Abstract
Among various microscopic techniques for characterizing protein structures and functions, high-speed atomic force microscopy (HS-AFM) is a unique technique in that it allows direct visualization of structural changes and molecular interactions of proteins without any labeling in a liquid environment. Since the development of the HS-AFM was first reported in 2001, it has been applied to analyze the dynamics of various types of proteins, including motor proteins, membrane proteins, DNA-binding proteins, amyloid proteins, and artificial proteins. This method has now become a versatile tool indispensable for biophysical research. This short review summarizes some bioimaging applications of HS-AFM reported in the last few years and novel applications of HS-AFM utilizing the unique ability of AFM to gain mechanical properties of samples in addition to structural information.
Keywords: High-speed atomic force microscopy, Single-molecule imaging, Conformational dynamics, Intermolecular interaction, Mechanical indentation
Introduction
Proteins inherently exert their unique physiological functions through a variety of dynamics, such as structural changes triggered by variations in the external environment, binding of substrates, and association and dissociation with other proteins. Hence, detailed information on various dynamic properties of proteins on the sub-second timescale obtained by directly filming them as movies can provide deep insights and understanding of essential mechanisms of their functional expression. The development of various types of bioimaging techniques based on optical microscopy has enabled us to track movements of proteins at the single molecular level in vitro and even in vivo and has contributed greatly to the progress of life science (Miller et al. 2018) (Joo et al. 2008). On the other hand, what is generally being imaged by optical microscopy techniques is not the protein itself, but the change in the locus of the fluorescent regions and the fluorescence intensity emitted by the labeled molecules bound to the protein. The behavior of the protein is indirectly determined from these observations. To understand intuitively proteins’ functions, a microscopy technique capable to resolve their higher order structure and high-speed imaging in solution is needed.
Atomic force microscopy (AFM) is part of the family of scanning probe microscopy techniques and was originally invented in 1986 for the surface structure analysis of solid materials. Because AFM can image the surface structure with nanometer resolution and can also map various physical properties (elasticity, viscoelasticity, charge distribution, etc.) of sample surfaces without any special pretreatment, it has become an analytical tool indispensable for nanoscience and nanotechnology. A major feature of AFM is that the operation principle is independent of the environment and, therefore, it has been applied to a wide range of biological samples from nucleic acids, proteins, and chromosomes to living cells in solution (Alessandrini and Facci 2005; Frederix et al. 2003). On the other hand, because the time resolution of conventional AFM varies from seconds to several minutes, dynamic imaging was impossible, and only a “dead” biological sample, which was firmly fixed to a solid substrate, could be imaged. Prof. Ando’s group at Kanazawa University has been working on improving the imaging speed of AFM since around 1993 in order to break through this limitation, and finally succeeded in capturing a protein with an imaging rate of 80 ms/frame in 2001 (Ando et al. 2001; Ando et al. 2008). Thanks to the success of the development of high-speed AFM (HS-AFM), it has become possible to capture movies of proteins at work and HS-AFM is now being applied to the imaging of conformational changes and intermolecular interactions of various proteins (Ando et al. 2014; Uchihashi and Scheuring 2018; Ando 2018).
This review provides overviews of several recent bioimaging applications achieved by HS-AFM, classified into imaging studies of conformational dynamics and protein-protein interactions. In addition, a functional extension of HS-AFM for molecular manipulation is described that takes advantage of one of the AFM’s significant features, the local force application. The examples in this review were chosen to showcase the latest applications of HS-AFM and newest developments to expand its functionality. The studies on conformational dynamics demonstrate the ability of HS-AFM to elucidate structure-function relationships, while the cases on intermolecular interaction show that also effects on the molecular level are accessible. The applications of force modulation highlight future directions of HS-AFM as a multimodal technique.
