A living cell is an autonomous molecular system surrounded by a biological membrane that possesses a soft structure composed of a lipid bilayer. Various biochemical reactions, which have crucial roles in regulating the molecular system, occur in the space surrounded by the cellular membrane or at the interface of the membrane. These reactions rely on various properties of the membrane, such as fluidity, surface charge, and protein interactions (Alberts et al. 2008). Unraveling phenomena specific to such “soft” compartments/interfaces will not only lead to a better understanding of living systems but will also allow the designing of more accurate artificial biosystems.
We thus organized a symposium entitled “Utilization of soft compartments/interfaces from nano to macroscale: Exploring the potential of living systems” at the 57th annual meeting of Biophysical Society of Japan (Fig. 1). This symposium focused on soft compartments and interfaces, i.e., the inner space of lipid vesicles and the surface of lipid bilayers, at several hierarchical levels, ranging from the molecular to the tissue level. We invited two speakers, Dr. Yuno Natsume and Dr. Kaori Kuribayashi-Shigetomi, and selected three speakers from among the conference participants. We, the organizers, also gave talks on the following papers: self-assembly of designed-nanostructures on the supported lipid bilayer interface (Suzuki et al. 2018); isothermal amplification of short single-stranded DNAs inside a giant unilamellar vesicle (GUV) (Sato et al. 2019); and the construction of cell-containing lipid vesicles (Morita et al. 2018). In this Commentary, we provide an overview of the topics presented by the invited/selected speakers and an outlook on the potential of furthering knowledge regarding soft compartments/interfaces.
Fig. 1.
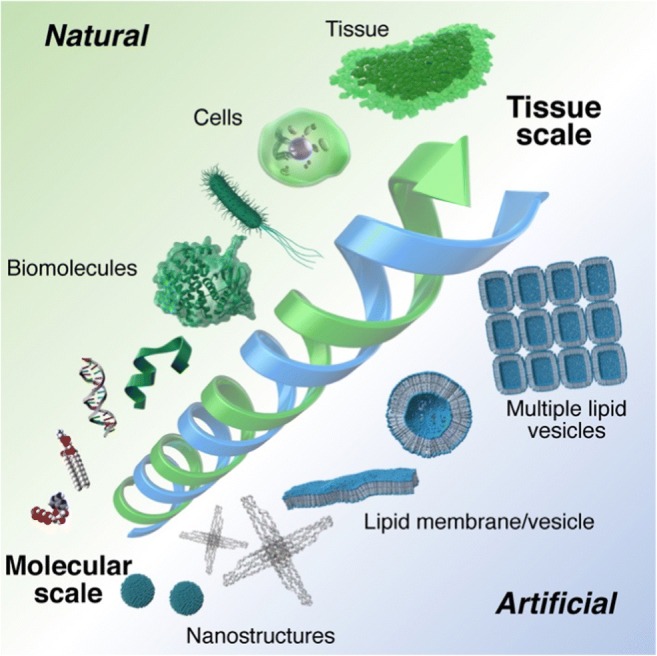
Conceptual representation of the session. Utilization of soft compartments/interfaces from molecular to tissue scales was discussed
Mr. Keiju Suda from Akiyama group in Kyushu University gave a talk about the molecular crowding effect on a biomembrane. They studied crystallization of bacteriorhodopsins using statistical mechanics theory. The results showed that molecular crowding could serve as a driving force for the self-assembly of bacteriorhodopsins. It seems that this mechanism is a general one for the assembly/formation of transmembrane proteins drifting on a membrane.
Dr. Yuno Natsume from Utsunomiya University focused on crowding of micrometer-sized objects in a confined space (GUV) from both a theoretical and experimental view (Natsume and Toyota 2016, Natsume et al. 2019). She showed that polystyrene beads encapsulated in a GUV could induce the deformation of the membrane upon osmotic stress treatment, and this could be explained by entropic interaction between the polystyrene beads and the inner membrane. Moreover, when beads of two different sizes (diameter 0.1 and 1 μm) were encapsulated into a GUV, the smaller beads preferentially localized on the inner membrane because of the depletion volume effect and their higher diffusivity. The theoretical estimation and the modeled experiments will help us understand how a variety of micrometer-scale cellular structures (organelles) are organized in living cells.
Dr. Naoto Nemoto from Saitama University gave a talk on the in vitro selection and evolution of pore-forming peptides. In his strategy, a cDNA method (Yamaguchi et al. 2009) was combined with GUVs and a fluorescence-activated cell sorting (FACS) technique. Peptides that have random sequences were displayed as cDNA-display molecules and mixed with GUVs along with two differently sized fluorescent molecules (large and small), which were used to assess the pore formation. Peptide function was evaluated by measuring the leakage of the fluorescent molecules from the GUVs. The GUVs that showed the higher leakage of fluorescent molecules were collected by using FACS. Then, cDNA-peptide conjugates collected from the GUVs were used in amplification by PCR and a further selection process. By repeating these cycles, the peptides capable of pore formation were obtained and their amino-acid sequences were identified. His work demonstrated the potential application of lipid vesicles in the de novo designing and engineering of peptides that function on the membrane.
