Abstract
Highly variable resistance rates to erythromycin and clindamycin have been reported in the β-hemolytic streptococcal species Streptococcus pyogenes, Streptococcus agalactiae, and Streptococcus dysgalactiae, depending on geographic and temporal context. In the present study we aimed to examine the longitudinal trends of antimicrobial resistance in these three species in a northern European setting. Furthermore, we used whole genome sequencing to identify resistance determinants and the mobile genetic elements involved in their dissemination, as well as elucidate phylogenetic relationships. All cases of invasive β-hemolytic streptococcal diseases in Health Region Bergen, western Norway, in the period 2004 to 2018 were retrospectively identified, comprising 271, 358, and 280 cases of S. pyogenes, S. agalactiae, and S. dysgalactiae, respectively. Antimicrobial susceptibility testing revealed a gradual but significant increase in erythromycin and clindamycin resistance for S. agalactiae and S. dysgalactiae during the study period. Whole genome sequencing of the erythromycin and clindamycin resistant bacterial population revealed a substantial phylogenetic diversity in S. agalactiae and S. dysgalactiae. However, the mobile genetic elements harboring the resistance determinants showed remarkable intra- and interspecies similarities, suggesting a dissemination of antimicrobial resistance predominantly through conjugative transfer rather than clonal expansion of resistant strains in these two species. Conversely, antimicrobial resistance in S. pyogenes remained low, apart from a transient outbreak of a clindamycin and erythromycin resistant emm11/ST403-clone in 2010–2012. Increased epidemiological attentiveness is warranted to monitor the emerging threat of antimicrobial resistance in β-hemolytic streptococci, particularly in S. agalactiae and S. dysgalactiae.
Keywords: β-hemolytic streptococci, antimicrobial resistance, Streptococcus dysgalactiae, Streptococcus pyogenes, Streptococcus agalactiae
Introduction
The major human pathogens among β-hemolytic streptococci (BHS) are Streptococcus pyogenes (Lancefield group A streptococcus, GAS), Streptococcus agalactiae (Lancefield group B streptococcus, GBS), and Streptococcus dysgalactiae (Lancefield group C and G streptococcus, SD). These phylogenetically closely related species produce overlapping clinical manifestations, and collectively they are responsible for substantial global disease burden (Carapetis et al., 2005; Le Doare and Heath, 2013; Rantala, 2014). Historically, SD was regarded as a rare cause of human disease, but the past decades a rapidly increasing incidence of invasive SD infections has been documented, and the rates have surpassed those of GAS and GBS in several geographic regions (Bramhachari et al., 2010; Oppegaard et al., 2015; Wajima et al., 2016).
Although Penicillin remains the drug of choice for treating infections caused by BHS, macrolides and clindamycin are important alternatives in β-lactam-intolerant patients. Furthermore, adjunctive clindamycin therapy reduces mortality in patients suffering from severe GAS disease manifestations (Linner et al., 2014), probably through the abrogation of bacterial toxin production (Mascini et al., 2001).
Resistance to the antimicrobial classes macrolides, lincosamides (such as clindamycin) and streptogramin B (MLSB) in BHS is primarily linked to the acquisition of target modification enzymes encoded by erm genes, mediating resistance to all three classes, or mef genes encoding efflux pumps targeting only macrolides (Varaldo et al., 2009). The genes lsa and lnu, causing lincosamide resistance, are also occasionally encountered (Hawkins et al., 2017). In GAS and GBS, all these resistance genes are associated with mobile genetic elements, and dissemination of these elements occur by either clonal expansion or horizontal genetic transfer (Varaldo et al., 2009; Palmieri et al., 2013; Hawkins et al., 2017; Zhou et al., 2017). The genetic environment of resistance determinants in SD is largely unknown, and the mechanisms involved in their dissemination have not been extensively explored. However, in vitro conjugal transfer of integrative conjugative elements (ICEs) harboring resistance genes has been demonstrated between these three species, and a common reservoir for resistance determinants is feasible (Palmieri et al., 2013).
There are considerable variations in reported resistance rates to MLSB antibiotics in BHS, and differences in susceptibility-testing methodology and ever-changing reference breakpoints make temporospatial comparisons difficult. Antimicrobial susceptibility rates in both GAS and GBS are under national surveillance in some countries, and whereas resistance to erythromycin and clindamycin appears to have a fluctuating course among GAS, gradually increasing rates of resistance has been noted in GBS (Norm/Norm-Vet, 2018; Public Health England, 2018; Swedres-Swarm, 2018) currently approximating 50% for both agents in the United States of America1. Consequently, the Infectious Diseases Society of America recently revised their national guidelines for antimicrobial treatment of GBS-infections (CDCgov, 2010). Long term epidemiologic trends of resistance rates in SD have not been previously studied in detail.
In the present study, we sought to examine the temporal trends of antimicrobial resistance in invasive BHS in western Norway, with a particular emphasis on SD. Moreover, we used whole genome sequencing to identify validated resistance genes and characterize their associated mobile genetic elements, as well as explore phylogenetic relationships and clonality in the resistant bacterial population.
Materials and Methods
Study Setting and Definitions
Health Region Bergen in western Norway has a catchment area of approximately 450,000 inhabitants, and comprises the tertiary care hospital Haukeland University Hospital, along with the two secondary care hospitals Haraldsplass Deaconess Hospital and Voss Hospital. All cases of invasive β-hemolytic streptococcal infections in this region presenting in the period 2004–2018 were retrospectively identified. Invasive disease was defined as isolation of BHS from normally sterile sites or from a non-sterile site in combination with surgically proven necrotizing soft tissue infection. Culture negative cases identified by sequencing of the 16S rRNA gene were excluded. To avoid issues regarding persistent and relapsing infections, only one incident per person was included.
Bacterial Isolates
All BHS have been identified based on large colony size (>0.5 mm in diameter) and β-hemolytic reaction on 5% sheep blood agar after incubation for 24 h, along with serogroup specificity using a rapid agglutination test (Oxoid, Basingstoke, Hampshire, United Kingdom). Species identity has been confirmed using Matrix Assisted Laser Desorption Ionization Time of Flight Mass Spectrometry (MALDI ToF MS) with the MALDI Biotyper database (Bruker Daltonik, Bremen, Germany).
Antimicrobial Susceptibility Testing
The isolates were tested for susceptibility to penicillin G, erythromycin, clindamycin, tetracycline, and trimethoprim-sulfamethoxazole by the disk diffusion method in accordance with the European Committee on Antimicrobial Susceptibility Testing (EUCAST) guidelines2. MLSB resistance phenotype was tested using the double disc diffusion method (D-test), placing erythromycin and clindamycin discs edge to edge 12 mm apart. Isolates resistant to erythromycin and/or clindamycin were categorized as having constitutive MLSB-resistance (cMLSB), inducible MLSB-resistance (iMLSB), macrolide resistance alone (M-phenotype), or isolated lincosamide resistance (L-phenotype). Streptococcus pneumoniae ATCC 49619 was used as control strain. EUCAST clinical breakpoints version 9.0 (2019) were used for categorization into susceptible, susceptible increased exposure and resistant phenotypes (see text footnote 2).