Recent bioimaging applications of HS-AFM
Although there are many applications of high-speed AFM for observing protein dynamics, the most important applications in the biophysical field would be visualization of conformational changes of single molecules associated to physiological functions and analyses of dynamic intermolecular interactions. Regarding conformational dynamics, the HS-AFM imaging of ATPases such as myosin V (Kodera et al. 2010) and F1-ATPase (Uchihashi et al. 2011) and membrane proteins such as bacteriorhodopsin (Shibata et al. 2010), a cyclic nucleotide-modulated channel (MloK1) (Rangl et al. 2016), and a glutamate transporter (GltPh) (Ruan et al. 2017) have been successfully demonstrated. As for the intermolecular interactions, diffusion and molecular association of membrane or membrane-associated proteins such as bacteriorhodopsin (Shibata et al. 2010), aquaporin-0 (Colom et al. 2012), OmpF (Casuso et al. 2012), and annexin-V (Miyagi et al. 2016) in lipid membranes have been reported. Here, an overview is presented of some examples on conformational dynamics and intermolecular interactions reported in the last 2 years.
Conformational dynamics of ring-shaped ATPases
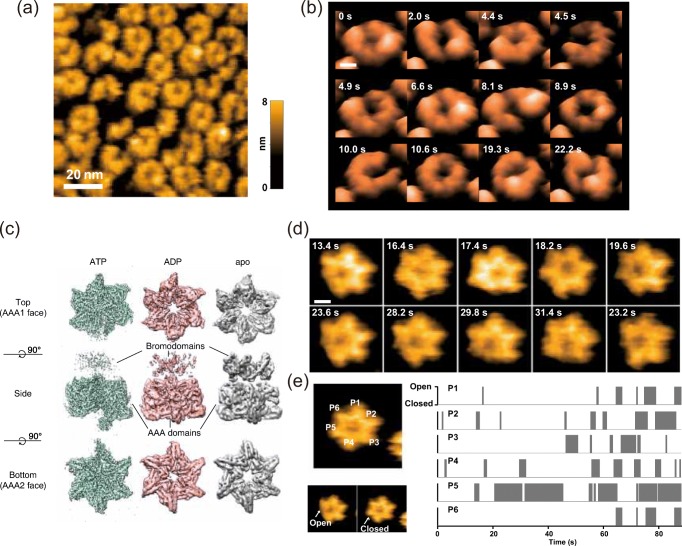
Saccharomyces cerevisiae Hsp104 and its bacterial homolog ClpB are AAA+ ATPases and called disaggregating molecular chaperones, which refold toxic protein aggregates into the native state in cooperation with the Hsp70 partner. Biochemical and electron microscopic (EM) analyses have revealed that they form a ring-shaped hexamer that unwinds the aggregated proteins by threading the peptides from the aggregates into the central pore with a conformational change driven by the energy of ATP hydrolysis (Watanabe et al. 2002; Nakazaki and Watanabe 2014; Lee et al. 2003). Also, a recent cryo-EM single-particle analysis has demonstrated that the hexamer has a helical structure rather than a symmetric ring structure (Deville et al. 2017), which had been believed previously. However, little is known about the structural dynamics of Hsp104 and ClpB related to the disaggregation activity. A HS-AFM image of the N-terminal deletion mutant of TClpB (ΔN-TClpB) weakly adsorbed on a mica substrate showed diverse oligomeric structures in the presence of ATP (Fig. 1a). These findings indicate that the ClpB hexamers are in heterogeneous states showing the round-symmetric ring, the asymmetric ring, and even the open ring. These various hexameric conformations randomly interchanged with time (Fig. 1b). The frequency of structural changes between the symmetric and the asymmetric forms including the open ring was dependent on ATP concentrations, suggesting that the large conformational changes observed are driven by the ATP hydrolysis. These results imply that the entire ClpB hexamer ring’s conformational dynamics are strongly pronounced. Further HS-AFM analyses of the oligomer forms of various ClpB mutants (ATP-binding/hydrolysis deficient, suppressed/enhanced disaggregation activity) provided a comprehensive picture of the role of ATP binding and hydrolysis in the formation of oligomers and the correlation between large-scale conformational dynamics and the disaggregation activity.
Fig. 1.
