Abstract
The structurally homologous Mtf1 and TFB2M proteins serve as transcription initiation factors of mitochondrial RNA polymerases in Saccharomyces cerevisiae and humans, respectively. These transcription factors directly interact with the nontemplate strand of the transcription bubble to drive promoter melting. Given the key roles of Mtf1 and TFB2M in promoter-specific transcription initiation, it can be expected that the DNA binding activity of the mitochondrial transcription factors is regulated to prevent DNA binding at inappropriate times. However, little information is available on how mitochondrial DNA transcription is regulated. While studying C-terminal (C-tail) deletion mutants of Mtf1 and TFB2M, we stumbled upon a finding that suggested that the flexible C-tail region of these factors autoregulates their DNA binding activity. Quantitative DNA binding studies with fluorescence anisotropy-based titrations revealed that Mtf1 with an intact C-tail has no affinity for DNA but deletion of the C-tail greatly increases Mtf1's DNA binding affinity. Similar observations were made with TFB2M, although autoinhibition by the C-tail of TFB2M was not as complete as in Mtf1. Analysis of available TFB2M structures disclosed that the C-tail engages in intramolecular interactions with the DNA binding groove in the free factor, which, we propose, inhibits its DNA binding activity. Further experiments showed that RNA polymerase relieves this autoinhibition by interacting with the C-tail and engaging it in complex formation. In conclusion, our biochemical and structural analyses reveal autoinhibitory and activation mechanisms of mitochondrial transcription factors that regulate their DNA binding activities and aid in specific assembly of transcription initiation complexes.
Keywords: mitochondria, transcriptional coactivator, RNA polymerase, fluorescence anisotropy, protein–DNA interaction, autoinhibition, mitochondrial RNA polymerase, mitochondrial transcription factors, Mtf1, TFB2M
Introduction
Mitochondrial RNA polymerases (RNAPs)2 are evolutionarily related to the single-subunit bacteriophage T7/T3 RNAP (1). However, unlike T7/T3 RNAP, the core subunit of the mitochondrial RNAP is unable to initiate promoter-specific transcription and requires assistance from accessory transcription initiation factors. To catalyze promoter-specific transcription initiation, the core subunit of Saccharomyces cerevisiae (yeast) mitochondrial RNAP, Rpo41, requires mitochondrial transcription factor 1 (Mtf1) (2, 3) and the human mitochondrial RNAP POLRMT requires transcription factor B2 mitochondrial (TFB2M) and transcription factor A mitochondrial (TFAM) (3–7). These factors play a key role in stabilizing the transcription initiation bubble.
Mtf1 and TFB2M proteins are evolutionarily related to bacterial rRNA methyltransferases (8, 9). Methylation activity is not required for transcription initiation; hence, Mtf1 has lost methyltransferase activity, but interestingly, TFB2M has residual methyltransferase activity (10). However, both factors have maintained their nucleic acid binding function, which is essential for promoter melting. The nucleic acid binding groove in the factors serves as a binding pocket for the nontemplate strand to stabilize the transcription initiation bubble (8, 11).
Transcription factors are regulated in various ways, including reversible protein phosphorylation, other accessory factors, and autoregulatory mechanisms (12–16). For example, bacterial σ factors are regulated by anti-σ factors or autoinhibited by the σ-1.1 domain (16). No such regulatory mechanism has been reported for mitochondrial transcription factors.
During our study of the C-terminal deletion mutants of Mtf1 (3), we stumbled upon a new finding that suggested that the flexible C-terminal tail of Mtf1 has an autoinhibitory function in DNA binding. Mtf1 and TFB2M contain a flexible C-tail region consisting of 16–20 aa that, as we showed recently, plays a critical role in template strand alignment, including supporting high-affinity binding of the initiating nucleotide for an efficient RNA priming reaction (3). The 16 aa of the Mtf1 C-tail were not resolved in the crystal structure (9), whereas in the crystal structures of TFB2M, the fully or partially resolved C-tail region was found to be in different conformations (8).
To investigate the regulatory role of the flexible C-tail region in DNA binding, we used fluorescence anisotropy–based titrations and measured the DNA Kd values of the WT and several C-tail deletion mutants of Mtf1 and TFB2M. We used promoter and nonpromoter sequences to test the specificity of the DNA binding site. Our studies show that DNA binding is nonspecific and that the C-tail region completely or partially suppresses the DNA binding activity of the factors. Inhibition is released, however, when the factors bind to RNAP and the C-tail is engaged with the RNAP. Based on our biochemical data and analysis of available structures, we propose a model that explains the autoinhibition and activation of DNA binding involving the C-tail of mitochondrial transcription factors. This model provides new insights into assembly and regulation of the mitochondrial transcription initiation complex.
