Abstract
COPD is a heterogeneous syndrome. Many COPD subtypes have been proposed, but there is not yet consensus on how many COPD subtypes there are and how they should be defined. The COPD Genetic Epidemiology Study (COPDGene), which has generated 10-year longitudinal chest imaging, spirometry, and molecular data, is a rich resource for relating COPD phenotypes to underlying genetic and molecular mechanisms. In this article, we place COPDGene clustering studies in context with other highly cited COPD clustering studies, and summarize the main COPD subtype findings from COPDGene. First, most manifestations of COPD occur along a continuum, which explains why continuous aspects of COPD or disease axes may be more accurate and reproducible than subtypes identified through clustering methods. Second, continuous COPD-related measures can be used to create subgroups through the use of predictive models to define cut-points, and we review COPDGene research on blood eosinophil count thresholds as a specific example. Third, COPD phenotypes identified or prioritized through machine learning methods have led to novel biological discoveries, including novel emphysema genetic risk variants and systemic inflammatory subtypes of COPD. Fourth, trajectory-based COPD subtyping captures differences in the longitudinal evolution of COPD, addressing a major limitation of clustering analyses that are confounded by disease severity. Ongoing longitudinal characterization of subjects in COPDGene will provide useful insights about the relationship between lung imaging parameters, molecular markers, and COPD progression that will enable the identification of subtypes based on underlying disease processes and distinct patterns of disease progression, with the potential to improve the clinical relevance and reproducibility of COPD subtypes.
Key Words: COPD, emphysema, machine learning
Abbreviations: GOLD, Global Initiative for Chronic Obstructive Lung Disease; ICGC, International COPD Genetics Consortium; TGF-β, transforming growth factor-β
COPD has many different clinical presentations, and COPD can be viewed as an umbrella syndrome that encompasses many distinct diseases.1 Despite recent efforts to expand the criteria for diagnosing and staging COPD, the definitions from expert panels2 do not fully capture the clinical heterogeneity of the disease.
The COPD Genetic Epidemiology Study (COPDGene) has generated detailed, longitudinal clinical phenotyping and genomic data for thousands of smokers, and these data are a rich resource for understanding the clinical and molecular heterogeneity of COPD. Machine learning methods can be used to identify new subtypes of COPD, defined by using patterns of clinical and molecular markers. Dozens of articles using COPDGene data have addressed this question, but there has been no comprehensive review of these scientific contributions.
The current article reviews the most relevant subtyping articles from COPDGene according to the broad questions they address: (1) How can clustering methods be used to discover novel subtypes, and are these subtypes reproducible? (2) What other machine learning methods besides clustering can be used to study COPD heterogeneity? (3) How can cut-points be defined in a data-driven way to turn continuous COPD measures into subtypes? (4) How can machine learning on chest CT data improve our ability to characterize COPD heterogeneity? (5) Are there distinct trajectories of lung function over the life course that correspond to molecular subtypes of COPD? In addition to this review, the contributions of COPDGene to COPD imaging,3 physiology,4 clinical epidemiology,5 genetics,6 and biomarker discovery7 have been covered in separate reviews.
The following sections provide a brief background on the study of COPD subtypes and the use of unsupervised machine learning methods for disease subtyping, and we summarize the most important published results in this area using COPDGene data.
Historical Perspective on COPD Subtypes
Clinicians and COPD researchers have long recognized that COPD encompasses multiple different disease processes. However, it has been difficult to precisely define the molecular underpinnings of the diverse phenotypic manifestations of COPD. As a result, the COPD field lacks the information required to develop a sufficiently detailed, comprehensive disease classification. The 1958 CIBA Symposium was a landmark event in COPD subtyping, and the summary of this symposium states that the lack of a precise COPD definition resulted in “confusion and misunderstanding between investigators working in different centers and in different branches of medicine” that limited the fundamental understanding of COPD.8 The CIBA Symposium framework remains influential today, particularly with respect to: (1) pathologic classification of emphysema based on the anatomy of the secondary pulmonary lobule; (2) the differentiation of reversible (asthma-related) from irreversible (COPD-related) pulmonary obstruction; and (3) the identification of chronic bronchitis and emphysema as the two primary clinical phenotypes of COPD.
In subsequent work, Charles Fletcher and Benjamin Burrows expanded on the concept of the chronic bronchitis and emphysema-predominant subtypes of COPD by using a variety of clinical measurements to define type A (emphysema-predominant) and type B (bronchial) COPD subtypes.9,10 Notably, this classification also included type X patients who did not meet criteria for either category, and although the authors provided general outlines for these subtypes, they concluded that “firm definitions of the syndromes would be premature” due to lack of understanding of the etiologic mechanisms of COPD. In subsequent years, multiple additional COPD subtypes were proposed, including the frequent exacerbator subtype,11,12 asthma-COPD overlap,13 and upper lobe-predominant emphysema.14
With the advent of larger datasets, machine learning methods were used for COPD subtype discovery,15, 16, 17, 18, 19, 20 and a selected list of such studies in included in Table 1. However, these clustering studies used different methods and variables, making it challenging to synthesize and interpret this literature.21
Table 1.
Selected Publications Using Machine Learning Methods to Identify Clusters or Disease Axes in COPD
| Category | PMID | Year | No. of Subjects | No. of Clusters/Axes | Method |
|---|---|---|---|---|---|
| Clustering | 18248806 | 2008 | 415 | 2 clusters | Fuzzy clustering |
| 19501190 | 2009 | 415 | 2 clusters | Multidimensional scaling and KHM clustering | |
| 20233420 | 2010 | 308 | 4 clusters | K-means | |
| 20075045 | 2010 | 322 | 4 clusters | Principal components analysis and hierarchical clustering | |
| 21177668 | 2011 | 342 | 3 clusters | K-means | |
| 22154126 | 2012 | 102 | 2 clusters | K-means | |
| 23236428 | 2012 | 527 | 3 clusters | Principal components analysis and hierarchical clustering | |
| 23392440 | 2013 | 213 | 5 clusters | Self-organizing maps | |
| 23613569 | 2013 | 1,543 | 3 clusters | Tree-based clustering | |
| 23536961 | 2013 | 157 | 4 clusters | Factor analysis and k-means | |
| 24563194 | 2014 | 8,288 | 4 clusters | K-means | |
| 25642832 | 2015 | 2,164 | 5 clusters | Factor analysis and random forests clustering | |
| 26773458 | 2016 | 364 | 4 clusters | Network-based stratification | |
| 28943279 | 2017 | 9,210 | 3 clusters | Random forests clustering | |
| 29097431 | 2017 | 6,060 | 5 clusters | Hierarchical clustering | |
| 28637835 | 2017 | 17,146 | Multiple solutions | Random forests and k-medoids clustering | |
| 29671603 | 2018 | 4,606 | 4 trajectories | Bayesian trajectory modeling | |
| Disease axes | 19480658 | 2009 | 127 | 4 disease axes | Principal components analysis |
| 29771274 | 2018 | 8,157 | 5 disease axes | Factor analysis | |
| 31189730 | 2019 | 4,726 | 6 disease axes | Weighted logistic regression |
In addition, there are fundamentally different perspectives on whether COPD is best described by using distinct subgroups or rather multiple overlapping disease processes. The term COPD “subtypes” has two common uses. It refers broadly to the study of COPD heterogeneity, but in its more specific meaning it refers to distinct, nonoverlapping subgroups of subjects. The term “endotype”22 refers to underlying molecular processes that define subtypes, similar to the concept of T-helper type 2-mediated airway inflammation in asthma.23 Unlike subtypes or endotypes, the term “treatable traits”24 was proposed as an alternative to the concept of subtypes in which rigid subgroup boundaries were replaced by a more flexible characterization based on overlapping traits, such as bronchodilator responsiveness, airway wall thickening, and sputum eosinophilia. In the treatable traits paradigm, subjects with COPD can have many overlapping disease processes that may vary in severity, rather than being classified into one and only one subtype. A similar concept has also been proposed in diabetes.25 Finally, the term “disease axis”26 refers specifically to continuous measures that are composed of many contributing variables. Disease axes are produced by a specific class of machine learning methods called dimension reduction algorithms, and they were proposed as an alternative to clustering algorithms for COPD subtyping.