HS-AFM images capturing conformational dynamics of ring-shaped ATPases. a Diverse oligomeric forms of ΔN-TClpB observed by HS-AFM in 0.5 mM ATP at 25 °C. Scale bar, 20 nm. b Clipped HS-AFM images of aΔN-TClpB hexamer captured at 10 fps in the presence of 10 μM ATP. Scale bar, 5 nm. c Cryo-EM maps of Abo1 in the ATP, ADP, and apo states. d Clipped HS-AFM images of an Abo1 hexamer undergoing conformational change in the presence of 2 mM ATP captured at 5 fps. Scale bar, 5 nm. e Analysis of the position of Abo1 ring opening (gray boxes) sorted by the protomers according to time shows random subunit activation
Abo1 is the homolog of fission yeast ATAD2 which are AAA+ ATPases and a family of histone chaperones that regulate nucleosome density and chromatin dynamics by histone H3–H4 loading or removal (Zou et al. 2007). Similar to other AAA+ ATPases, Abo1 is considered to form a ring-shaped hexameric oligomer and it functions to pull substrates through the ring’s central pore. Cryo-EM analysis revealed the distinct conformations of the hexamer for three different nucleotide states (ATP-, ADP-, and apo-state) (Fig. 1c) (Cho et al. 2019). The Abo1 hexamer in the apo- and ADP states showed a similar symmetric planar ring structure, whereas, in the ATP-state, the protomers were staggered in height and shifted towards the center, forming an asymmetric spiral with a smaller pore. HS-AFM was applied to investigate how the hexamer conformations change over time. In the presence of 2 mM ATP, real-time HS-AFM images revealed striking symmetry-breaking events, where individual blades of the hexameric ring seemed to disappear due to a decrease in the height of a subunit (Fig. 1d) (Cho et al. 2019). In most cases of asymmetric states, only one blade disappeared, but there were also rare transient cases where two blades of the ring disappeared simultaneously. Interestingly, the Abo1 Walker B mutant, which lost its ATP-hydrolysis activity, displayed only the single symmetry breaking structure in the presence of ATP, indicating that ATP can bind to a single protomer in the hexamer. A tracking analysis of the ring opening positions showed that the ring opening takes place randomly with no ordered sequence (Fig. 1e), suggesting that Abo1 subunits hydrolyze ATP stochastically at least under basal conditions without substrates added.
Intermolecular interaction
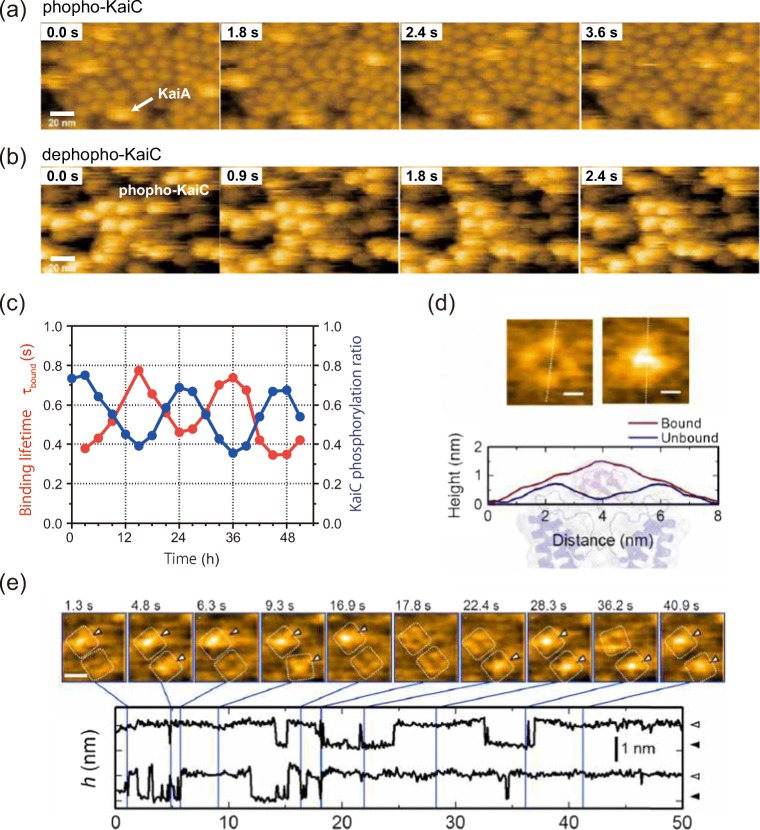
Here, two typical examples are shown for dynamic intermolecular interactions between two different proteins monitored by HS-AFM. The first example is a binding analysis for the bacterial circadian clock proteins. The cyanobacterial circadian clock system consists of oscillators composed of three Kai proteins (KaiA, KaiB, KaiC) encoded by the Kai genes. The unique feature of the Kai protein system is that the phosphorylation state of KaiC oscillates autonomously over a long period of time simply by mixing KaiA, KaiB, and KaiC with ATP in vitro in a circadian cycle (Nakajima et al. 2005). KaiC, the centerpiece of circadian rhythms, forms a doughnut-shaped homohexamer consisting of two stacked rings, the CI ring (N-terminal side) and the CII ring (C-terminal side). It has autokinase and autophosphatase activities, and the circadian rhythm is regulated by switching these activities. KaiA and KaiB are responsible for switching the activity of KaiC. KaiA is a dimer that binds to the tail structure on the CII ring side of KaiC to promote its phosphorylation, while KaiB is a monomer that binds to the CI ring side of KaiC and promotes the dephosphorylation of KaiC by sequestering KaiA. HS-AFM was applied to visualize the binding interaction of KaiA to KaiC on sub-second timescales (Mori et al. 2018). When the hexamer