methods described herein represent just one such approach, but they are expected to be helpful to predict and understand protein structures and functions.
Abbreviations
- MSA
multiple sequence alignment
- MD
molecular dynamics
- RNN
residual neural network
- GNN
graphic neural network
- CNN
convolutional neural network
- DIO
differences between the input and output
- NMR
nuclear magnetic resonance
Funding information
This research was partially supported as a Platform Project for Supporting Drug Discovery and Life Science Research (Basis for Supporting Innovative Drug Discovery and Life Science Research (BINDS)) from AMED under Grant number JP19am0101110.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Adhikari B, Bhattacharya D, Cao R, Cheng J. CONFOLD: residue-residue contact-guided ab initio protein folding. Proteins. 2015;83:1436–1449. doi: 10.1002/prot.24829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahdanau D et al (2014) Neural machine translation by jointly learning to align and translate. In: International Conference on Learning Representations
- Chen H, Engkvist O, Wang Y, Olivecrona M, Blaschke T. The rise of deep learning in drug discovery. Drug Discov Today. 2018;23(6):1241–1250. doi: 10.1016/j.drudis.2018.01.039. [DOI] [PubMed] [Google Scholar]
- Cooper A, Dryden DTF. Allostery without conformational change - a plausible model. Eur Biophys J. 1984;11:103–109. doi: 10.1007/BF00276625. [DOI] [PubMed] [Google Scholar]
- Costa F, De Grave K (2010) Fast neighborhood subgraph pairwise distance kernel. In: International Conference on Machine Learning
- El-Gebali S, et al. The Pfam protein families database in 2019. Nucleic Acids Res. 2019;47:D427–D432. doi: 10.1093/nar/gky995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuentes EJ, Der CJ, Lee AL. Ligand-dependent dynamics and intramolecular signaling in a PDZ domain. J Mol Biol. 2004;335:1105–1115. doi: 10.1016/j.jmb.2003.11.010. [DOI] [PubMed] [Google Scholar]
- Fukuda H, Tomii K. DeepECA: an end-to-end learning framework for protein contact prediction from a multiple sequence alignment. BMC Bioinformatics. 2020;21:10. doi: 10.1186/s12859-019-3190-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Günther S, et al. Supertarget and matador: resources for exploring drug-target relationships. Nucleic Acids Res. 2008;36:D919–D922. doi: 10.1093/nar/gkm862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandathil SM, Greener JG, Jones DT. Recent developments in deep learning applied to protein structure prediction. Proteins. 2019;87:1179–1189. doi: 10.1002/prot.25824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karsch-Mizrachi I, et al. The international nucleotide sequence database collaboration. Nucleic Acids Res. 2018;46:D48–D51. doi: 10.1093/nar/gkx1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521:435–444. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- Lemke T, Peter C. EncoderMap: dimensionality reduction and generation of molecule conformations. J Chem Theory Comput. 2019;15:1209–1215. doi: 10.1021/acs.jctc.8b00975. [DOI] [PubMed] [Google Scholar]
- Liu J, Nussinov R. Allostery: an overview of its history, concepts, methods, and applications. PLoS Comput Biol. 2016;12:e1004966. doi: 10.1371/journal.pcbi.1004966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Sun J, Guan J, Zheng J, Zhou S. Improving compound-protein interaction prediction by building up highly credible negative samples. Bioinformatics. 2015;31:i221–i229. doi: 10.1093/bioinformatics/btv256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monastyrskyy B, D'Andrea D, Fidelis K, Tramontano A, Kryshtafovych A. New encouraging developments in contact prediction: assessment of the CASP11 results. Proteins. 2016;84(Suppl 1):131–144. doi: 10.1002/prot.24943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mysinger MM, Carchia M, Irwin JJ, Shoichet BK. Directory of useful decoys, enhanced (DUD-E): better ligands and decoys for better benchmarking. J Med Chem. 2012;55:6582–6594. doi: 10.1021/jm300687e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team (2018) R: a language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria
- Rosenblatt F. The perceptron: a probabilistic model for information storage and organization in the brain. Psychol Rev. 1958;65:386–408. doi: 10.1037/h0042519. [DOI] [PubMed] [Google Scholar]
- Schaarschmidt J, Monastyrskyy B, Kryshtafovych A, Bonvin AMJJ. Assessment of contact predictions in CASP12: co-evolution and deep learning coming of age. Proteins. 2018;86(Suppl 1):51–66. doi: 10.1002/prot.25407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senior AW, et al. Improved protein structure prediction using potentials from deep learning. Nature. 2020;577:706–710. doi: 10.1038/s41586-019-1923-7. [DOI] [PubMed] [Google Scholar]
- Shi Q, Chen W, Huang S, Wang Y, Xue Z (2019) Deep learning for mining protein data. Brief Bioinform. 10.1093/bib/bbz156 [DOI] [PubMed]
- Shimagaki K, Weigt M. Selection of sequence motifs and generative Hopfield-Potts models for protein families. Phys Rev E. 2019;100:032128. doi: 10.1103/PhysRevE.100.032128. [DOI] [PubMed] [Google Scholar]
- Sutskever I, Vinyals O, Le QV. Advances in Neural Information Processing Systems. 2014. Sequence to sequence learning with neural networks; pp. 3104–3112. [Google Scholar]
- Toxvaerd S, Heilmann OJ, Dyre JC. Energy conservation in molecular dynamics simulations of classical systems. J Chem Phys. 2012;136:224106. doi: 10.1063/1.4726728. [DOI] [PubMed] [Google Scholar]
- Tsubaki M, Tomii K, Sese J. Compound–protein interaction prediction with end-to-end learning of neural networks for graphs and sequences. Bioinformatics. 2019;35:309–318. doi: 10.1093/bioinformatics/bty535. [DOI] [PubMed] [Google Scholar]
- Tsuchiya Y, Taneishi K, Yonezawa Y. Autoencoder-based detection of dynamic allostery triggered by ligand binding based on molecular dynamics. J Chem Inf Model. 2019;59:4043–4051. doi: 10.1021/acs.jcim.9b00426. [DOI] [PubMed] [Google Scholar]
- Wishart DS, et al. Drugbank: a knowledgebase for drugs, drug actions and drug targets. Nucleic Acids Res. 2008;36:D901–D906. doi: 10.1093/nar/gkm958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Sapienza PJ, Ke H, Chang A, Hengel SR, Wang H, Phillips GN, Lee AL. Crystallographic and nuclear magnetic resonance evaluation of the impact of peptide binding to the second PDZ domain of protein tyrosine phosphatase 1E. Biochemistry. 2010;49:9280–9291. doi: 10.1021/bi101131f. [DOI] [PMC free article] [PubMed] [Google Scholar]