Abstract
Optogenetics is a growing technique which allows manipulation of biological events simply by illumination. The technique is appreciated especially in the neuroscience field because of its availability in controlling neuronal functions. A light-gated cation channel, Cr_ChR2 from Chlamydomonas reinhardtii, is the first and mostly applied to optogenetics for activating neuronal excitability. In addition, the molecular mechanism of Cr_ChR2 has been intensively studied by electrophysiology, spectroscopy, X-ray structural studies, etc. Novel cation channelrhodopsins from Guillardia theta, namely, Gt_CCR1–4, were discovered in 2016 and 2017. These channelrhodopsins are more homologous to haloarchaeal rhodopsins, particularly the proton pumps. Thus these cryptophyte-type light-gated cation channels are structurally and mechanistically distinct from chlorophyte channelrhodopsin such as Cr_ChR2. We here compared the photocurrent properties, cation selectivity, and kinetics between well-known Cr_ChR2 and Gt_CCR4. The light sensitivity of Gt_CCR4 is significantly higher than that of Cr_ChR2, while the channel open lifetime is in the same range as that of Cr_ChR2. Gt_CCR4 shows high Na+ selectivity in which the selectivity ratio for Na+ was 37-fold larger than that for Cr_ChR2, which primarily conducts H+. On the other hand, Gt_CCR4 conducted almost no H+ and no Ca2+ under physiological conditions. Other unique features and the applicability of Gt_CCR4 for optogenetics were discussed.
Keywords: Optogenetics, Microbial rhodopsin, Channelrhodopsin, Electrophysiology, Ion channel
Introduction
Microbial-type rhodopsins (type I rhodopsins), found in archaea, bacteria, eukaryota (such as fungi and algae), and viruses, are consist of seven or eight transmembrane helices with a covalently bound all-trans retinal as the chromophore (Ernst et al. 2014; Govorunova et al. 2017). They are physiologically responsible for energy production and the phototaxis reaction in the native organisms. Molecular functions of microbial rhodopsin involve ion transporters, sensors, and light-regulated enzymes. The ion-transporting rhodopsins are categorized into ion pumps and channels. Bacteriorhodopsin (BR) was the first identified outward-directed proton-pumping rhodopsin (Oesterhelt and Stoeckenius 1971). The discovery of a Cl− pump, a Na+ pump, and inward-directed proton pumps has been reported, even until recently (Matsuno-Yagi and Mukohata 1977; Inoue et al. 2013)(Inoue et al. 2016; Polovinkin et al. 2017). Structure-based and spectroscopic studies, when combined with electrophysiology and molecular dynamics studies, revealed the detailed molecular mechanism of these molecules.
In 2002 and 2003, light-gated ion channels, channelrhodopsin-1 and channelrhodopsin-2 (Cr_ChR1 and Cr_ChR2) have been discovered from green (chlorophyte) algae Chlamydomonas reinhardtii (Nagel et al. 2002, 2003). Anion-channel rhodopsins have been discovered in 2015 (Govorunova et al. 2015). New abundant group of microbial-type rhodopsin, heliorhodopsins, with converted membrane topology has been reported in 2018 (Pushkarev et al. 2018). Although their molecular function is still unknown, they are distributed in archaea, bacteria, eukarya, and viruses. Thus, the functions of rhodopsins are still spreading.
Channelrhodopsins and optogenetics
Cation channelrhodopsins, such as Cr_ChR1 and Cr_ChR2, permeate cations non-selectively for H+, Na+, K+, Ca2+, etc. Extensive studies by spectroscopy and electrophysiology have been performed on CrCCR2, revealing the opening/closing dynamics (Schneider et al. 2015). High-resolution X-ray structures of ChRs revealed details of their molecular architecture and provided insight into their photoactivation and ion conduction (Kato et al. 2012; Volkov et al. 2017). Light-induced difference infrared spectra revealed that structural changes of ChR are necessary for its function (Ritter et al. 2008; Lorenz-Fonfria et al. 2013; Ito et al. 2014).