Challenges and Applications of Unsupervised Machine Learning Methods in COPD Subtyping
Machine learning refers to the design, development, and analysis of computational algorithms that automatically “learn” from experience (data) to achieve a specific task. In COPD, unsupervised learning algorithms have been used to discover novel subtypes by mining complex datasets. Two major classes of unsupervised machine learning algorithms are clustering and dimension reduction. Clustering algorithms such as k-means or hierarchical clustering seek to assign subjects into groups by some measure of similarity. In this sense, clustering methods simplify data along the subject dimension by compressing a large number of subjects into a smaller number of groups or clusters. When the data do not intrinsically have distinct clusters, the choice of cluster number can be arbitrary, and highly dataset- and method-dependent.
Dimension reduction methods simplify datasets along the variable dimension by combining measured variables into a smaller number of composite variables that contain as much of the original information as possible. Dimension reduction is most useful when there is strong correlation structure in a dataset, because much of the information can be “compressed” into a smaller number of composite variables, thereby reducing the dimension of the original dataset.
With the increasing availability of data-rich measurements such as CT images and genomic datasets in thousands of subjects with COPD, machine learning has the potential to discover novel connections between the physiologic manifestations of COPD and their underlying biological processes. However, the application of machine learning to COPD subtyping faces many challenges. Machine learning algorithms are complex and do not always produce results that are reliable or readily interpretable. Effective applications of machine learning often still rely on human expertise to extract the proper meaning from noisy variables and to evaluate between multiple possible outputs from the same algorithm. The current article illustrates the limitations of machine learning in COPD subtyping and some of the successes to date.
COPDGene Contributions to the Identification of COPD Subtypes and Disease Axes
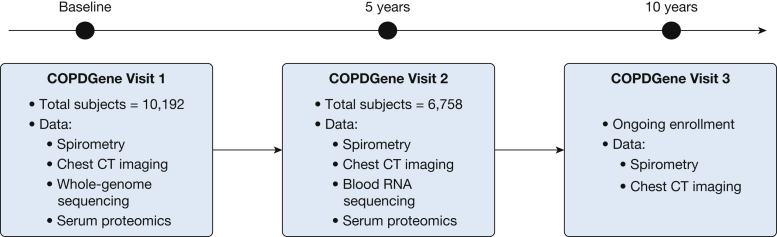
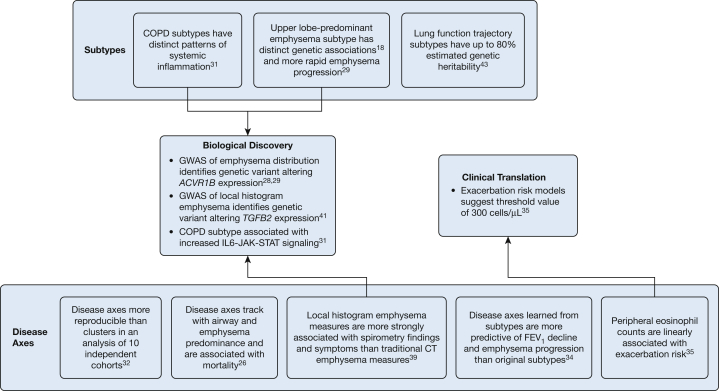
COPDGene enrolled a total of 10,192 current and former smokers across the full spectrum of lung function at 21 different centers across the United States.27 At baseline, 43% of subjects had normal spirometry findings, and 36% were in Global Initiative for Chronic Obstructive Lung Disease (GOLD) stage 2, 3, or 4. Two-thirds of the subjects were non-Hispanic white, and one-third were African American. Forty-seven percent were women, and the average age of subjects at enrollment was 60 years. Nearly all study subjects underwent spirometry, questionnaire assessments, standardized inspiratory and expiratory chest CT imaging, and genome-wide genotyping. Five-year follow-up data were obtained for 6,758 subjects, and 10-year visits are currently being conducted. Figure 1 provides an overview of the number of subjects and data types currently available for each of the three COPDGene visits, and Figure 2 provides an overview of the major findings from machine learning analyses of COPD subtypes and disease axes in COPDGene data.
Figure 1.
Overview of data gathered at the baseline, 5-year, and 10-year visits of the COPD Genetic Epidemiology Study (COPDGene).
Figure 2.
Summary of contributions from COPDGene to machine learning approaches to COPD subtyping. GWAS = genome-wide association study. See Figure 1 legend for expansion of other abbreviation.
How Can Clustering Methods Be Used to Discover Novel Subtypes, and Are These Subtypes Reproducible?
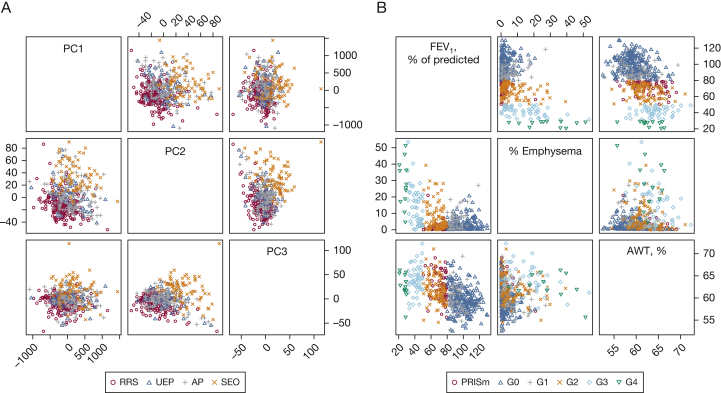
To identify COPD subtypes using clinical variables, Castaldi et al18 analyzed spirometric and imaging variables using k-means clustering to identify four clusters of phenotypically distinct subjects in COPDGene. These clusters were: (1) relatively resistant to smoking; (2) mild upper lobe emphysema-predominant; (3) airway-predominant COPD; and (4) severe airflow obstruction and emphysema. Although the average characteristics of the four clusters were distinct, when we visualized the clusters, there was little separability between the groups (Fig 3A), indicating that the subjects in COPDGene are distributed along a continuous spectrum of phenotypic variability, rather than forming clearly distinct clusters.