of the mutant KaiC mimicking the phosphorylated state was adsorbed on the chemically modified mica substrate with the CII ring side facing up, it was found that KaiA dimers repeated the binding and dissociation to KaiC hexamers on a timescale in seconds (Fig. 2a). On the other hand, KaiA bound strongly to the dephosphorylated-state mutant of KaiC and formed a complex over several seconds. By examining the relationship between the phosphorylation state of wild-type KaiC and the binding time of KaiA, τbound, during the circadian oscillation, it was found that the affinity of KaiA and KaiC oscillates in a circadian rhythm in synchronization with the phosphorylation state of KaiC (Fig. 2c). Combining the HS-AFM experimental results with a mathematical model further revealed that the KaiC phosphoform-dependent differential affinity (PDDA) broadens the range of Kai protein stoichiometries that allow rhythmicity, explaining how the oscillation is resilient in an in vivo milieu that includes noise.
Fig. 2.
HS-AFM monitoring of intermolecular interactions. Binding of KaiA molecules to the CI side of a the KaiC phospho-mimics and b the KaiC dephospho-mimics. Frame rate, 10 fps. Scale bar, 20 nm. c KaiA’s bound state lifetime (τbound) depends on KaiCWT phosphostatus over a 51 h time course of the in vitro cycle of phosphorylation. d Typical AFM images of a KscA channel reconstituted in a DMPC bilayer with (right) or without (left) AgTx2. Height profiles along the white dotted lines in the AFM images are shown below the images. The background illustration behind the height profiles indicates the corresponding structures of the channel and AgTx2. e Time-lapse images of AgTx2 binding to and dissociation from the KcsA channels (top) and time courses of the averaged height h (nm) around the center of the extracellular surface of two corresponding K+ channels (bottom). White dotted squares represent regions of interest for visualization of the tetrameric channels. AgTx2 bindings onto the channels are indicated by white arrowheads on the AFM images. Frame rate, 10 fps. Scale bar, 5 nm
As the second example for the observation of intermolecular interactions, an HS-AFM analysis of the binding dynamics of agitoxin-2 to a K+ channel is described (Sumino et al. 2019). Agitoxin-2 (AgTx2) from scorpion venom is a potent inhibitor of K+ channels. It is known that AgTx2 is a 38 amino acid peptide that binds to the extracellular surface of K+ channels and blocks the passage of ions. However, it has not been uncovered whether the binding dynamics can be explained by a simple two-state model or a more complicated mechanism such as induced fit or conformational selection. Here, single-molecule observation to monitor the binding dynamics of AgTx2 to a K+ channel, KcsA, was carried out using HS-AFM. Since KcsA forms a tetramer arranged in a square, the binding of AgTx2 to the extracellular side of the tetramer bulges the central pore of the channel through which K+ ions pass, elevating the height of the tetramer (Fig. 2d). The analysis of the time course of the height change showed the repeated binding and dissociation of AgTx2 to the KcsA tetramer (Fig. 2e). The analysis of the time course of the height change showed that an increase of the concentration of AgTx2 in the solution leads to an increase in the probability of the bound state. Event-oriented, detailed single-molecule analyses revealed that the affinity of the channel for AgTx2 increased during persistent binding and decreased during persistent dissociation. From these observations, an induced fit model can be proposed which includes four states with at least two high- and low-affinity states of the channel for both, the binding and dissociation states.
