Abstract
The inside of living cells is highly crowded with biological macromolecules. It has long been considered that the properties of nucleic acids and proteins, such as their structures, dynamics, interactions, and enzymatic activities, in intracellular environments are different from those under in vitro dilute conditions. In-cell NMR is a robust and powerful method used in the direct measurement of those properties in living cells. However, until 2 years ago, in-cell NMR was limited to Xenopus laevis oocytes due to technical challenges of incorporating exogenous nucleic acids. In the last 2 years, in-cell NMR spectra of nucleic acid introduced into living human cells have been reported. By use of the in-cell NMR spectra of nucleic acids in living human cells, the formation of hairpin structures with Watson–Crick base pairs, and i-motif and G-quadruplex structures with non-Watson–Crick base pairs was demonstrated. Others investigated the mRNA–antisense drug interactions and DNA–small compound interactions. In this article, we review these studies to underscore the potential of in-cell NMR for addressing the structures, dynamics, and interactions of nucleic acids in living human cells.
Keywords: In-cell NMR, DNA, RNA, Human cell, Intracellular structure, Streptolysin O (SLO)
Introduction
Genome-wide surveys have indicated that only ~ 2% of the human genome comprises protein-coding regions (Dunham et al. 2012). On the other hand, ~ 80% of the human genome is thought to be involved in gene regulation (Elkon and Agami 2017). The non-coding genome regions may include cis-regulatory elements or regions that are transcribed into non-coding RNAs (Elkon and Agami 2017). More than 70% of the human genome is reportedly transcribed into RNAs (Djebali et al. 2012). These findings suggest that nucleic acids themselves regulate various biological events. Understanding the molecular mechanisms by which these nucleic acids exert their functional roles is a necessary task. These functional nucleic acids are present inside living cells, whose internal environments are highly crowded with biological macromolecules such as nucleic acids and proteins. It is known that the biophysical characteristics, such as macromolecular interactions, molecular crowding, water activity, and viscosity, within living cells differ considerably from dilute solutions generally used in vitro (Ellis 2001; Zhou et al. 2008; Sarkar et al. 2013; Hänsel et al. 2014). Due to these differences, the structures, dynamics, and interactions of nucleic acids are assumed to be different between in vitro and living cells. Hydrophilic polymers, such as polyethylene glycol, have been used to examine the structure of nucleic acids in vitro to simulate intracellular molecular crowding. These studies also suggested that the structural and functional properties of nucleic acids are sensitive to environmental conditions even under in vitro conditions (Nakano et al. 2014). Therefore, it is essential to clearly define how the structure, dynamics, and interactions of nucleic acids within live cells differ from in vitro conditions. To meet this need, methodologies for analyzing nucleic acids inside living cells are continuously being developed (Giassa et al. 2018; Dzatko et al. 2020).
To investigate the structural features of biological macromolecules in cells, in-cell NMR is one of the prominent methods. In-cell NMR is an application of NMR spectroscopy involving a living cell suspension as a sample, where the biomolecule of interest is introduced into the cells. Due to the non-invasive property of NMR spectroscopy, the NMR signal of the target biomolecule in living cells can be obtained. The measurement of in-cell NMR spectra was firstly reported for proteins in E. coli in 2001 by Dötsch and coworkers (Serber et al. 2001), and development of the methodology is continuing (Luchinat and Banci 2017; Nishida et al. 2020). Various cell types have been used for in-cell NMR analysis of proteins (Stadmiller and Pielak 2018), including E. coli (Serber et al. 2001; Sakakibara et al. 2009), yeast (Bertrand et al. 2012), Xenopus laevis oocytes (Selenko et al. 2006), insect cells (Tanaka et al. 2019), and human cells (Inomata et al. 2009; Ogino et al. 2009; Banci et al. 2013; Theillet et al. 2016).
On the other hand, for nucleic acids, the first report was published in 2009 by Trantirek, Schwalbe, and Dötsch, who used Xenopus laevis oocytes (Hänsel et al. 2009). Compared with proteins, in-cell NMR studies on nucleic acids have long been limited to Xenopus laevis oocytes (Hänsel et al. 2013; Salgado et al. 2015; Bao et al. 2017; Ishizuka et al. 2017; Manna et al. 2018) due to the limitation of the method for introducing adequate amounts of nucleic acids into huge numbers of cells. The structure, dynamics, and intermolecular interactions of nucleic acids should be characterized in the physiological environment, including pH, redox potential, viscosity, and the presence of interactive partners, such as proteins, ions, metabolites, and so on. In particular, the expression patterns of the endogenous proteins that interact with nucleic acids should be different between Xenopus laevis oocyte and other cells. Our group was the first to use cells other than Xenopus laevis oocytes, and we succeeded in obtaining NMR signals of nucleic acids in living human cells in 2018 (Yamaoki et al. 2018). In this article, we review the rapidly developing in-cell NMR methodologies for nucleic acids in living human cells (Dzatko et al. 2018; Bao et al. 2019; Schlagnitweit et al. 2019; Krafcikova et al. 2019).