Whole Genome Sequencing and Bioinformatics
All isolates displaying reduced susceptibility to erythromycin or clindamycin were subjected to whole genome sequencing. Bacterial DNA was extracted using MagNA Pure (Roche Life science, Basel, Switzerland) and quantified using Qubit (Thermo Fisher Scientific, United States). Genomic libraries were constructed using the Illumina Nextera XT kit (Illumina, Essex, United Kingdom), and 150 base pair paired end sequencing was performed on a MiSeq platform (Illumina).
Paired end reads were assessed for quality using FastQC3, trimmed with Trimmomatic (Bolger et al., 2014), assembled by Spades (Nurk et al., 2013), and subsequently annotated using RAST (Aziz et al., 2008).
The emm-types of GAS and SD isolates were verified in-silico by BLAST-search against the emm-type database4. Capsule-types in GBS isolates were also checked using a pipeline developed by Metcalf (Metcalf et al., 2017). Multilocus sequence typing (MLST) profiles were identified using the Center for Genomic Epidemiology (CGE) website (Larsen et al., 2012).
Relevant resistance genes were identified by BLAST-searches in Geneious (Kearse et al., 2012), using the ResFinder and CARD databases as references (Zankari et al., 2012; Alcock et al., 2020). The genomic context of any identified resistance determinants was subsequently manually explored in Geneious for the characterization of their association with mobile genetic elements. Verification of potential ICEs and bacteriophages was performed using ICEfinder (Liu et al., 2019) and PHASTER (Arndt et al., 2016), in combination with meticulous literature review and BLAST-searches. Mauve was used for comparison of ICEs (Darling et al., 2004).
Phylogenetic relationships and clonality among the resistant isolates were evaluated by analysis of single nucleotide polymorphisms (SNPs) using CSI Phylogeny version 1.4 on the CGE website at default settings. One resistant isolate for each species were randomly picked as reference genome. Phylogenetic trees were constructed in Geneious using Geneious tree builder with the Tamura-Nei Genetic distance model and Neighbor-joining method without bootstrapping.
Statistics
Temporal trends were evaluated for statistical significance by Poisson regression analysis. Data were analyzed using SPSS PASW STATISTICS, version 23.0 (IBM SPSS Statistics for Windows, Armonk, NY, United States: IBM Corp.). A two-tailed p-value <0.05 was considered significant.
Ethical Statement
The study underwent institutional ethics review and approval (2010/1406 Regional Ethics Committee West, Norway). The study was exempted from written consent as it does not involve patient information or human biological material.
Results
We identified a total of 995 culture positive cases of invasive BHS disease, affecting 957 patients in the period 2004–2018. Recurrent infections were predominantly caused by GBS and SD (17 cases each). Clinical isolates were available for antimicrobial susceptibility testing for 909 out of the 957 unique cases (95%), comprising 271 GAS, 358 GBS, and 280 SD.
Antimicrobial Susceptibility
All isolates were fully susceptible to penicillin G, and only one isolate each of GAS and SD displayed phenotypic resistance to trimethoprim-sulfamethoxazole. Reduced susceptibility to tetracycline was observed in 14%, 67%, and 40% of GAS, GBS, and SD isolates, respectively. A total of 87 isolates were resistant to MLSB antibiotics, comprising 15 GAS, 53 GBS, and 19 SD. The distribution of the zone diameters for all the tested antimicrobial agents is presented in Table 1.
TABLE 1.
Distribution of antimicrobial resistance testing zone diameters.
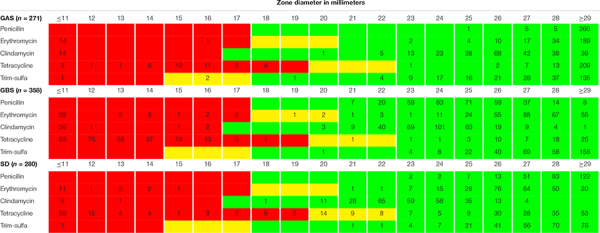 |
The distribution is presented as the number (n) of isolates in each zone diameter category. Isolates have been categorized as susceptible (green), susceptible increased exposure (yellow), or resistant (red) using EUCAST breakpoints v9.0 (2019). GAS, Streptococcus pyogenes; GBS, Streptococcus agalactiae; SD, Streptococcus dysgalactiae; Trim-sulfa, trimethoprim-sulfamethoxazole.