Adequate expression of Cr_ChR2 in neurons allowed the optical manipulation of the action potential (Boyden et al. 2005; Ishizuka et al. 2006). This technique, so called optogenetics, revolutionized the neuroscience research field. Optical stimulation has been widely applied in cultured neurons, tissues, and even freely moving animals, while optical inhibition was realized by light-driven ion pumps such as NpHR (Cl− pump) AR3 (proton pump) or anion channelrhodopsins (ACRs)(Zhang et al. 2007; Chow et al. 2010; Govorunova et al. 2015). After ChR2 has become a standard optogenetics tool, the number of variant molecules has been engineered to improve or to modify the functionality of ChRs. Simultaneously, homologous ChRs have been explored in related organisms (Schneider et al. 2015). One of the important aspect was color tuning of ChRs. Cr_ChR2 displays an action spectrum maximum at 470 nm (Nagel et al. 2003). ChR variants such as C1V1, which is the chimeric version of ChR1 from Chlamydomonas reinhardtii and Volvox carteri, or C1C2 (a green receiver), the chimeric version of Cr_ChR1 and Cr_ChR2, absorb light at around 530~545 nm (Tsunoda and Hegemann 2009; Wen et al. 2010; Prigge et al. 2012). Another red-shifted ChR, Chrimson from Chlamydomonas noctigama, exhibits an absorption maximum at 590 nm, which allows reliable neuronal stimulation by light exceeding 600 nm (Klapoetke et al. 2014). On the other hand, TsChR or PsChR absorb a shorter wavelength, making it possible to excite neurons at 440 nm (Govorunova et al. 2013). The palette of color-tuned ChRs covers almost the entire visible range, which enables to manipulate neuronal excitability by any choice of visible light.
The lifetime of channel opening can be extended by mutations at C128 and D156 (DC pair) which form a hydrogen bond bridge in Cr_ChR2. Mutations at C128 slowed the kinetics of channel closing up to 1000-fold (Berndt et al. 2009). Cr_ChR2 C128S/D156A displayed an even stronger effect, namely, prolonged lifetime of the open channel by as much as 30 min (Yizhar et al. 2011).
DTD channelrhodopsins
The amino acid motive in TM3 is the functional determinant in microbial rhodopsins. For the intramolecular proton transport, BR has a proton acceptor and donor attached to the protonated Schiff base, which are D85 and D96 located at TM3, respectively (Fig. 1b) (Ernst et al. 2014). T89 forms a hydrogen bond with D85 in BR, and these three residues are highly conserved among archaeal light-driven proton-pumping rhodopsins.
Fig. 1.
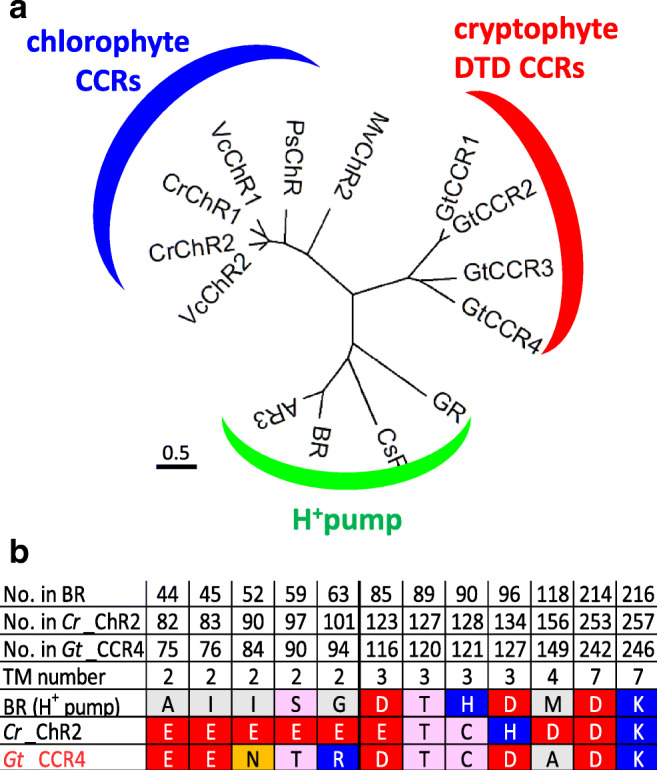
a A phylogenetic tree of two families of cation channelrhodopsins (CCRs) and proton pumps. Chlorophyte CCRs shown are from Chlamydomonas reinhardtii (Cr_ChR1, Cr_ChR2), Volvox carteri (Vc_ChR1, Vc_ChR2), Mesostigma viride (Mv_ChR2), and Platymonas subcordiformis (Ps_ChR). Cryptophyte DTD CCRs are from Guillardia theta (Gt_CCR1–4). H+ pumps shown are from Gloeobacter violaceus (GR), Coccomyxa subellipsoidea C-169 (CsR), Halobacterium salinarum (BR), and Halorubrum sodomense (AR3). b Amino acid alignments of bacteriorhodopsin (BR), Cr_ChR2, and Gt_CCR4. The characteristic of amino acids in BR, Cr_ChR2, and Gt_CCR4 were listed. In addition, amino acid numbers of each protein and transmembrane helix (TM) numbers are indicated
Recently, light-driven Na+ and Cl−-pumping rhodopsins are found from marine bacteria (Inoue et al. 2013; Yoshizawa et al. 2014), which contain residues of NDQ and NTQ, at the corresponding position of D85, T89, and D96 in BR, respectively. Therefore, the DTD, NDQ, and NTQ motifs are the characteristic of the light-driven archaeal proton pump, the eubacterial Na+ pump, and the eubacterial Cl− pump, respectively. Eubacterial proton pump proteorhodopsin (PR) possesses the DTE motif, while archaeal Cl− pump halorhodopsin (HR) possesses the TSA motif (Kandori 2015). These motifs substantially characterize the function of ion pump rhodopsins, namely, the proton pump exhibits DTD or DTE, where the first and third residues are carboxylates acting for proton transfer. In contrast, Na+ pump displays NDQ, where only the second residue is a carboxylate, and Cl− pump contains only neutral residues such as NTQ and TSA.
