Biological molecular motors can operate robustly and efficiently in highly fluctuating nano-scale environments. How these molecules achieve such remarkable functions is an intriguing question that requires the understanding of the general principles of structural design, enzymatic kinetics, and nonequilibrium dynamics of the biological machineries. By bringing together both experimental and theoretical experts from interdisciplinary fields (Fig. 1), this symposium aimed to explore (A) the novel experimental techniques in probing the energetic efficiencies at the single-molecule level; (B) the recent theoretical methods in explaining the biophysical principles of molecular energetics; and (C) the common strategies shared among different bio-molecular machines in achieving high efficiencies.
Fig. 1.
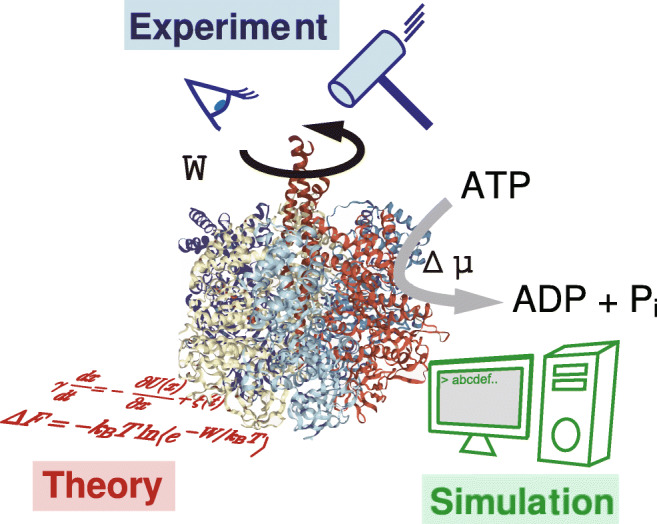
This symposium brings together experts in experiment, simulation, and theory of the field to explore the nonequilibrium energetics of biological molecular motors
This symposium was motivated and co-sponsored by the collaborative project “Molecular Engine” (a Grant-in Aid for Scientific Research on Innovative Areas, JSPS KAKENHI Grant No. JP18H05427) led by Kazushi Kinbara (Tokyo Inst. of Tech.). The goal of the project is to design molecules with autonomous functions resembling those in biological molecular motors, especially focusing on the functions associated with energy conversions.
The symposium started with a brief remark by Chun-Biu Li (Stockholm Univ.). Shoichi Toyabe (Tohoku Univ.) introduced basic concepts of energetics in the context of molecular motors, including efficiencies and precision.
Takayuki Ariga (Yamaguchi Univ.) introduced their recent single-molecule assay of energetics in kinesin molecules, which combined a force-clamp experiment with high-speed feedback and a nonequilibrium relation to determine the heat dissipation (Ariga et al. 2018). They found that kinesin has a large internal dissipation, implying a low energetic efficiency.
Toshiharu Suzuki (Tokyo Inst. of Tech.) showed their works on the ATP synthase with a novel crystallographic technique. They visualized the structural rearrangements in elementary chemical steps, leading to an understanding of the highly efficient energy conversion by the ATP synthase (Soga et al. 2017; Guo et al. 2019).
Holger Flechsig (Kanazawa Univ.) presented their theoretical model describing the walking motions of myosin V along actin filaments to understand its dynamics and energetics. Their model successfully reproduced and explained the recent observations from interactive high-speed AFM experiments, showing that stimulations by the AFM tip can trigger the myosin walking in the absence of chemical energy supply from ATP.
Jin Yu (UC Irvine) discussed the energetics and structural dynamics of a viral RNA polymerase to understand its mechano-chemical coupling and fidelity (Long et al. 2019). Their massive molecular-dynamics simulations quantitatively characterized the Brownian-ratchet working scenario of the polymerase.
Simone Pigolotti (Okinawa Inst of Sci. and Tech.) discussed their theoretical finding on the trade-off between the error and speed in the biopolymer synthesis process (Sartori and Pigolotti 2013). He also discussed the intriguing correlation between fluctuations in the error and speed (Chiuchiú et al. 2019).
Kei-ichi Okazaki (Inst. for Mol. Sci.) presented a novel approach to extract the energy landscape of a linear motor chitinase from single-molecule trajectories based on hidden Markov model to estimate parameters of a diffusion model on discrete potential surfaces (Okazaki et al. 2019). Their work revealed the dynamic burnt-bridge Brownian-ratchet mechanism of the chitinase motor.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Shoichi Toyabe, Email: toyabe@tohoku.ac.jp.
Chun-Biu Li, Email: cbli@math.su.se.
Kazushi Kinbara, Email: kkinbara@bio.titech.ac.jp.
References
- Ariga T, Tomishige M, Mizuno D. Nonequilibrium energetics of molecular motor kinesin. Phys Rev Lett. 2018;121:218,101. doi: 10.1103/PhysRevLett.121.218101. [DOI] [PubMed] [Google Scholar]
- Chiuchiú D, Tu Y, Pigolotti S. Error-speed correlations in biopolymer synthesis. Phys Rev Lett. 2019;123:0381,013. doi: 10.1103/PhysRevLett.123.038101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Suzuki T, Rubinstein JL. Structure of a bacterial ATP synthase. eLife. 2019;8:e43128. doi: 10.7554/eLife.43128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long C, Ee C, Da L-T, Yu J. A viral t7 RNA polymerase ratcheting along DNA with fidelity control. Comp Str Biotech J. 2019;17:638–644. doi: 10.1016/j.csbj.2019.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazaki K, Nakamura A, Iino R (2019) Dynamic free energy landscape of a linear motor chitinase from single-particle tracking trajectories. biorXiv 10.1101/655878
- Sartori P, Pigolotti S. Kinetic versus energetic discrimination in biological copying. Phys Rev Lett. 2013;110:188,101. doi: 10.1103/PhysRevLett.110.188101. [DOI] [PubMed] [Google Scholar]
- Soga N, Kimura K, Kinosita K, Yoshida M, Suzuki T. Perfect chemomechanical coupling of FoF1-ATP synthase. Proc Nat Acad Sci. 2017;114:4960–4965. doi: 10.1073/pnas.1700801114. [DOI] [PMC free article] [PubMed] [Google Scholar]