Results
The C-tail inhibits the DNA binding activity of Mtf1
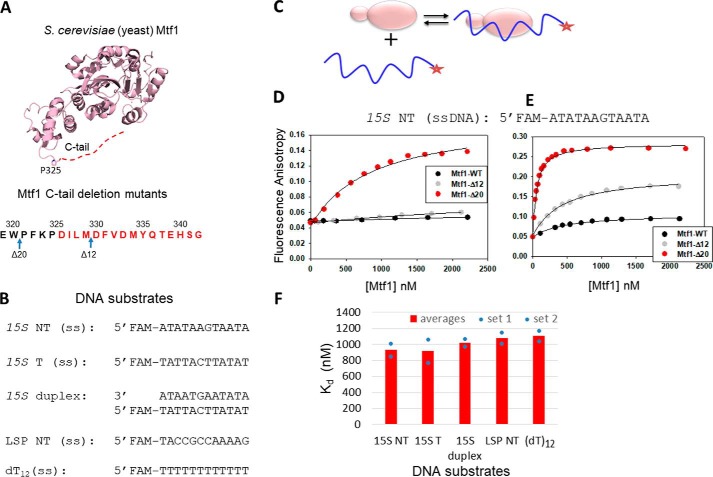
In this study, we used two Mtf1 C-tail deletion mutants that were used in our previous study (3). Mtf1-Δ20 lacks the entire C-tail region (20 aa), and the partially deleted C-tail mutant Mtf1-Δ12 lacks only the terminal 12 aa (Fig. 1A). Because Mtf1 binds to the nontemplate strand of the transcription bubble in the initiation complex (11), we synthesized a 12-nt single-stranded DNA (ssDNA) that contained the −8 to +4 sequence of the nontemplate strand of the yeast 15S rRNA promoter (Fig. 1B). This DNA included the −4 to +2 region that forms a transcription bubble (17) and interacts with Mtf1 in the initiation complex (11).
Figure 1.
The C-tail of Mtf1 drastically autoinhibits the DNA binding activity of Mtf1. A, structure of the yeast mitochondrial transcription factor Mtf1 (rose, PDB code 1I4W). The missing 16 aa of the C-tail of Mtf1 in the crystal structure are shown as a red dotted line and are also marked in red in the amino acid sequence of the C-tail of Mtf1. B, DNA sequences of the substrates used for the Mtf1 DNA binding studies. C, cartoon showing the basic scheme of the fluorescence anisotropy assays to monitor protein–DNA binding. D, representative binding curves showing the fluorescence anisotropy changes resulting from titration of the 15S NT DNA with Mtf1. 15S NT DNA (5 nm) was titrated with Mtf1-WT (black circles), Mtf1-Δ12 (gray circles), and Mtf1-Δ20 (red circles) in buffer A (see “Experimental procedures”). E, 15S NT (5 nm) was titrated with Mtf1-WT (black circles), Mtf1-Δ12 (gray circles), and Mtf1-Δ20 (red circles) in buffer A without potassium glutamate. The solid lines represent fit to the hyperbolic Equation 1 with Kd values as follows: Mtf1-WT = 447 ± 60 nm (amplitude, 0.059), Mtf1-Δ12 = 426 ± 33 nm (amplitude, 0.156), Mtf1-Δ20 = 51 ± 2.8 nm (amplitude: 0.24). F, the average DNA Kd values of Mtf1-Δ20 are shown for the DNA substrates in B. The blue dots are the individual values for set 1 and set 2 titration data, which are shown in Fig. S1.
We used fluorescence anisotropy–based titrations to measure the DNA Kd values (Fig. 1C) (18). Fluorescein-labeled 15S NT ssDNA was titrated with increasing concentrations of Mtf1 protein. We observed negligible changes in fluorescence anisotropy with Mtf1-WT and Mtf1-Δ12 even after adding 2 μm protein (Fig. 1D). On the other hand, Mtf1-Δ20, with a larger deletion of the C-tail, showed a significant increase in fluorescence anisotropy in titration experiments. The binding data of Mtf1-Δ20 fit well to a hyperbola and provided a Kd of ∼930 (±114) nm for the 15S NT complex. These results indicate that the presence of the full or even partial C-tail region inhibits the DNA binding activity of Mtf1. Thus, complete deletion of the C-tail is needed to activate the DNA binding activity of Mtf1.
The DNA binding buffer in the above experiments contained 50 mm potassium glutamate. To determine whether the DNA binding interactions were salt-sensitive, we eliminated potassium glutamate from the buffer. Under less stringent salt conditions, we observed some amount of DNA binding to the Mtf1-WT and Mtf1-Δ12, although the amplitudes of fluorescence anisotropy change remained lower than with Mtf1-Δ20 (Fig. 1E). Interestingly, there was a clear C-tail length–dependent effect on the DNA binding amplitudes. Mtf1-WT with an intact C-tail showed the lowest amount of DNA binding, followed by Mtf1-Δ12 with an intermediate level of binding and Mtf1-Δ20 showing the highest amount of DNA binding. We do not know the exact reason for the different plateau in the anisotropy values for the different mutants. It could be due to higher-order interactions of the protein–DNA complexes in the absence of salt. Removal of the salt increased the DNA binding affinity of Mtf1-Δ20 by 18-fold. These results point to an ionic nature of interaction in the DNA binding groove and suggest that Mtf1 makes contact with the charged DNA phosphate backbone. Thus, competition with salt explains the lower DNA binding affinity of Mtf1-Δ12.
To investigate whether the DNA binding groove of Mtf1 has a preference for binding a particular DNA sequence or structure, we compared the DNA Kd values of promoter versus nonpromoter sequences and ssDNA versus dsDNA. We used the template and nontemplate ssDNA of the yeast 15S promoter as DNA substrates, the duplex 15S promoter, an unrelated ssDNA sequence of the human mitochondrial promoter (LSP NT), and dT12 (Fig. 1B). Mtf1-WT did not bind to any of these DNA substrates. Mtf1-Δ20, on the other hand, bound to all DNA substrates (Fig. 1F and Fig. S1) with very similar Kd values (750–1000 nm). These results indicate that the DNA binding groove of Mtf1 has no sequence or structural preference for the DNA. This means that the C-tail prevents Mtf1 from binding to specific and nonspecific DNAs. In summary, the DNA binding studies described above provide first evidence that the C-tail region has a role in regulating the DNA binding activity of Mtf1 prior to transcription initiation.