Figure 3.
Scatterplot matrices show the distribution of clustering-defined subtypes (Castaldi et al18) in principal component space for 500 subjects from COPDGene (A), and the same subjects projected along the dimensions of FEV1 % predicted, CT quantitative emphysema, and CT airway wall thickness with points colored by Global Initiative for Chronic Obstructive Lung Disease spirometric categories (B). AP = airway predominant; AWT, % = airway wall thickness as a percentage of total luminal area for segmental airways; G0-G4 = Global Initiative for Chronic Obstructive Lung Disease spirometric stages 0 to 4; PC = principal component; PRISm = preserved ratio impaired spirometry (ie, FEV1 < 80% of predicted, FEV1/FVC > 0.7); RRS = relatively resistant smokers; SEO = severe emphysema and obstruction; UEP = upper lobe emphysema predominant. See Figure 1 legend for expansion of other abbreviation.
When genetic association testing was performed for these clusters, the severe obstruction/emphysema and the upper lobe-predominant groups exhibited a strong association with several known COPD-associated variants, whereas the airway-predominant groups had a much weaker pattern of genetic association. The observation of strong genetic associations to the mild upper lobe-predominant groups led to subsequent articles examining the genetic basis of apico-basal emphysema distribution. A genome-wide association study for emphysema distribution identified five genome-wide significant associations,28 and subsequent cell-based functional studies identified an emphysema-associated functional variant altering the expression of ACVR1B, a signaling receptor in the transforming growth factor-β (TGF-β) superfamily.29 In a separate clustering analysis focused specifically on measures of emphysema distribution, the upper lobe-predominant group was observed to have more rapid 5-year progression of emphysema in both unadjusted and multivariate adjusted analyses.30
To determine whether blood gene expression data can be used to stratify smokers according to systemic inflammation state, Chang et al31 applied a network-based stratification method to gene expression data from subjects from COPDGene and Evaluation of COPD Longitudinally to Identify Predictive Surrogate Endpoints (ECLIPSE) studies. This analysis identified reproducible gene expression signatures that distinguished four subtypes of smokers. These signatures distinguished subjects with moderate airflow obstruction from those without obstruction; in addition, the signatures of some subgroups were enriched for inflammatory pathways such as IL6-JAK-STAT signaling, in which the expression of this pathway was increased in the cluster with the lowest average FEV1 relative to the cluster with the highest FEV1. Other gene functional categories such as lymphocyte activation, wound healing, and protein catabolism were also associated with subtype signatures. When we compared the overlap between the clustering assignments for 120 COPDGene subjects included in both the Castaldi et al18 and Chang et al31 articles, the clusterings were different (Table 2).
Table 2.
Comparison of K-Means Clustering and NBS Clustering Results Shows Little Overlap
| Variable | NBS1 | NBS2 | NBS3 | NBS4 |
|---|---|---|---|---|
| Relatively resistant smokers | 20 | 10 | 3 | 2 |
| Upper lobe predominant emphysema | 5 | 9 | 1 | 1 |
| Airway predominant | 15 | 11 | 0 | 3 |
| Severe COPD | 15 | 13 | 1 | 11 |
For 120 COPDGene subjects analyzed in both the Castaldi et al18 phenotype clustering article and the Chang et al31 gene expression clustering article, the overlap between clustering assignments was modest. Network-based stratification (NBS) clusters are ordered as in the original manuscript by average level of FEV1 (ie, NBS1 has highest average FEV1).
Because the studies by Castaldi et al18 and Chang et al31 used different input variables, it is not surprising that the clusters differed. However, for studies evaluating similar variables, one would expect that clustering studies across different cohorts would produce similar results. In fact, when comparing the average characteristics of clusters, the subtypes identified by Castaldi et al do show some similarity to those reported in other studies. In 342 subjects with COPD hospitalized for respiratory exacerbation,16 three clusters were identified, two of which resembled the airway-predominant COPD and severe airflow and emphysema clusters. The upper lobe-predominant emphysema cluster was not identified in this study, which was expected because it did not include CT-quantified emphysema. In another study of 415 subjects with COPD recruited from outpatient clinics,15 two clusters were identified that again resembled the airway-predominant COPD and severe airflow and emphysema groups. In the only systematic review conducted of COPD clustering studies, Pinto et al21 found two recurring clusters that seem to share characteristics with the airway-predominant COPD and severe airflow and emphysema clusters. However, Pinto et al also noted that it was not possible to perform a quantitative comparison of clustering results because the methods and variables used across studies were dissimilar, and thus quantitative assessment of the reproducibility of clustering results could not be performed.
To directly address the question of the reproducibility of clustering in COPD, a collaborative study in the International COPD Genetics Consortium (ICGC) was performed to assess the subject-level similarity of clustering results from multiple methods applied across multiple cohorts.32 This study showed that clustering results were only modestly reproducible. However, the principal component axes derived from these same datasets were very stable. This suggests that, for the set of variables studied, the COPD “phenotypic space” is a continuum rather than a group of discrete clusters. Figure 3B shows the continuous nature of the COPD phenotypic space for different sets of variables in COPDGene. This continuous phenotypic space is more amenable to dimension reduction than clustering.
A subsequent clustering reproducibility study33 from the COPD Cohorts Collaborative International Assessment (3CIA) reported greater agreement, although the metrics of reproducibility differed between the two studies. In the ICGC study,32 clustering reproducibility was assessed by comparing the results of clustering analyses performed de novo in each of the participating cohorts. In the 3CIA study, clustering was performed in a single cohort, and the reproducibility of cluster-specific mortality rates was assessed across multiple cohorts by using this single clustering solution. Although both studies are valid, the definition of clustering reproducibility is not the same. The ICGC results provide information on the reproducibility of the clustering process itself, whereas the 3CIA study reports the reproducibility of average characteristics and event rates of a single clustering solution.
In summary, these studies highlight that clustering is useful for identifying novel connections between clinical phenotype and molecular measures. However, the reproducibility of clustering across datasets may not be high, because COPD clinical datasets often do not have a strong clustering structure. Thus, for any clustering result, demonstration of reproducibility of the clustering process itself is essential for any claims about the generalizability or clinical translation of the cluster assignments.
What Other Machine Learning Methods Besides Clustering Can Be Used to Study COPD Heterogeneity?