Introduction of nucleic acids into living human cells by the SLO method
The first in-cell NMR study on nucleic acids was reported 10 years ago. Firstly, in-cell NMR of nucleic acids was established using Xenopus lavis oocytes (Hänsel et al. 2009). In-cell NMR measurement requires the introduction of adequate amounts of nucleic acids into living cells. For Xenopus laevis oocytes, the microinjection method was successfully used and about 200 oocytes into which nucleic acids had been injected were subjected to in-cell NMR measurements. However, in the case of human cell lines, such as HeLa cell, 2 × 107 cells are required for in-cell NMR measurements, with adequate amounts of target nucleic acids. Consequently, the microinjection method is not feasible in these cell types. We applied a bacterial toxic protein, streptolysin O (SLO), that can reversibly form pores on cell membranes (Fig. 1). The SLO method had originally been developed to introduce various kinds of molecules in large amounts into living human cells (Kano et al. 2012, 2017; Kunishige et al. 2020). In terms of in-cell NMR application, this technique was firstly used for proteins (Ogino et al. 2009; Kubo et al. 2013; Mochizuki et al. 2018).
Fig. 1.
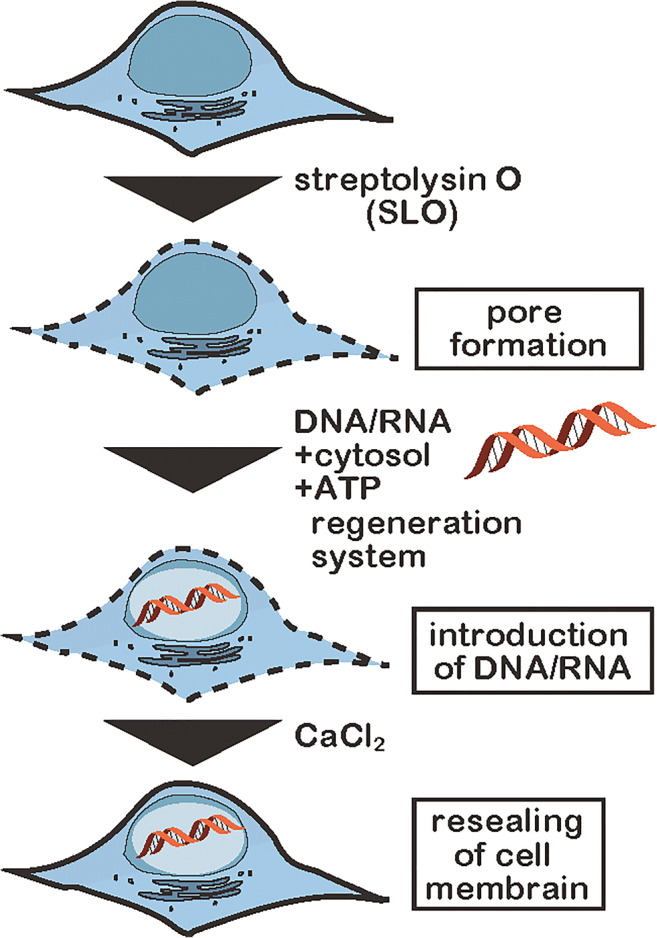
Schematic illustration of introduction of nucleic acids into living human cells by the modified streptolysin O (SLO) method
We applied this SLO method with some modifications to introduce nucleic acids into living human cells for measurement of in-cell NMR (Yamaoki et al. 2018). SLO oligomerizes into a ring structure and produces transient pores with a diameter of 25–30 nm on the cell membrane. Addition of calcium ions induces the deformation of SLO oligomers, by which the pores are resealed. Nucleic acids of interest are allowed to enter SLO-permeabilized cells, after which pores are resealed (Fig. 1). We determined that a key modification to the SLO method was the inclusion of a cytosolic extract and an ATP-regenerating system (ATP, creatine kinase, and creatine phosphate). These additives supply intracellular components, and support the survival and recovery of the resealed cells. This resulted in efficient introduction of nucleic acids and resealing of the permeabilized cells. The intracellular localization of both FAM-labeled DNA and RNA introduced by the SLO method was examined by confocal fluorescence microscopy. They were found to be localized mainly in cell nuclei (Yamaoki et al. 2018). The SLO-treated HeLa cells including the target DNA or RNA were employed as samples for in-cell NMR measurements (Fig. 1).