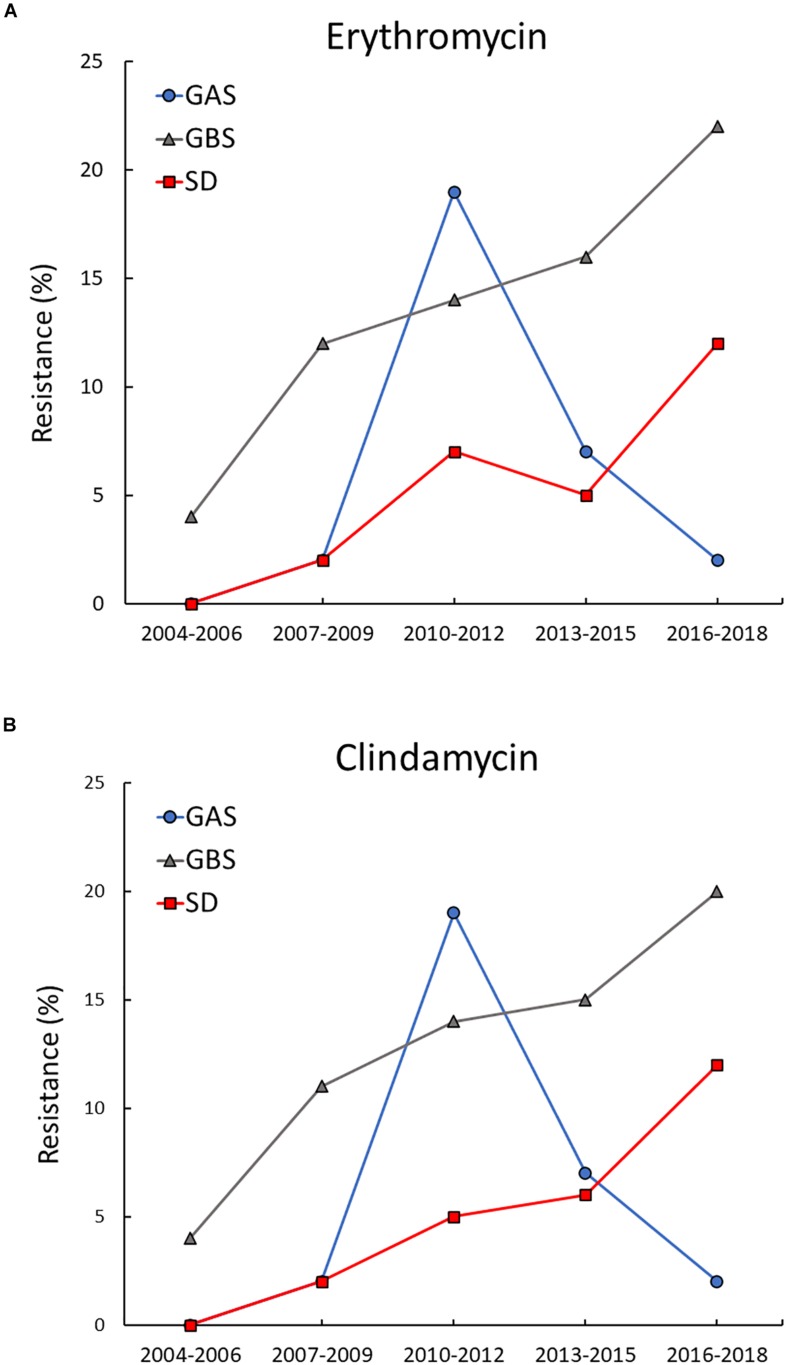
Distinct temporal trends for erythromycin and clindamycin resistance were revealed for the three BHS species (Figure 1). For GAS, the overall prevalence of resistance was very low, apart from a transient increase in resistance to erythromycin and clindamycin in 2010–2012. During this period almost 20% of the invasive GAS isolates belonged to a cMLSB-resistant emm11-lineage, but the outbreak quickly abated. The rates of resistance in GBS displayed a gradual and significant increase in the study period, rising from 4% during 2004–2006 to 22% in the period 2016–2018 for erythromycin (p = 0.004), and from 4 to 20% for clindamycin (p = 0.013). A similar upward trend was observed for SD, where no resistant isolates where detected until 2009. Subsequently, the incidence of erythromycin and clindamycin resistance rose significantly to 12% for both agents by 2016–2018 (p = 0.012).
FIGURE 1.
Temporal trends of erythromycin and clindamycin resistance in BHS. Resistance to erythromycin (A) and clindamycin (B) increased significantly in Streptococcus agalactiae (GBS) and Streptococcus dysgalactiae (SD) during the study period. In Streptococcus pyogenes (GAS) resistance to erythromycin and clindamycin remained low, apart from a transient outbreak of resistance to both agents in the period 2010–2012.
Detection of Resistance Genes
All 87 BHS isolates displaying reduced susceptibility to MLSB antibiotics were subjected to whole genome sequencing, and in 86 of these validated resistance genes were detected (Table 2). Chromosomal mutations in ribosomal genes were not investigated. Almost all isolates displaying the iMLSB phenotype harbored the erm(A)-gene, but erm(T) was found in one GBS isolate. The cMLSB phenotype was predominantly associated with the erm(B)-gene, mef(A) was linked to the M phenotype and the lsa(C)-gene was present in three isolates with the L phenotype. Presence of additional clindamycin-resistance genes was found in three erm(B)-positive GBS isolates; one harbored both lsa(E) and lnu(B), and in two isolates lsa(C) was detected.
TABLE 2.
Antimicrobial resistance phenotype and genotype of the macrolide and lincosamide resistant β-hemolytic streptococcal isolates.
| Microbe | n | Phenotype | (n) | Genotype (n) |
| S. pyogenes | 15 | iMLSB | (1) | erm(A) (1) |
| cMLSB | (14) | erm(B) (14) | ||
| M | (0) | |||
| L | (0) | |||
| Tet R | (14) | tet(M) (14) | ||
| S. agalactiae | 53 | iMLSB | (7) | erm(A) (6), erm(T) (1) |
| cMLSB | (38) | erm(A) (8), erm(B) (26), erm(B) + lsa(C) (2), erm(B) + lsa(E) + lnu(B) (1), ND (1) | ||
| M | (6) | mef(A) (6) | ||
| L | (2) | lsa(C) (2) | ||
| Tet R | (50) | tet(M) (31), tet(O) (17), tet(M) + tet(O) (2) | ||
| S. dysgalactiae | 19 | iMLSB | (11) | erm(A) (11) |
| cMLSB | (6) | erm(A) (3), erm(B) (3) | ||
| M | (1) | mef(A) (1) | ||
| L | (1) | lsa(C) (1) | ||
| Tet R | (6) | tet(M) (4), tet(O) (1), tet(M) + tet(O) (1) |
n, number of isolates; MLSB, macrolide, lincosamide, streptogramin B; iMLSB, inducible MLSB-resistance; cMLSB, constitutive MLSB-resistance, M, macrolide resistance alone; L, lincosamide resistance alone; Tet R, tetracycline resistance; ND, no resistance gene detected.
Co-resistance to tetracycline was evident in almost all GAS (14/15) and GBS (50/53) with reduced susceptibility to MLSB antibiotics, whereas only 31% (6/19) of the SD isolates displayed dual resistance. tet(M) was the most common resistance determinant in all three species, but tet(O) was also frequent in GBS (Table 2).
The 14 cMLSB and tetracycline resistant GAS isolates were discovered also to encode a cassette of genes mediating resistance to aminoglycosides and streptothricins, comprising aadE, aphA3, and sat4. A similar element was detected in 14 cMLSB and tetracycline resistant GBS isolates. The single GAS and SD isolates resistant to trimethoprim-sulfamethoxazole were also whole genome sequenced and found to contain the dfr(A)-gene.
Phylogenetic Analysis of the BHS Population Resistant to Erythromycin and Clindamycin
To facilitate temporospatial comparisons of clonal distribution of macrolide and clindamycin resistance, an analysis of the combination of emm-type, MLST-type and MLSB-resistance gene has often been used as a rough marker for resistant clones in GAS (Perez-Trallero et al., 2007; Willems et al., 2011). In the present study we have extrapolated this approach to SD, and substituted emm-type with capsule-type for GBS. An overview of the spectrum of resistant clones for the three species is presented in Table 3.
TABLE 3.