Mechanical manipulation and indentation on single molecules with HS-AFM
Since AFM is a mechanically sensitive surface probe, it has been used as a microscopic tool for evaluating the mechanical properties such as stiffness, elastic modulus, and viscosity of sample surfaces in addition to imaging topography (Dufrêne et al. 2013; Kasas and Dietler 2008). Positive utilization of the mechanical contact between the probe and the sample also allows local mechanical indentation and structural manipulation of the sample. Here, recent research examples are introduced in which the characteristics of AFM mechanical measurements are utilized in HS-AFM.
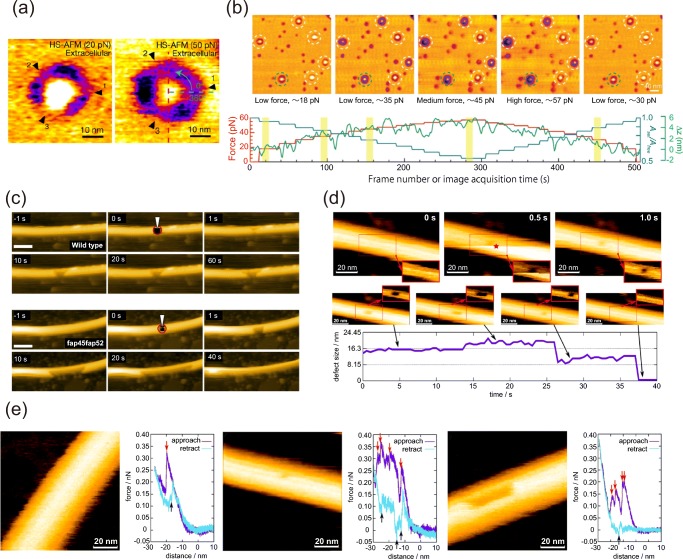
For its operation, HS-AFM employs tapping mode, in which the AFM cantilever is oscillated at its resonant frequency. The probe-surface distance is controlled by a feedback loop that keeps the oscillation amplitude constant. By changing the reference value (cantilever amplitude) of the feedback control during the HS-AFM imaging, the force applied between probe and sample can be continuously varied, and the structural change of a sample can be simultaneously monitored. An excellent application of the force-controlled imaging was demonstrated for the mechanosensitive channel PIEZO1 which converts mechanical force to chemical signals (Lin et al. 2019). PIEZO1 forms a triskelion-shaped homotrimer with a central pore module in a lipid bilayer and the extrasellar surface of the trimer was imaged by HS-AFM under different applied forces (Fig. 3a, left panel). The conformation of the PIEZO1 is also continuously modulated by increasing the probe-surface force (Fig. 3b). Furthermore, the applied force can be rapidly increased at a local point during the imaging and the following events on the sample can be monitored. This technique was applied to assess the mechanical stability of doublet microtubules with or without inner lumen proteins (Fig. 3c) (Owa et al. 2019).
Fig. 3.