The first observation of NMR signals of nucleic acids in living human cells by in-cell NMR
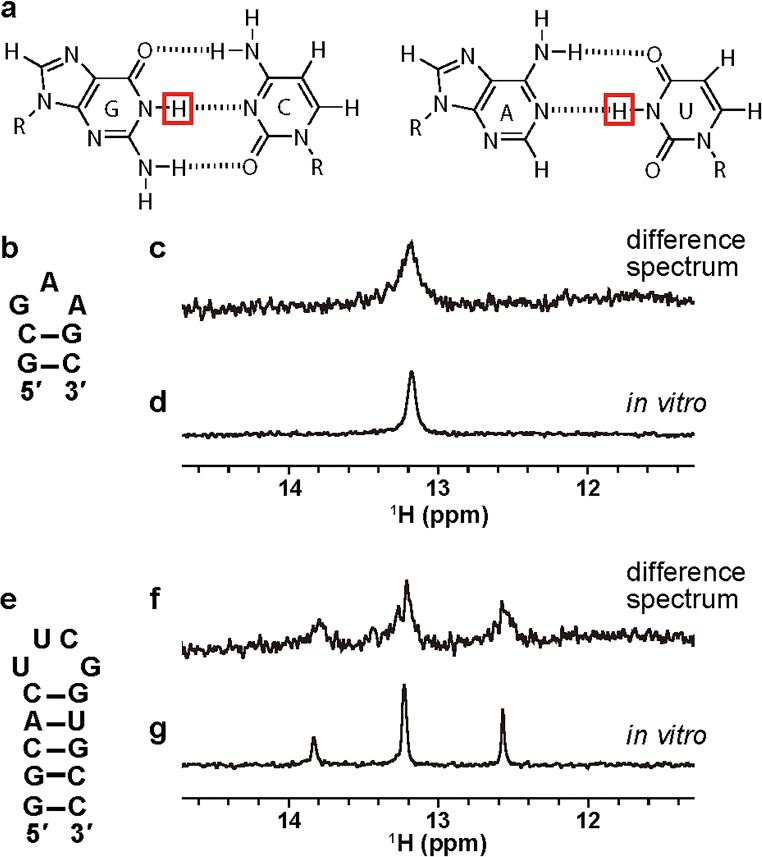
As the first examples, oligo DNA (d[GCGAAGC]) and RNA (r[GGCACUUCGGUGCC]) that form hairpin structures (Fig. 2b, e) with Watson–Crick base pairs (Fig. 2a) under in vitro dilute solution conditions were introduced into HeLa cells. This so-called hairpin DNA and RNA has been well-studied as highly stable motifs. The hairpin structures of DNAs are considered to be formed through the triplet repeat expansion associated with genetic diseases. Also, the hairpin structures of RNAs are expectedly included in various kinds of non-coding RNAs such as rRNAs, tRNAs, pre-miRNAs, and untranslated regions of mRNAs.
Fig. 2.
In-cell NMR spectra of nucleic acids in living human cells. a Watson–Crick base pairs. Imino protons are highlighted. b, e The hairpin structures of DNA and RNA, respectively. c, f The difference NMR spectra (pure in-cell spectra) of DNA and RNA, respectively. d, g The in vitro NMR spectra of DNA and RNA, respectively. (b–g Adapted from Yamaoki et al. 2018 with permission from the PCCP Owner Societies)
For in-cell NMR of nucleic acids, the imino proton signals are the first choice to monitor nucleic acids in living human cells. First of all, the imino proton signals provide much information on the mode of base-pairing of nucleic acids, as explained below. The imino region of an NMR spectrum is isolated from the regions of other signals of endogenous proteins. Additionally, endogenous nucleic acids such as genome DNAs, rRNAs, and mRNAs are too large for detection of the NMR signals of their imino protons. In the case of small RNAs, such as miRNAs, the concentrations are so low that their imino signals cannot be observed. In fact, there is no signal in the imino region of NMR spectra of intact human cells into which no exogenous DNAs and RNAs had been introduced. This means that we can obtain in-cell NMR spectra having almost no background for the imino region even without any isotopic labeling such as with 15N and 13C for identification. Using SLO-treated cells into which hairpin DNA and RNA had been introduced, we observed the NMR signals of nucleic acids in living human cells for the first time (Yamaoki et al. 2018). To prove that the signals were derived from the nucleic acids in the living cells, we removed the cells by centrifugation and obtained an NMR spectrum of the supernatant after in-cell NMR measurements. In the spectrum of the supernatant, weak imino proton signals derived from the nucleic acids that had leaked from dead cells were detected. By subtracting the spectrum of the supernatant from the original in-cell NMR spectrum, we successfully obtained a pure in-cell NMR spectrum of the nucleic acids in living human cells for the first time (Yamaoki et al. 2018) (Fig. 2c, f).
