Abstract
Effective treatments for coronavirus disease 2019 (COVID-19) are urgently needed to control this current pandemic, caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Replication of SARS-CoV-2 depends on the viral RNA-dependent RNA polymerase (RdRp), which is the likely target of the investigational nucleotide analogue remdesivir (RDV). RDV shows broad-spectrum antiviral activity against RNA viruses, and previous studies with RdRps from Ebola virus and Middle East respiratory syndrome coronavirus (MERS-CoV) have revealed that delayed chain termination is RDV's plausible mechanism of action. Here, we expressed and purified active SARS-CoV-2 RdRp composed of the nonstructural proteins nsp8 and nsp12. Enzyme kinetics indicated that this RdRp efficiently incorporates the active triphosphate form of RDV (RDV-TP) into RNA. Incorporation of RDV-TP at position i caused termination of RNA synthesis at position i+3. We obtained almost identical results with SARS-CoV, MERS-CoV, and SARS-CoV-2 RdRps. A unique property of RDV-TP is its high selectivity over incorporation of its natural nucleotide counterpart ATP. In this regard, the triphosphate forms of 2′-C-methylated compounds, including sofosbuvir, approved for the management of hepatitis C virus infection, and the broad-acting antivirals favipiravir and ribavirin, exhibited significant deficits. Furthermore, we provide evidence for the target specificity of RDV, as RDV-TP was less efficiently incorporated by the distantly related Lassa virus RdRp, and termination of RNA synthesis was not observed. These results collectively provide a unifying, refined mechanism of RDV-mediated RNA synthesis inhibition in coronaviruses and define this nucleotide analogue as a direct-acting antiviral.
Keywords: RNA polymerase, plus-stranded RNA virus, drug action, drug development, drug discovery, COVID-19, coronavirus (CoV), MERS, SARS, SARS-CoV-2, Ebola virus, Lassa virus, RNA-dependent RNA polymerase (RdRp), replication, remdesivir, sofosbuvir, favipiravir, ribavirin
Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)3 is a positive-sense RNA virus and the causative agent of coronavirus disease 2019 (COVID-19) (1, 2). The initial outbreak in December 2019 in Wuhan, China, was declared a pandemic by the World Health Organization on March 10, 2020 (3). Antiviral treatments are urgently needed to relieve the burden on healthcare systems worldwide. Effective therapeutics are expected to reduce mortality and hospitalizations. In the absence of a vaccine, antiviral therapeutics could also be utilized prophylactically to protect vulnerable populations, including those who are frequently exposed to the virus. Despite the earlier outbreaks of SARS in 2003 and Middle East respiratory syndrome (MERS) in 2012, coronavirus-specific antivirals have yet to advance into clinical trials. At this point, the focus is on compounds with a demonstrated broad spectrum of antiviral activities and on drugs developed for other therapeutic purposes with evidence of acting also against coronaviruses. Several of these compounds are currently being assessed in randomized controlled clinical trials, including remdesivir (RDV; formerly GS-5734) (4).
RDV is a phosphoramidate prodrug of a 1′-cyano-substituted nucleotide analogue (5). Its triphosphate form (RDV-TP) resembles ATP and is used as a substrate of several viral RNA-dependent RNA polymerase (RdRp) enzymes or complexes (6–9). The compound has shown broad-spectrum in vitro and in vivo antiviral activity against nonsegmented negative-sense RNA viruses of the Filoviridae (e.g. Ebola virus (EBOV)) (5, 8) and Paramyxoviridae (e.g. Nipah virus (NiV)) families (7, 10, 11), as well as in vitro activity against viruses in the Pneumoviridae (e.g. respiratory syncytial virus (RSV)) family (10). Antiviral activity against a broad spectrum of coronaviruses, including SARS-CoV and MERS-CoV, was subsequently demonstrated both in vitro and in animal models (6, 12–15). No in vitro inhibition was reported for several segmented negative-sense RNA viruses of the Arenaviridae family (e.g. Lassa virus (LASV)) and the Bunyavirals order (formerly the family Bunyaviridae; e.g. Crimean Congo hemorrhagic fever virus) (10). RDV was also recently tested in a randomized controlled trial during the 2019 Ebola outbreak in the Democratic Republic of the Congo (16). Although two antibody-based treatments showed superior efficacy, mortality in the RDV arm was lower than the overall mortality rate of the outbreak, and human safety data are now available (16). Inhibition of MERS-CoV replication and therapeutic efficacy of RDV was also demonstrated in mouse and rhesus macaque models (13, 15).