Genetic characterization of the resistant BHS population.
| Clone | n | Phenotype | ICE-family | Ref | Other resistance genes | ||
| GAS | |||||||
| emm | MLST | res-gene | |||||
| emm11 | ST403 | erm(B) | 14 | cMLSB | Tn6003 | A | tet(M) (14), MAS (14) |
| emm58 | ST176 | erm(A) | 1 | iMLSB | ICEsp2905 | B | |
| GBS | |||||||
| Capsule | MLST | res-gene | |||||
| Ib | ST12 | erm(B) | 3 | cMLSB | ICESa2603 | C | tet(O) (3), MAS (3), lsa(E) + lnu(B) (1) |
| II | ST1 | erm(A) | 1 | iMLSB | Unknown | tet(M) | |
| II | ST12 | erm(B) | 1 | cMLSB | ICESa2603 | C | tet(O) |
| II | ST22 | erm(B) | 1 | cMLSB | Tn3872 | A | tet(M) |
| III | ST17 | erm(B) | 9 | cMLSB | ICESa2603 | C | tet(O) (9), tet(M) (2), MAS (8) |
| III | ST1434 | erm(B) | 1 | cMLSB | ICESa2603 | C | tet(O), MAS |
| III | ST19 | erm(B) | 4 | cMLSB | ICESa2603 | C | tet(O) (2), tet(M) (2), lsa(C) (2), MAS (2) |
| III | ST27 | erm(B) | 1 | cMLSB | ICESa2603 | C | tet(O) |
| III | ST1432 | erm(B) | 1 | cMLSB | Tn3872 | A | tet(M), cat(pc194) |
| III | ST335 | erm(A) | 2 | iMLSB | ICEsp2905 | B | tet(M) (2) |
| III | ST107 | erm(A) | 1 | cMLSB | ICEsp2905 | B | |
| IV | ST1433 | erm(B) | 1 | cMLSB | ICESa2603 | C | tet(O) |
| IV | ST459 | erm(A) | 6 | cMLSB | ICEsp2905 | B | tet(M) (5) |
| V | ST1 | erm(A) | 4 | iMLSB | Unknown | tet(M) (4) | |
| V | ST1 | erm(B) | 7 | cMLSB | Tn3872 | A | tet(M) (7) |
| III | ST17 | erm(T) | 1 | iMLSB | pRW35 | A | tet(M) |
| Ia | ST23 | mef | 6 | M | Phage1207.3 | A | tet(M) (6) |
| III | ST1399 | lsa(C) | 1 | L | IMESagUCN70 | D | tet(M) |
| III | ST1167 | lsa(C) | 1 | L | IMESagUCN70 | D | tet(O) |
| Ia | ST7 | ND | 1 | cMLSB | ND | ||
| SD | |||||||
| emm | MLST | res-gene | |||||
| stC74a | ST17 | erm(A) | 4 | iMLSB | ICEsp2905 | B | |
| stC74a | ST29 | erm(A) | 1 | iMLSB | ICEsp2905 | B | tet(M) |
| stG480 | ST8 | erm(A) | 2 | iMLSB | ICEsp2905 | B | |
| stG245 | ST29 | erm(A) | 1 | cMLSB | ICEsp2905 | B | |
| stG62647 | ST319 | erm(A) | 1 | iMLSB | ICEsp2905 | B | |
| stG2574 | ST23 | erm(A) | 1 | cMLSB | ICEsp2905 | B | tet(M) |
| stG485 | ST55 | erm(A) | 1 | iMLSB | ICEsp2905 | B | tet(M) + tet(O) |
| stG485 | ST17 | erm(A) | 1 | cMLSB | ICEsp2905 | B | |
| stG485 | ST29 | erm(B) | 1 | cMLSB | Unknown | ||
| stG2078 | ST473 | erm(A) | 1 | iMLSB | ICEsp2905 | B | |
| stG2078 | ST17 | erm(A) | 1 | iMLSB | ICEsp2905 | B | |
| stG2078 | ST17 | erm(B) | 1 | cMLSB | Tn6218 | * | |
| stG840 | ST269 | erm(B) | 1 | cMLSB | Tn3872 | A | tet(M) |
| stG485 | ST128 | mef(A) | 1 | M | Phage1207.3 | A | tet(M) |
| stG643 | ST12 | lsa(C) | 1 | L | ICEsp2905 | B | tet(O) |
A (Varaldo et al., 2009); B (Giovanetti et al., 2012); C (Hawkins et al., 2017), and D (Zhou et al., 2017). *Unpublished, GenBank accession HG002388. n, number of isolates; ICE, integrative conjugative element; MLST, multilocus sequence typing; res-gene, resistance gene; GAS, Streptococcus pyogenes; GBS, Streptococcus agalactiae; SD, Streptococcus dysgalactiae; MLSB, macrolide, lincosamide, streptogramin B; iMLSB, inducible MLSB-resistance; cMLSB, constitutive MLSB-resistance, M, macrolide resistance alone; L, lincosamide resistance alone; ND, no resistance gene detected. MAS, macrolide-aminoglycoside-streptothricin resistance element containing the aminoglycoside and streptothricin resistance genes aadE, aphA3, and sat4 in addition to the macrolide resistance determinant ermB.