By using the in-cell NMR technique, structural information on nucleic acids in living human cells can be obtained. The NMR signal of an imino proton can be observed only when the imino proton is involved in hydrogen bonding or protected from exchange with the bulk water (Fig. 2a). This is because the imino protons of guanine, thymine, and uracil are rapidly exchanged with the protons of bulk water. Therefore, the observation of imino proton signals indicates the formation of base pairs. Moreover, the chemical shifts of imino proton signals are sensitive to the type of base pair such as Watson–Crick and non-Watson–Crick ones. We demonstrated that the chemical shifts of imino proton signals derived from the hairpin DNA and RNA under cellular conditions are similar to those under in vitro conditions (Fig. 2c, d, f, g). These results indicated that the DNA and RNA formed Watson–Crick base pairs in the living human cells as well as in vitro. Thus, the in-cell NMR study revealed that DNA and RNA form quite similar hairpin structures in living human cells as well as under in vitro conditions (Yamaoki et al. 2018).
As described above, the imino proton rapidly exchanges with water proton. Therefore, to observe the NMR signal of imino proton, the pulse sequence with pre-saturation for water suppression should be avoided. Our in-cell NMR spectra were recorded by the Selective Optimized-Flip-Angle Short-Transient (SOFAST) technique with PC9 and rSNOB pulses for water suppression (Schanda et al. 2005).
Application of in-cell NMR to detect non-canonical structures of nucleic acids in living human cells
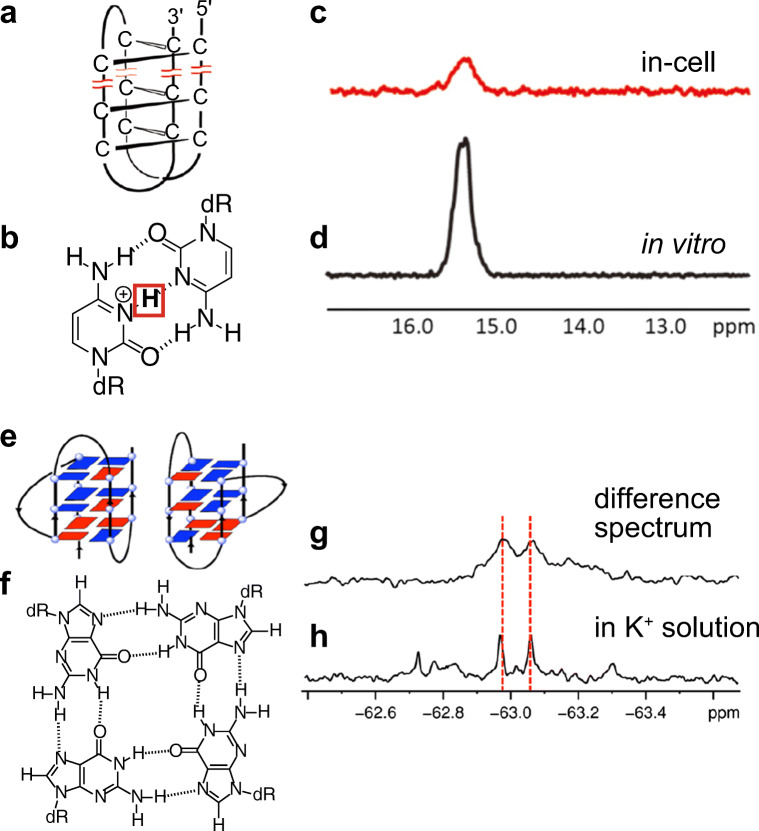
Following our report, Trantirek and coworkers also reported an in-cell NMR study on DNA in living human cells (Dzatko et al. 2018). In their study, the electroporation method was used for the introduction of nucleic acids into living human cells. The electroporation method also reversibly forms pores on cell membrane by electric pulse. As of now, the intracellular concentrations of nucleic acids that were introduced from outside the cell are comparable, 5–20 μM for both SLO and electroporation methods (Dzatko et al. 2018; Yamaoki et al. 2018). Theoretically, both methods using SLO and electroporation are applicable to various kinds of cells. They used various cytosine-rich oligo DNAs that form one of the non-canonical structures, the i-motif, formed through the intercalation of hemi-protonated cytosine–cytosine base pairs (C–C+) in vitro (Fig. 3a, b). The i-motif structure is currently considered a regulatory motif for gene transcription (Kendrick et al. 2014; Sedghi Masoud and Nagasawa 2018; Sedghi Masoud et al. 2018). They detected imino proton signals in the 15–16 ppm region of in-cell NMR spectra, which strongly indicated the formation of C–C+ base pairs and i-motif structures even in living human cells as well as in vitro (Fig. 3c, d). Furthermore, temperature variable in-cell NMR measurements showed that some i-motif structures are more stable under intracellular conditions than under in vitro conditions (Dzatko et al. 2018).