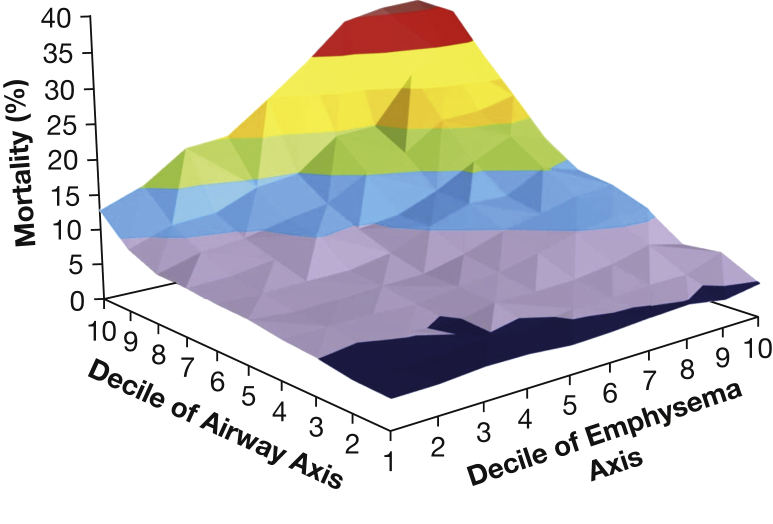
As an extension of the finding that disease axes were more reproducible than clusters, Kinney et al26 applied another dimension reduction method (factor analysis) to 28 chest CT and pulmonary function measures in COPDGene to identify COPD disease axes. In factor analysis, the contribution of the original variables to each factor can be quantified through the factor loadings for each axis. Pulmonary function measures contributed strongly to the first two factors: the first was labeled as the emphysema disease axis based on contributions from multiple CT emphysema measures, and the second was labeled as the airway disease axis due to contributions of CT measures of the thickness of the segmental airway walls. Three other factors were identified: two represented both gas trapping and hyperinflation, and one captured CT measurement variability associated with BMI. These factors were then incorporated into predictive models of mortality and clinical outcomes in COPD. Both the airway and emphysema disease axes were related to mortality, with a statistically significant, synergistic interaction between the airway and emphysema disease axes (Fig 4).
Figure 4.
The y-axis represents the predicted probability of all-cause mortality ranging from 4% (shown in dark blue), 5% to 10% (shown in purple), 10% to 15% (shown in blue), 15% to 20% (shown in green), 20% to 25% (shown in orange), 25% to 30% (shown in yellow), 30% to 35% (shown in red), to > 35% (shown in dark red) for each decile of loading score for factors 1 (Emphysema Axis) and 2 (Airway Axis) in a Cox proportional hazards model including age, sex, current smoking, pack years of smoking, BMI, high BP, each of the five factors, the interaction between factors 1 and 2, and a quadratic term for factor 2. The x and z axes represent deciles of each axis, ranging from 1 (representing a small loading score) to 10 (representing a large loading score).
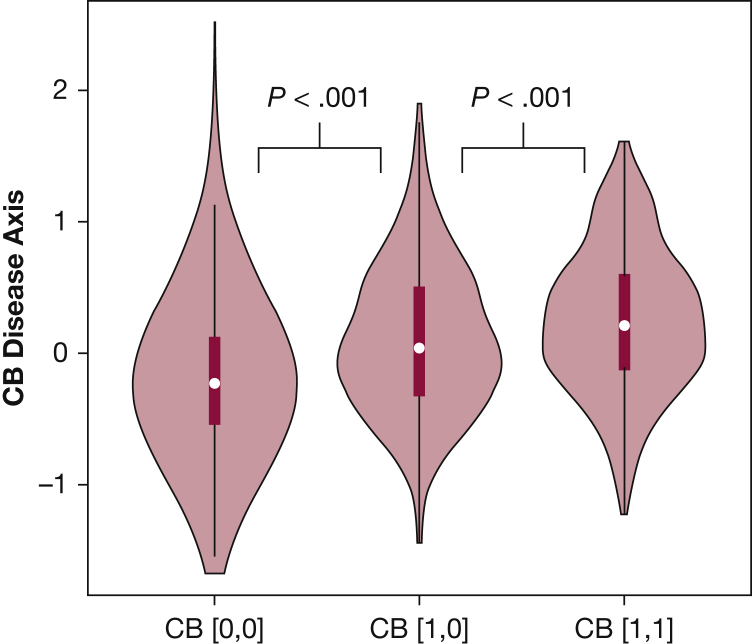
Chen et al34 developed an approach to generate more clinically interpretable disease axes that would allow users to have a greater level of control in determining the orientation of a disease axis. The concept of this method is to create disease axes that are oriented or “anchored” at either end by known COPD subtypes. In practice, this is done by building a logistic regression model to discriminate between the two subtypes or subgroups, with the predicted values from this model constituting a subtype-defined disease axis. We applied this method to build a chronic bronchitis disease axis. We observed that, relative to the presence or absence of chronic bronchitis at baseline, the disease axis provided better prediction for 5-year change in FEV1 (6.4% vs 6.0% variance explained) and emphysema (12.8% vs 7.5% variance explained), and disease axis values at baseline were predictive of persistent chronic bronchitis symptoms at the COPDGene 5-year follow-up visit (Fig 5).
Figure 5.
Distribution of chronic bronchitis disease axis values at the COPDGene baseline visit according to presence of chronic bronchitis symptoms at the baseline and 5-year study visit. Subjects with persistent chronic bronchitis symptoms (ie, present at both visits, CB [1,1]) had disease axis values that were higher than subjects without chronic bronchitis (CB [0,0]) and subjects with intermittent symptoms (CB [1,0] for chronic bronchitis at baseline but not at the 5-year visit). P values were calculated by using the Mann-Whitney U test. See Figure 1 legend for expansion of abbreviation.
In summary, COPD clinical variability is typically distributed along a continuum, and continuous disease axes generated by dimension reduction methods are more natural representations of this continuum that are also more likely to be reproducible than clusters. A direct comparison between subtypes and disease axes showed that disease axes often provide more accurate prediction of future COPD-related events.
How Can Cut-Points Be Defined in a Data-Driven Way to Turn Continuous COPD Measures Into Subtypes?
If continuous disease axes are more accurate and reproducible than clusters, how could such continuous phenotypes be used to help make clinical decisions? Yun et al35 addressed this question by examining the relation between peripheral blood eosinophil measurements and risk of COPD exacerbations. In COPDGene subjects in GOLD spirometric stages 2, 3, or 4, the number of respiratory exacerbations was linearly related to the number of eosinophils in the peripheral blood, and this relation was stronger with absolute eosinophil counts rather than with eosinophil percentage. To determine a reasonable cutoff, prediction models for exacerbations were made using a range of cutoffs on absolute eosinophil count, with a value of 300 cells/μL having the best performance. These models were validated in subjects from the ECLIPSE study. These findings are consistent with other reports, including an analysis of 7,225 subjects with COPD in the Copenhagen General Population Study, which also found that absolute eosinophil counts provided superior prediction of respiratory exacerbations relative to eosinophil percentages.36 This study used a similar count threshold of 340 cells/μL. Another analysis of 7,245 subjects with COPD confirmed that a cutoff of 300 cells/μL was associated with exacerbation rate in multivariate models, and the exacerbation rate increased with higher cutoff thresholds.37
The study by Yun et al35 provides a roadmap for how to turn continuous COPD phenotypes (in this case, peripheral eosinophilia) into clinically relevant subtypes according to criteria based on assessment of risk for COPD-related outcomes. This article shows how predictive models can be used to identify specific subtype cutoffs, although this method also raises the possibility of having different sets of COPD subtypes corresponding to different clinical outcomes.