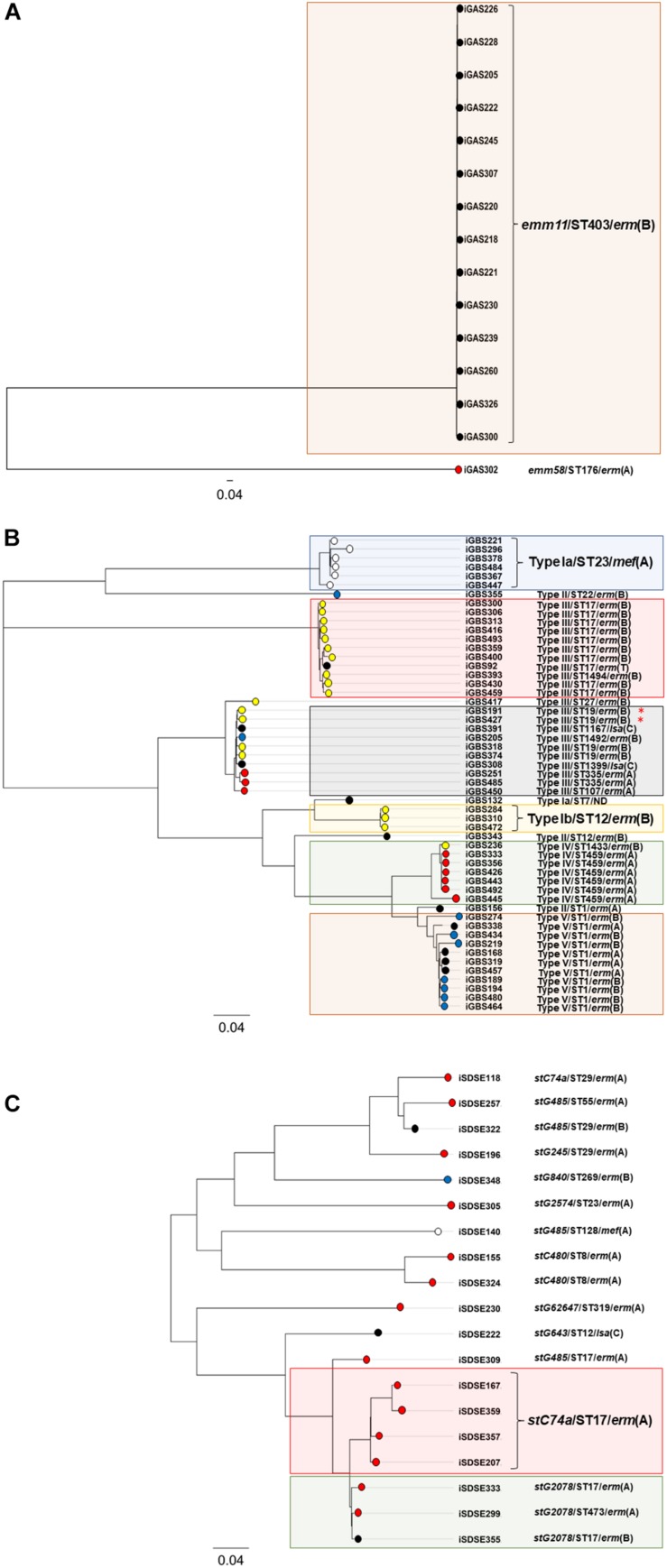
For GAS, the emm11/ST403/erm(B) lineage dominated, and only one isolate was found to belong to a different clade, emm58/ST176/erm(A). Differently, 20 different clones were identified in resistant GBS isolates, the predominant being III/ST17/erm(B) (n = 9), V/ST1/erm(B) (n = 7), IV/ST459/erm(A) (n = 6), and Ia/ST23/mef(A) (n = 6). A similar diversity was observed in SD, where 15 different lineages were observed among the 19 isolates resistant to MLSB antibiotics. Only stC74a/ST17/erm(A) (n = 4) and stG480/ST8/erm(A) (n = 2) were associated with more than one isolate. All sequenced SD isolates belonged to the subspecies S. dysgalactiae subspecies equisimilis.
Phylogeny of the erythromycin and clindamycin resistant BHS population was also examined by SNP-analysis. The isolates iGAS300, iGBS300, and iSDSE357 were randomly chosen as reference genomes, yielding a core genome size of 1.7 Mbp (94% of reference genome), 1.7 Mbp (82%), and 1.8 Mbp (83%) for GAS, GBS, and SD, respectively. In GAS, 7,550 valid SNP-positions were found, and the isolates differed by a range from 4 to 7,528 SNPs. However, the isolates belonging to the emm11/ST403/erm(B) lineage were only separated by a maximum of 18 SNPs, indicating a highly clonal group. A total of 19,645 valid SNPs was detected in GBS, ranging from 3 to 8,979 SNP variations between the isolates, whereas in SD 15,524 SNPs were found (range 82–5,865). Phylogenetic trees based on the SNP-analysis are depicted in Figure 2.
FIGURE 2.
Phylogenetic trees of the resistant BHS – population. The phylogenetic trees are based on core-genome SNP-analysis of S. pyogenes (A), S. agalactiae (B), and S. dysgalactiae (C) isolates displaying reduced susceptibility to erythromycin and/or clindamycin. The scale indicates substitutions per site. Isolates clustering with >97% similarity have been highlighted with colored rectangular boxes. The circular node tips have been assigned color coding according to the mobile genetic element harboring the MLSB-resistance gene in the respective isolates. Red indicates ICEsp2907, blue color has been assigned to Tn3872, yellow indicates ICESa2603, isolates carrying the bacteriophage 1207.3 are white, and all other elements are depicted in black. The red asterisk indicates the two isolates co-harboring lsa(C).
Characterization of Mobile Genetic Elements Associated With Macrolide and Lincosamide Resistance
A wide variety of mobile genetic elements carrying MLSB-resistance genes were detected in the BHS isolates (Table 3). mef(A) was the only resistance gene associated with a bacteriophage (Phage 1207.3), and erm(T) was carried by the small plasmid, pRW35. The remaining resistance genes were carried by ICEs. The emm11/ST403/erm(B) GAS lineage was equipped with ICETn6003, a composite element derived from the fusion of a multi resistance macrolide, aminoglycoside, streptothricin (MAS) element into orf20 of the tet(M) tetracycline resistance gene carrying Tn916 element. Tn3872, another Tn916 based element with erm(B) integrated in orf9, was harbored by nine GBS isolates and one SD isolate. Mobile genetic elements belonging to the ICESa2603 family were predominant among GBS, detected in 20 erm(B) positive isolates. Fourteen of these ICESa2603 elements also contained a MAS-like fragment, eighteen carried tet(O), whereas two co-harbored tet(M).
Interestingly, the composite ICEsp2905, with its embedded integrative mobilizable element IMEsp2907 carrying erm(A), was detected in all three species, and was the dominant cause of resistance to MLSB antibiotics in SD (14/19 isolates). Comparison of IMEsp2907 from different isolates revealed a high degree of similarity (Figure 3A). On a similar note, highly homologous ICEs were found in phylogenetically distantly related GBS isolates, as judged by core-genome SNP-analysis. Although the isolates iGBS310 and iGBS313 differ by 8632 SNPs, they contained ICESa2603 elements displaying 99.5% whole sequence homology (65,877/66,185 bp identity). In the equally phylogenetically distanced isolates iGBS274 and iGBS355, we detected Tn3872 elements showing 99.9% similarity (23,102/23,113 bp identity) (Figure 3B). Moreover, comparison to the Tn3872 element in iSDSE305 also revealed a high genetic resemblance (96.1%).
FIGURE 3.
Genetic composition of the mobile genetic element IMEsp2907 in three BHS. Mauve was used for the sequence alignment of the mobile genetic element. Panel (A) depicts a comparison of IMEsp2907 detected in the invasive BHS isolates iGAS302 (S. pyogenes), iGBS485 (S. agalactiae), and iSDSE357 (S. dysgalactiae). The integrative mobilizable element displayed a conserved genetic architecture and highly similar nucleotide sequences in these three species. Genes involved in mobilization and integration accounted for the majority of the discrepancies. The gene immediately downstream from erm(A) is annotated by RAST as a spectinomycin resistance gene based on sequence similarities, but has not been experimentally verified. Panel (B) displays an alignment of Tn3872 detected in two GBS isolates and one SD isolate. The ICE Tn3872 is composed of an erm(B) module (highlighted in a pink box) integrated into a Tn916 element.
