Abstract
The cystine/glutamate transporter system xc− consists of the light-chain subunit xCT (SLC7A11) and the heavy-chain subunit CD98 (4F2hc or SLC3A2) and exchanges extracellular cystine for intracellular glutamate at the plasma membrane. The imported cystine is reduced to cysteine and used for synthesis of GSH, one of the most important antioxidants in cancer cells. Because cancer cells have increased levels of reactive oxygen species, xCT, responsible for cystine–glutamate exchange, is overexpressed in many cancers, including glioblastoma. However, under glucose-limited conditions, xCT overexpression induces reactive oxygen species accumulation and cell death. Here we report that cell survival under glucose deprivation depends on cell density. We found that high cell density (HD) down-regulates xCT levels and increases cell viability under glucose deprivation. We also found that growth of glioblastoma cells at HD inactivates mTOR and that treatment of cells grown at low density with the mTOR inhibitor Torin 1 down-regulates xCT and inhibits glucose deprivation-induced cell death. The lysosome inhibitor bafilomycin A1 suppressed xCT down-regulation in HD-cultured glioblastoma cells and in Torin 1–treated cells grown at low density. Additionally, bafilomycin A1 exposure or ectopic xCT expression restored glucose deprivation–induced cell death at HD. These results suggest that HD inactivates mTOR and promotes lysosomal degradation of xCT, leading to improved glioblastoma cell viability under glucose-limited conditions. Our findings provide evidence that control of xCT protein expression via lysosomal degradation is an important mechanism for metabolic adaptation in glioblastoma cells.
Keywords: amino acid transport, cell death, cancer biology, cell biology, lysosome
Introduction
It is well-known that many cancer cells depend on glucose for proliferation and survival. They up-regulate glucose transporters and enzymes involved in glucose metabolism. The increased glucose metabolism is utilized to supply sufficient energy and biosynthetic intermediates. In addition, cancer cells use glucose to generate NADPH, which is required for GSH and thioredoxin systems, the major antioxidant systems in cancer cells (1–4). Therefore, to survive under glucose-insufficient conditions, cancer cells need to adapt to metabolic stress or migrate toward more favorable environments.
The system xc− is composed of the light chain subunit xCT (SLC7A11) and the heavy chain subunit CD98 (4F2hc, SLC3A2) and mediates the exchange of extracellular cystine and intracellular glutamate across the plasma membrane. The amino acid specificity and transport activity of system xc− depend on xCT, and increased expression of xCT results in increased cystine/glutamate exchange. Expression of xCT is often up-regulated in cancer cells, including glioblastoma cells, and its expression correlates with tumor growth and poor survival (5–7). When cystine is imported and intracellularly reduced to cysteine, it is used to synthesize reduced GSH. GSH is required for optimal activity of GSH peroxidase 4, a key regulator of ferroptosis. Ferroptosis is a form of cell death induced by phospholipid peroxidation, and treatment with pharmacological inhibitors of xCT or depletion of GSH in several types of cancer cells causes ferroptosis (8–11). Thus, intracellular transport of cystine is important to avoid oxidative stress and cell death in cancer cells. On the other hand, overexpression of xCT in cancer cells induces cell death under glucose deprivation (12–15). This mechanism involves depletion of intracellular glutamate because of xCT-mediated export and production of reactive oxygen species induced by xCT-mediated cystine uptake. The transcription factors NRF2 and ATF4 up-regulate xCT expression and function in the glucose dependence of cancer cells (12, 13). However, how the activity of xCT is regulated in cancer cells is not fully understood.
mTOR is a central regulator of cell growth and proliferation. The activity of mTOR is regulated in response to growth factors and environmental nutrient conditions (16, 17). mTOR signaling is frequently activated in cancer cells and functions in tumor growth and progression (16, 18). Activation of mTOR promotes anabolic processes (synthesis of proteins, lipids, and nucleotides) and suppression of catabolic processes (the autophagy–lysosome system and ubiquitin–proteasome system) (19–22). mTOR signaling also plays a role in metabolic reprograming in cancer cells by regulating the expression and activity of key enzymes involved in glucose, amino acid, and fatty acid metabolism (18). In this study, we demonstrate that cell density regulates xCT protein stability through an mTOR-dependent pathway in glioblastoma cells.
Results
A high cell density down-regulates xCT and suppresses glucose deprivation–induced cell death in glioblastoma cells
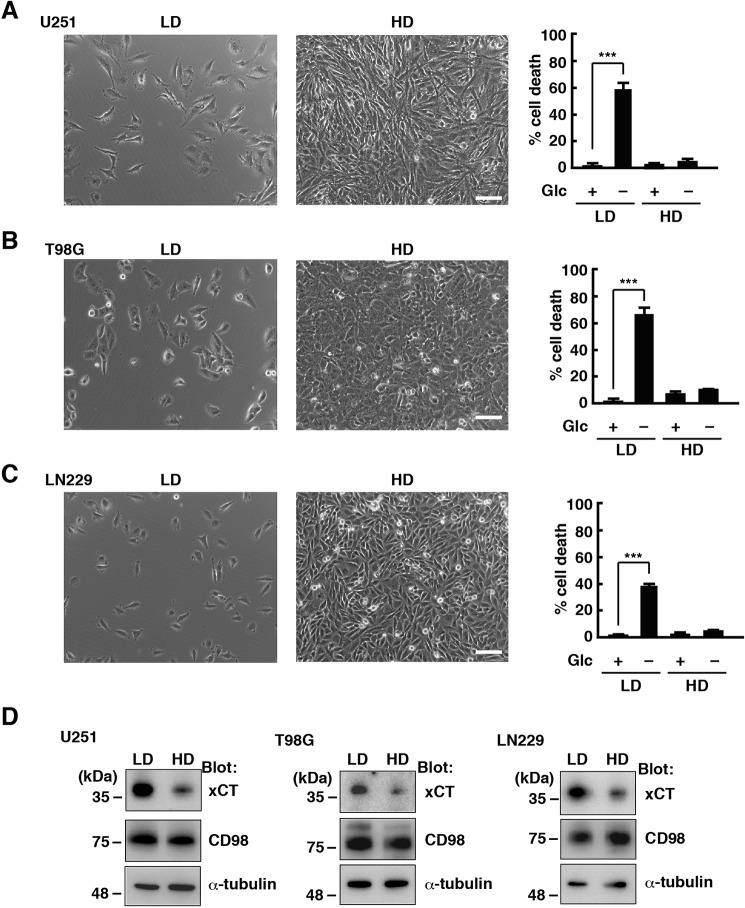
In glioblastoma cells, glucose deprivation from the medium rapidly induced cell death. We found that this depends on cell density. We plated U251 glioblastoma cells at a low cell density (LD;2 1 × 104 cells/cm2) and high cell density (HD, 1 × 105 cells/cm2) and cultured them for 48 h. Then the culture medium was collected 24 h after deprivation of glucose, and the amount of lactate dehydrogenase (LDH) released in the medium was measured to quantify cell death. Glucose deprivation significantly increased cell death at LD. However, most cells were alive when they were plated at HD (Fig. 1A). We used other glioblastoma cell lines, T98G and LN229, and obtained similar results (Fig. 1, B and C). We previously reported that the cystine/glutamate antiporter xCT mediates glucose deprivation–induced cell death in glioblastoma cells (14). Therefore, we next examined whether xCT or its binding partner CD98 is regulated by cell density. The protein levels of xCT were down-regulated in U251, T98G, and LN229 cells cultured at HD for 48 h (Fig. 1D). On the other hand, cell density had little effect on CD98 expression. These results suggest that HD reduces the protein levels of xCT and suppresses glucose deprivation–induced cell death in glioblastoma cells.