Structural basis of C-tail–mediated inhibition of DNA binding
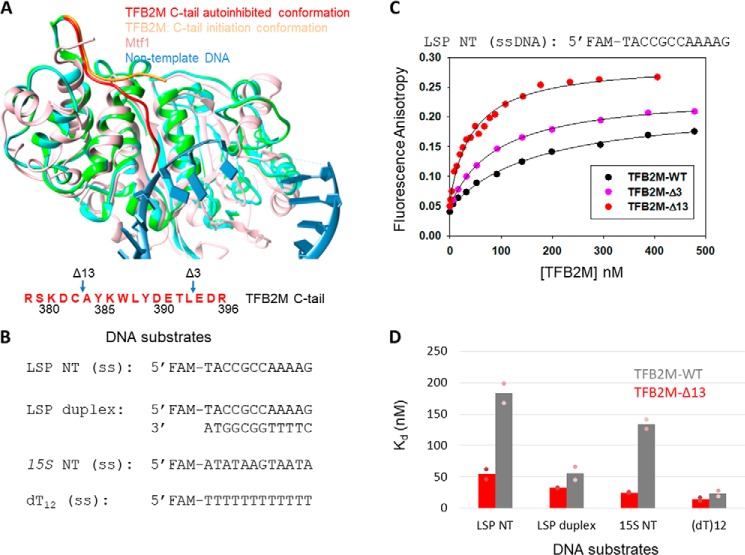
To understand how the C-tail region of Mtf1 inhibits DNA binding, we analyzed the structures of the homologous human TFB2M protein (8). The C-tail region in free TFB2M is fully resolved in chain A of the molecule (Fig. 2A, TFB2M in green and C-tail in red). In chain A, the C-tail makes intramolecular interactions with the DNA binding groove (Fig. S2). In the initiation structure, the DNA binding groove is engaged with the nontemplate strand (Fig. 2A, sky blue), and therefore, the C-tail (Fig. 2A, orange) is projected in a different direction away from the DNA binding groove. Thus, analysis of the TFB2M structures suggests that the conformation of the flexible C-tail in free TFB2M is sterically incompatible with DNA binding. Although Mtf1 and TFB2M share only 12% sequence identity, there is a high degree of structural homology between the two proteins (Fig. 2A, Mtf1 in rose and TFB2M in green). We hypothesized that the DNA binding activity of Mtf1 is similarly autoinhibited by C-tail interactions with the DNA binding groove.
Figure 2.
The C-tail of TFB2M mildly autoinhibits the DNA binding activity of TFB2M. A, the aligned structures of free human TFB2M (PDB code 6ERO, cyan), TFB2M in the initiation complex (PDB code 6ERP, green), and yeast Mtf1 (PDB code 1I4W, rose) are shown. The relative positions of the C-tail in all three structures are shown. The amino acid sequence of the C-tail of TFB2M is shown below in red. B, DNA substrates used for the TFB2M DNA binding studies. C, representative binding curves show the fluorescence anisotropy change resulting from titration of LSP NT (5 nm) with TFB2M-WT (black circles), TFB2M-Δ3 (pink circles), and TFB2M-Δ13 (red circles). The data were fit to the hyperbolic Equation 1 to obtain the following Kd values: TFB2M-WT = 169 ± 18 nm (amplitude, 0.17), TFB2M-Δ3 = 92 ± 3.3 nm (amplitude, 0.19), TFB2M-Δ13 = 46 ± 5.2 nm (amplitude, 0.23). D, the gray and red columns show the DNA Kd values of TFB2M-WT and TFB2M-Δ13, respectively, for the various DNA substrates shown in B. The pink dots represent individual values for set 1 and set 2 titration data, which are shown in Fig. S3.
The autoinhibitory role of the C-tail is conserved in the human homolog TFB2M
The above structural analysis predicts that the C-tail will inhibit the DNA binding activity of TFB2M. To directly test the autoinhibitory role of the C-tail in TFB2M, we measured the DNA Kd values of TFB2M-WT and its two C-tail deletion mutants, TFB2M-Δ3 and TFB2M-Δ13, which lack three and 13 aa of the C-tail, respectively (Fig. 2A) (3). In our initial DNA binding studies, we used the nontemplate ssDNA of the human mitochondrial light strand promoter (LSP NT) as the substrate, which contains the DNA sequence that binds to TFB2M in the initiation complex. Additionally, we synthesized the LSP duplex and unrelated 15S NT and dT12 ssDNAs to determine whether TFB2M has a preference for binding to promoter sequences (Fig. 2B).
TFB2M-WT binds to LSP NT ssDNA with a Kd of about 180 (±22) nm, and deletion of 13 aa of the TFB2M C-tail increases the DNA binding affinity of LSP NT by about 4-fold (Fig. 2C and Fig. S3). Deletion of 3 aa of the C-tail results in an intermediate level of DNA binding (Fig. 2C and Fig. S3). This shows a C-tail length–dependent effect on DNA binding activity, similar to Mtf1.
Studies with other DNA sequences indicate that TFB2M binds to all DNAs but with different Kd values (Fig. 2D). The nonpromoter 15S NT ssDNA binds with a Kd of 130 (±11) nm, which is similar to the Kd of LSP NT. Thus, the DNA binding activity of free TFB2M appears to be nonspecific. The LSP duplex has a 3-fold higher affinity than LSP NT ssDNA. For reasons unknown, TFB2M has a high affinity for dT12 DNA. In all cases, however, C-tail deletion increases the DNA binding affinity of TFB2M.