In GBS, all four lsa(C) genes were found on the small integrative mobilizable element IMESagUCN70, whereas the lincosamide resistance determinants lsa(E) and lnu(B) co-resided on a mobile genetic element previously described in a GBS isolate (SGB76) (Hawkins et al., 2017). Differently, the lsa(C) gene detected in SD was directly integrated into an ICE of the ICEsp2905-family.
Two resistance gene related elements, associated with erm(A) and erm(B), respectively, could not be identified through BLAST-search or literature review, and will be further investigated. Details on the repertoire of resistance genes and associated ICEs for each individual isolate is detailed in Supplementary Table S1.
Discussion
In the present study, we have conducted a comprehensive analysis of macrolide and clindamycin resistance in invasive BHS isolates over a 15-year period, and to the best of our knowledge, this is the first study investigating the temporal trends of such resistance in SD isolates.
Alarmingly, we found significantly increasing resistance rates in our health region for SD, rising incrementally from the start of the current decade. Whole genome sequencing of the resistant bacterial population revealed a substantial phylogenetic diversity, but the mobile genetic elements harboring the resistance genes displayed a high degree of genetic similarity. Taken together this indicates dissemination of MLSB-resistance genes in SD predominantly through conjugative transfer rather than clonal dissemination.
Although several studies have described a high diversity of emm-types among resistant SD isolates, knowledge on MLST-profiles in this population is scarce (Wajima et al., 2016; Zheng et al., 2017; Kim et al., 2018). Kim et al. (2018) recently characterized contemporary SD isolates from Japan and Korea, and reported a polyclonal distribution, with 13 emm/ST profiles among 20 isolates and 6 emm/ST profiles among 24 isolates in the two countries, respectively. Strikingly, despite the temporal and geographic proximity they only detected two mutual resistant clones, stG485/ST128/mef(A) and stG840/ST269/erm(B). Those clones were also present in our community, but no further similarities were evident. This underlines the highly polyclonal distribution of SD isolates, but could also infer the existence of some globally successful lineages. However, the paucity of data precludes firm conclusions.
Conversely, we found erythromycin and clindamycin resistance in GAS to be almost exclusively linked to a transient outbreak of an emm11/ST403/erm(B) lineage during 2010–2012. The emergence of this lineage has previously been documented in several other countries, though a few years prior to its arrival in Norway (Perez-Trallero et al., 2007; Silva-Costa et al., 2008). Although such clonal outbreaks can have a rapid and major impact on resistance rates, surging from 2 to 20% in 1 year in our region, the long-term implications are often negligible. With the gradual acquisition of specific immunity in the general population, the outbreak usually subsides (Montes et al., 2014).
Substantiating this, longitudinal epidemiologic surveillance in Spain and Portugal from the mid-90s revealed the erythromycin resistant population in GAS to be highly clonal (Perez-Trallero et al., 2007; Silva-Costa et al., 2012; Montes et al., 2014). The emergence and disappearance of new dominant clones continuously renders the phylogenetic landscape and is reflected in the fluctuating course of the resistance rates (Silva-Costa et al., 2008; Montes et al., 2014). From erythromycin resistance rates approximating 30% in the late 90s, the problem has almost vanished, and is currently below 5% in Portugal (Friaes et al., 2019). National surveillance reports from the Nordic countries during the past five years have all documented low resistance rates to MLSB antibiotics in GAS, indicating that the emergence of new resistant GAS clones is a rare event in northern Europe (DANMAP, 2017; Norm/Norm-Vet, 2018; Swedres-Swarm, 2018).
In the present study, a significant upward trend for resistance to erythromycin and clindamycin was detected in GBS, exceeding 20% for both agents by the end of the study period. A similar development and level of resistance has been documented in our neighboring countries (DANMAP, 2017; Public Health England, 2018; Swedres-Swarm, 2018). The United States of America appeared to start down this path some years earlier, and the resistance rates approximated 50% by 20155.
Like in SD, the gradually increasing resistance to erythromycin and clindamycin in GBS in our region was predominantly caused by a polyclonal expansion of the resistant population. A comparable diversity has been reported by others, including a similar distribution of capsule-types and MLST-profiles (Bergseng et al., 2008; Rojo-Bezares et al., 2016). However, given the limited number of capsule-types as compared to GAS and SD emm-types, the genotype/ST-profile/resistance-gene annotation provides a much lower phylogenetic resolution in GBS, making cross-study comparisons more difficult.
The past decades SD has emerged as an important cause of human infectious disease, and in Finland, it was the fifth most frequent pathogen detected in blood cultures in 2016 (Finland National Institute for Health and Welfare, 2016). Whereas GAS and GBS are under national surveillance in several countries, almost none monitor the epidemiology of SD on a national level. Public Health England report annual resistance rates for BHS, but present data for group C and group G streptococci separately, without identification to the species level (Public Health England, 2018). Furthermore, susceptibility data is only available for 60–70% of the isolates, posing a risk of reporting bias. Nonetheless, the major increase in resistance observed in the United Kingdom over the past decades is alarming, rising from 13 to 40% and 7 to 30% in group G streptococci for erythromycin and clindamycin, respectively. Moreover, even higher rates have been reported from Asia (Zheng et al., 2017). Although the level of resistance in SD in our community is still low in this context, the general direction of the temporal trend along with the polyclonal nature of the resistant isolates are concerning. Antimicrobial resistance spreading by conjugative transfer is less affected by herd immunity in the host population, and thus likely more challenging to combat (Lerminiaux and Cameron, 2019).
The erm(A) carrying integrative mobilizable element IMEsp2907 was initially discovered in GAS as a part of a larger composite ICEsp2905, also harboring a separate Tet(O) element (Giovanetti et al., 2012). All these modules could be transferred independently or collectively to new GAS recipients, revealing the intricate yet highly versatile transfer dynamics of horizontal genetic exchange between streptococcal species. Interestingly, we found homologs of IMEsp2907 in all three species of BHS, but in varying genomic contexts. On a similar note, the distribution of the different resistance determinant associated ICEs in S. agalactiae and S. dysgalactiae did not appear to intimately correlate with phylogenetic origin as judged by core-genome SNP-analysis. Taken together, this could infer a pivotal role for conjugative transfer in the dissemination of antimicrobial resistance among BHS. Furthermore, by searching the NCBI database, an IMEsp2907 homolog can also be detected in Streptococcus anginosus and Anaerococcus species, raising the question of the origin of these genetic elements. Hence, the emphasis of future research on resistance determinants in BHS should be to identify their major reservoirs, and the investigate the factors that influence the frequency of conjugative spread.
