Abstract
Elucidation of the molecular mechanism underlying the metabolic adaptation is critical to understand homeostasis of life. This review provides the concept, experimental and computational methods, to deal with this issue using “trans-omics” approaches.
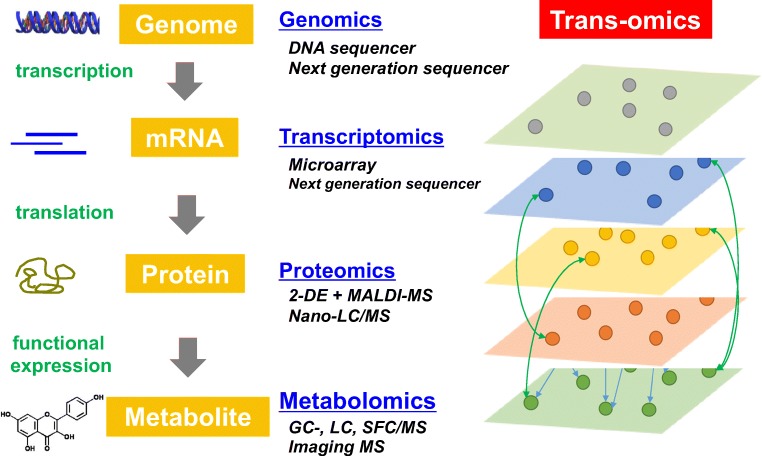
Life is maintained by dynamic flow and fine tuning of metabolism. In this circumstance, rewiring of the metabolic networks is considered to be the result of whole-body metabolic adaptation to its environment. To understand the molecular mechanism underlying the metabolic adaptation, measuring and analyzing “trans-omics” networks which are defined as the interactions occurring among molecules across multi-omic layers, such as genome, transcriptome, proteome, and metabolome, are necessary (Yugi et al. 2016; Yugi and Kuroda 2018) (Fig. 1). Therefore, the development of technologies associated with quantifying each “omics” molecule and integrating the information gained from the measurement is critical. To deal with those issues, we held the trans-omics symposium at the 57th BSJ Meeting in Miyazaki, Japan, to shed light on strategies in obtaining and integrating such multiple omics data for establishing trans-omics approaches, and also to promote discussions among cutting-edge researchers in the area of omics research. This article reviews the researches presented by the session speakers in the 2019 BSJ Meeting and a researcher in the trans-omics research group in Japan to highlight the metabolic adaptation.
Fig. 1.
Trans-omics approaches to measure and analyze the information flow in life
The session was opened by Takeshi Bamba from Kyushu University. He has been working on the development of various metabolomics technologies (Takeda et al. 2018, 2019). In his presentation, the development of quantitative technology of metabolomic analysis was discussed. His group has recently developed a semi-quantitative metabolome analysis based on gas chromatography/mass spectrometry (GC/MS) and a targeted quantitative lipidomics methodology involving supercritical fluid chromatography/triple-quadrupole mass spectrometry (SFC/QqQMS). This method enables the acquisition of quantitative metabolome data, which has been difficult to obtain up to now, and will be an advantageous tool in trans-omics research.
Next generation sequencing (NGS) methods are a major technology in the omics field and have become a standard experimental tool for capturing detailed information of DNA/RNA and histone modification status. The next speaker, Takashi Ito from Kyushu University, has been developing bisulfate sequencing methods to identify DNA methylation and has recently developed an ultra-sensitive method called post-bisulfite adaptor tagging (PBAT) that is an improved low-input whole-genome bisulfite sequencing method (Miura et al. 2019) which can be applied for single-cell sequencing. His methods have been widely utilized in research community to accelerate various methylome analyses.
Hozumi Motohashi, from Tohoku University, discussed about the impact of metabolic adaption of cancer cells by focusing on the NRF2 transcription factor. NRF2 was originally identified as a key regulator of sulfur-employing defense mechanism against oxidative and electrophilic stress and was later found to contribute to metabolic reprogramming in different types of cancer cells. The NRF2-adapted cancer cell shows glutamine dependency and less dependency on catabolic activity in mitochondria (Mitsuishi et al. 2012). She raised a possibility of cancer treatment by targeting metabolic liabilities unique to NRF2-adapted cancer cells.