Compared to ion pumps, the motif is more diverged for channelrhodopsins (ChRs). In fact, ChR1 (CrCCR1) and 2 (CrCCR2) from Chlamydomonas reinhardtii possess an ETH motif, while ChR1 from Chlamydomonas augustae (CaCCR1) and Mesostigma viride (MvCCR1) contain ECH and ETA motifs, respectively (Nagel et al. 2002; Hou et al. 2012; Watanabe et al. 2016). Anion channelrhodopsin 1 (Gt_ACR1) and 2 (Gt_ACR2) from Guillardia theta contain STL and STQ motifs, respectively (Govorunova et al. 2015). A novel cation channelrhodopsin was reported in 2016 and 2017 from Guillardia theta, namely, Gt_CCR1–4 (Govorunova et al. 2016; Yamauchi et al. 2017), though DTD has been unique to light-driven archaeal proton pumps. Guillardia theta, a cryptophyte, encodes more than 40 microbial rhodopsins, including anion channels, Gt_ACR1 and Gt_ACR2 (Curtis et al. 2012). Patch-clamp measurements showed that three DTD rhodopsins from G. theta (Gt_CCR1, Gt_CCR2, Gt_CCR3, and Gt_CCR4) function as cation channels, suggesting that channel function in rhodopsins has evolved via multiple routes. Amino acid sequences of these cation channelrhodopsins (CCRs) are more homologous to haloarchaeal rhodopsins, such as BR, than to chlorophyte CCRs including Cr_ChR2. In fact, the phylogenetic tree in Fig. 1a shows that Gt_CCR1–4 are well separated from the cluster of chlorophyte CCR and that they are closer to H+-pumping rhodopsins. Gt_CCRs conserve the characteristic amino-acid residues involved in unidirectional proton transfer, including the proton acceptor D85 and the proton donor D96 in BR, while D96 in BR is replaced with a positively charged histidine residue (H134 in Cr_ChR2) (Fig. 1b).
On the other hand, a characteristic glutamic acid in TM2 (E90 in Cr_ChR2) which is crucial for channel gating and ion selectivity in Cr_ChR2 is not conserved in Gt_CCRs (Asn in Gt_CCR4) (Sugiyama et al. 2009; Wietek et al. 2014)(Fig. 1b). Cr_ChR2 possesses a so-called D-C pair (C128 and D156 in Cr_ChR2), which is responsible for the channel open lifetime (Berndt et al. 2009; Nack et al. 2010; Dawydow et al. 2014). But such pair is not found in Gt_CCRs. Thus, overall sequence patterns separate these cryptophyte CCRs form chlorophyte channels. The molecular mechanisms, such as the channel gating mechanism and ion selectivity, might be distinct in chlorophyte CCRs. It was already revealed that the retinal Schiff base (SB) in Gt_CCR2 rapidly deprotonates to the D85 homolog, as in BR, upon photoisomerization. Channel opening requires deprotonation of the D96 homolog (Sineshchekov et al. 2017). Reprotonation of SB was achieved by the direct return of a proton from the D85 homolog. This step (M decay) corresponds to the ion channel closing, implicating tight coupling between retinal dynamics and channel function.
However, the M decay of Cr_ChR2 does not functionally correspond to the channel closing. For the reprotonation of SB, E156 in TM4 provides the proton in Cr_ChR2 (Lorenz-Fonfria et al. 2013). It has been also reported that the secondary structural change in the primary reaction in Gt_CCR4 was much smaller than in Cr_ChR2 (Yamauchi et al. 2017). These differences in the molecular mechanism place the cryptophyte CCR in a new family of channelrhodopsins, which we described as “DTD channelrhodopsins” or “BR-like cation channelrhodopsins”(Govorunova et al. 2016; Yamauchi et al. 2017; Shigemura et al. 2019). After the emergence of the DTD ChR, its molecular properties are intriguing in comparison with other ChRs. In the following chapter, we compare ion channel properties of Gt_CCR4 and Cr_ChR2.