This study is limited by the low number of resistant isolates. However, in 2018 all three BHS species were included in the Norwegian Surveillance Program for Antimicrobial Resistance (NORM) for the first time, confirming that our findings reflect the national incidence of resistance to erythromycin and clindamycin (Norm/Norm-Vet, 2018). The transferability of our data to regions of high-level resistance remains to be elucidated, and further molecular studies on the resistant BHS population are warranted, especially for SD.
Conclusion
Resistance to erythromycin and clindamycin in GBS and SD is increasing in western Norway. The resistant bacterial population is highly polyclonal, but a limited number of ICEs appear to mediate the dissemination of resistance genes. This could indicate a spread through conjugal transfer rather than clonal expansion, but further studies are needed to elucidate the mechanisms of transmission.
Data Availability Statement
Whole genome data for all sequenced isolates has been deposited at DDBJ/ENA/GenBank under the Bioproject PRJNA600655, and the NCBI accession numbers JAAABO000000000–JAAAEW000000000.
Author Contributions
OO and BK conceived the study. SS and HM contributed to the study design. OO performed statistical analysis and bioinformatics, and drafted the manuscript. All authors read and approved the final manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the staff at the microbiological department at Haukeland University Hospital for excellent technical assistance and for providing access to their laboratory facilities.
Funding. This study received funding from the Western Norway Regional Health Authority (grant number 912231), and the Norwegian Surveillance System for Antimicrobial Drug Resistance. The funders were not involved in the study design, interpretation of data, the writing of this article or the decision to submit it for publication.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.00797/full#supplementary-material
Overview of resistance genes and their associated mobile genetic elements.
References
- Alcock B. P., Raphenya A. R., Lau T. T. Y., Tsang K. K., Bouchard M., Edalatmand A., et al. (2020). CARD 2020: antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 48 D517–D525. 10.1093/nar/gkz935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arndt D., Grant J. R., Marcu A., Sajed T., Pon A., Liang Y., et al. (2016). PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res. 44 W16–W21. 10.1093/nar/gkw387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aziz R. K., Bartels D., Best A. A., DeJongh M., Disz T., Edwards R. A., et al. (2008). The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9:75. 10.1186/1471-2164-9-75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergseng H., Rygg M., Bevanger L., Bergh K. (2008). Invasive group B Streptococcus (GBS) disease in Norway 1996-2006. Eur. J. Clin. Microbiol. Infect. Dis. 27 1193–1199. 10.1007/s10096-008-0565-8 [DOI] [PubMed] [Google Scholar]
- Bolger A. M., Lohse M., Usadel B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30 2114–2120. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bramhachari P. V., Kaul S. Y., McMillan D. J., Shaila M. S., Karmarkar M. G., Sriprakash K. S. (2010). Disease burden due to Streptococcus dysgalactiae subsp. equisimilis (group G and C streptococcus) is higher than that due to Streptococcus pyogenes among Mumbai school children. J. Med. Microbiol. 59(Pt 2), 220–223. 10.1099/jmm.0.015644-0 [DOI] [PubMed] [Google Scholar]
- Carapetis J. R., Steer A. C., Mulholland E. K., Weber M. (2005). The global burden of group A streptococcal diseases. Lancet Infect. Dis. 5 685–694. 10.1016/S1473-3099(05)70267-X [DOI] [PubMed] [Google Scholar]
- CDCgov (2010). Prevention of perinatal group B streptococcal disease, revised guidelines from CDC. MMWR Morb. Mortal. Wkly. Rep. 59 1–32. [PubMed] [Google Scholar]
- DANMAP (2017). Use of Antimicrobial Agents and Occurrence of Antimicrobial Resistance in Bacteria from Food Animals, Food and Humans in Denmark. Lyngby: Technical University of Denmark. [Google Scholar]
- Darling A. C., Mau B., Blattner F. R., Perna N. T. (2004). Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 14 1394–1403. 10.1101/gr.2289704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finland National Institute for Health and Welfare (2016). Infectious Diseases in Finland 2016. Helsinki: Finland National Institute for Health and Welfare. [Google Scholar]
- Friaes A., Melo-Cristino J., Ramirez M. Portuguese Group for the Study of Streptococcal (2019). Changes in emm types and superantigen gene content of Streptococcus pyogenes causing invasive infections in Portugal. Sci. Rep. 9:18051. 10.1038/s41598-019-54409-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovanetti E., Brenciani A., Tiberi E., Bacciaglia A., Varaldo P. E. (2012). ICESp2905, the erm(TR)-tet(O) element of Streptococcus pyogenes, is formed by two independent integrative and conjugative elements. Antimicrob. Agents Chemother. 56 591–594. 10.1128/AAC.05352-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins P. A., Law C. S., Metcalf B. J., Chochua S., Jackson D. M., Westblade L. F., et al. (2017). Cross-resistance to lincosamides, streptogramins A and pleuromutilins in Streptococcus agalactiae isolates from the USA. J. Antimicrob. Chemother. 72 1886–1892. 10.1093/jac/dkx077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse M., Moir R., Wilson A., Stones-Havas S., Cheung M., Sturrock S., et al. (2012). Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28 1647–1649. 10.1093/bioinformatics/bts199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Byun J. H., Park H., Lee J., Lee H. S., Yoshida H., et al. (2018). Molecular epidemiological features and antibiotic susceptibility patterns of Streptococcus dysgalactiae subsp. equisimilis Isolates from Korea and Japan. Ann. Lab. Med. 38 212–219. 10.3343/alm.2018.38.3.212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen M. V., Cosentino S., Rasmussen S., Friis C., Hasman H., Marvig R. L., et al. (2012). Multilocus sequence typing of total-genome-sequenced bacteria. J. Clin. Microbiol. 50 1355–1361. 10.1128/JCM.06094-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Doare K., Heath P. T. (2013). An overview of global GBS epidemiology. Vaccine 31(Suppl. 4), D7–D12. 10.1016/j.vaccine.2013.01.009 [DOI] [PubMed] [Google Scholar]
- Lerminiaux N. A., Cameron A. D. S. (2019). Horizontal transfer of antibiotic resistance genes in clinical environments. Can. J. Microbiol. 65 34–44. 10.1139/cjm-2018-0275 [DOI] [PubMed] [Google Scholar]