Fig. 3.
Evaluation of non-canonical structure formation of nucleic acids by in-cell NMR. a, b Schematic illustrations of i-motif and C–C+ base pair, respectively. Imino proton is highlighted. c, d In-cell and in vitro NMR spectra of C-rich oligo DNA, respectively. e, f Schematic illustrations of hybrid-type G4 structure and guanine quartet, respectively. Blue and red rectangules represent guanine bases in the anti and syn conformations, respectively. g, h In-cell and in vitro 19F-NMR spectra of telomeric DNA, respectively. (a–d Adapted from Dzatko et al. 2018. e–h Adapted from Bao et al. 2019)
Xu and coworkers also obtained structural information on another type of non-canonical structure, the guanine-quadruplex (G4 structure, Fig. 3e), in living human cells using the 19F-labeling technique (Bao et al. 2019). The G4 structure is formed through the stacking of planar guanine quartets consisting of four guanines that are hydrogen-bonded to the two adjacent guanines (Fig. 3f). The G4 structure is considered a regulatory unit for various biological events such as DNA replication, RNA transcription, protein translation, and telomere elongation (Tian et al. 2018). They introduced telomeric DNAs into living human cells by the SLO method and obtained one-dimensional 19F-NMR spectra. They revealed oligo telomeric DNAs formed the hybrid-type G4 structure (for d[AGGG(TTAGGG)3]) and the two-tetrad antiparallel G4 structure (for d[GGG(TTAGGG)3T]) in living human cells based on the chemical shifts of the 3,5-bis(trifluoromethyl)phenyl moiety (Fig. 3g, h) (Bao et al. 2019). Studies by Trantirek, Xu, and coworkers showed that in-cell NMR is a powerful tool for the structural analysis of nucleic acids in living human cells.
Application of in-cell NMR to evaluate intermolecular interactions of nucleic acids
Another application of in-cell NMR as to nucleic acids is the observation of the intermolecular interactions of nucleic acids in living human cells. The in-cell NMR in human cells allows early assessment of drug potency for human by evaluating the intracellular binding specificity of drug candidates (Luchinat et al. 2020).
Petzold and coworkers demonstrated that an antisense nucleic acid drug can interact with a target mRNA in living human cells (Schlagnitweit et al. 2019). Notably, they applied dynamic nuclear polarization (DNP)–assisted solid-state NMR for in-cell NMR of nucleic acids in living human cells (Schlagnitweit et al. 2019). The DNP technique improved the sensitivity of NMR measurements.
Trantirek and coworkers performed in-cell NMR measurements for DNA–ligand complexes. They used three ligand compounds that can bind to double-stranded DNA including T-T mismatches with similar affinity under in vitro conditions. Interestingly, although two of them can bind to the DNA even in living human cells, the third ligand cannot interact with the DNA due to its low specificity (Krafcikova et al. 2019). Studies by Petzold, Trantirek, and coworkers demonstrated the potential of in-cell NMR of nucleic acids for the development of nucleic acid drugs and drug compounds targeting nucleic acids.
Future perspective
One of the next challenges for in-cell NMR as to nucleic acids in living human cells is an observation of interactions with biologically relevant ligands such as endogenous proteins and metabolites. The intermolecular interactions of nucleic acids with such specific binding partners should be different from those under in vitro dilute conditions. So far, the intracellular concentrations of delivered nucleic acids are 5–20 μM. These concentrations are higher than physiological concentrations of RNAs such as tRNA (0.5–1 μM in Xenopus laevis oocyte). However, the concentrations of delivered nucleic acids are still close to physiological concentrations of endogenous proteins that are candidates for specific binding partners of nucleic acids of interest. For example, the intracellular concentrations of hnRNP A1, human superoxide dismutase 1 (SOD1), and human α-synuclein are 13–42 μM (Maharana et al. 2018), 10 μM (Banci et al. 2013), and 5–50 μM (Theillet et al. 2016), respectively. Therefore, the interactions of nucleic acids of interest with endogenous proteins could be observed by in-cell NMR using human cells.