Figure 1.
HD down-regulates xCT and suppresses glucose deprivation–induced cell death in glioblastoma cells. A–C, U251 (A), T98G (B), and LN229 (C) cells were cultured at LD (1 × 104 cells/cm2) or HD (1 × 105 cells/cm2) for 48 h and placed in medium with or without glucose (Glc, 5 mm) for 24 h. Scale bar = 100 μm. Quantification of cell death was performed using an LDH release assay. Cells treated with 0.1% Tween 20 were used to calculate 100% cell death. Error bars represent S.D. (n = 3). ***, p < 0.001, calculated by one-way ANOVA with Tukey's post hoc test. D, immunoblot analysis of U251, T98G, and LN229 cells cultured at LD or HD for 48 h.
The activity of mTOR is required for xCT expression and glucose deprivation–induced cell death
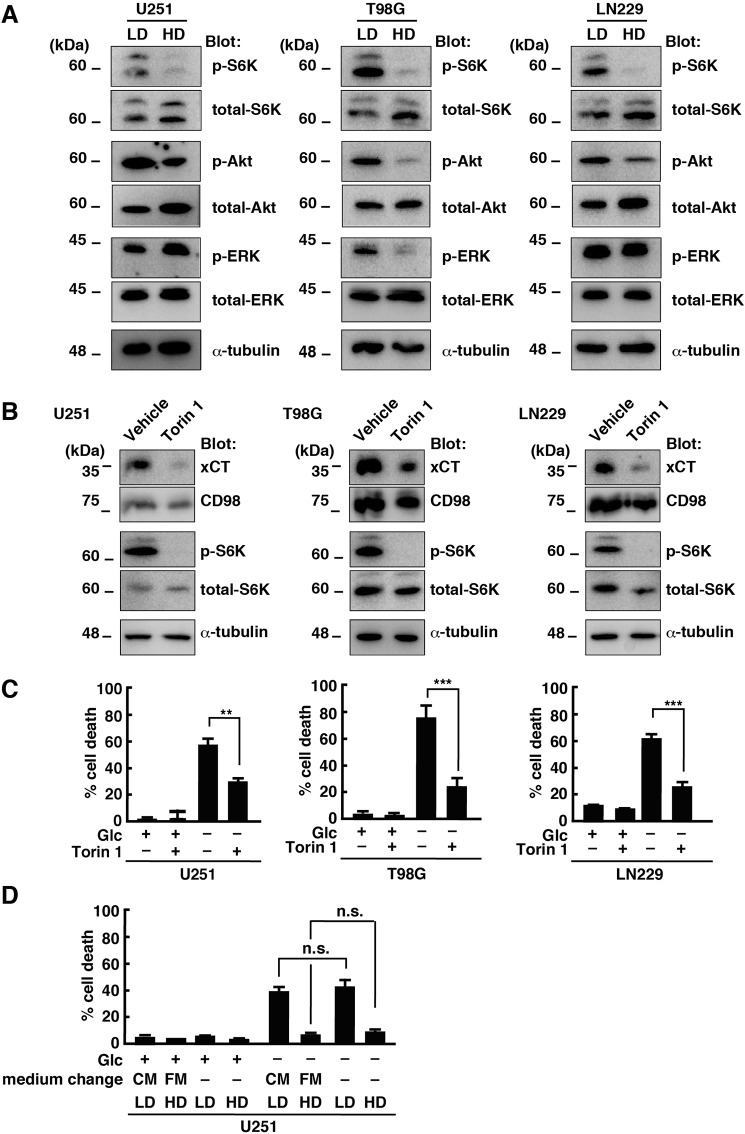
We next examined the mechanism involved in cell density–dependent regulation of xCT expression in glioblastoma cells. Previous studies reported that the mTOR signaling pathway is inhibited at HD (23, 24). Consistent with this, the phosphorylation levels of p70 S6 kinase (p-S6K), a downstream target of mTOR, were suppressed in U251, T98G, and LN229 cells when they were cultured at HD (Fig. 2A). The levels of Akt phosphorylation (threonine 308) were also slightly decreased, but cell density had little effect on the levels of extracellular signal-regulated kinase phosphorylation. Therefore, we focused on the effects of the mTOR inhibitor Torin 1 on xCT expression. Treatment of glioblastoma cells with Torin 1 at LD reduced the protein levels of xCT in U251, T98G, and LN229 cells (Fig. 2B). CD98 expression was also slightly affected by Torin 1. Furthermore, Torin 1 treatment significantly suppressed glucose deprivation–induced cell death in U251, T98G, and LN229 cells cultured at LD (Fig. 2C). These results suggest that mTOR activation plays a key role in xCT expression and cell viability under glucose deprivation.
Figure 2.
Inhibition of mTOR activity down-regulates xCT and suppresses glucose deprivation-induced cell death. A, immunoblot analysis of U251, T98G, and LN229 cells cultured at LD or HD for 48 h. ERK, extracellular signal-regulated kinase. B, immunoblot analysis of U251, T98G, and LN229 cells treated with Torin 1 (250 nm) at LD for 24 h. C, U251, T98G, and LN229 cells were cultured at LD with or without Torin 1 for 48 h and placed in medium with or without glucose (Glc, 5 mm) for 24 h. D, U251 cells cultured at LD or HD were replaced in fresh medium (FM) or conditioned medium (CM) collected from HD culture after 48 h. Cells were then placed in medium with or without glucose (5 mm) for 24 h. Cell death 24 h after medium change was quantified using an LDH release assay. Cells treated with 0.1% Tween 20 were used for calculating 100% cell death. Error bars represent S.D. (n = 3). ***, p < 0.001; **, p < 0.01; n.s., not significant; calculated by one-way ANOVA with Tukey's post hoc test.
mTOR is inactivated after exhaustion of cell medium in normal epithelial cells (23). Therefore, we next examined whether the medium from HD culture can inhibit glucose deprivation–induced cell death at LD. However, the medium from U251 cell culture at HD (conditioned medium) had little effect on glucose deprivation-induced cell death at LD (Fig. 2D). In addition, replacement of the medium with fresh medium also had no effect on cell viability in U251 cells under glucose deprivation at HD. Therefore, it is likely that inactivation of mTOR activity at HD was not due to exhaustion of the medium under our experimental conditions.