How Can Machine Learning on Chest CT Data Improve Our Ability to Characterize COPD Heterogeneity?
Semi-automated classification of emphysema patterns and airway wall thickness from thousands of COPDGene CT scans has improved our ability to divide COPD into distinct subgroups. Mendoza et al38 used k-nearest neighbor clustering to quantify distinct CT emphysema patterns by comparing local lung density histograms vs a set of manually curated reference patterns of pathologic emphysema in > 9,000 CT scans from COPDGene. The resulting local histogram emphysema quantifications had stronger associations to a range of spirometric and functional measures than standard measures of CT emphysema,39 and genome-wide association study of these measures identified known and novel genetic associations.40 One of the genetic regions identified by the genome-wide association study was subsequently shown using CRISPR gene editing to contain a fibroblast-specific enhancer element that increases the expression of TGFB2 in fibroblasts; this finding provides additional genetic evidence of the link between emphysema and TGF-β signaling in human COPD.41
Are There Distinct Trajectories of Lung Function Over the Life Course That Correspond to Molecular Subtypes of COPD?
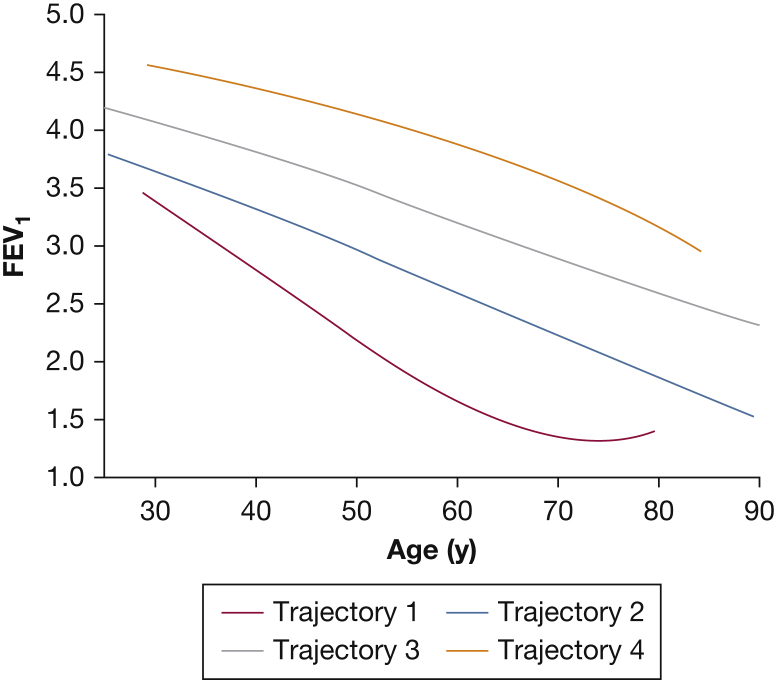
COPD subtypes are usually defined based on cross-sectional data, but subtypes learned in this manner can be confounded by differences in disease severity. For certain tasks, such as the identification of genetic associations to COPD, it is desirable to identify distinct patterns of disease progression that are not confounded by these severity differences. To address this need, Ross et al42 developed a Bayesian modeling approach that incorporates the concept of disease trajectories into COPD subtype identification. This study used decades-long longitudinal spirometric data in the Normative Aging Study (NAS) to identify and model four distinct patterns of FEV1 decline. Interestingly, the trajectory with the most rapid rate of decline in mid-life was also characterized by the lowest maximal FEV1 attained, suggesting this was a low lung growth/rapid decline trajectory (Fig 6).
Figure 6.
Four lung function trajectories learned from analyzing 1,060 men followed up for > 20 years in the Normative Aging Study. Trajectory 1 was characterized by both a lower maximal FEV1 attained as well as a more rapid rate of lung function loss in mid-life. The other trajectories differed primarily in maximal FEV1 attained but not in rate of decline. See Figure 1 legend for expansion of abbreviation.
These models were then applied to a subset of COPDGene subjects to infer their lung function trajectory assignment. In COPDGene, subjects with severe COPD were overrepresented in the low growth/rapid decline trajectory. This trajectory seems to be strongly associated with genetic differences based on a higher rate of parental COPD and the high genetic contribution to trajectories identified from heritability-based analysis.43 These findings are consistent with the results of other trajectory-based analyses of COPD,44, 45, 46 and this is a promising approach for integrating information between studies that have varying amounts of longitudinal follow-up available.
Discussion and Future Directions
The main findings from the studies covered in this review are as follows: (1) clustering is most useful for exploratory analyses of COPD subtypes; (2) continuous disease axes more accurately represent COPD heterogeneity than clusters; (3) chest CT phenotypes obtained through machine learning algorithms have improved our ability to quantify COPD heterogeneity and have led to novel biological discoveries, including in the TGF-β pathway; and (4) trajectories of lung growth and decline show strong genetic influences and may enable more powerful biological discoveries in COPD.
Although the use of machine learning with rich COPD datasets is promising, a strict replication analysis in 10 cohorts found that clustering results were poorly reproducible. The conclusion from this study is that, in some instances, clustering is poorly suited for COPD data that are distributed along a continuum without distinct subgroups.32 Because of these issues of reproducibility, greater focus has been placed on the identification of continuous measures of COPD-related disease processes, such as treatable traits and disease axes. Disease axes have been shown to be more reproducible than clusters32 and more predictive of 5-year changes in FEV1 and emphysema.34
Clinical translation of disease axes and treatable traits requires that clinically relevant cutoffs be identified for these continuous measures. The research by Yun et al35 in peripheral eosinophilia shows how support for cutoff values can be derived from predictive risk models. By relating eosinophilia to exacerbation risk, standard statistical methods provided support for a cutoff of 300 cells/μL. Based on many additional studies of stability of blood eosinophil counts and retrospective analysis of clinical trial data, the GOLD 2019 criteria also included the 300 eosinophils/μL threshold for considering first-line inhaled corticosteroids in subjects with group D COPD.2 Thus, peripheral eosinophilia is a concrete example of how a continuous COPD phenotype can be translated into subtypes for clinical practice through the development and replication of risk models for a COPD-related outcome. This implies that different cutoffs and subgroups may need to be defined for different outcomes. Thus, rather than asking “What are the subtypes of COPD?” it may be better to determine which subtypes are the most useful for a specific clinical purpose.