Thus, our results show that the C-tail of TFB2M autoinhibits the DNA binding activity but that autoinhibition by the C-tail of TFB2M is not absolute as observed in Mtf1. The general trend is the same; that is, the presence of the C-tail inhibits the DNA binding activity. Thus, the C-tail has a conserved role in autoinhibiting the DNA binding activity of the free mitochondrial transcription factors.
The C-tail of Mtf1 is required for stable complex formation with the RNAP subunit
Next we asked how the DNA binding activity of Mtf1 and TFB2M is activated for transcription initiation. Structural analysis shows that, in the initiation complex, the partially resolved C-tail of TFB2M interacts with the RNAP subunit POLRMT (Fig. S4) (8). This suggests that RNAP may release the autoinhibited state by engaging the C-tail in an altered conformation.
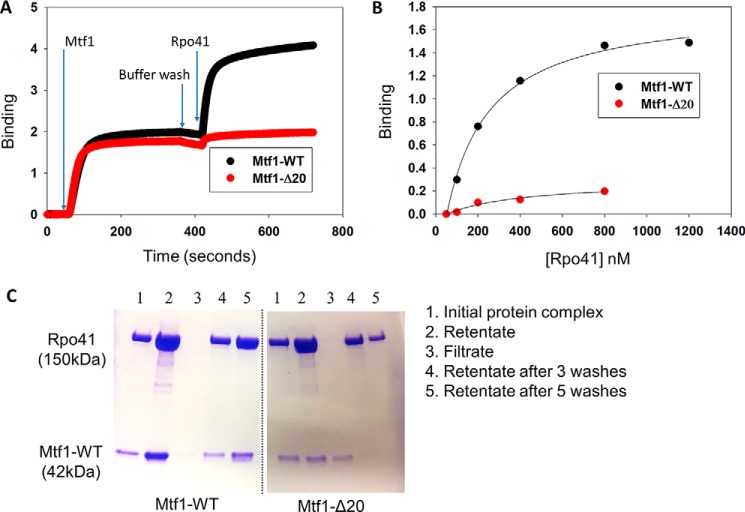
To investigate the RNAP-mediated mechanism of activation, we investigated whether the C-tail has a role in complex formation with the RNAP subunit. We used biolayer interferometry (BLI) and ultrafiltration approaches and measured protein–protein interactions between Mtf1 and the RNAP subunit Rpo41. His-tagged Mtf1-WT and Mtf1-Δ20 proteins were immobilized on the anti-His BLI biosensors, and binding to Rpo41 was assessed by measuring the time-dependent increase in the light interference signal (Fig. 3A and Fig. S5). The amplitude of light interference corresponding to the amount of Rpo41–Mtf1 complex was recorded and plotted against increasing concentrations of Rpo41 (Fig. 3B and S5). The resulting binding curves indicated robust binding of Mtf1-WT to Rpo41 and relatively little binding of Mtf1-Δ20 to Rpo41.
Figure 3.
The C-tail of Mtf1 mediates complex formation with Rpo41. A, representative binding plots showing complex formation between Mtf1 and Rpo41 using biolayer interferometry assays. The first 60 s represent the baseline. Over the next 300 s, the biosensor HIS1K was treated with 0.4 μm His-tagged Mtf1-WT (black line) or Mtf1-Δ20 (red line) protein, followed by washing with buffer for 60 s. The probes were then dipped in Rpo41 (0.5 μm) for 300 s, followed by washing for 60 s. B, the degree of binding (y axis) was calculated from the difference in light interference values before and after adding Rpo41 in A. C, an equimolar complex of Rpo41 and Mtf1-WT or Mtf1-Δ20 at a final concentration 2 μm (lane 1) was filtered through a 100-kDa molecular mass cutoff Microcon centrifugal filter unit. Lane 2 is the retentates, and lane 3 is the filtrates. The retentate was washed with 500 μl of buffer three times (lane 4). The retentate was washed two more times (lane 5). Samples of the initial protein complex, retentate, filtrate, and retentate samples after washing were run on a 4%–20% SDS-PAGE gel.
Ultrafiltration is an alternative solution-based method that has been used previously to assay complex formation between Rpo41 and Mtf1 (19). Application of this method also showed efficient complex formation between Rpo41 and Mtf1-WT, as evident from retention of both proteins in the retentate (even after several washes) and almost no protein in the filtrate (Fig. 3C, Mtf1-WT, lanes 2 and 3). A stable complex was not observed with Mtf1-Δ20 (Fig. 3C, Mtf1-Δ20, lanes 2 and 3). Rpo41 was present in the retentate, and most of the Mtf1-Δ20 was in the filtrate. The above methods of analysis of protein–protein interactions provide consistent results indicating that the C-tail of Mtf1 is involved in complex formation with the RNAP subunit.
The C-tail of TFB2M promotes a stable initiation complex with POLRMT
We tried measuring complex formation between TFB2M and POLRMT in the absence of DNA using BLI and ultrafiltration, but these methods failed to provide consistent results because TFB2M binds nonspecifically to the biosensor probe and the filters. Hence, we used transcription assays and followed runoff products to monitor complex formation between TFB2M and POLRMT in the initiation complex.