HD promotes lysosomal degradation of xCT in glioblastoma cells
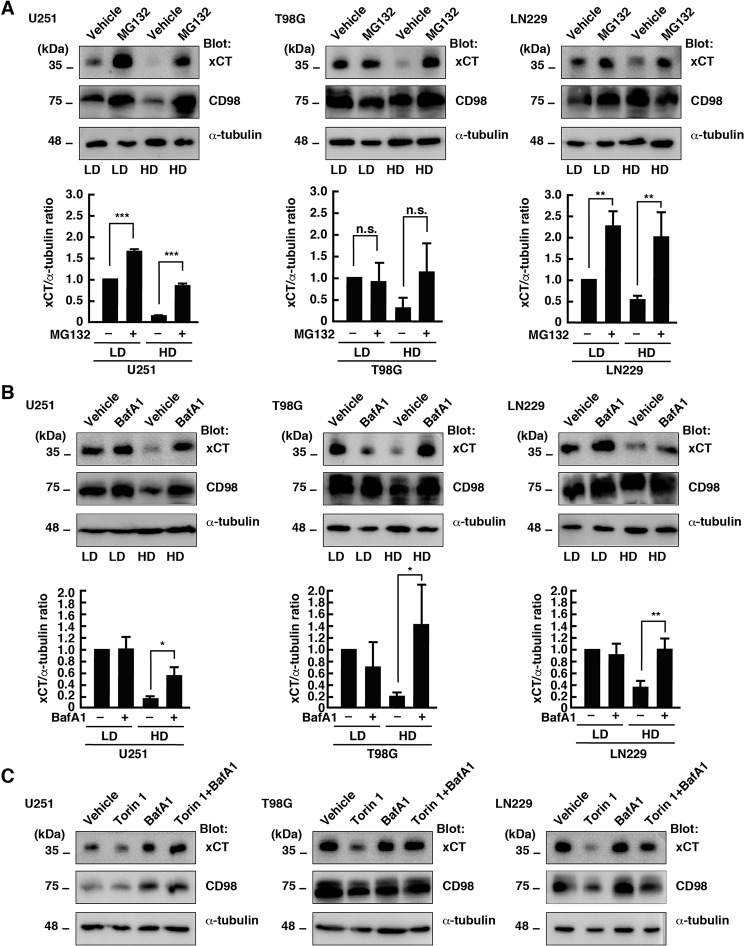
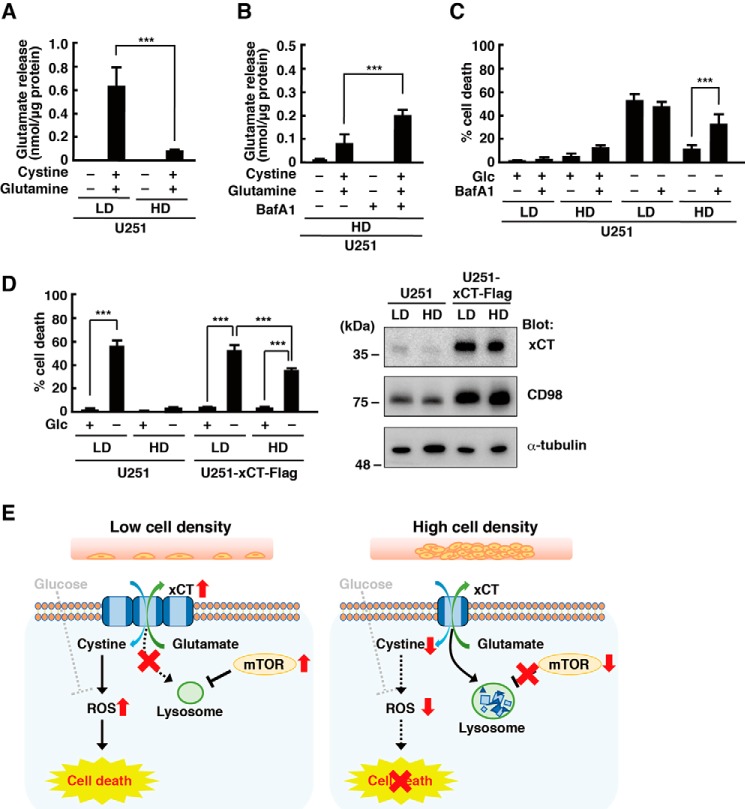
A previous study reported that the protein stability of xCT is regulated by the deubiquitinase OTUB1 in a proteasome-dependent manner (25). The proteasome inhibitor MG132 increased the protein levels of xCT in U251 cells at LD and HD (Fig. 3A). On the other hand, treatment with the lysosome inhibitor Bafilomycin A1 (BafA1) increased xCT expression in U251 cells at HD but had little effect on the protein level of xCT at LD (Fig. 3B). Similar results were obtained in T98G and LN229 cells, but MG132 did not increase the xCT protein level in T98G cells at LD. These results suggest that HD promotes degradation of xCT, at least in part, in a lysosome-dependent manner. On the other hand, the steady state levels of xCT in U251 and LN229 cells are regulated by proteasome-dependent degradation. In U251 cells, HD slightly decreased the protein level of CD98, suggesting that cell density regulates CD98 protein expression in a cell type–specific manner.
Figure 3.
HD promotes lysosomal degradation of xCT. A, immunoblot analysis of U251, T98G, and LN229 cells cultured at LD or HD with or without MG132 (20 μm) for 24 h. B, immunoblot analysis of U251, T98G, and LN229 cells cultured at LD or HD with or without BafA1 (1 μm) for 24 h. The xCT/α-tubulin ratio relative to that of vehicle treatment at LD is shown (mean ± S.D. of three independent experiments). C, immunoblot analysis of U251, T98G, and LN229 cells cultured at LD with or without BafA1 (1 μm) or Torin 1 (250 nm) for 24 h. ***, p < 0.001; **, p < 0.01; *, p < 0.05; n.s., not significant, calculated by one-way ANOVA with Tukey's post hoc test.
Previous studies reported that activation of mTOR induces suppression of catabolic processes, including lysosomal degradation (26, 27). Therefore, we next examined whether lysosomal degradation functions in Torin 1–induced down-regulation of xCT in glioblastoma cells at LD. Treatment with BafA1 restored the levels of xCT protein in Torin 1–treated U251, T98G, and LN229 cells at LD (Fig. 3C). These results suggest that inhibition of mTOR activity is sufficient for lysosomal degradation of xCT in glioblastoma cells.