Electrophysiological properties of channelrhodopsins, Gt_CCR4 and Cr_ChR2
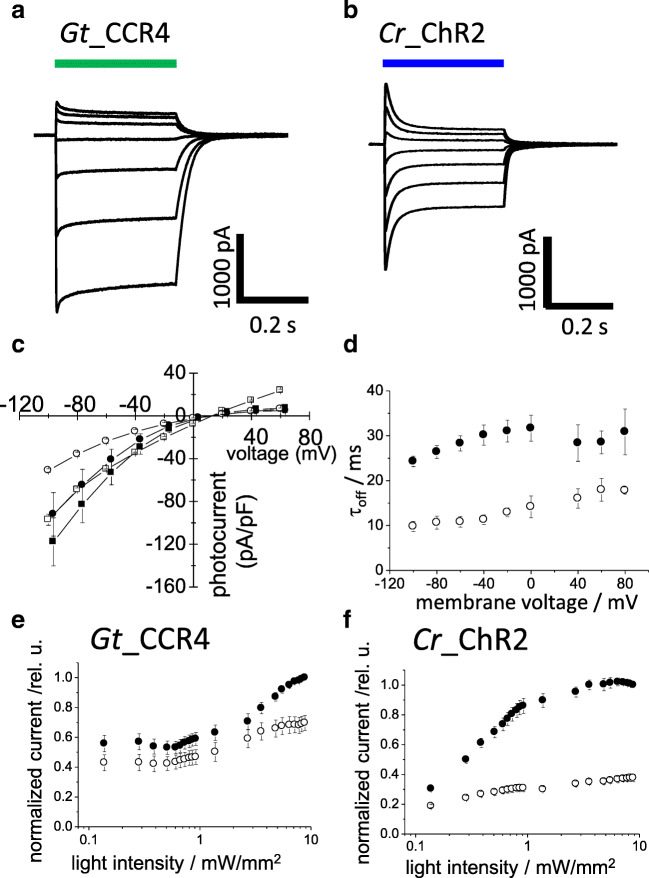
Light-induced ion conductance (photocurrents) has been characterized by electrical recordings from ChR-expressing cells mainly under a voltage-clamp configuration. Figures 2 a and b show representative photocurrents of Gt_CCR4 and Cr_ChR2 measured in ND7/23 cells (Shigemura et al. 2019). Upon illumination with a bright continuous light (530 nm, 6.8 mW/mm2), large photocurrent was observed from the Gt_CCR4-expressing cells (Fig. 2a). As shown on the I-V plot in Fig. 2b, the current amplitude depends on the applied voltage. At negative membrane voltage, Gt_CCR4 exhibits inward-directed photocurrent carried by cations. The photocurrent reached − 2 nA at − 60 mV (Fig. 2a). Around 0~+ 20 mV, the current direction is reversed from negative to positive direction, indicating outward-directed cation flux. As for the current shape, it reaches its peak within a few milliseconds upon illumination, followed by a slight decay into a steady-state level. The current amplitude of the steady state still retained about 80% of the transient peak component. After shutting off light, the photocurrent decayed into zero level with a time constant of 20–30 ms (Fig. 2d).
Fig. 2.
Basic ion channel properties of Gt_CCR4 and Cr_ChR2. a and b Typical photocurrent of two CCRs under voltage clamp configuration. Each cation channelrhodopsin expressed in ND7/23 cells was stimulated by green (530 nm) or blue (470 nm) light. Membrane potentials were clamped from − 60 mV to + 60 mV in + 20 mV steps. c Current-voltage relationship (I-V plot) of Gt_CCR4 (filled symbol) and Cr_ChR2 (empty symbol). Current peak component (square) and steady-state amplitude (circle) of two channels are depicted. d Current-decay kinetics of Gt_CCR4 and Cr_ChR2. τoff is plotted as a function of membrane voltage. Filled circle, Gt_CCR4; empty circle, Cr_ChR2. e and f Light power dependency of photocurrents from Gt_CCR4 to Cr_ChR2. Each channel was stimulated by 530 nm (Gt_CCR4) and 470 nm (Cr_ChR2). Current peak component (filled circle) and steady-state amplitude (empty circle) are depicted. For experimental details, refer Shigemura et al. 2019
The photocurrent from Cr_ChR2 showed a large peak component which reached about − 2 nA at − 60 mV (Fig. 2b). However, the current shape is obviously different from that of Gt_CCR4. The initial peak decayed into a lower level by about 50%. This means that Cr_ChR2 exhibits a markedly large inactivation compared to Gt_CCR4. Figure 2 c depicts the current-voltage relationship of photocurrent (I-V plot) from Gt_CCR4 and Cr_ChR2. Both peak component (Ip) and steady-state current (Iss) are plotted. The reversal potentials of two channels are around + 10 mV. The shape of the I-V plot from Gt_CCR4 indicates strong inward rectification. Iss of Cr_ChR2 displayed similarly inward rectification, while Ip was weakly rectified, in which a markedly outward-directed current was observed at positive membrane voltages (Fig. 2a and b).