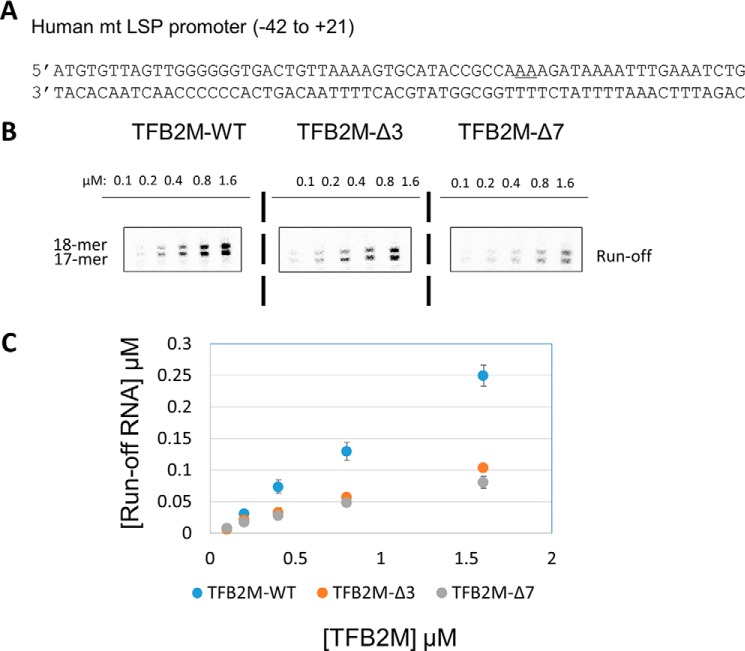
We used TFB2M-Δ3 and TFB2M-Δ7 in the runoff assays because TFB2M-Δ13 does not support transcription (3). Runoff synthesis was measured using an in vitro reconstituted complex of POLRMT, TFB2M, TFAM, and the LSP promoter (Fig. 4A). Transcription initiation from the LSP DNA produces short abortive RNAs and two runoff products 17- and 18-mer in length (Fig. 4B and Fig. S6). The two runoff products result from the two reported start sites on LSP (underlined in the LSP sequence in Fig. 4A) (3, 7).
Figure 4.
The C-tail deletion in TFB2M affects initiation complex formation. A, the promoter fragment LSP used in this study. The two start sites are underlined. B, runoff transcription profiles of TFB2M-WT and the C-tail deletion mutants TFB2M-Δ3 and TFB2M-Δ7 on the LSP promoter (the full gel profile is shown in Fig. S6). Reactions were carried out with 0.6 μm POLRMT, TFAM, and promoter duplex and increasing concentrations of TFB2M-WT or C-tail mutant and 250 μm ATP, UTP, GTP, and γ[32P]ATP for 15 min at 25 °C in transcription buffer. C, plot showing quantitation of grouped runoff products for each reaction. The error bars represent errors calculated from two independent experiments.
To measure the stability of the initiation complexes containing TFB2M-WT or C-tail deletion mutants, the transcription reactions were carried out at increasing TFB2M concentrations (Fig. 4, B and C, and Fig. S6). TFB2M-WT reactions showed a steeper increase in runoff RNA products with increasing TFB2M concentration compared with TFB2M-Δ3 and TFB2M-Δ7. Close to 4-fold greater amounts of mutant proteins were required to observe the same amount of runoff products as with TFB2M-WT. These results indicate that initiation complexes with C-tail deletion mutants are weaker compared with TFB2M-WT. The results are consistent with the model where the TFB2M C-tail is involved in complex formation with the RNAP subunit. Thus, the results with Mtf1 and TFB2M suggest a consistent mechanism of activation of the DNA binding activity of mitochondrial transcription factors.
Discussion
The mitochondrial transcription factors Mtf1 and TFB2M of S. cerevisiae and humans, respectively, have a well-established role in promoter melting during transcription initiation (8, 11, 17, 20). These homologous transcription factors bind to the nontemplate strand of the transcription bubble to drive the promoter melting step. In this study, we show that the DNA binding activity of the transcription factors is autoinhibited by their C-terminal tail regions prior to their association with the RNAP subunit. Mtf1 and TFB2M contain a flexible C-terminal tail region of 16–20 aa that, as we showed recently, plays important roles in transcription initiation (3). This study shows that the C-tail has additional roles in regulating the DNA binding activity of the free factors.
Parallel studies of the yeast and human mitochondrial transcription factors provided an opportunity to compare and contrast the mechanism of DNA regulation by the C-tail region. We find that the C-tail regions of Mtf1 and TFB2M have a conserved role in autoinhibiting the DNA binding activity of the free factors. Quantitation of DNA binding activity, however, shows that the C-tail of Mtf1 drastically inhibits DNA binding, whereas the C-tail of TFB2M only partially inhibits DNA binding. Moreover, it appears that the DNA binding activity of the free factors is nonspecific. The activated factors bind to single-stranded DNA and dsDNA. Based on our results, we propose that Mtf1 is unlikely to associate with nonspecific DNA regions prior to transcription initiation. On the other hand, unless there are alternative means of inhibiting the DNA binding activity, free TFB2M will associate with DNA prior to initiation.