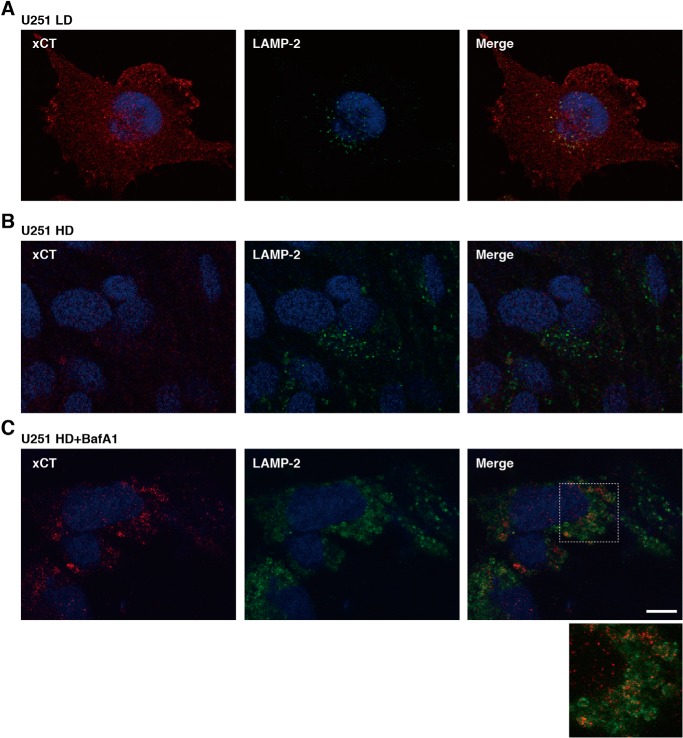
We next examined the localization of xCT in U251 cells cultured at LD and HD to confirm that xCT is degraded in lysosomes at HD. In U251 cells at LD, xCT was observed throughout the cells (Fig. 4A). In U251 cells at HD, xCT labeling was very low, consistent with Western blot data (Fig. 4B). However, xCT was observed in vesicular structures, some of which were stained by an antibody against LAMP2, a marker for lysosomes, in U251 cells treated with BafA1 at HD (Fig. 4C). As reported previously (28, 29), the LAMP2-labeled membranes were enlarged in BafA1-treated cells, probably because of accumulation of undegraded material in lysosomes. These results suggest that xCT is recruited to lysosomes for degradation in glioblastoma cells at HD.
Figure 4.
Localization of xCT in U251 cells at LD and HD. A–C, confocal images of U251 cells cultured at LD (A) and HD (B) or treated with BafA1 (1 μm) at HD for 48 h (C). Cells were stained with anti-xCT (red) and anti-LAMP2 (green) antibodies. Nuclei were also stained with Hoechst 33258 (blue). Scale bar = 10 μm.
Inhibition of lysosomal activity in glioblastoma cells at HD promotes glucose deprivation–induced cell death
We measured glutamate release into the medium to examine whether the increased xCT protein level by lysosomal inhibition at HD leads to increased cystine–glutamate exchange. As we reported previously (30), glutamate release was detected when U251 cells were placed in amino acid–free medium supplemented with cystine and glutamine at LD. However, it was greatly reduced in U251 cells at HD (Fig. 5A). Treatment with BafA1 partially restored glutamate release (Fig. 5B). These results suggest that inhibition of lysosomal activity in U251 cells at HD increases the level of functional xCT protein. We next examined the effects of lysosomal inhibition on glucose deprivation–induced cell death and found that BafA1 increased the level of cell death in U251 cells at HD (Fig. 5C). To confirm that the increased level of xCT was responsible for HD-induced promotion of cell viability under glucose deprivation, we established U251 cells stably expressing xCT, in which high protein levels of xCT were observed even at HD. Overexpression of xCT restored glucose deprivation–induced cell death in U251 cells cultured at HD (Fig. 5D), suggesting that down-regulation of xCT by lysosomal degradation at HD improves cell viability under glucose deprivation in glioblastoma cells. In xCT-transfected cells, we observed a higher CD98 signal than in untransfected cells. The reason for this is unknown, but CD98 functions as a chaperone-like protein to direct xCT to the plasma membrane (5, 7), and increased xCT expression may require increased protein levels of CD98.
Figure 5.
Inhibition of lysosomal activity promotes glucose deprivation–induced cell death. A, U251 cells cultured at LD or HD for 48 h were placed in amino acid–free medium with or without cystine (0.2 mm) and glutamine (2 mm) for 4 h, and glutamate released into the medium was measured. B, U251 cells were cultured at HD with or without BafA1 (1 μm) for 48 h. The cells were placed in amino acid–free medium with or without cystine (0.2 mm) and glutamine (2 mm) for 4 h, and glutamate released into the medium was measured. C, U251 cells were cultured at LD or HD with or without BafA1 (1 μm) for 48 h and placed in medium with or without glucose (Glc, 5 mm) for 24 h. D, U251 cells expressing FLAG-tagged xCT were cultured at LD or HD for 48 h and placed in medium with or without glucose (5 mm) for 24 h. Cell death 24 h after medium change was quantified using an LDH release assay. Cells treated with 0.1% Tween 20 were used for calculating 100% cell death. E, model of cell density–dependent glioblastoma cell death under glucose deprivation. Error bars represent S.D. (n = 3). ***, p < 0.001, calculated by one-way ANOVA with Tukey's post hoc test.
Discussion
The viability of cancer cells is regulated by numerous factors, including environmental conditions. In this study, we found that cell density influences the viability of glioblastoma cells under glucose-limited conditions. In glioblastoma cells, lysosomal activity is negatively regulated by mTOR, and HD inhibits mTOR activity, which results in promotion of lysosomal degradation of xCT. Glioblastoma cells expressing high levels of xCT undergo cell death when exposed to glucose deprivation. Therefore, reduced protein levels of xCT lead to improved cell viability of glioblastoma cells under glucose deprivation. Thus, our results suggest that cell density controls sensitivity to low glucose in glioblastoma cells and that this mechanism involves lysosomal degradation of xCT (Fig. 5E). As increased xCT expression in HD by lysosomal inhibition or ectopic expression of xCT did not fully restore glucose deprivation–induced cell death, we did not exclude the possibility that another mechanism is involved in promotion of cell viability in glioblastoma cells at HD. There is a positive correlation between xCT expression and tumor growth and poor survival. However, when glioblastoma cells are exposed to glucose deprivation at LD, higher expression of xCT conversely causes oxidative stress and cell death. Therefore, inhibition of glucose metabolism may be an attractive therapeutic strategy for glioblastoma, with higher expression of xCT at LD.