Kinetics of the photocurrent decay after shutting off the light is shown in Fig. 2d. The time constant of Gt_CCR4 is about 25–35 ms under a membrane voltage between − 100 and 80 mV, while Cr_ChR2 showed faster kinetics by about 10–20 ms. Light sensitivity in the photocurrent of two channels are compared (Figs. 2e and 2f). The photocurrent amplitude from Cr_ChR2 grows as a typical sigmoidal curve for both the initial peak and the steady-state components (Fig. 2f). EC50 was determined as 0.8 mW/mm2 for Ip and 0.35 mW/mm2 for Iss. On the other hand, the Gt_CCR4 current showed unique growth in terms of light dependency with two apparent phases in which the current first saturated at 0.1 mW/mm2 at about 50% of full activation, followed by the second phase of growth from 1 to 10 mW/mm2 (Fig. 2e). The EC50 was determined as 0.13 mW/mm2 for Ip and 0.18 mW/mm2 for Iss. Thus, Gt_CCR4 is more sensitive to light with respect to channel activation.
Ion selectivity
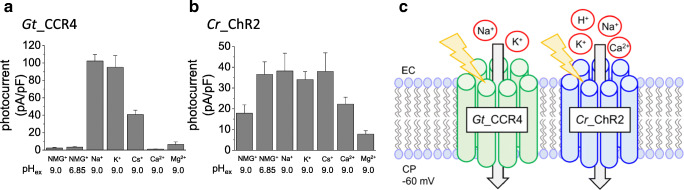
In electrophysiological studies, ion selectivity is usually investigated by reversal potential shift (Hille 2001). Cr_ChR2 conducts not only protons but also monovalent cations such as Na+ and K+ and divalent cations such as Ca2+ (Kleinlogel et al. 2011). On the other hand, Gt_CCR1-4 are highly selective for monovalent cation such as Na+ and K+ and less selective for H+(Govorunova et al. 2016; Shigemura et al. 2019). Actually, H+ permeability for Cr_ChR2 is about 1 × 106 (Nagel et al. 2003), while that for Gt_CCR4 is 2.1 × 104, indicating about 37-fold less permeability for H+ in Gt_CCR4 (Shigemura et al. 2019).
The photocurrent amplitude of Gt_CCR4 and Cr_ChR2 at − 60 mV under each condition is summarized in Fig. 3a and b. The current amplitude of Gt_CCR4 was significantly large in the presence of Na+ and K+, close to – 100 pA/pF, and in the presence of Cs+ at about – 40 pA/pF. In contrast, only a negligible current was observed at low pH or in the presence of Ca2+ or Mg2+. This supports the notion that Gt_CCR4 is more selective for monovalent metal cations and less selective for H+ and divalent cations. Interestingly, photocurrents by Gt_CCR4 were suppressed at a higher Ca2+ concentration (Shigemura et al. 2019). This implies that cation flow is blocked by Ca2+. In contrast, photocurrent amplitudes from Cr_ChR2 in various ionic conditions do not show such dramatic change, which is due to low ion selectivity in Cr_ChR2 (Fig. 3b).
Fig. 3.
Ion selectivity of Gt_CCR4 and Cr_ChR2. a and b Comparison of photocurrent density in the presence of various cations at − 60 mV. Experimental details were described in Shigemura et al. 2019. c Schematic image of ion conductance of Gt_CCR4 and Cr_ChR2
Summary
In this review, we aimed to elucidate the ion channel properties of a recently discovered light-gated cation channel Gt_CCR4 from a cryptophyte and compare it to the well-known Cr_ChR2 from Chlamydomonas reinhardtii. In ND7/23 cells, Gt_CCR4 showed a large current density (Fig. 2c). Inactivation of the photocurrent obtained from Gt_CCR4 was smaller than in Cr_ChR2. In other words, a large current was observed under constant light (Fig. 2a and b). These characteristics promise stable and reproducible stimulation of neuronal excitability by Gt_CCR4.