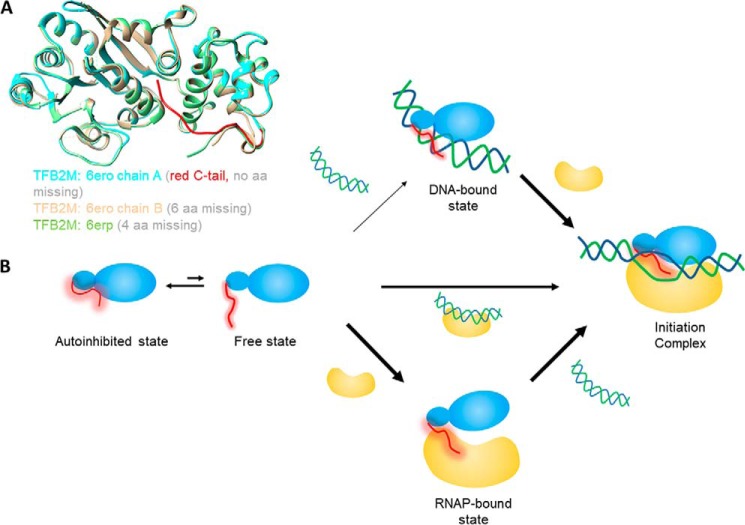
Based on the crystal structure of TFB2M, we propose that the flexible C-tail can exist in two conformations: an autoinhibited state and a free state (Fig. 5B). The crystal structure of free TFB2M shows two conformations of the C-tail in the two molecules of the asymmetric unit (Fig. 5A, cyan and sand). In chain A, the C-tail (Fig. 5A, red) is in the autoinhibited state making intramolecular interactions with the DNA binding groove and masking the DNA binding site. In chain B, the partially resolved C-tail (Fig. 5A, sand) projects away from the DNA binding groove and is more in line with the C-tail in the initiation complex (Fig. 5A, green).
Figure 5.
Model to explain the autoregulatory role of the C-tail of TFB2M and Mtf1 in assembly of the initiation complex. A, crystal structures of TFB2M showing different states of the C-tail in free and DNA-bound states. Chain A of TFB2M in free state is shown in cyan and the C-tail in red. Chain B of TFB2M in the free state is shown in sand, and TFB2M in the initiation complex is shown in green. B, the flexible C-tail (red) of the mitochondrial transcription factors Mtf1 and TFB2M is in equilibrium between an autoinhibited state and free state. For Mtf1, the equilibrium is toward the autoinhibited state, whereas for TFB2M, both states exist. Therefore, Mtf1 needs to bind to RNAP or the RNAP–DNA complex to generate the initiation complex. On the other hand, TFB2M can take all three pathways to form the initiation complex. Under cellular conditions, the DNA binding activity of TFB2M might be regulated by other mechanisms.
We propose that the equilibrium constants of the free and autoinhibited states is different in Mtf1 and TFB2M. The Mtf1 C-tail is mostly in the autoinhibited state, whereas a significant portion of the TFB2M C-tail is in the free state. It is not known whether TFB2M binds DNA under cellular conditions because it might be regulated by additional mechanisms. For example, it is known that TFB2M is posttranslationally modified and that some of the modifications are in the C-tail region (13). Thus, the C-tail autoinhibition mechanism can be potentially reinforced by C-tail modifications. However, further studies are needed to understand these alternative mechanisms of TFB2M regulation.
This study also provides insights into how the transcription factors are activated for promoter-specific transcription initiation. It turns out that the C-tail region is essential for forming a stable complex with RNAP. Thus, it appears that the RNAP subunit can activate the factors by engaging the C-tail with itself, releasing it from the autoinhibited state. The structure of TFB2M shows that the C-tail relocates from the DNA binding groove and is engaged with the thumb domain and the intercalating hairpin of RNAP in the initiation complex (Fig. S4) (8). The C-tail engagement with RNAP would expose the DNA binding groove, which can bind to the nontemplate strand and stabilize the transcription bubble in the initiation complex.
Autoregulatory mechanisms are widely found in biological processes, including DNA transcription. Bacterial and eukaryotic transcription factors, such as σ-70 and Ets-1, have a built-in regulatory domain that modulates their DNA binding activity prior to transcription. The DNA binding activity of σ-70 is autoinhibited by the σ-1.1 domain, and, similar to mitochondrial transcription factors, autoinhibition is relieved when σ-70 forms a complex with the RNAP subunit (16). Autoinhibition by a flexible C-terminal tail is also found in various DNA binding proteins. For example, the DNA binding activity of gp2.5 protein of bacteriophage T7 and bacterial single-stranded DNA-binding protein is autoinhibited by their acidic C-tails (14, 15), which, as we propose for the mitochondrial transcription factors, competes for the basic DNA binding cleft with the DNA.
Thus, this study identifies a previously unknown and conserved role of the C-tail in regulating the DNA binding activity of yeast and human mitochondrial transcription factors. Such autoregulatory mechanisms increase the specificity of the transcription reaction and prevent transcription from occurring at nonpromoter sites.
Experimental procedures
Nucleic acid substrates
Oligodeoxynucleotides were custom-synthesized with a 5′-end fluorescein modification and purified by HPLC (Integrated DNA Technologies, Coralville, IA). DNA concentration was determined from absorbance at 260 nm and the corresponding molar extinction coefficients. Complementary ssDNAs were mixed at a 1:1 ratio, annealed at 95 °C for 1 min, and cooled over 1 h to room temperature to construct the duplex DNA molecules.
Protein purification
Expression and purification of Mtf1, TFB2M, and the respective C-tail deletion mutant proteins were carried out as reported previously (3, 7, 21). The yeast proteins were stored in 50% glycerol and the human proteins were stored in 10% glycerol at −80 °C. The molar concentrations of the proteins were determined in guanidium HCl buffer from absorbance measurements at 280 nm and the respective molar extinction coefficients.