Conclusions
Ten years has passed since the first in-cell NMR study on nucleic acids. Now we can obtain in-cell NMR spectra of nucleic acids in living human cells. In the last 2 years, studies involving living human cells have rapidly increased. This methodology is a powerful and promising tool for analyzing and understanding the behavior of functional DNAs and RNAs, such as gene regulation element DNAs, siRNAs, microRNAs, long non-coding RNAs, and DNA/RNA aptamers, in living human cells.
Funding information
This work was supported by MEXT, Japan [19K16054 and 17K14515 to Y.Y.; 17K07307 and 17H05878 to T.N.; and 18H04550 and 18K19397 to M.K.].
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflicts of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Banci L, Barbieri L, Bertini I, Luchinat E, Secci E, Zhao Y, Aricescu AR. Atomic-resolution monitoring of protein maturation in live human cells by NMR. Nat Chem Biol. 2013;9:297–299. doi: 10.1038/nchembio.1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao HL, Ishizuka T, Sakamoto T, Fujimoto K, Uechi T, Kenmochi N, Xu Y. Characterization of human telomere RNA G-quadruplex structures in vitro and in living cells using 19F NMR spectroscopy. Nucleic Acids Res. 2017;45:5501–5511. doi: 10.1093/nar/gkx109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao HL, Liu HS, Xu Y. Hybrid-type and two-tetrad antiparallel telomere DNA G-quadruplex structures in living human cells. Nucleic Acids Res. 2019;47:4940–4947. doi: 10.1093/nar/gkz276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand K, Reverdatto S, Burz DS, Zitomer R, Shekhtman A. Structure of proteins in eukaryotic compartments. J Am Chem Soc. 2012;134:12798–12806. doi: 10.1021/ja304809s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djebali S, Davis C, Merkel A, et al. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunham I, Kundaje A, Aldred S, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzatko S, Krafcikova M, Hänsel-Hertsch R, Fessl T, Fiala R, Loja T, Krafcik D, Mergny JL, Foldynova-Trantirkova S, Trantirek L. Evaluation of the stability of DNA i-motifs in the nuclei of living mammalian cells. Angew Chem Int Ed Engl. 2018;57:2165–2169. doi: 10.1002/anie.201712284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzatko Simon, Fiala Radovan, Hänsel-Hertsch Robert, Foldynova-Trantirkova Silvie, Trantirek Lukas. New Developments in NMR. Cambridge: Royal Society of Chemistry; 2019. Chapter 16. In-cell NMR Spectroscopy of Nucleic Acids; pp. 272–297. [Google Scholar]
- Elkon R, Agami R. Characterization of noncoding regulatory DNA in the human genome. Nat Biotechnol. 2017;35:732–746. doi: 10.1038/nbt.3863. [DOI] [PubMed] [Google Scholar]
- Ellis RJ. Macromolecular crowding: obvious but underappreciated. Trends Biochem Sci. 2001;26:597–604. doi: 10.1016/s0968-0004(01)01938-7. [DOI] [PubMed] [Google Scholar]
- Giassa IC, Rynes J, Fessl T, Foldynova-Trantirkova S, Trantirek L. Advances in the cellular structural biology of nucleic acids. FEBS Lett. 2018;592:1997–2011. doi: 10.1002/1873-3468.13054. [DOI] [PubMed] [Google Scholar]
- Hänsel R, Foldynová-Trantírková S, Löhr F, Buck J, Bongartz E, Bamberg E, Schwalbe H, Dötsch V, Trantírek L. Evaluation of parameters critical for observing nucleic acids inside living Xenopus laevis oocytes by in-cell NMR spectroscopy. J Am Chem Soc. 2009;131:15761–15768. doi: 10.1021/ja9052027. [DOI] [PubMed] [Google Scholar]
- Hänsel R, Löhr F, Trantirek L, Dötsch V. High-resolution insight into G-overhang architecture. J Am Chem Soc. 2013;135:2816–2824. doi: 10.1021/ja312403b. [DOI] [PubMed] [Google Scholar]
- Hänsel R, Luh LM, Corbeski I, Trantirek L, Dötsch V. In-cell NMR and EPR spectroscopy of biomacromolecules. Angew Chem Int Ed Engl. 2014;53:10300–10314. doi: 10.1002/anie.201311320. [DOI] [PubMed] [Google Scholar]
- Inomata K, Ohno A, Tochio H, Isogai S, Tenno T, Nakase I, Takeuchi T, Futaki S, Ito Y, Hiroaki H, Shirakawa M. High-resolution multi-dimensional NMR spectroscopy of proteins in human cells. Nature. 2009;458:106–109. doi: 10.1038/nature07839. [DOI] [PubMed] [Google Scholar]