Different cell conditions often affect cell viability. In particular, contact inhibition or HD controls cell survival and proliferation. One important mechanism underlying contact inhibition is the Hippo signaling pathway. When cells are plated at HD, the Hippo signaling pathway is activated, which results in distribution of Yes-associated protein/transcriptional coactivator with PDZ-binding motif from the nucleus to the cytosol and their inactivation, leading to cessation of cell proliferation (31–33). Although cancer cells lose contact inhibition and induce dysregulated proliferation, it has been reported that cell density also affects viability in cancer cells. For example, HD causes drug resistance and increases cell viability in many cancer cells, including glioblastoma (34–36). HD also protects cancer cells from ferroptosis (37, 38), although the precise mechanisms remain to be elucidated. On the other hand, HD inhibits mTOR and prevents cellular senescence (23). In this study, we found that HD promotes glioblastoma cell viability under glucose deprivation via mTOR inactivation and degradation of xCT. As cystine uptake through xCT functions in GSH synthesis in many cancer cells, cell density may affect the amount of GSH and the antioxidant ability of cancer cells.
Expression of xCT is regulated transcriptionally and posttranslationally in response to different stimuli. It is well-known that NRF2 and ATF4 transcription factors induce xCT mRNA expression under oxidative stress conditions, whereas xCT expression is negatively regulated by the p53 tumor suppressor protein (5, 7, 12, 13, 39). The stability of xCT protein is also regulated by its binding partners. A variant of CD44, an adhesion molecule for the extracellular matrix, binds to xCT and stabilizes it at the plasma membrane, promoting GSH synthesis and suppressing reactive oxygen species generation in cancer cells (40). Epidermal growth factor receptors also promote surface expression of xCT (41). In addition, interaction with CD44 promotes interaction of xCT with OTUB1, an ovarian tumor family deubiquitinase, and increases xCT protein stability (25). We demonstrated that mTOR activation increases xCT protein levels by suppressing lysosomal degradation in glioblastoma cells. Therefore, expression of xCT is regulated by multiple mechanisms in response to different stimuli. It has been suggested that there is cross-talk between the lysosomal and proteasomal degradation systems (42, 43). For example, proteasomal inhibitors induce compensatory actions to cause autophagy and lysosomal degradation (44–46). On the other hand, lysosomal inhibition has been found to increase or reduce proteasomal activity (47, 48). Thus, the relationship between the lysosomal and proteasomal pathways is complicated, and we cannot exclude the possibility that the proteasomal pathway is involved in xCT degradation at HD. On the other hand, several studies previously reported regulatory mechanisms of xCT activity. The component of the class III phosphatidylinositol 3-kinase complex BECN1 binds to xCT when it is phosphorylated by AMP-activated protein kinase and negatively regulates xCT activity (49). The activity of xCT is also negatively regulated by phosphorylation on serine 26 in the N-terminal cytosolic region (50, 51), although it is unclear whether this affects xCT protein stability.
Regulation of mTOR activity is important for cancer cells to proliferate and survive. The activity of mTOR is controlled by growth factors, nutrients, cellular energy levels, and extracellular oxygen levels (16, 18, 52, 53). When glioblastoma cells are cultured at HD, the activity of mTOR is suppressed, consistent with previous studies using other types of cancer cells (23, 24). As conditional medium from glioblastoma cells cultured at HD had little effect on xCT expression, it is unlikely that factors secreted from the cells or exhaustion of the medium plays a role in mTOR inactivation or degradation of xCT. How mTOR is regulated in response to cell density in glioblastoma cells should be investigated in future studies. On the other hand, there is a close relationship between mTOR activity and lysosomal function (26, 54). The basic helix–loop–helix transcription factor EB induces expression of genes encoding proteins involved in lysosomal function (55), and mTOR phosphorylates and sequesters transcription factor EB in the cytoplasm, leading to inhibition of lysosomal gene expression (56, 57). Activation of mTOR also controls lysosomal functions through ATP-sensitive Na+ channels (27). Thus, regulation of mTOR activity is essential for lysosomal activity and protein degradation. Last, it is important for future studies to investigate how xCT is delivered to lysosomes in response to HD.
Experimental procedures
Plasmids and reagents
The expression plasmid pCXN2 vector was a generous gift from Dr. J. Miyazaki (Osaka University). The coding sequence for human xCT was amplified by RT-PCR from HeLa cells and subcloned into pCXN2 with a FLAG tag sequence at the C terminus. Medium without glucose or amino acids was prepared as described previously (14). Inhibitors were used at the following concentrations: Torin 1 (Merck, 250 nm), BafA1 (Cayman Chemical, 1 μm), and MG132 (Wako Pure Chemical Industries, 10 μm).
Cell culture and transfection
U251 cells were obtained from the European Collection of Authenticated Cell Cultures (EC09063001). T98G cells were provided by the RIKEN BRC through the National Bio-Resource Project of the MEXT, Japan (RCB1954). LN229 cells were obtained from the ATCC (CRL-2611). They were grown in DMEM containing 10% FBS, 4 mm glutamine, 100 units/ml penicillin, and 0.1 mg/ml streptomycin in humidified air containing 5% CO2 at 37 °C. Transfection of U251 cells was performed using Lipofectamine 2000 (Life Technologies). All cell lines were tested and found to be negative for mycoplasma contamination using the EZ-PCR Mycoplasma Test Kit (Biological Industries). To generate U251 cells stably expressing xCT-FLAG, U251 cells were seeded in a 6-cm dish (250,000 cells/dish) and transfected with pCXN2-xCT-FLAG 24 h later. Two days after transfection, the cells were collected and seeded in two 10-cm dishes in medium containing 250 μg/ml G418 (Wako) to eliminate untransfected cells. Ten days after selection, colonies grown from single cells were isolated. These clones were expanded and screened by immunoblotting with anti-xCT and anti-FLAG antibodies.
Glucose deprivation conditions and cell death experiments
Cells were plated in a 48-well plate (Greiner Bio-One, 677180) at LD (1 × 104 cells/well) or HD (1 × 105 cells/well). 48 h after plating, cells were rinsed twice with PBS, and the medium was replaced with glucose-free medium containing 10% dialyzed FBS (HyClone) for 24 h. Cell death was measured by LDH release assay using the MTX LDH kit (Kyokuto Pharmaceutical Industrial) according to the manufacturer's instructions. The optical density was measured at 595 nm using a microplate reader (Tecan, GENious). The value of LDH release after treatment with 0.1% Tween 20 was defined as 100% cell death.
Immunoblotting and antibodies
Cell lysates cultured at LD or HD for 48 h were analyzed by immunoblotting as described previously (28). The signals were captured with an Amersham Biosciences Imager 600 (GE Healthcare/Life Sciences). Densitometric analysis was performed using Amersham Biosciences Imager 600 analysis software. The following antibodies were used in this study: antibodies against xCT/SLC7A11 (12691), CD98/4F2hc (47213), phospho-p70 S6 kinase (Thr-389, 9234), p70 S6 kinase (9202), Thr-202/Tyr-204 phospho-extracellular signal-regulated kinase (4370), p44/42 mitogen-activated protein kinase (4695), Thr-308 phospho-Akt (2965), and Akt (9272) (Cell Signaling Technology); anti-α-tubulin antibody (T5168) and anti-FLAG antibody (M2, F1804, Sigma-Aldrich); and secondary antibodies against mouse IgG (P0447) and rabbit IgG (P0448) conjugated to horseradish peroxidase (DAKO).