Shinsuke Uda from Kyushu University is a mathematician and he introduced the topic of information theory into the session. Shannon’s information theory is a powerful tool to analyze information processing in cells (Uda et al. 2013). In this theory, measures of information are mainly defined and formulated by the context of communication between a sender and receiver, and can be also utilized to infer the network structure within the omics data. However, sample size and computational burden often become a bottleneck in the analysis. He gave an overview of information theory in biological application.
Teppei Shimamura is a statistician from Nagoya University. In his research, he applies a Bayesian framework to the analysis of multi-modal biological data (Minoura et al. 2019). Bayesian framework is a mathematical scheme to describe a generation process of these data by a set of probabilistic models. He applied the method to time-series cytometry data in order to infer cell population dynamics. He also applies the method to single-cell transcriptome data in order to infer cell-to-cell communications and their associations with environmental factors using a ligand-receptor matrix.
For the last topic of the session, Mariko Okada from Osaka University presented her work on B cell gene regulation mediated by the NF-κB transcription factor. She introduced a simple mathematical model that predicts gene expression quantity from epigenome data. The analysis showed that the longer the distance of H3K27ac modification together with the degree of open chromatin determines the extent of increased gene expression. Such regions also generate more transcriptional noise than the short regions in single cell transcriptome, indicating a potential mechanism of cell-to-cell variability (Michida, et al. in revision).
Within this special issue are included detailed review articles by two speakers from the session, Hozumi Motohashi (Tohoku University) and Shinsuke Uda (Kyushu University). Takaaki Horinouchi (RIKEN), who studies the in vitro evolution of bacteria, has also kindly contributed a review article on their own research in the field of trans-omics.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Minoura K, Abe K, Maeda Y, Nishikawa H, Shimamura T. Model-based cell clustering and population tracking for time-series flow cytometry data. BMC Bioinformatics. 2019;20(Suppl 23):633. doi: 10.1186/s12859-019-3294-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuishi Y, Taguchi K, Kawatani Y, Shibata T, Nukiwa T, Aburatani H, Yamamoto M, Motohashi H. Nrf2 redirects glucose and glutamine into anabolic pathways in metabolic reprogramming. Cancer Cell. 2012;22(1):66–79. doi: 10.1016/j.ccr.2012.05.016. [DOI] [PubMed] [Google Scholar]
- Miura F, Shibata Y, Miura M, Sangatsuda Y, Hisano O, Araki H, Ito T. Highly efficient single-stranded DNA ligation technique improves low-input whole-genome bisulfite sequencing by post-bisulfite adaptor tagging. Nucleic Acids Res. 2019;47(15):e85. doi: 10.1093/nar/gkz435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda H, Izumi Y, Takahashi M, Paxton T, Tamura S, Koike T, Yu Y, Kato N, Nagase K, Shiomi M, Bamba T. Widely-targeted quantitative lipidomics method by supercritical fluid chromatography triple quadrupole mass spectrometry. J Lipid Res. 2018;59(7):1283–1293. doi: 10.1194/jlr.D083014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda Hiroaki, Takahashi Masatomo, Hara Takeshi, Izumi Yoshihiro, Bamba Takeshi. Improved quantitation of lipid classes using supercritical fluid chromatography with a charged aerosol detector. Journal of Lipid Research. 2019;60(8):1465–1474. doi: 10.1194/jlr.D094516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uda S, Saito TH, Kudo T, Kokaji T, Tsuchiya T, Kubota H, Komori Y, Ozaki Y, Kuroda S. Robustness and compensation of information transmission of signaling pathways. Science. 2013;341(6145):558–561. doi: 10.1126/science.1234511. [DOI] [PubMed] [Google Scholar]
- Yugi K, Kubota H, Hatano A, Kuroda S. Trans-omics: how to reconstruct biochemical networks across multiple ‘omic’ layers. Trends Biotechnol. 2016;34(4):276–290. doi: 10.1016/j.tibtech.2015. [DOI] [PubMed] [Google Scholar]
- Yugi K, Kuroda S. Metabolism as a signal generator across trans-omic networks at distinct time scales. Current Opinion in Systems Biology. 2018;8:59–66. doi: 10.1016/j.coisb.2017.12.002. [DOI] [Google Scholar]