As we discover more COPD-related biomarkers, we can expect that COPD subtypes will increasingly be defined by using a combination of clinical features, imaging characteristics, and molecular markers. As our knowledge of genetic associations to COPD steadily increases,47 and the quality of COPD phenotypes improves, updated COPD subtype definitions will better capture the clinical and biological heterogeneity of COPD.
What are the key areas in which we anticipate additional contributions from COPDGene? First, when 10-year follow-up data are available, associations to disease progression will be more apparent, and more detailed descriptions of lung function trajectories will be possible. Second, the large-scale generation of DNA sequencing, RNA sequencing, DNA methylation, and proteomic data from blood samples at the 5- and 10-year visits will identify key molecular biomarkers of COPD progression that will lead to improved definitions of COPD molecular subtypes. Third, updated analyses of disease progression can identify the minimal sets of variables necessary for accurate risk stratification, making subtyping more broadly applicable in a clinical setting. Finally, advances in machine learning methods may lead to a more detailed understanding of the relation between COPD heterogeneity and disease progression.
Acknowledgements
Financial/nonfinancial disclosures: The authors have reported to CHEST the following: P. J. C. has received research support and consulting fees from GlaxoSmithKline and Novartis. E. K. S. received honoraria from Novartis for Continuing Medical Education Seminars; and grant and travel support from GlaxoSmithKline. C. P. H. reports personal fees from Mylan, AstraZeneca, Concert Pharmaceuticals, and 23andMe; and grants from Novartis and Boehringer Ingelheim. G. W. reports grants and other support from Boehringer Ingelheim, PulmonX, BTG Interventional Medicine, Janssen Pharmaceuticals, and GlaxoSmithKline. R. S. J. E. reports personal fees from Boehringer Ingelheim, Eolo Medical, and Toshiba. M. H. C. reports grants from GlaxoSmithKline; and personal fees from Genentech. None declared (A. B., J. Y., J. C. R., G. L. K., K. A. Y., E. A. R., D. A. L., G. J. C., J. G. D., S. I. R., R. C., B. J. M., J. C., J. E. H.).
*COPDGene Investigators, Core Units: Administrative Center: James D. Crapo, MD (Principal Investigator); Edwin K. Silverman, MD, PhD (Principal Investigator); Barry J. Make, MD; and Elizabeth A. Regan, MD, PhD. Genetic Analysis Center: Terri Beaty, PhD; Ferdouse Begum, PhD; Peter J. Castaldi, MD; Michael Cho, MD; Dawn L. DeMeo, MD, MPH; Adel R. Boueiz, MD; Marilyn G. Foreman, MD, MS; Eitan Halper-Stromberg, MD, PhD; Lystra P. Hayden, MD; Craig P. Hersh, MD, MPH; Jacqueline Hetmanski, MS, MPH; Brian D. Hobbs, MD; John E. Hokanson, MPH, PhD; Nan Laird, PhD; Christoph Lange, PhD; Sharon M. Lutz, PhD; Merry-Lynn McDonald, PhD; Margaret M. Parker, PhD; Dmitry Prokopenko, PhD; Dandi Qiao, PhD; Elizabeth A. Regan, MD, PhD; Phuwanat Sakornsakolpat, MD; Edwin K. Silverman, MD, PhD; Emily S. Wan, MD; and Sungho Won, PhD. Imaging Center: Juan Pablo Centeno, MSc; Jean-Paul Charbonnier, PhD; Harvey O. Coxson, PhD; Craig J. Galban, PhD; MeiLan K. Han, MD; Eric A. Hoffman, PhD; Stephen Humphries, PhD; Francine L. Jacobson, MD, MPH; Philip F. Judy, PhD; Ella A. Kazerooni, MD; Alex Kluiber, BA; David A. Lynch, MB; Pietro Nardelli, PhD; John D. Newell Jr, MD; Aleena Notary, MS; Andrea Oh, MD; Elizabeth A. Regan, MD, PhD; James C. Ross, PhD; Raul San Jose Estepar, PhD; Joyce Schroeder, MD; Jered Sieren, MHA; Berend C. Stoel, PhD; Juerg Tschirren, PhD; Edwin Van Beek, MD, PhD; Bram van Ginneken, PhD; Eva van Rikxoort, PhD; Gonzalo Vegas Sanchez-Ferrero, PhD; Lucas Veitel, BA; George R. Washko, MD; and Carla G. Wilson, MS. PFT QA Center, Salt Lake City, UT: Robert Jensen, PhD. Data Coordinating Center and Biostatistics, National Jewish Health, Denver, CO: Douglas Everett, PhD; Jim Crooks, PhD; Katherine Pratte, PhD; Matt Strand, PhD; and Carla G. Wilson, MS. Epidemiology Core, University of Colorado Anschutz Medical Campus, Aurora, CO: John E. Hokanson, MPH, PhD; Gregory Kinney, MPH, PhD; Sharon M. Lutz, PhD; and Kendra A. Young, PhD. Mortality Adjudication Core: Surya P. Bhatt, MD; Jessica Bon, MD; Alejandro A. Diaz, MD, MPH; MeiLan K. Han, MD; Barry Make, MD; Susan Murray, ScD; Elizabeth Regan, MD; Xavier Soler, MD; and Carla G. Wilson, MS. Biomarker Core: Russell P. Bowler, MD, PhD; Katerina Kechris, PhD; and Farnoush Banaei-Kashani, PhD.
Other contributions: The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Heart, Lung, and Blood Institute or the National Institutes of Health.
Footnotes
FUNDING/SUPPORT: The project described was supported by the National Heart, Lung, and Blood Institute [Awards U01 HL089897, U01 HL089856, and R01 HL124233]. The COPD Genetic Epidemiology Study (COPDGene) is also supported by the COPD Foundation through contributions made to an Industry Advisory Board comprising AstraZeneca, Boehringer-Ingelheim, Genentech, GlaxoSmithKline, Novartis, and Sunovion.
Contributor Information
Peter J. Castaldi, Email: repjc@channing.harvard.edu.