Immunofluorescence microscopy
Cells cultured at LD or HD on coverslips for 48 h were fixed with 4% paraformaldehyde in PBS for 20 min and washed with PBS five times. Cells were permeabilized with 0.2% Triton X-100 in PBS for 10 min and incubated with 10% FBS in PBS for 30 min to block nonspecific antibody binding. Then cells were incubated with anti-xCT antibody (1:1000) and anti-LAMP-2 antibody (1:200, sc-18822, Santa Cruz Biotechnology) in PBS for 24 h. After washing with PBS, cells were incubated with anti-mouse IgG conjugated with Alexa Fluor 488 (A11029, Thermo Fisher Scientific), anti-rabbit IgG conjugated with Alexa 594 (A11037, Thermo Fisher Scientific), and Hoechst 33258 (H1398, Thermo Fisher Scientific, 250 ng/ml) in PBS for 1 h. After washing with PBS, cells were mounted in 90% glycerol containing 0.1% p-phenylenediamine dihydrochloride in PBS. Images were captured using a confocal microscope (FV3000) with a ×60 PLAPON objective (Olympus Co. Ltd.).
Glutamate release assay
U251 cells plated in a 48-well plate at LD or HD were cultured for 48 h. They were then rinsed twice with PBS, and the medium was replaced with amino acid–free medium supplemented with or without 0.2 mm cystine and 2 mm glutamine for 4 h. Glutamate release into the medium was measured using The L-Glutamate Assay Kit YAMASA NEO (Yamasa Corp.) according to the manufacturer's instructions. To normalize the cell number, protein concentration in each well was measured using the Protein Assay BCA Kit (Nacalai Tesque).
Data analysis
Data were analyzed using analysis of variance (ANOVA) with Tukey honestly significant difference post hoc test. p < 0.05 was considered significant. Statistical analyses were performed using KaleidaGraph (Synergy Software).
Data Availability
All data are contained within the manuscript.
Author contributions
I. Y. and H. K. conceptualization; I. Y. and H. K. data curation; I. Y., S. H. Y., and H. K. formal analysis; I. Y., S. H. Y., and H. K. investigation; I. Y., S. H. Y., and H. K. visualization; I. Y. and H. K. methodology; I. Y., S. H. Y., and H. K. writing-review and editing; S. H. Y. and H. K. resources; H. K. funding acquisition; H. K. writing-original draft; H. K. project administration.
Acknowledgment
We thank Dr. Junichi Miyazaki (Osaka University, Osaka, Japan) for the CAG promoter–containing vector.
This work was supported in part by Grant-in-Aid for Scientific Research from the Japan Society for the Promotion of Science 18K06215 and Princess Takamatsu Cancer Research Fund Grant 18-25006. The authors declare that they have no conflicts of interest with the contents of this article.
- LD
- low cell density
- HD
- high cell density
- LDH
- lactate dehydrogenase
- BafA1
- Bafilomycin A1
- ANOVA
- analysis of variance.
References
- 1. Hanahan D., and Weinberg R. A. (2011) Hallmarks of cancer: the next generation. Cell 144, 646–674 10.1016/j.cell.2011.02.013 [DOI] [PubMed] [Google Scholar]
- 2. Ward P. S., and Thompson C. B. (2012) Metabolic reprogramming: a cancer hallmarks even Warburg did not anticipate. Cancer Cell 21, 297–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hay N. (2016) Reprogramming glucose metabolism in cancer: can it be exploited for cancer therapy? Nat. Rev. Cancer 16, 635–649 10.1038/nrc.2016.77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Galadari S., Rahman A., Pallichankandy S., and Thayyullathil F. (2017) Reactive oxygen species and cancer paradox: to promote or to suppress? Free Radic. Biol. Med. 104, 144–164 10.1016/j.freeradbiomed.2017.01.004 [DOI] [PubMed] [Google Scholar]
- 5. Conrad M., and Sato H. (2012) The oxidative stress-inducible cystine/glutamate antiporter, system X(c)(−): cystine supplier and beyond. Amino Acids 42, 231–246 10.1007/s00726-011-0867-5 [DOI] [PubMed] [Google Scholar]
- 6. Takeuchi S., Wada K., Toyooka T., Shinomiya N., Shimazaki H., Nakanishi K., Nagatani K., Otani N., Osada H., Uozumi Y., Matsuo H., and Nawashiro H. (2013) Increased xCT expression correlates with tumor invasion and outcome in patients with glioblastoma. Neurosurgery 72, 33–41 10.1227/NEU.0b013e318276b2de [DOI] [PubMed] [Google Scholar]
- 7. Lewerenz J., Hewett S. J., Huang Y., Lambros M., Gout P. W., Kalivas P. W., Massie A., Smolders I., Methner A., Pergande M., Smith S. B., Ganapathy V., and Maher P. (2013) The cystine/glutamate antiporter system X(c)(−) in health and disease: from molecular mechanisms to novel therapeutic opportunities. Antioxid. Redox Signal. 18, 522–555 10.1089/ars.2011.4391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dixon S. J., Lemberg K. M., Lamprecht M. R., Skouta R., Zaitsev E. M., Gleason C. E., Patel D. N., Bauer A. J., Cantley A. M., Yang W. S., Morrison B. 3rd, and Stockwell B. R. (2012) Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell 149, 1060–1072 10.1016/j.cell.2012.03.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yang W. S., SriRamaratnam R., Welsch M. E., Shimada K., Skouta R., Viswanathan V. S., Cheah J. H., Clemons P. A., Shamji A. F., Clish C. B., Brown L. M., Girotti A. W., Cornish V. W., Schreiber S. L., and Stockwell B. R. (2014) Regulation of ferroptotic cancer death by GPX4. Cell 156, 317–331 10.1016/j.cell.2013.12.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hassannia B., Vandenabeele P., and Vanden Berghe T. (2019) Targeting ferroptosis to iron out cancer. Cancer Cell 35, 830–849 10.1016/j.ccell.2019.04.002 [DOI] [PubMed] [Google Scholar]
- 11. Friedmann Angeli J. P., Krysko D. V., and Conrad M. (2019) Ferroptosis at the crossroads of cancer-acquired drug resistance and immune evasion. Nat. Rev. Cancer 19, 405–414 10.1038/s41568-019-0149-1 [DOI] [PubMed] [Google Scholar]