- Linner A., Darenberg J., Sjolin J., Henriques-Normark B., Norrby-Teglund A. (2014). Clinical efficacy of polyspecific intravenous immunoglobulin therapy in patients with streptococcal toxic shock syndrome: a comparative observational study. Clin. Infect. Dis 59 851–857. 10.1093/cid/ciu449 [DOI] [PubMed] [Google Scholar]
- Liu M., Li X., Xie Y., Bi D., Sun J., Li J., et al. (2019). ICEberg 2.0: an updated database of bacterial integrative and conjugative elements. Nucleic Acids Res. 47 D660–D665. 10.1093/nar/gky1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascini E. M., Jansze M., Schouls L. M., Verhoef J., Van Dijk H. (2001). Penicillin and clindamycin differentially inhibit the production of pyrogenic exotoxins A and B by group A streptococci. Int. J. Antimicrob. Agents 18 395–398. 10.1016/s0924-8579(01)00413-7 [DOI] [PubMed] [Google Scholar]
- Metcalf B. J., Chochua S., Gertz R. E., Jr., Hawkins P. A., Ricaldi J., Li Z., et al. (2017). Short-read whole genome sequencing for determination of antimicrobial resistance mechanisms and capsular serotypes of current invasive Streptococcus agalactiae recovered in the USA. Clin. Microbiol. Infect 23 574.e14–577.e14. 10.1016/j.cmi.2017.02.021 [DOI] [PubMed] [Google Scholar]
- Montes M., Tamayo E., Mojica C., Garcia-Arenzana J. M., Esnal O., Perez-Trallero E. (2014). What causes decreased erythromycin resistance in Streptococcus pyogenes? Dynamics of four clones in a southern European region from 2005 to 2012. J. Antimicrob. Chemother. 69 1474–1482. 10.1093/jac/dku039 [DOI] [PubMed] [Google Scholar]
- Norm/Norm-Vet (2018). Usage og Antimicrobial Agents and Occurrence of Antimicrobial Resistance in Norway. Available online at: https://unn.no/fag-og-forskning/norm-norsk-overvakingssystem-for-antibiotikaresistens-hos-mikrober (accessed March 26, 2020). [Google Scholar]
- Nurk S., Bankevich A., Antipov D., Gurevich A. A., Korobeynikov A., Lapidus A., et al. (2013). Assembling single-cell genomes and mini-metagenomes from chimeric MDA products. J. Comput. Biol. 20 714–737. 10.1089/cmb.2013.0084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oppegaard O., Mylvaganam H., Kittang B. R. (2015). Beta-haemolytic group A, C and G streptococcal infections in Western Norway: a 15-year retrospective survey. Clin. Microbiol. Infect. 21 171–178. 10.1016/j.cmi.2014.08.019 [DOI] [PubMed] [Google Scholar]
- Palmieri C., Magi G., Creti R., Baldassarri L., Imperi M., Gherardi G., et al. (2013). Interspecies mobilization of an ermT-carrying plasmid of Streptococcus dysgalactiae subsp. equisimilis by a coresident ICE of the ICESa2603 family. J. Antimicrob. Chemother. 68 23–26. 10.1093/jac/dks352 [DOI] [PubMed] [Google Scholar]
- Perez-Trallero E., Montes M., Orden B., Tamayo E., Garcia-Arenzana J. M., Marimon J. M. (2007). Phenotypic and genotypic characterization of Streptococcus pyogenes isolates displaying the MLSB phenotype of macrolide resistance in Spain, 1999 to 2005. Antimicrob. Agents Chemother. 51 1228–1233. 10.1128/AAC.01054-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Public Health England (2018). Laboratory surveillance of pyogenic and non-pyogenic streptococcal bacteraemia in England, Wales and Northern Ireland: 2017. Health Protect. Rep. 12:41. [Google Scholar]
- Rantala S. (2014). Streptococcus dysgalactiae subsp. equisimilis bacteremia: an emerging infection. Eur. J. Clin. Microbiol. Infect. Dis. 33 1303–1310. 10.1007/s10096-014-2092-0 [DOI] [PubMed] [Google Scholar]
- Rojo-Bezares B., Azcona-Gutierrez J. M., Martin C., Jareno M. S., Torres C., Saenz Y. (2016). Streptococcus agalactiae from pregnant women: antibiotic and heavy-metal resistance mechanisms and molecular typing. Epidemiol. Infect. 144 3205–3214. 10.1017/S0950268816001692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva-Costa C., Friaes A., Ramirez M., Melo-Cristino J., Strin I. (2012). Differences between macrolide-resistant and -susceptible Streptococcus pyogenes: importance of clonal properties in addition to antibiotic consumption. Antimicrob. Agents Chemother. 56 5661–5666. 10.1128/AAC.01133-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva-Costa C., Pinto F. R., Ramirez M., Melo-Cristino J., Portuguese P. (2008). Decrease in macrolide resistance and clonal instability among Streptococcus pyogenes in Portugal. Clin. Microbiol. Infect. 14 1152–1159. 10.1111/j.1469-0691.2008.02104.x [DOI] [PubMed] [Google Scholar]
- Swedres-Swarm (2018). Consumption of antibiotics and occurrence of resistance in Sweden. Solna/Uppsala ISSN1650-6332. [Google Scholar]
- Varaldo P. E., Montanari M. P., Giovanetti E. (2009). Genetic elements responsible for erythromycin resistance in streptococci. Antimicrob. Agents Chemother. 53 343–353. 10.1128/AAC.00781-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wajima T., Morozumi M., Hanada S., Sunaoshi K., Chiba N., Iwata S., et al. (2016). Molecular characterization of invasive Streptococcus dysgalactiae subsp. equisimilis, Japan. Emerg. Infect Dis. 22 247–254. 10.3201/eid2202.141732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willems R. J., Hanage W. P., Bessen D. E., Feil E. J. (2011). Population biology of Gram-positive pathogens: high-risk clones for dissemination of antibiotic resistance. FEMS Microbiol. Rev. 35 872–900. 10.1111/j.1574-6976.2011.00284.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zankari E., Hasman H., Cosentino S., Vestergaard M., Rasmussen S., Lund O., et al. (2012). Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 67 2640–2644. 10.1093/jac/dks261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng P. X., Chan Y. C., Chiou C. S., Hsieh C. L., Chiang-Ni C., Wu J. J. (2017). Highly prevalent emmSTG840.0 and emmSTC839.0 types of erythromycin non-susceptible group G Streptococcus isolated from bacteremia in southern Taiwan. J. Microbiol. Immunol. Infect. 50 831–838. 10.1016/j.jmii.2016.12.010 [DOI] [PubMed] [Google Scholar]
- Zhou K., Xie L., Han L., Guo X., Wang Y., Sun J. (2017). ICESag37, a novel integrative and conjugative element carrying antimicrobial resistance genes and potential virulence factors in Streptococcus agalactiae. Front. Microbiol. 8:1921. 10.3389/fmicb.2017.01921 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Overview of resistance genes and their associated mobile genetic elements.
Data Availability Statement
Whole genome data for all sequenced isolates has been deposited at DDBJ/ENA/GenBank under the Bioproject PRJNA600655, and the NCBI accession numbers JAAABO000000000–JAAAEW000000000.