In the work presented by Dr. Yutetsu Kuruma, from Japan Agency for Marine-Earth Science and Technology, lipid vesicles were used as the body of an artificial cell containing organelle-like structures (Berhanu et al. 2019). Purified FoF1-ATP synthases and bacteriorhodopsins were embedded in nano-sized lipid vesicles and then these vesicles were further encapsulated in a GUV together with a cell-free protein synthesis system (PURE system). These artificial cells were capable of synthesizing proteins by consuming self-sufficient energy (ATP) that is produced by FoF1-ATP synthases through light irradiation. Notably, the self-production of the key components in his artificial photosynthetic cells was achieved. This work, in which membrane proteins and transcription and translation machinery proteins were integrated into a GUV to form a system capable of synthesizing its own constituents, is an important milestone in the field of synthetic biology.
In this symposium, navigating the assembly of living cells was also focused on to obtain guidelines on the construction of artificial biosystems at higher hierarchical levels. Dr. Kaori Kuribayashi-Shigetomi from Hokkaido University presented a three-dimensional (3D) cell structure that was created by a unique technique, termed “cell origami” (Kuribayashi-Shigetomi et al. 2012). In this technique, living cells are cultured on patterned micro-sized plates with hinges. Contractile force generated by the cells resulted in the folding of the plate like origami, the traditional Japanese art of paper folding. By rationally designing the plate’s geometry, the cells were folded into various 3D shapes, such as cubes, tetrahedron, dodecahedrons, and cylindrical helical tubes. In addition, 3D cell co-culture microstructures were constructed using the cell-origami technique (He et al. 2018). This technique paves the way to organize cultured cells into tissue-like higher order structures using arbitrary designed shapes. Applying the folding technique to artificial cells would also be a fascinating direction for constructing “artificial tissues.”
Lipid membranes and vesicles have been mainly adopted as alternative tools to investigate the functions/phenomena that occur on cellular membranes. The studies presented in this symposium demonstrated a variety of applications of these tools, including crystallization of membrane proteins, in vitro selection of pore-forming peptides, and construction of artificial cells as well as models of organelles and cells. These successful studies encourage us to envision more sophisticated artificial biosystems that possess adaptability and autonomy, which are characteristics of living organisms.
Acknowledgments
The authors thank all invited and contributed speakers for giving exciting talks. The Authors also thank participants for contributing constructive discussions.
Funding information
This work was supported by the Japan Society for the Promotion of Science (JSPS) Grant-in-Aid for Scientific Research (KAKENHI) (grant numbers JP18J00720 to Yusuke Sato; and 18K19831 and 19H04201 to Yuki Suzuki).
Compliance with ethical standards
Conflict of interest
The authors declare that there is no conflict of interest.
Ethical approval
This article does not contain any studies on human or animals performed by any of the authors.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Yusuke Sato, Email: sato.y.cf@m.titech.ac.jp.
Masamune Morita, Email: morita.m9@aist.go.jp.
Yuki Suzuki, Email: ysuzuki79r@molbot.mech.tohoku.ac.jp.
References
- Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P. Membrane structure. In: Willson J, Hunt T, editors. Molecular biology of the cell. 5. New York: Garland Science; 2008. pp. 617–650. [Google Scholar]
- Berhanu S, Ueda T, Kuruma Y. Artificial photosynthetic cell producing energy for protein synthesis. Nat Commun. 2019;10:1325. doi: 10.1038/s41467-019-09147-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Q, Okajima T, Onoe H, Subagyo A, Sueoka K, Kuribayashi-Shigetomi K. Origami-based self-folding of co-cultured NIH/3T3 and HepG2 cells into 3D microstructures. Sci Rep. 2018;8:4556. doi: 10.1038/s41598-018-22598-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuribayashi-Shigetomi K, Onoe H, Takeuchi S. Cell origami: self-folding of three-dimensional cell-laden microstructures driven by cell traction force. PLoS One. 2012;7:e51085. doi: 10.1371/journal.pone.0051085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita M, Katoh K, Noda N. Direct observation of bacterial growth in giant unilamellar vesicles: a novel tool for bacterial cultures. Chem Open. 2018;7:845–849. doi: 10.1002/open.201800126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natsume Y, Toyota T. Asymmetrical polyhedral configuration of giant vesicles induced by orderly array of encapsulated colloidal particles. PLoS One. 2016;11:e0146683. doi: 10.1371/journal.pone.0146683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natsume Y, Noguchi E, Kurihara K. Spontaneous localization of particles in giant vesicles owing to depletion force. J Phys Soc Jpn. 2019;88:033001. doi: 10.7566/JPSJ.88.033001. [DOI] [Google Scholar]
- Sato Y, Komiya K, Kawamata I, Murata S, Nomura SM. Isothermal amplification of specific DNA molecules inside giant unilamellar vesicles. Chem Commun. 2019;55:9084–9087. doi: 10.1039/C9CC03277K. [DOI] [PubMed] [Google Scholar]
- Suzuki Y, Sugiyama H, Endo M. Complexing DNA origami frameworks through sequential self-assembly based on directed docking. Angew Chem Int Ed. 2018;57:7061–7065. doi: 10.1002/anie.201801983. [DOI] [PubMed] [Google Scholar]
- Yamaguchi J, Naimuddin M, Biyani M, Sasaki T, Machida M, Kubo T, Funatsu T, Husimi Y, Nemoto N. cDNA display: a novel screening method for functional disulfide-rich peptides by solid-phase synthesis and stabilization of Mrna-protein fusions. Nucleic Acids Res. 2009;37:e108. doi: 10.1093/nar/gkp514. [DOI] [PMC free article] [PubMed] [Google Scholar]