COPDGene Investigators:
James D. Crapo, Edwin K. Silverman, Barry J. Make, Elizabeth A. Regan, Terri Beaty, Ferdouse Begum, Peter J. Castaldi, Michael Cho, Dawn L. DeMeo, Adel R. Boueiz, Marilyn G. Foreman, Eitan Halper-Stromberg, Lystra P. Hayden, Craig P. Hersh, Jacqueline Hetmanski, Brian D. Hobbs, John E. Hokanson, Nan Laird, Christoph Lange, Sharon M. Lutz, Merry-Lynn McDonald, Margaret M. Parker, Dmitry Prokopenko, Dandi Qiao, Elizabeth A. Regan, Phuwanat Sakornsakolpat, Edwin K. Silverman, Emily S. Wan, Sungho Won, Juan Pablo Centeno, Jean-Paul Charbonnier, Harvey O. Coxson, Craig J. Galban, MeiLan K. Han, Eric A. Hoffman, Stephen Humphries, Francine L. Jacobson, Philip F. Judy, Ella A. Kazerooni, Alex Kluiber, David A. Lynch, Pietro Nardelli, John D. Newell, Jr., Aleena Notary, Andrea Oh, Elizabeth A. Regan, James C. Ross, Raul San Jose Estepar, Joyce Schroeder, Jered Sieren, Berend C. Stoel, Juerg Tschirren, Edwin Van Beek, Bram van Ginneken, Eva van Rikxoort, Gonzalo Vegas Sanchez-Ferrero, Lucas Veitel, George R. Washko, Carla G. Wilson, Robert Jensen, Douglas Everett, Jim Crooks, Katherine Pratte, Matt Strand, Carla G. Wilson, John E. Hokanson, Gregory Kinney, Sharon M. Lutz, Kendra A. Young, Surya P. Bhatt, Jessica Bon, Alejandro A. Diaz, MeiLan K. Han, Barry Make, Susan Murray, Elizabeth Regan, Xavier Soler, Carla G. Wilson, Russell P. Bowler, Katerina Kechris, and Farnoush Banaei-Kashani
References
- 1.Rennard S.I., Vestbo J. The many “small COPDs.”. Chest. 2008;134(3):623. doi: 10.1378/chest.07-3059. [DOI] [PubMed] [Google Scholar]
- 2.Singh D., Agustí A.G.N., Anzueto A. Global strategy for the diagnosis, management, and prevention of chronic obstructive lung disease: the GOLD science committee report 2019. Eur Respir J. 2019;53(5):1900164. doi: 10.1183/13993003.00164-2019. [DOI] [PubMed] [Google Scholar]
- 3.Bhatt S.P., Washko G.R., Hoffman E.A. Imaging advances in chronic obstructive pulmonary disease. Insights from the Genetic Epidemiology of Chronic Obstructive Pulmonary Disease (COPDGene) Study. Am J Respir Crit Care Med. 2019;199(3):286–301. doi: 10.1164/rccm.201807-1351SO. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stringer W.W., Porsasz J., Bhatt S.P., McCormack M.C., Make B.J., Casaburi R. Physiologic insights from the COPDGene study. Journal of the COPD Foundation. 2019;6(3):256–266. doi: 10.15326/jcopdf.6.3.2019.0128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maselli D.J., Bhatt S.P., Anzueto A. Clinical epidemiology of COPD: insights from 10 years of the COPDGene study. Chest. 2019;156(2):228–238. doi: 10.1016/j.chest.2019.04.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ragland M.F., Benway C.J., Lutz S.M. Genetic advances in chronic obstructive pulmonary disease. Insights from COPDGene. Am J Respir Crit Care Med. 2019;200(6):677–690. doi: 10.1164/rccm.201808-1455SO. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Regan E.A., Hersh C.P., Castaldi P.J. Omics and the search for blood biomarkers in chronic obstructive pulmonary disease. Insights from COPDGene. Am J Respir Cell Mol Biol. 2019;61(2):143–149. doi: 10.1165/rcmb.2018-0245PS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fletcher C.M., Gilson J.G., Hugh-Jones P., Scadding J.G. Terminology, definitions, and classification of chronic pulmonary emphysema and related conditions: a report of the conclusions of a Ciba Guest Symposium. Thorax. 1959;14(4):286–299. [Google Scholar]
- 9.Burrows B., Niden A.H., Fletcher C.M., Jones N.L. Clinical types of chronic obstructive lung disease in London and in Chicago. A study of one hundred patients. Am Rev Respir Dis. 1964;90:14–27. doi: 10.1164/arrd.1964.90.1.14. [DOI] [PubMed] [Google Scholar]
- 10.Burrows B., Fletcher C.M., Heard B.E., Jones N.L., Wootliff J.S. The emphysematous and bronchial types of chronic airways obstruction. A clinicopathological study of patients in London and Chicago. Lancet. 1966;1(7442):830–835. doi: 10.1016/s0140-6736(66)90181-4. [DOI] [PubMed] [Google Scholar]
- 11.Donaldson G.C., Seemungal T.A.R., Bhowmik A., Wedzicha J.A. Relationship between exacerbation frequency and lung function decline in chronic obstructive pulmonary disease. Thorax. 2002;57(10):847–852. doi: 10.1136/thorax.57.10.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hurst J.R., Vestbo J., Anzueto A. Susceptibility to exacerbation in chronic obstructive pulmonary disease. N Engl J Med. 2010;363(12):1128–1138. doi: 10.1056/NEJMoa0909883. [DOI] [PubMed] [Google Scholar]
- 13.Gibson P.G., Simpson J.L. The overlap syndrome of asthma and COPD: what are its features and how important is it? Thorax. 2009;64(8):728–735. doi: 10.1136/thx.2008.108027. [DOI] [PubMed] [Google Scholar]
- 14.Fishman A., Martinez F., Naunheim K. A randomized trial comparing lung-volume-reduction surgery with medical therapy for severe emphysema. N Engl J Med. 2003;348(21):2059–2073. doi: 10.1056/NEJMoa030287. [DOI] [PubMed] [Google Scholar]
- 15.Pistolesi M., Camiciottoli G., Paoletti M. Identification of a predominant COPD phenotype in clinical practice. Respir Med. 2008;102(3):367–376. doi: 10.1016/j.rmed.2007.10.019. [DOI] [PubMed] [Google Scholar]
- 16.Garcia-Aymerich J., Gómez F.P., Benet M. Identification and prospective validation of clinically relevant chronic obstructive pulmonary disease (COPD) subtypes. Thorax. 2011;66(5):430–437. doi: 10.1136/thx.2010.154484. [DOI] [PubMed] [Google Scholar]
- 17.Cho M., Washko G.R., Hoffmann T.J. Cluster analysis in severe emphysema subjects using phenotype and genotype data: an exploratory investigation. Respir Res. 2010;11:30. doi: 10.1186/1465-9921-11-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Castaldi P.J., Dy J.G., Ross J. Cluster analysis in the COPDGene study identifies subtypes of smokers with distinct patterns of airway disease and emphysema. Thorax. 2014;69(5):415–422. doi: 10.1136/thoraxjnl-2013-203601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vanfleteren L., Spruit M., Groenen M. Clusters of comorbidities based on validated objective measurements and systemic inflammation in patients with chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2013;187(7):728–735. doi: 10.1164/rccm.201209-1665OC. [DOI] [PubMed] [Google Scholar]
- 20.Burgel P.R., Paillasseur J.L., Caillaud D. Clinical COPD phenotypes: a novel approach using principal component and cluster analyses. Eur Respir J. 2010;36(3):531–539. doi: 10.1183/09031936.00175109. [DOI] [PubMed] [Google Scholar]