- 12. Shin C. S., Mishra P., Watrous J. D., Carelli V., D'Aurelio M., Jain M., and Chan D. C. (2017) The glutamate/cystine xCT antiporter antagonizes glutamine metabolism and reduces nutrient flexibility. Nat. Commun. 8, 15074 10.1038/ncomms15074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Koppula P., Zhang Y., Shi J., Li W., and Gan B. (2017) The glutamate/cystine antiporter SLC7A11/xCT enhances cancer cell dependency on glucose by exporting glutamate. J. Biol. Chem. 292, 14240–14249 10.1074/jbc.M117.798405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Goji T., Takahara K., Negishi M., and Katoh H. (2017) Cystine uptake through the cystine/glutamate antiporter xCT triggers glioblastoma cell death under glucose deprivation. J. Biol. Chem. 292, 19721–19732 10.1074/jbc.M117.814392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Koppula P., Zhang Y., Zhuang L., and Gan B. (2018) Amino acid transporter SLC7A11/xCT at the crossroads of regulating redox homeostasis and nutrient dependency of cancer. Cancer Commun. (Lond.) 38, 12 10.1186/s40880-018-0288-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Saxton R. A., and Sabatini D. M. (2017) mTOR signaling in growth, metabolism, and disease. Cell 168, 960–976 10.1016/j.cell.2017.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lawrence R. E., and Zoncu R. (2019) The lysosome as a cellular centre for signalling, metabolism and quality control. Nat. Cell Biol. 21, 133–142 10.1038/s41556-018-0244-7 [DOI] [PubMed] [Google Scholar]
- 18. Mossmann D., Park S., and Hall M. N. (2018) mTOR signalling and cellular metabolism are mutual determinants in cancer. Nat. Rev. Cancer 18, 744–757 10.1038/s41568-018-0074-8 [DOI] [PubMed] [Google Scholar]
- 19. Jung C. H., Ro S. H., Cao J., Otto N. M., and Kim D. H. (2010) mTOR regulation of autophagy. FEBS Lett. 584, 1287–1295 10.1016/j.febslet.2010.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kim J., Kundu M., Viollet B., and Guan K. L. (2011) AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat. Cell Biol. 13, 132–141 10.1038/ncb2152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhao J., Zhai B., Gygi S. P., and Goldberg A. L. (2015) mTOR inhibition activates overall protein degradation by the ubiquitin proteasome system as well as by autophagy. Proc. Natl. Acad. Sci. U.S.A. 112, 15790–15797 10.1073/pnas.1521919112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rousseau A., and Bertolotti A. (2016) An evolutionarily conserved pathway controls proteasome homeostasis. Nature 536, 184–189 10.1038/nature18943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Leontieva O. V., Demidenko Z. N., and Blagosklonny M. V. (2014) Contact inhibition and high cell density deactivate the mammalian target of rapamycin pathway, thus suppressing the senescence program. Proc. Natl. Acad. Sci. U.S.A. 111, 8832–8837 10.1073/pnas.1405723111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Trajkovic K., Valdez C., Ysselstein D., and Krainc D. (2019) Fluctuations in cell density alter protein markers of multiple cellular compartments, confounding experimental outcomes. PLoS ONE 14, e0211727 10.1371/journal.pone.0211727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Liu T., Jiang L., Tavana O., and Gu W. (2019) The deubiquitylase OTUB1 mediates ferroptosis via stabilization of SLC7A11. Cancer Res. 79, 1913–1924 10.1158/0008-5472.CAN-18-3037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Puertollano R. (2014) mTOR and lysosome regulation. F1000Prime Rep. 6, 52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cang C., Zhou Y., Navarro B., Seo Y. J., Aranda K., Shi L., Battaglia-Hsu S., Nissim I., Clapham D. E., and Ren D. (2013) mTOR regulates lysosomal ATP-sensitive two-pore Na+ channels to adapt to metabolic state. Cell 152, 778–790 10.1016/j.cell.2013.01.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mousavi S. A., Kjeken R., Berg T. O., Seglen P. O., Berg T., and Brech A. (2001) Effects of inhibitors of the vacuolar proton pump on hepatic heterophagy and autophagy. Biochim. Biophys. Acta 1510, 243–257 10.1016/S0005-2736(00)00354-0 [DOI] [PubMed] [Google Scholar]
- 29. Fass E., Shvets E., Degani I., Hirschberg K., and Elazar Z. (2006) Microtubules support production of starvation-induced autophagosomes but not their targeting and fusion with lysosomes. J. Biol. Chem. 281, 36303–36316 10.1074/jbc.M607031200 [DOI] [PubMed] [Google Scholar]
- 30. Teramoto K., and Katoh H. (2019) The cystine/glutamate antiporter xCT is a key regulator of EphA2 S897 phosphorylation under glucose-limited conditions. Cell. Signal. 62, 109329 10.1016/j.cellsig.2019.05.014 [DOI] [PubMed] [Google Scholar]
- 31. Zhao B., Wei X., Li W., Udan R. S., Yang Q., Kim J., Xie J., Ikenoue T., Yu J., Li L., Zheng P., Ye K., Chinnaiyan A., Halder G., Lai Z. C., and Guan K. L. (2007) Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 21, 2747–2761 10.1101/gad.1602907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Reuven N., Adler J., Meltser V., and Shaul Y. (2013) The Hippo pathway kinase Lats2 prevents DNA damage-induced apoptosis through inhibition of the tyrosine kinase c-Abl. Cell Death Differ. 20, 1330–1340 10.1038/cdd.2013.83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pavel M., Renna M., Park S. J., Menzies F. M., Ricketts T., Füllgrabe J., Ashkenazi A., Frake R. A., Lombarte A. C., Bento C. F., Franze K., and Rubinsztein D. C. (2018) Contact inhibition controls cell survival and proliferation via YAP/TAZ-autophagy axis. Nat. Commun. 9, 2961 10.1038/s41467-018-05388-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kobayashi H., Takemura Y., and Ohnuma T. (1992) Relationship between tumor cell density and drug concentration and the cytotoxic effects of doxorubicin or vincristine: mechanism of inoculum effects. Cancer Chemother. Pharmacol. 31, 6–10 10.1007/BF00695987 [DOI] [PubMed] [Google Scholar]
- 35. Westhoff M. A., Zhou S., Bachem M. G., Debatin K. M., and Fulda S. (2008) Identification of a novel switch in the dominant forms of cell adhesion-mediated drug resistance in glioblastoma cells. Oncogene 27, 5169–5181 10.1038/onc.2008.148 [DOI] [PubMed] [Google Scholar]