- Ishizuka T, Zhao PY, Bao HL, Xu Y. A multi-functional guanine derivative for studying the DNA G-quadruplex structure. Analyst. 2017;142:4083–4088. doi: 10.1039/c7an00941k. [DOI] [PubMed] [Google Scholar]
- Kano F, Nakatsu D, Noguchi Y, Yamamoto A, Murata M. A resealed-cell system for analyzing pathogenic intracellular events: perturbation of endocytic pathways under diabetic conditions. PLoS One. 2012;7:e44127. doi: 10.1371/journal.pone.0044127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kano F, Noguchi Y, Murata M. Establishment and phenotyping of disease model cells created by cell-resealing technique. Sci Rep. 2017;7:15167. doi: 10.1038/s41598-017-15443-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendrick S, Kang HJ, Alam MP, Madathil MM, Agrawal P, Gokhale V, Yang D, Hecht SM, Hurley LH. The dynamic character of the BCL2 promoter i-motif provides a mechanism for modulation of gene expression by compounds that bind selectively to the alternative DNA hairpin structure. J Am Chem Soc. 2014;136:4161–4171. doi: 10.1021/ja410934b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krafcikova M, Dzatko S, Caron C, Granzhan A, Fiala R, Loja T, Teulade-Fichou MP, Fessl T, Hänsel-Hertsch R, Mergny JL, Foldynova-Trantirkova S, Trantirek L. Monitoring DNA-ligand interactions in living human cells using NMR spectroscopy. J Am Chem Soc. 2019;141:13281–13285. doi: 10.1021/jacs.9b03031. [DOI] [PubMed] [Google Scholar]
- Kubo S, Nishida N, Udagawa Y, Takarada O, Ogino S, Shimada I. A gel-encapsulated bioreactor system for NMR studies of protein-protein interactions in living mammalian cells. Angew Chem Int Ed Engl. 2013;52:1208–1211. doi: 10.1002/anie.201207243. [DOI] [PubMed] [Google Scholar]
- Kunishige R, Kano F, Murata M. The cell resealing technique for manipulating, visualizing, and elucidating molecular functions in living cells. Biochim Biophys Acta Gen Subj. 2020;1864:129329. doi: 10.1016/j.bbagen.2019.03.015. [DOI] [PubMed] [Google Scholar]
- Luchinat E, Banci L. In-cell NMR: a topical review. IUCrJ. 2017;4:108–118. doi: 10.1107/S2052252516020625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luchinat Enrico, Barbieri Letizia, Cremonini Matteo, Nocentini Alessio, Supuran Claudiu T., Banci Lucia. Drug Screening in Human Cells by NMR Spectroscopy Allows the Early Assessment of Drug Potency. Angewandte Chemie International Edition. 2020;59(16):6535–6539. doi: 10.1002/anie.201913436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maharana S, Wang J, Papadopoulos DK, Richter D, Pozniakovsky A, Poser I, Bickle M, Rizk S, Guillén-Boixet J, Franzmann TM, Jahnel M, Marrone L, Chang YT, Sterneckert J, Tomancak P, Hyman AA, Alberti S. RNA buffers the phase separation behavior of prion-like RNA binding proteins. Science. 2018;360:918–921. doi: 10.1126/science.aar7366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manna S, Sarkar D, Srivatsan SG. A dual-app nucleoside probe provides structural insights into the human telomeric overhang in live cells. J Am Chem Soc. 2018;140:12622–12633. doi: 10.1021/jacs.8b08436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki A, Saso A, Zhao Q, Kubo S, Nishida N, Shimada I. Balanced regulation of redox status of intracellular thioredoxin revealed by in-cell NMR. J Am Chem Soc. 2018;140:3784–3790. doi: 10.1021/jacs.8b00426. [DOI] [PubMed] [Google Scholar]
- Nakano S, Miyoshi D, Sugimoto N. Effects of molecular crowding on the structures, interactions, and functions of nucleic acids. Chemical Rev. 2014;114:2733–2758. doi: 10.1021/cr400113m. [DOI] [PubMed] [Google Scholar]
- Nishida N, Ito Y, Shimada I. In situ structural biology using in-cell NMR. Biochim Biophys Acta Gen Subj. 2020;1864:129364. doi: 10.1016/j.bbagen.2019.05.007. [DOI] [PubMed] [Google Scholar]