Mechanical indentation with HS-AFM. a HS-AFM images of the extracellular side of PIEZO1 in a lipid bilayer observed at specific applied force (left, about 20 pN; right, about 50 pN). b Top: HS-AFM images of PIEZO1 (dashed circles) during the stepwise increase in the loading force. Bottom: loading force (red), Aset/Afree ratio (blue) from the feedback control and z-piezo displacement (green) as function of frame acquisition time. c Time-lapse HS-AFM images of doublet microtubules with (upper panels) and without (lower panels) inner lumen proteins after increasing the force at a local area indicated by the red circles. In the doublet microtubules with inner lumen proteins present, enlargement of the hole usually stopped within a few seconds, whereas most of the B-tubules without the inner lumen proteins were broken by 40 s. Frame rate, 1 fps. Scale bar, 100 nm. d Creation (upper panels) of a defect in a microtubule using in-line-force-curve HS-AFM and the subsequent growth and complete healing of the defect (lower panels). The insets in the images show the region of the dashed rectangle with a pronounced contrast for the defect. The red star indicates the frame and the position of force application. e HS-AFM images after the local mechanical indentation and force curves during the indentation for different situations (left, fully reversible deformation; middle, single dimer defect; right, 12 dimer defect). The red arrows indicate the downward jumps during approach and the black arrows indicate the upward jumps during retraction
A disadvantage of the previously described local force application by HS-AFM is the difficulty to quantify the interaction force due to the oscillating cantilever. To enable an accurate quantitative force evaluation, the in-line force curve mode was developed. In this mode, the cantilever oscillation is stopped at a target position during the scanning and the force curve is acquired within 40 ms (Ganser and Uchihashi 2019). One can quantitatively evaluate the interaction force between the probe and the sample from the force curve. Also, the events directly following the force indentation can be captured. The in-line force curve mode was demonstrated to create single tubulin-dimer defects on a microtubule and monitor the expanding and healing processes of the defects (Fig. 3d). Furthermore, the quantitative evaluation of the energy dissipation depending on the irreversible deformation of microtubules and the size of created defects has been demonstrated (Fig. 3e).
Conclusion
HS-AFM is currently the only microscopic tool capable of visualizing dynamic behaviors of biomolecules in solution on the nanometer scale, in real time, which could previously be observed only as ensemble averages or static images. It has become possible to verify facts and hypotheses using a variety of measurement methods as clear visual evidence. On the other hand, there are still technical difficulties that need to be improved. For one, disturbances caused by direct contact of the probe with delicate proteins and their influence to the structures and physiological functions during imaging are often not negligible. Therefore, further technical developments to minimize the invasiveness are required. In addition, most of the samples to which HS-AFM has been applied are isolated and purified proteins, and it is still difficult to apply the HS-AFM directly to complicated molecular systems involving several kinds of different molecules. One solution for the latter problem, a correlated combination of HS-AFM and single-molecule optical microscopy, has already been developed (Fukuda et al. 2013; Umakoshi et al. 2020).
One future direction to utilize the features of the AFM technique that are not feasible with other microscopic methodologies would be to enhance mechanical characterization. At present, quantitative force measurement can be performed only at a few points during the imaging to achieve a fast image rate. If the fast mapping of mechanical properties becomes possible, we could examine the correlation between structural dynamics and local mechanical properties of proteins and the influences of mechanical properties on protein functions.
Acknowledgments
We thank Prof. Toshio Ando for his long-term dedication to the development of HS-AFM and its application.
Funding information
This work was partly supported by JST/CREST (#JPMJCR13M1); KAKENHI from the Ministry of Education, Culture, Sports, Science and Technology, Japan (19H05389, 18H04512 and 18H01837 to TU, and 19K23737 to CG); and the Joint Research of the Exploratory Research Center on Life and Living Systems (ExCELLS) (ExCELLS program No. 18-101 to T.U.).