Fluorescence anisotropy experiments to determine Kd values of protein binding to DNA
Fluorescence anisotropy-based titration experiments were carried out on a Fluoro-Max-4 spectrofluorometer (Jobin Yvon-Spex Instruments S.A., Inc.) at 25 °C. Fluorescein-labeled DNA (5 nm) was titrated with Mtf1-WT, TFB2M, or the C-tail deletion mutants. Reaction buffer A (50 mm Tris acetate (pH 7.5), 50 mm potassium glutamate, 10 mm magnesium acetate, 1 mm DTT, and 0.05% Tween 20) was used in studies of Mtf1, and reaction buffer B (50 mm Tris acetate (pH 7.5), 100 mm sodium glutamate, 10 mm magnesium acetate, 10 mm DTT, and 0.01% Tween 20) was used in studies of the TFB2M. Anisotropy values (robs) were recorded with excitation at 490 nm (4-nm bandwidth) and emission at 514 nm (8-nm bandwidth). The robs were plotted against total protein concentration ([P]) and fit to Equation 1 to obtain the Kd. Here, rmax is the fluorescence anisotropy amplitude, and rf is the initial fluorescence anisotropy of the DNA before protein addition.
| (Eq. 1) |
Biolayer interferometry
The BLItz binding assays were performed with the Fortebio BLItz system with a Dip and Read Penta-HIS (HIS1K) biosensor (ForteBio) using the tube method to measure complex formation between Rpo41 and Mtf1. The probes were equilibrated in water overnight and then in buffer A for 30 min prior to use. The biosensors were washed in buffer A for 60 s to obtain the baseline. Mtf1 (400 nm) was immobilized on HIS1K biosensors for 300 s, washed in buffer A for 60 s, dipped in increasing concentrations of Rpo41 solution (50–800 nm), and finally washed again in buffer A for 60 s. The wavelength shift was recorded in real time with ForteBio software.
Ultracentrifugation assays
An equimolar complex of Rpo41 and Mtf1 (2 μm each) was mixed in reaction buffer C (50 mm Tris acetate (pH 7.5), 100 mm potassium glutamate, 10 mm magnesium acetate, 5 mm fresh DTT, 0.01% protein-grade Tween 20, and 5% glycerol) in a final volume of 500 μl. The mixture was incubated at 25 °C for 15 min (initial protein complex) before filtering through a 100-kDa molecular mass cutoff Microcon centrifugal filter unit until the volume of the first retentate was about 50 μl (1/10 of initial mixture). The retentate was diluted to 500 μl with buffer C and filtered again. This washing step was repeated, and a sample was taken after three and five washes. Samples consisting of initial protein complex, first retentate, filtrate, and three and five retentate samples were collected and run on a 4%–20% SDS-PAGE gel.
Transcription reactions
Transcription reactions were carried out at 25 °C using 1 μm POLRMT, 1 μm TFAM, 1 μm TFB2M-WT or deletion mutants, and 1 μm promoter DNA in transcription buffer (50 mm Tris acetate (pH 7.5), 100 mm sodium glutamate, 10 mm magnesium acetate, 10 mm DTT, and 0.01% Tween 20). For runoff RNA synthesis, we used 250 μm ATP, UTP, or GTP spiked with [γ-32P]ATP. Reactions were stopped after 15 min using 400 mm EDTA and formamide dye (98% formamide, 0.025% bromphenol blue, and 10 mm EDTA). Samples were heated to 95 °C for 2 min and chilled on ice, and the RNA products were resolved on a 24% sequencing gel containing 4 m urea. The gel was exposed to a phosphor screen overnight and scanned on a Typhoon 9410 PhosphorImager instrument (Amersham Biosciences). The free ATP and RNA bands were quantified using ImageQuant, and molar amounts of RNA synthesized were calculated according to Equation 2:
| (Eq. 2) |
where R and A are the band intensities of RNA products and free ATP, respectively, and [ATP] is the molar concentration of ATP added to the reaction.
Data availability
All data are in the manuscript.
Author contributions
U. B. and S. S. P. conceptualization; U. B., N. M., M. F., J. S., and L. C. J. data curation; U. B., J. S., and S. S. P. formal analysis; U. B. and S. S. P. supervision; U. B. and S. S. P. funding acquisition; U. B., N. M., M. F., and J. S. investigation; U. B., N. M., M. F., and J. S. methodology; U. B. writing-original draft; U. B., N. M., M. F., J. S., L. C. J., and S. S. P. writing-review and editing; N. M., M. F., J. S., L. C. J., and S. S. P. validation; J. S. resources; S. S. P. visualization; S. S. P. project administration.
Supplementary Material
Acknowledgments
We thank the Patel laboratory members for advice and suggestions regarding this work.
This work was supported by American Heart Association Grant 16PRE30400001 and University and Louis Bevier dissertation completion fellowship from Rutgers University (to U. B.) and NIGMS, National Institutes of Health Grant GM118086 (to S. S. P.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1–S6.
- RNAP
- RNA polymerase
- aa
- amino acids
- ssDNA
- single-stranded DNA
- LSP
- light strand promoter
- BLI
- biolayer interferometry
- NT
- non-template
- nt
- nucleotide
- POLRMT
- Polymerase mitochondrial.