The light sensitivity of Gt_CCR4 is higher than that of Cr_ChR2, with a particular steady-state component (Iss) (Fig. 2e and f). ChR variants with high light sensitivity have already been developed (Berndt et al. 2009; Yizhar et al. 2011; Dawydow et al. 2014). However, those have a long channel life time with at least two orders of magnitude or even much longer. Therefore, these are inappropriate for high-frequency light stimulation. In contrast, Gt_CCR4 has a short open life time of 25–30 ms, which is about the same range as Cr_ChR2, i.e., 10–15 ms (Fig. 1d). Together, Gt_CCR4 is light sensitive and useful as an optogenetics tool with high time resolution. Optical irradiation causes heat and elevates temperature by 0.2~2 °C, especially in cranial nerve experiments (Owen et al. 2019). Moreover, it has been demonstrated that the rise in temperature suppressed neuronal spiking in multiple brain regions, serving as a warning of the use of strong light for neuronal stimulation. Such an undesirable artifact has to be avoided by lowering light intensity or reducing duration of illumination, while effective depolarization has to be stably maintained. Gt_CCR4 has the potential for overcoming this problem.
H+ permeability is high for Cr_ChR2 (Nagel et al. 2003). Not only for monovalent cations such as Na+ and K+, permeability for Ca2+ has been reported (Nagel et al. 2003; Kleinlogel et al. 2011). On the other hand, Gt_CCR4 showed high selectivity in monovalent metal cations and low H+ permeability (Fig. 3c). The permeability of a divalent cation such as Ca2+ seems to be very low or negligible. The position that is important to ion selectivity has already been identified in Cr_ChR2. E90 in the central gate is crucial for cation/anion selection (Wietek et al. 2014). The outer gate in Chrimson (E139) on the extracellular side is important for Na+ extrusion (Vierock et al. 2017). Both glutamic acids are not conserved in Gt_CCR4, implying that a different ion selection property resides in DTD channels. It would be necessary to study selectivity based on variant analysis and structural information in the future. Considering its application in optogenetics, Gt_CCR4 would not cause a significant change to pH in the cell membrane because of its very low H+ permeability, which could be advantageous when an unknown effect by pH needs to be prevented. To enable optical stimulation without improper calcium signaling, Gt_CCR4 might work better than Cr_ChR2.
In conclusion, Gt_CCR4 shows characteristic channel properties, such as high conductance and high cation selectivity without significant inactivation, which would be an appropriate set of features for optogenetics applications. We are currently assessing the feasibility of Gt_CCR4 as an optical stimulator in cultured neurons.
Funding information
This work was supported by the Japanese Ministry of Education, Culture, Sports, Science and Technology (25104009, 15H02391 to H.K and 18 K06109 to S.P.T.), a JST CREST grant (JPMJCR1753 to H.K.), and a JST PRESTO grant (JPMJPR1688 to S.P.T). S.H. is a Research Fellow of the Japan Society for the Promotion of Science (JSPS Research Fellow).
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Berndt A, Yizhar O, Gunaydin LA, et al. Bi-stable neural state switches. Nat Neurosci. 2009;12:229–234. doi: 10.1038/nn.2247. [DOI] [PubMed] [Google Scholar]
- Boyden ES, Zhang F, Bamberg E, et al. Millisecond-timescale, genetically targeted optical control of neural activity. Nat Neurosci. 2005;8:1263–1268. doi: 10.1038/nn1525. [DOI] [PubMed] [Google Scholar]
- Chow BY, Han X, Dobry AS, et al. High-performance genetically targetable optical neural silencing by light-driven proton pumps. Nature. 2010;463:98–102. doi: 10.1038/nature08652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis BA, Tanifuji G, Maruyama S, et al. Algal genomes reveal evolutionary mosaicism and the fate of nucleomorphs. Nature. 2012;492:59–65. doi: 10.1038/nature11681. [DOI] [PubMed] [Google Scholar]