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Alessandrini A, Facci P. AFM: a versatile tool in biophysics. Meas Sci Technol. 2005;16:R65–R92. doi: 10.1088/0957-0233/16/6/R01. [DOI] [Google Scholar]
- Ando T. High-speed atomic force microscopy and its future prospects. Biophys Rev. 2018;10:285–292. doi: 10.1007/s12551-017-0356-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ando T, Kodera N, Takai E, et al. A high-speed atomic force microscope for studying biological macromolecules. Proc Natl Acad Sci. 2001;98:12468–12472. doi: 10.1073/pnas.211400898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ando T, Uchihashi T, Fukuma T. High-speed atomic force microscopy for nano-visualization of dynamic biomolecular processes. Prog Surf Sci. 2008;83:337–437. doi: 10.1016/j.progsurf.2008.09.001. [DOI] [Google Scholar]
- Ando T, Uchihashi T, Scheuring S. Filming biomolecular processes by high-speed atomic force microscopy. Chem Rev. 2014;114:3120–3188. doi: 10.1021/cr4003837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casuso I, Khao J, Chami M, et al. Characterization of the motion of membrane proteins using high-speed atomic force microscopy. Nat Nanotechnol. 2012;7:525–529. doi: 10.1038/nnano.2012.109. [DOI] [PubMed] [Google Scholar]
- Cho C, Jang J, Kang Y, et al. Structural basis of nucleosome assembly by the Abo1 AAA+ ATPase histone chaperone. Nat Commun. 2019;10:5764. doi: 10.1038/s41467-019-13743-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colom A, Casuso I, Boudier T, Scheuring S. High-speed atomic force microscopy: cooperative adhesion and dynamic equilibrium of Junctional microdomain membrane proteins. J Mol Biol. 2012;423:249–256. doi: 10.1016/j.jmb.2012.07.004. [DOI] [PubMed] [Google Scholar]
- Deville C, Carroni M, Franke KB, et al. Structural pathway of regulated substrate transfer and threading through an Hsp100 disaggregase. Sci Adv. 2017;3:e1701726. doi: 10.1126/sciadv.1701726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufrêne YF, Martínez-Martín D, Medalsy I, et al. Multiparametric imaging of biological systems by force-distance curve–based AFM. Nat Methods. 2013;10:847–854. doi: 10.1038/nmeth.2602. [DOI] [PubMed] [Google Scholar]
- Frederix PLTM, Akiyama T, Staufer U, et al. Atomic force bio-analytics. Curr Opin Chem Biol. 2003;7:641–647. doi: 10.1016/j.cbpa.2003.08.010. [DOI] [PubMed] [Google Scholar]
- Fukuda S, Uchihashi T, Iino R, et al. High-speed atomic force microscope combined with single-molecule fluorescence microscope. Rev Sci Instrum. 2013;84:073706. doi: 10.1063/1.4813280. [DOI] [PubMed] [Google Scholar]
- Ganser C, Uchihashi T. Microtubule self-healing and defect creation investigated by in-line force measurements during high-speed atomic force microscopy imaging. Nanoscale. 2019;11:125–135. doi: 10.1039/C8NR07392A. [DOI] [PubMed] [Google Scholar]
- Joo C, Balci H, Ishitsuka Y, et al. Advances in single-molecule fluorescence methods for molecular biology. Annu Rev Biochem. 2008;77:51–76. doi: 10.1146/annurev.biochem.77.070606.101543. [DOI] [PubMed] [Google Scholar]
- Kasas S, Dietler G. Probing nanomechanical properties from biomolecules to living cells. Pflugers Arch - Eur J Physiol. 2008;456:13–27. doi: 10.1007/s00424-008-0448-y. [DOI] [PubMed] [Google Scholar]
- Kodera N, Yamamoto D, Ishikawa R, Ando T. Video imaging of walking myosin V by high-speed atomic force microscopy. Nature. 2010;468:72–76. doi: 10.1038/nature09450. [DOI] [PubMed] [Google Scholar]
- Lee S, Sowa ME, Watanabe Y, et al. The structure of ClpB. Cell. 2003;115:229–240. doi: 10.1016/S0092-8674(03)00807-9. [DOI] [PubMed] [Google Scholar]