- 36. Gujral T. S., and Kirschner M. W. (2017) Hippo pathway mediates resistance to cytotoxic drugs. Proc. Natl. Acad. Sci. U.S.A. 114, E3729–E3738 10.1073/pnas.1703096114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Brown C. W., Amante J. J., and Mercurio A. M. (2018) Cell clustering mediated by the adhesion protein PVRL4 is necessary for α6β4 integrin-promoted ferroptosis resistance in matrix-detached cells. J. Biol. Chem. 293, 12741–12748 10.1074/jbc.RA118.003017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wenz C., Faust D., Linz B., Turmann C., Nikolova T., and Dietrich C. (2019) Cell-cell contacts protect against t-BuOOH-induced cellular damage and ferroptosis in vitro. Arch. Toxicol. 93, 1265–1279 10.1007/s00204-019-02413-w [DOI] [PubMed] [Google Scholar]
- 39. Jiang L., Kon N., Li T., Wang S. J., Su T., Hibshoosh H., Baer R., and Gu W. (2015) Ferroptosis as a p53-mediated activity during tumour suppression. Nature 520, 57–62 10.1038/nature14344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ishimoto T., Nagano O., Yae T., Tamada M., Motohara T., Oshima H., Oshima M., Ikeda T., Asaba R., Yagi H., Masuko T., Shimizu T., Ishikawa T., Kai K., Takahashi E., et al. (2011) CD44 variant regulates redox status in cancer cells by stabilizing the xCT subunit of system xc− and thereby promotes tumor growth. Cancer Cell 19, 387–400 10.1016/j.ccr.2011.01.038 [DOI] [PubMed] [Google Scholar]
- 41. Tsuchihashi K., Okazaki S., Ohmura M., Ishikawa M., Sampetrean O., Onishi N., Wakimoto H., Yoshikawa M., Seishima R., Iwasaki Y., Morikawa T., Abe S., Takao A., Shimizu M., Masuko T., et al. (2016) The EGF receptor promotes the malignant potential of glioma by regulating amino acid transport system X(c)(−). Cancer Res. 76, 2954–2963 10.1158/0008-5472.CAN-15-2121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Dikic I. (2017) Proteasomal and autophagic degradation systems. Annu. Rev. Biochem. 86, 193–224 10.1146/annurev-biochem-061516-044908 [DOI] [PubMed] [Google Scholar]
- 43. Ji C. H., and Kwon Y. T. (2017) Crosstalk and interplay between the ubiquitin-proteasome system and autophagy. Mol. Cells 40, 441–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Matsumoto G., Wada K., Okuno M., Kurosawa M., and Nukina N. (2011) Serine 403 phosphorylation of p62/SQSTM1 regulates selective autophagic clearance of ubiquitinated proteins. Mol. Cell 44, 279–289 10.1016/j.molcel.2011.07.039 [DOI] [PubMed] [Google Scholar]
- 45. Gu D., Wang S., Kuiatse I., Wang H., He J., Dai Y., Jones R. J., Bjorklund C. C., Yang J., Grant S., and Orlowski R. Z. (2014) Inhibition of the MDM2 E3 ligase induces apoptosis and autophagy in wild-type and mutant p53 models of multiple myeloma, and acts synergistically with ABT-737. PLoS ONE 9, e103015 10.1371/journal.pone.0103015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lagunas-Martínez A., García-Villa E., Arellano-Gaytán M., Contreras-Ochoa C. O., Dimas-González J., López-Arellano M. E., Madrid-Marina V., and Gariglio P. (2017) MG132 plus apoptosis antigen-1 (APO-1) antibody cooperate to restore p53 activity inducing autophagy and p53-dependent apoptosis in HPV16 E6-expressing keratinocytes. Apoptosis 22, 27–40 10.1007/s10495-016-1299-1 [DOI] [PubMed] [Google Scholar]
- 47. Wang X. J., Yu J., Wong S. H., Cheng A. S. L., Chan F. K., Ng S. S. M., Cho C. H., Sung J. J., and Wu W. K. (2013) A novel crosstalk between two major protein degradation systems: regulation of proteasomal activity by autophagy. Autophagy 9, 1500–1508 10.4161/auto.25573 [DOI] [PubMed] [Google Scholar]
- 48. Qiao L., and Zhang J. (2009) Inhibition of lysosomal functions reduces proteasomal activity. Neurosci. Lett. 456, 15–19 10.1016/j.neulet.2009.03.085 [DOI] [PubMed] [Google Scholar]
- 49. Song X., Zhu S., Chen P., Hou W., Wen Q., Liu J., Xie Y., Liu J., Klionsky D. J., Kroemer G., Lotze M. T., Zeh H. J., Kang R., and Tang D. (2018) AMPK-mediated BECN1 phosphorylation promotes ferroptosis by directly blocking system X(c)(−) activity. Curr. Biol. 28, 2388–2399 10.1016/j.cub.2018.05.094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Gu Y., Albuquerque C. P., Braas D., Zhang W., Villa G. R., Bi J., Ikegami S., Masui K., Gini B., Yang H., Gahman T. C., Shiau A. K., Cloughesy T. F., Christofk H. R., Zhou H., et al. (2017) mTORC2 regulates amino acid metabolism in cancer by phosphorylation of the cystine-glutamate antiporter xCT. Mol. Cell 67, 128–138.e7 10.1016/j.molcel.2017.05.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lien E. C., Ghisolfi L., Geck R. C., Asara J. M., and Toker A. (2017) Oncogene PI3K promotes methionine dependency in breast cancer cells through the cystine-glutamate antiporter xCT. Sci. Signal. 10, eaao6604 10.1126/scisignal.aao6604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Inoki K., Zhu T., and Guan K. L. (2003) TSC2 mediates cellular energy response to control cell growth and survival. Cell 115, 577–590 10.1016/S0092-8674(03)00929-2 [DOI] [PubMed] [Google Scholar]
- 53. Liu L., Cash T. P., Jones R. G., Keith B., Thompson C. B., and Simon M. C. (2006) Hypoxia-induced energy stress regulates mRNA translation and cell growth. Mol. Cell 21, 521–531 10.1016/j.molcel.2006.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Lamming D. W., and Bar-Peled L. (2019) Lysosome: the metabolic signaling hub. Traffic 20, 27–38 10.1111/tra.12617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Sardiello M., Palmieri M., di Ronza A., Medina D. L., Valenza M., Gennarino V. A., Di Malta C., Donaudy F., Embrione V., Polishchuk R. S., Banfi S., Parenti G., Cattaneo E., and Ballabio A. (2009) A gene network regulating lysosomal biogenesis and function. Science 325, 473–477 10.1126/science.1174447 [DOI] [PubMed] [Google Scholar]
- 56. Settembre C., Zoncu R., Medina D. L., Vetrini F., Erdin S., Erdin S., Huynh T., Ferron M., Karsenty G., Vellard M. C., Facchinetti V., Sabatini D. M., and Ballabio A. (2012) A lysosome-to nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. EMBO J. 31, 1095–1108 10.1038/emboj.2012.32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Roczniak-Ferguson A., Petit C. S., Froehlich F., Qian S., Ky J., Angarola B., Walther T. C., and Ferguson S. M. (2012) The transcription factor TFEB links mTORC1 signaling to transcriptional control of lysosome homeostasis. Sci. Signal. 5, ra42 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data are contained within the manuscript.