References
- 1. Cermakian N., Ikeda T. M., Miramontes P., Lang B. F., Gray M. W., and Cedergren R. (1997) On the evolution of the single-subunit RNA polymerases. J. Mol. Evol. 45, 671–681 10.1007/PL00006271 [DOI] [PubMed] [Google Scholar]
- 2. Jang S. H., and Jaehning J. A. (1991) The yeast mitochondrial RNA polymerase specificity factor, MTF1, is similar to bacterial sigma factors. J. Biol. Chem. 266, 22671–22677 [PubMed] [Google Scholar]
- 3. Basu U., Lee S. W., Deshpande A., Shen J., Sohn B. K., Cho H., Kim H., and Patel S. S. (2020) The C-terminal tail of the yeast mitochondrial transcription factor Mtf1 coordinates template strand alignment, DNA scrunching and timely transition into elongation. Nucleic Acids Res. 48, 2604–2620 10.1093/nar/gkaa040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Falkenberg M., Gaspari M., Rantanen A., Trifunovic A., Larsson N. G., and Gustafsson C. M. (2002) Mitochondrial transcription factors B1 and B2 activate transcription of human mtDNA. Nat. Genet. 31, 289–294 10.1038/ng909 [DOI] [PubMed] [Google Scholar]
- 5. Litonin D., Sologub M., Shi Y., Savkina M., Anikin M., Falkenberg M., Gustafsson C. M., and Temiakov D. (2010) Human mitochondrial transcription revisited: only TFAM and TFB2M are required for transcription of the mitochondrial genes in vitro. J. Biol. Chem. 285, 18129–18133 10.1074/jbc.C110.128918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Shutt T. E., Bestwick M., and Shadel G. S. (2011) The core human mitochondrial transcription initiation complex: it only takes two to tango. Transcription 2, 55–59 10.4161/trns.2.2.14296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ramachandran A., Basu U., Sultana S., Nandakumar D., and Patel S. S. (2017) Human mitochondrial transcription factors TFAM and TFB2M work synergistically in promoter melting during transcription initiation. Nucleic Acids Res. 45, 861–874 10.1093/nar/gkw1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hillen H. S., Morozov Y. I., Sarfallah A., Temiakov D., and Cramer P. (2017) Structural basis of mitochondrial transcription initiation. Cell 171, 1072–1081.e10 10.1016/j.cell.2017.10.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Schubot F. D., Chen C. J., Rose J. P., Dailey T. A., Dailey H. A., and Wang B. C. (2001) Crystal structure of the transcription factor sc-mtTFB offers insights into mitochondrial transcription. Protein Sci. 10, 1980–1988 10.1110/ps.11201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cotney J., and Shadel G. S. (2006) Evidence for an early gene duplication event in the evolution of the mitochondrial transcription factor B family and maintenance of rRNA methyltransferase activity in human mtTFB1 and mtTFB2. J. Mol. Evol. 63, 707–717 10.1007/s00239-006-0075-1 [DOI] [PubMed] [Google Scholar]
- 11. Paratkar S., and Patel S. S. (2010) Mitochondrial transcription factor Mtf1 traps the unwound non-template strand to facilitate open complex formation. J. Biol. Chem. 285, 3949–3956 10.1074/jbc.M109.050732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Guccione E., and Richard S. (2019) The regulation, functions and clinical relevance of arginine methylation. Nat. Rev. Mol. Cell Biol. 20, 642–657 10.1038/s41580-019-0155-x [DOI] [PubMed] [Google Scholar]
- 13. Hornbeck P. V., Zhang B., Murray B., Kornhauser J. M., Latham V., and Skrzypek E. (2015) PhosphoSitePlus, 2014: mutations, PTMs and recalibrations. Nucleic Acids Res. 43, D512–520 10.1093/nar/gku1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kozlov A. G., Cox M. M., and Lohman T. M. (2010) Regulation of single-stranded DNA binding by the C termini of Escherichia coli single-stranded DNA-binding (SSB) protein. J. Biol. Chem. 285, 17246–17252 10.1074/jbc.M110.118273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Marintcheva B., Marintchev A., Wagner G., and Richardson C. C. (2008) Acidic C-terminal tail of the ssDNA-binding protein of bacteriophage T7 and ssDNA compete for the same binding surface. Proc. Natl. Acad. Sci. U.S.A. 105, 1855–1860 10.1073/pnas.0711919105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Schwartz E. C., Shekhtman A., Dutta K., Pratt M. R., Cowburn D., Darst S., and Muir T. W. (2008) A full-length group 1 bacterial σ factor adopts a compact structure incompatible with DNA binding. Chem. Biol. 15, 1091–1103 10.1016/j.chembiol.2008.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tang G. Q., Paratkar S., and Patel S. S. (2009) Fluorescence mapping of the open complex of yeast mitochondrial RNA polymerase. J. Biol. Chem. 284, 5514–5522 10.1074/jbc.M807880200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tang G. Q., Deshpande A. P., and Patel S. S. (2011) Transcription factor-dependent DNA bending governs promoter recognition by the mitochondrial RNA polymerase. J. Biol. Chem. 286, 38805–38813 10.1074/jbc.M111.261966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Velazquez G., Sousa R., and Brieba L. G. (2015) The thumb subdomain of yeast mitochondrial RNA polymerase is involved in processivity, transcript fidelity and mitochondrial transcription factor binding. RNA Biol. 12, 514–524 10.1080/15476286.2015.1014283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Posse V., and Gustafsson C. M. (2017) Human mitochondrial transcription factor B2 Is required for promoter melting during initiation of transcription. J. Biol. Chem. 292, 2637–2645 10.1074/jbc.M116.751008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bird J. G., Basu U., Kuster D., Ramachandran A., Grudzien-Nogalska E., Towheed A., Wallace D. C., Kiledjian M., Temiakov D., Patel S. S., Ebright R. H., and Nickels B. E. (2018) Highly efficient 5′ capping of mitochondrial RNA with NAD(+) and NADH by yeast and human mitochondrial RNA polymerase. Elife 10.7554/eLife.42179 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are in the manuscript.