- Lin Y-C, Guo YR, Miyagi A, et al. Force-induced conformational changes in PIEZO1. Nature. 2019;573:230–234. doi: 10.1038/s41586-019-1499-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller H, Zhou Z, Shepherd J, et al. Single-molecule techniques in biophysics: a review of the progress in methods and applications. Rep Prog Phys. 2018;81:024601. doi: 10.1088/1361-6633/aa8a02. [DOI] [PubMed] [Google Scholar]
- Miyagi A, Chipot C, Rangl M, Scheuring S. High-speed atomic force microscopy shows that annexin V stabilizes membranes on the second timescale. Nat Nanotechnol. 2016;11:783–790. doi: 10.1038/nnano.2016.89. [DOI] [PubMed] [Google Scholar]
- Mori T, Sugiyama S, Byrne M, et al. Revealing circadian mechanisms of integration and resilience by visualizing clock proteins working in real time. Nat Commun. 2018;9:3245. doi: 10.1038/s41467-018-05438-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima M, Imai K, Ito H, et al. Reconstitution of circadian oscillation of cyanobacterial KaiC phosphorylation in vitro. Science (80-) 2005;308:414–415. doi: 10.1126/science.1108451. [DOI] [PubMed] [Google Scholar]
- Nakazaki Y, Watanabe Y-H. ClpB chaperone passively threads soluble denatured proteins through its central pore. Genes Cells. 2014;19:891–900. doi: 10.1111/gtc.12188. [DOI] [PubMed] [Google Scholar]
- Owa M, Uchihashi T, Yanagisawa H, et al. Inner lumen proteins stabilize doublet microtubules in cilia and flagella. Nat Commun. 2019;10:1143. doi: 10.1038/s41467-019-09051-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangl M, Miyagi A, Kowal J, et al. Real-time visualization of conformational changes within single MloK1 cyclic nucleotide-modulated channels. Nat Commun. 2016;7:12789. doi: 10.1038/ncomms12789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan Y, Miyagi A, Wang X, et al. Direct visualization of glutamate transporter elevator mechanism by high-speed AFM. Proc Natl Acad Sci. 2017;114:1584–1588. doi: 10.1073/pnas.1616413114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata M, Yamashita H, Uchihashi T, et al. High-speed atomic force microscopy shows dynamic molecular processes in photoactivated bacteriorhodopsin. Nat Nanotechnol. 2010;5:208–212. doi: 10.1038/nnano.2010.7. [DOI] [PubMed] [Google Scholar]
- Sumino A, Sumikama T, Uchihashi T, Oiki S. High-speed AFM reveals accelerated binding of agitoxin-2 to a K + channel by induced fit. Sci Adv. 2019;5:eaax0495. doi: 10.1126/sciadv.aax0495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchihashi T, Iino R, Ando T, Noji H. High-speed atomic force microscopy reveals rotary catalysis of rotorless F 1-ATPase. Science (80-) 2011;333:755–758. doi: 10.1126/science.1205510. [DOI] [PubMed] [Google Scholar]
- Uchihashi T, Scheuring S. Applications of high-speed atomic force microscopy to real-time visualization of dynamic biomolecular processes. Biochim Biophys Acta, Gen Subj. 2018;1862:229–240. doi: 10.1016/j.bbagen.2017.07.010. [DOI] [PubMed] [Google Scholar]
- Umakoshi T, Fukuda S, Iino R, et al. High-speed near-field fluorescence microscopy combined with high-speed atomic force microscopy for biological studies. Biochim Biophys Acta, Gen Subj. 2020;1864:S0304-4165(19)30061-3. doi: 10.1016/j.bbagen.2019.03.011. [DOI] [PubMed] [Google Scholar]
- Watanabe Y, Motohashi K, Yoshida M. Roles of the two ATP binding sites of ClpB from Thermus thermophilus. J Biol Chem. 2002;277:5804–5809. doi: 10.1074/jbc.M109349200. [DOI] [PubMed] [Google Scholar]
- Zou JX, Revenko AS, Li LB, et al. ANCCA, an estrogen-regulated AAA+ ATPase coactivator for ER , is required for coregulator occupancy and chromatin modification. Proc Natl Acad Sci. 2007;104:18067–18072. doi: 10.1073/pnas.0705814104. [DOI] [PMC free article] [PubMed] [Google Scholar]