This is a session commentary describing the “Current status and issues of protein solution biophysics—Session 1SDP” of the Annual Meeting of the Biophysical Society of Japan (Miyazaki 2019).
The advance of biophysico-chemical methods is enabling the quantitative and systematic analysis of biomolecules, including their interactions and conformations. In this symposium, the speakers introduced the current progress in biophysico-chemical measurement techniques.
To start the session, recent developments in the quantitative assessments of protein-mediated intermolecular solution interactions were summarized by Dr. Susumu Uchiyama, encompassing the progress in analytical ultracentrifugation (Uchiyama et al. 2018). Dr. Satoko Akashi (Grad. Sch. Med. Life Science, Yokohama City Univ.) overviewed the use of native mass spectrometry to analyze biomolecular complexes, and its development for analysis even under heterogeneous environments such as cell extracts (Akashi 2006). Dr. Satoru Nagatoishi (School of Engineering, University of Tokyo) summarized the thermodynamics of protein interactions for various therapies and diagnoses. Precise thermodynamic measurements for drug development are now possible for very weak but specific interactions of biomolecules (Nagatoishi et al. 2018). Mr. William E. Arter (Department of Chemistry, University of Cambridge) introduced biophysical analyses by microchip electrophoresis, focusing on alpha-synuclein oligomers (Arter et al. 2018).
High-speed AFM and cryo-EM are now two of the most powerful tools for the visualization of molecular events in solution. Dr. Hiroki Watanabe (Exploratory Research Center on Life and Living Systems, National Institutes of Natural Sciences) presented the dynamic structures and interactions of antibodies under physiologically relevant conditions using high-speed AFM (Yogo et al. 2019). Dr. Hiroshi Imai discussed a newly developed negative stain EM method for protein complexes at high protein concentrations (Imai et al. 2015). The broad theme to emerge from these talks was that combining several structure analysis methods is effective for precisely delineating molecular events in solution. Finally, Dr. Saeko Yanaka discussed the analysis of antibodies’ dynamic structures and their interactions under physiologically relevant conditions using nuclear magnetic resonance coupled with solution state scattering with MD simulations (Yanaka et al. 2019).
Due to the high sensitivity, specificity, and accuracy of these measurement approaches, even in situ measurements are becoming possible. In the discussion of the session, the future possibilities of these methods in realistic environments were discussed. We wish to share the fruits of these developments in future symposiums.
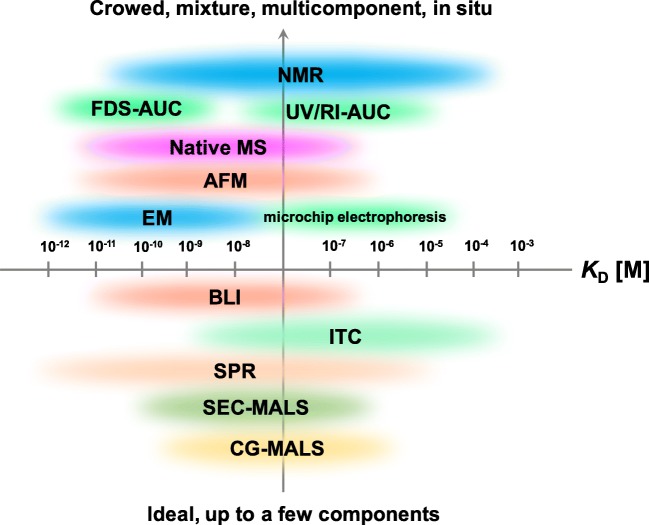
Figure: Current status and issues of protein solution biophysics. Possible quantitative assessments of intermolecular protein-mediated interactions. NMR, nuclear magnetic resonance; FDS-AUC, fluorescence detection system analytical ultracentrifugation; UV/IR-AUC, ultraviolet/infrared; Native MS, native mass spectroscopy; AFM, atomic force microscopy; EM, electron microscopy; BLI, bio-layer interferometry; ITC, isothermal titration calorimetry; SPR, surface plasmon resonance; SEC-MALS, size exclusion chromatography multi-angle light scattering; CG-MALS, composition-gradient multi-angle light scattering
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Akashi S. Investigation of molecular interaction within biological macromolecular complexes by mass spectrometry. Med Res Rev. 2006;26(3):339–368. doi: 10.1002/med.20051. [DOI] [PubMed] [Google Scholar]
- Arter WE, Charmet J, Kong J, Saar KL, Herling TW, Muller T, Keyser UF, Knowles TPJ. Combining affinity selection and specific ion mobility for microchip protein sensing. Anal Chem. 2018;90(17):10302–10310. doi: 10.1021/acs.analchem.8b02051. [DOI] [PubMed] [Google Scholar]
- Imai H, Shima T, Sutoh K, Walker ML, Knight PJ, Kon T, Burgess SA (2015) Direct observation shows superposition and large scale flexibility within cytoplasmic dynein motors moving along microtubules. Nat Commun 6 [DOI] [PMC free article] [PubMed]
- Nagatoishi S, Caaveiro JMM, Tsumoto K. Biophysical analysis of the protein-small molecule interactions to develop small molecule drug discovery. Yakugaku Zasshi. 2018;138(8):1033–1041. doi: 10.1248/yakushi.17-00211-2. [DOI] [PubMed] [Google Scholar]
- Uchiyama S, Noda M, Krayukhina E. Sedimentation velocity analytical ultracentrifugation for characterization of therapeutic antibodies. Biophys Rev. 2018;10(2):259–269. doi: 10.1007/s12551-017-0374-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanaka S, Yogo R, Inoue R, Sugiyama M, Itoh SG, Okumura H, Miyanoiri Y, Yagi H, Satoh T, Yamaguchi T, Kato K. Dynamic views of the Fc region of immunoglobulin G provided by experimental and computational observations. Antibodies. 2019;8(3):39. doi: 10.3390/antib8030039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yogo R, Yamaguchi Y, Watanabe H, Yagi H, Satoh T, Nakanishi M, Onitsuka M, Omasa T, Shimada M, Maruno T, Torisu T, Watanabe S, Higo D, Uchihashi T, Yanaka S, Uchiyama S, Kato K. The Fab portion of immunoglobulin G contributes to its binding to Fcγ receptor III. Sci Rep. 2019;9:11957. doi: 10.1038/s41598-019-48323-w. [DOI] [PMC free article] [PubMed] [Google Scholar]