- Dawydow A, Gueta R, Ljaschenko D, et al. Channelrhodopsin-2-XXL, a powerful optogenetic tool for low-light applications. Proc Natl Acad Sci U S A. 2014;111:13972–13977. doi: 10.1073/pnas.1408269111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst OP, Lodowski DT, Elstner M, et al. Microbial and animal rhodopsins: structures, functions, and molecular mechanisms. Chem Rev. 2014;114:126–163. doi: 10.1021/cr4003769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govorunova EG, Sineshchekov OA, Li H, et al. Characterization of a highly efficient blue-shifted channelrhodopsin from the marine alga Platymonas subcordiformis. J Biol Chem. 2013;288:29911–29922. doi: 10.1074/jbc.M113.505495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govorunova EG, Sineshchekov OA, Janz R, et al. Neuroscience. Natural light-gated anion channels: a family of microbial rhodopsins for advanced optogenetics. Science (80- ) 2015;349:647–650. doi: 10.1126/science.aaa7484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govorunova EG, Sineshchekov OA, Spudich JL. Structurally distinct cation channelrhodopsins from cryptophyte algae. Biophys J. 2016;110:2302–2304. doi: 10.1016/j.bpj.2016.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govorunova EG, Sineshchekov OA, Li H, Spudich JL. Microbial rhodopsins: diversity, mechanisms, and optogenetic applications. Annu Rev Biochem. 2017;86:845–872. doi: 10.1146/annurev-biochem-101910-144233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hille B. Ion Channels of Excitable Membranes. 2001. Chapter 1 introduction. [Google Scholar]
- Hou SY, Govorunova EG, Ntefidou M, et al. Diversity of Chlamydomonas channelrhodopsins. Photochem Photobiol. 2012;88:119–128. doi: 10.1111/j.1751-1097.2011.01027.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue K, Ono H, Abe-Yoshizumi R, et al. A light-driven sodium ion pump in marine bacteria. Nat Commun. 2013;4:1678. doi: 10.1038/ncomms2689. [DOI] [PubMed] [Google Scholar]
- Inoue K, Ito S, Kato Y, et al. A natural light-driven inward proton pump. Nat Commun. 2016;7:13415. doi: 10.1038/ncomms13415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizuka T, Kakuda M, Araki R, Yawo H. Kinetic evaluation of photosensitivity in genetically engineered neurons expressing green algae light-gated channels. Neurosci Res. 2006;54:85–94. doi: 10.1016/j.neures.2005.10.009. [DOI] [PubMed] [Google Scholar]
- Ito S, Kato HE, Taniguchi R, et al. Water-containing hydrogen-bonding network in the active center of channelrhodopsin. J Am Chem Soc. 2014;136:3475–3482. doi: 10.1021/ja410836g. [DOI] [PubMed] [Google Scholar]
- Kandori H. Ion-pumping microbial rhodopsins. Front Mol Biosci. 2015;2:52. doi: 10.3389/fmolb.2015.00052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato HE, Zhang F, Yizhar O, et al. Crystal structure of the channelrhodopsin light-gated cation channel. Nature. 2012;482:369–374. doi: 10.1038/nature10870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klapoetke NC, Murata Y, Kim SS, et al. Independent optical excitation of distinct neural populations. Nat Methods. 2014;11:338–346. doi: 10.1038/nmeth.2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinlogel S, Feldbauer K, Dempski RE, et al. Ultra light-sensitive and fast neuronal activation with the Ca2+−permeable channelrhodopsin catCh. Nat Neurosci. 2011;14:513–518. doi: 10.1038/nn.2776. [DOI] [PubMed] [Google Scholar]
- Lorenz-Fonfria VA, Resler T, Krause N, et al. Transient protonation changes in channelrhodopsin-2 and their relevance to channel gating. Proc Natl Acad Sci. 2013;110:E1273–E1281. doi: 10.1073/pnas.1219502110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuno-Yagi A, Mukohata Y. Two possible roles of bacteriorhodopsin; a comparative study of strains of Halobacz’eril’m halobium differing in pigmentation. Biochem Biophys Res Commun. 1977;78:237–243. doi: 10.1016/0006-291X(77)91245-1. [DOI] [PubMed] [Google Scholar]
- Nack M, Radu I, Gossing M, et al. The DC gate in channelrhodopsin-2: crucial hydrogen bonding interaction between C128 and D156. Photochem Photobiol Sci. 2010;9:194–198. doi: 10.1039/b9pp00157c. [DOI] [PubMed] [Google Scholar]
- Nagel G, Ollig D, Fuhrmann M, et al. Channelrhodopsin-1: a light-gated proton channel in green algae. Science (80- ) 2002;296:2395–2398. doi: 10.1126/science.1072068. [DOI] [PubMed] [Google Scholar]
- Nagel G, Szellas T, Huhn W, et al. Channelrhodopsin-2, a directly light-gated cation-selective membrane channel. Proc Natl Acad Sci U S A. 2003;100:13940–13945. doi: 10.1073/pnas.1936192100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oesterhelt D, Stoeckenius W. Rhodopsin-like protein from the purple membrane of Halobacterium halobium. Nat New Biol. 1971;233:149–152. doi: 10.1038/10.1038/newbio233149a0. [DOI] [PubMed] [Google Scholar]