- Ogino S, Kubo S, Umemoto R, Huang S, Nishida N, Shimada I. Observation of NMR signals from proteins introduced into living mammalian cells by reversible membrane permeabilization using a pore-forming toxin, streptolysin O. J Am Chem Soc. 2009;131:10834–10835. doi: 10.1021/ja904407w. [DOI] [PubMed] [Google Scholar]
- Sakakibara D, Sasaki A, Ikeya T, Hamatsu J, Hanashima T, Mishima M, Yoshimasu M, Hayashi N, Mikawa T, Wälchli M, Smith BO, Shirakawa M, Güntert P, Ito Y. Protein structure determination in living cells by in-cell NMR spectroscopy. Nature. 2009;458:102–105. doi: 10.1038/nature07814. [DOI] [PubMed] [Google Scholar]
- Salgado GF, Cazenave C, Kerkour A, Mergny JL. G-quadruplex DNA and ligand interaction in living cells using NMR spectroscopy. Chem Sci. 2015;6:3314–3320. doi: 10.1039/c4sc03853c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar M, Li C, Pielak GJ. Soft interactions and crowding. Biophys Rev. 2013;5:187–194. doi: 10.1007/s12551-013-0104-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schanda P, Kupce E, Brutscher B. SOFAST-HMQC experiments for recording two-dimensional heteronuclear correlation spectra of proteins within a few seconds. J Biomol NMR. 2005;33:199–211. doi: 10.1007/s10858-005-4425-x. [DOI] [PubMed] [Google Scholar]
- Schlagnitweit J, Sandoz SF, Jaworski A, Guzzetti I, Aussenac F, Carbajo RJ, Chiarparin E, Pell AJ, Petzold K. Observing an antisense drug complex in intact human cells by in-cell NMR spectroscopy. ChemBioChem. 2019;20:2474–2478. doi: 10.1002/cbic.201900297. [DOI] [PubMed] [Google Scholar]
- Sedghi Masoud S, Nagasawa K. I-motif-binding ligands and their effects on the structure and biological functions of i-motif. Chem Pharm Bull (Tokyo) 2018;66:1091–1103. doi: 10.1248/cpb.c18-00720. [DOI] [PubMed] [Google Scholar]
- Sedghi Masoud S, Yamaoki Y, Ma Y, Marchand A, Winnerdy FR, Gabelica V, Phan AT, Katahira M, Nagasawa K. Analysis of interactions between telomeric i-motif DNA and a cyclic tetraoxazole compound. ChemBioChem. 2018;19:2268–2272. doi: 10.1002/cbic.201800425. [DOI] [PubMed] [Google Scholar]
- Selenko P, Serber Z, Gadea B, Ruderman J, Wagner G. Quantitative NMR analysis of the protein G B1 domain in Xenopus laevis egg extracts and intact oocytes. Proc Natl Acad Sci U S A. 2006;103:11904–11909. doi: 10.1073/pnas.0604667103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serber Z, Ledwidge R, Miller SM, Dötsch V. Evaluation of parameters critical to observing proteins inside living Escherichia coli by in-cell NMR spectroscopy. J Am Chem Soc. 2001;123:8895–8901. doi: 10.1021/ja0112846. [DOI] [PubMed] [Google Scholar]
- Stadmiller SS, Pielak GJ. The expanding zoo of in-cell protein NMR. Biophys J. 2018;115:1628–1629. doi: 10.1016/j.bpj.2018.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T, Ikeya T, Kamoshida H, Suemoto Y, Mishima M, Shirakawa M, Güntert P, Ito Y. High-resolution protein 3D structure determination in living eukaryotic cells. Angew Chem Int Ed Engl. 2019;58:7284–7288. doi: 10.1002/anie.201900840. [DOI] [PubMed] [Google Scholar]
- Theillet FX, Binolfi A, Bekei B, Martorana A, Rose HM, Stuiver M, Verzini S, Lorenz D, van Rossum M, Goldfarb D, Selenko P. Structural disorder of monomeric α-synuclein persists in mammalian cells. Nature. 2016;530:45–50. doi: 10.1038/nature16531. [DOI] [PubMed] [Google Scholar]
- Tian T, Chen YQ, Wang SR, Zhou X. G-Quadruplex: a regulator of gene expression and its chemical targeting. Chem. 2018;4:1314–1344. doi: 10.1016/j.chempr.2018.02.014. [DOI] [Google Scholar]
- Yamaoki Y, Kiyoishi A, Miyake M, Kano F, Murata M, Nagata T, Katahira M. The first successful observation of in-cell NMR signals of DNA and RNA in living human cells. Phys Chem Chem Phys. 2018;20:2982–2985. doi: 10.1039/c7cp05188c. [DOI] [PubMed] [Google Scholar]
- Zhou HX, Rivas G, Minton AP. Macromolecular crowding and confinement: biochemical, biophysical, and potential physiological consequences. Annu Rev Biophys. 2008;37:375–397. doi: 10.1146/annurev.biophys.37.032807.125817. [DOI] [PMC free article] [PubMed] [Google Scholar]