- 21.Pinto L.M., Alghamdi M., Benedetti A., Zaihra T., Landry T., Bourbeau J. Derivation and validation of clinical phenotypes for COPD: a systematic review. Respir Res. 2015;16(1):50. doi: 10.1186/s12931-015-0208-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Woodruff P.G., Agustí A.G.N., Roche N., Singh D., Martinez F.J. Current concepts in targeting chronic obstructive pulmonary disease pharmacotherapy: making progress towards personalised management. Lancet. 2015;385(9979):1789–1798. doi: 10.1016/S0140-6736(15)60693-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Woodruff P.G., Modrek B., Choy D.F. T-helper type 2-driven inflammation defines major subphenotypes of asthma. Am J Respir Crit Care Med. 2009;180(5):388–395. doi: 10.1164/rccm.200903-0392OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Agustí A.G.N., Bel E., Thomas M. Treatable traits: toward precision medicine of chronic airway diseases. Eur Respir J. 2016;47(2):410–419. doi: 10.1183/13993003.01359-2015. [DOI] [PubMed] [Google Scholar]
- 25.McCarthy M.I. Painting a new picture of personalised medicine for diabetes. Diabetologia. 2017;60(5):793–799. doi: 10.1007/s00125-017-4210-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kinney G.L., Santorico S.A., Young K.A. Identification of chronic obstructive pulmonary disease axes that predict all-cause mortality: the COPDGene study. Am J Epidemiol. 2018;187(10):2109–2116. doi: 10.1093/aje/kwy087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Regan E.A., Hokanson J.E., Murphy J.R. Genetic epidemiology of COPD (COPDGene) study design. COPD. 2010;7(1):32–43. doi: 10.3109/15412550903499522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boueiz A., Lutz S.M., Cho M. Genome-wide association study of the genetic determinants of emphysema distribution. Am J Respir Crit Care Med. 2017;195(6):757–771. doi: 10.1164/rccm.201605-0997OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boueiz A., Pham B., Chase R. Integrative genomics analysis identifies ACVR1B as a candidate causal gene of emphysema distribution. Am J Respir Cell Mol Biol. 2019;60(4):388–398. doi: 10.1165/rcmb.2018-0110OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boueiz A., Chang Y., Cho M. Lobar Emphysema distribution is associated with 5-year radiological disease progression. Chest. 2017;153(1):65–76. doi: 10.1016/j.chest.2017.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chang Y., Glass K., Liu Y.Y. COPD subtypes identified by network-based clustering of blood gene expression. Genomics. 2016;107(2-3):51–58. doi: 10.1016/j.ygeno.2016.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Castaldi P.J., Benet M., Petersen H. Do COPD subtypes really exist? COPD heterogeneity and clustering in 10 independent cohorts. Thorax. 2017;72(11):998–1006. doi: 10.1136/thoraxjnl-2016-209846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Burgel P.R., Paillasseur J.L., Janssens W. A simple algorithm for the identification of clinical COPD phenotypes. Eur Respir J. 2017;50(5):1701034. doi: 10.1183/13993003.01034-2017. [DOI] [PubMed] [Google Scholar]
- 34.Chen J., Cho M., Silverman E.K. Turning subtypes into disease axes to improve prediction of COPD progression. Thorax. 2019;74(9):906–909. doi: 10.1136/thoraxjnl-2018-213005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yun J.H., Lamb A., Chase R. Blood eosinophil count thresholds and exacerbations in patients with chronic obstructive pulmonary disease. J Allergy Clin Immunol. 2018;141(6):2037–2047.e10. doi: 10.1016/j.jaci.2018.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vedel-Krogh S., Nielsen S.F., Lange P., Vestbo J., Nordestgaard B.G. Blood eosinophils and exacerbations in chronic obstructive pulmonary disease. The Copenhagen General Population Study. Am J Respir Crit Care Med. 2016;193(9):965–974. doi: 10.1164/rccm.201509-1869OC. [DOI] [PubMed] [Google Scholar]
- 37.Zeiger R.S., Tran T.N., Butler R.K. Relationship of blood eosinophil count to exacerbations in chronic obstructive pulmonary disease. J Allergy Clin Immunol Pract. 2018;6(3):944–954.e5. doi: 10.1016/j.jaip.2017.10.004. [DOI] [PubMed] [Google Scholar]
- 38.Mendoza C.S., Washko G.R., Crapo J.D. Emphysema quantification in a multi-scanner HRCT cohort using local intensity distributions. Proc IEEE Int Symp Biomed Imaging. 2012:474–477. doi: 10.1109/ISBI.2012.6235587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Castaldi P.J., San José Estépar R., Mendoza C.S. Distinct quantitative CT emphysema patterns are associated with physiology and function in smokers. Am J Respir Crit Care Med. 2013;188(9):1083–1090. doi: 10.1164/rccm.201305-0873OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Castaldi P.J., Cho M., San José Estépar R. Genome-wide association identifies regulatory loci associated with distinct local histogram emphysema patterns. Am J Respir Crit Care Med. 2014;190(4):399–409. doi: 10.1164/rccm.201403-0569OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Parker M.M., Hao Y., Guo F. Identification of an emphysema-associated genetic variant near TGFB2 with regulatory effects in lung fibroblasts. Elife. 2019;8 doi: 10.7554/eLife.42720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ross J.C., Castaldi P.J., Cho M. A Bayesian nonparametric model for disease subtyping: application to emphysema phenotypes. IEEE Trans Med Imaging. 2017;36(1):343–354. doi: 10.1109/TMI.2016.2608782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ross J.C., Castaldi P.J., Cho M. Longitudinal modeling of lung function trajectories in smokers with and without chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2018;198(8):1033–1042. doi: 10.1164/rccm.201707-1405OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lange P., Celli B., Agustí A.G.N. Lung-function trajectories leading to chronic obstructive pulmonary disease. N Engl J Med. 2015;373(2):111–122. doi: 10.1056/NEJMoa1411532. [DOI] [PubMed] [Google Scholar]
- 45.Bui D.S., Lodge C.J., Burgess J.A. Childhood predictors of lung function trajectories and future COPD risk: a prospective cohort study from the first to the sixth decade of life. Lancet Respir Med. 2018;6(7):535–544. doi: 10.1016/S2213-2600(18)30100-0. [DOI] [PubMed] [Google Scholar]
- 46.Agustí A.G.N., Faner R. Lung function trajectories in health and disease. Lancet Respir Med. 2019;7(4):358–364. doi: 10.1016/S2213-2600(18)30529-0. [DOI] [PubMed] [Google Scholar]
- 47.Sakornsakolpat P., Prokopenko D., Lamontagne M. Genetic landscape of chronic obstructive pulmonary disease identifies heterogeneous cell-type and phenotype associations. Nature Genetics. 2019;51(3):494–505. doi: 10.1038/s41588-018-0342-2. [DOI] [PMC free article] [PubMed] [Google Scholar]