- Owen SF, Liu MH, Kreitzer AC. Thermal constraints on in vivo optogenetic manipulations. Nat Neurosci. 2019;22:1061–1065. doi: 10.1038/s41593-019-0422-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polovinkin V, Rodriguez-Valera F, Bueldt G, et al. Inward H + pump xenorhodopsin: mechanism and alternative optogenetic approach. Sci Adv. 2017;3:e1603187. doi: 10.1126/sciadv.1603187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prigge M, Schneider F, Tsunoda SP, et al. Color-tuned channelrhodopsins for multiwavelength optogenetics. J Biol Chem. 2012;287:31804–31812. doi: 10.1074/jbc.M112.391185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pushkarev Alina, Inoue Keiichi, Larom Shirley, Flores-Uribe José, Singh Manish, Konno Masae, Tomida Sahoko, Ito Shota, Nakamura Ryoko, Tsunoda Satoshi P., Philosof Alon, Sharon Itai, Yutin Natalya, Koonin Eugene V., Kandori Hideki, Béjà Oded. A distinct abundant group of microbial rhodopsins discovered using functional metagenomics. Nature. 2018;558(7711):595–599. doi: 10.1038/s41586-018-0225-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritter E, Stehfest K, Berndt A, et al. Monitoring light-induced structural changes of channelrhodopsin-2 by UV-visible and Fourier transform infrared spectroscopy. J Biol Chem. 2008;283:35033–35041. doi: 10.1074/jbc.M806353200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider F, Grimm C, Hegemann P. Biophysics of channelrhodopsin. Annu Rev Biophys. 2015;44:167–186. doi: 10.1146/annurev-biophys-060414-034014. [DOI] [PubMed] [Google Scholar]
- Shigemura Shunta, Hososhima Shoko, Kandori Hideki, Tsunoda Satoshi P. Ion Channel Properties of a Cation Channelrhodopsin, Gt_CCR4. Applied Sciences. 2019;9(17):3440. doi: 10.3390/app9173440. [DOI] [Google Scholar]
- Sineshchekov OA, Govorunova EG, Li H, Spudich JL. Bacteriorhodopsin-like channelrhodopsins: alternative mechanism for control of cation conductance. Proc Natl Acad Sci. 2017;114:E9512–E9519. doi: 10.1073/pnas.1710702114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama Y, Wang H, Hikima T, et al. Photocurrent attenuation by a single polar-to-nonpolar point mutation of channelrhodopsin-2. Photochem Photobiol Sci. 2009;8:328–336. doi: 10.1039/b815762f. [DOI] [PubMed] [Google Scholar]
- Tsunoda SP, Hegemann P. Glu 87 of channelrhodopsin-1 causes pH-dependent color tuning and fast photocurrent inactivation. In: Photochemistry and Photobiology; 2009. pp. 564–569. [DOI] [PubMed] [Google Scholar]
- Vierock J, Grimm C, Nitzan N, Hegemann P. Molecular determinants of proton selectivity and gating in the red-light activated channelrhodopsin Chrimson. Sci Rep. 2017;7:1–15. doi: 10.1038/s41598-017-09600-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkov Oleksandr, Kovalev Kirill, Polovinkin Vitaly, Borshchevskiy Valentin, Bamann Christian, Astashkin Roman, Marin Egor, Popov Alexander, Balandin Taras, Willbold Dieter, Büldt Georg, Bamberg Ernst, Gordeliy Valentin. Structural insights into ion conduction by channelrhodopsin 2. Science. 2017;358(6366):eaan8862. doi: 10.1126/science.aan8862. [DOI] [PubMed] [Google Scholar]
- Watanabe S, Ishizuka T, Hososhima S, et al. The regulatory mechanism of ion permeation through a channelrhodopsin derived from Mesostigma viride (MvChR1) Photochem Photobiol Sci. 2016;15:365–374. doi: 10.1039/c5pp00290g. [DOI] [PubMed] [Google Scholar]
- Wen Lei, Wang Hongxia, Tanimoto Saki, Egawa Ryo, Matsuzaka Yoshiya, Mushiake Hajime, Ishizuka Toru, Yawo Hiromu. Opto-Current-Clamp Actuation of Cortical Neurons Using a Strategically Designed Channelrhodopsin. PLoS ONE. 2010;5(9):e12893. doi: 10.1371/journal.pone.0012893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wietek J, Wiegert JS, Adeishvili N, et al. Conversion of channelrhodopsin into a light-gated chloride channel. Science. 2014;344:409–412. doi: 10.1126/science.1249375. [DOI] [PubMed] [Google Scholar]
- Yamauchi Y, Konno M, Ito S, et al. Molecular properties of a DTD channelrhodopsin from Guillardia theta. Biophys Physicobiology. 2017;14:57–66. doi: 10.2142/biophysico.14.0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yizhar O, Fenno LE, Prigge M, et al. Neocortical excitation/inhibition balance in information processing and social dysfunction. Nature. 2011;477:171–178. doi: 10.1038/nature10360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshizawa S, Kumagai Y, Kim H, et al. Functional characterization of flavobacteria rhodopsins reveals a unique class of light-driven chloride pump in bacteria. Proc Natl Acad Sci U S A. 2014;111:6732–6737. doi: 10.1073/pnas.1403051111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Wang L-P, Brauner M, et al. Multimodal fast optical interrogation of neural circuitry. Nature. 2007;446:633–639. doi: 10.1038/nature05744. [DOI] [PubMed] [Google Scholar]