Progress has been made in elucidating the mechanism of action of RDV-TP. RDV-TP competes with ATP for incorporation by the EBOV RdRp complex composed of the L protein and VP35 (9). Steady-state kinetics reveal that incorporation of ATP is slightly more efficient compared with RDV-TP. In contrast to classic chain terminators, inhibition is not seen immediately following the incorporated RDV-TP, and the existence of a 3′-OH group allows the nucleophilic attack on the next incoming nucleotide. Studies with EBOV RdRp, RSV RdRp, and NiV RdRp have indicated that RNA synthesis is terminated after a few more nucleotide incorporation events (7, 9). RDV-TP incorporation at position i commonly yields delayed chain termination between positions i+3 and i+5. We have recently expressed and purified an active complex composed of MERS nonstructural proteins nsp8 and nsp12 (17). With the limitations of steady-state kinetic measurements, we showed that incorporation of RDV-TP is more efficient than ATP, and delayed chain termination is observed specifically at position i+3. Potential inhibitory effects of RDV-TP on SARS-CoV RdRp or on SARS-CoV-2 RdRp have not been studied. Active SARS-CoV RdRp was shown to form a complex composed of nsp7, nsp8, and nsp12, whereby nsp7 and nsp8 were connected with a linker (18). Structures of complexes with the three subunits were recently determined using cryo-EM (19).
Here, we expressed SARS-CoV and SARS-CoV-2 RdRp complexes in insect cells and monitored RNA synthesis on short model primer/templates during elongation. We demonstrate that RDV-TP inhibits SARS-CoV RdRp and SARS-CoV-2 RdRp with the same potency and mechanism of action. The ability to efficiently compete with the natural counterpart ATP is a favorable property of RDV-TP. We have tested in the same context several other nucleotide analogues and commonly observe that incorporation of the natural nucleotide is considerably preferred. Favorable selectivity for the RDV-TP over ATP and delayed chain termination at position i+3 are key elements of a refined mechanism of inhibition observed with SARS-CoV, MERS-CoV, and SARS-CoV-2 RdRp complexes.
Results
Expression of SARS-CoV and SARS-CoV-2 RdRp complexes
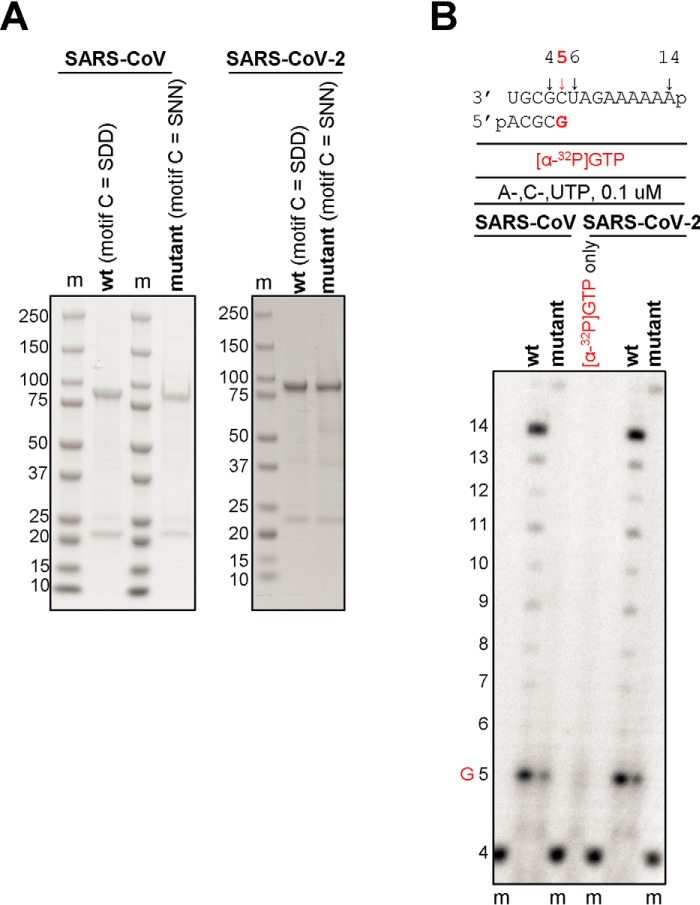
We have recently generated recombinant MERS-CoV RdRp from a bacmid containing nsp5, -7, -8, and -12 (17). Expression of this construct in insect cells resulted in the nsp5-7-8-12 polyprotein processed by the nsp5 protease. Nickel-nitrilotriacetic acid affinity chromatography yielded an active binary nsp8/12 RdRp complex. We therefore expressed SARS-CoV and SARS-CoV-2 RdRp from bacmids containing the equivalent nsp5, -7, -8, and -12 sequences. This approach yielded binary complexes with nsp8 and nsp12, as described previously for MERS-CoV (17). We also expressed mutant enzymes with amino acid substitutions in the conserved motif C of nsp12 (SDD to SNN) to inactivate the catalytic site (Fig. 1A). WT and mutant enzymes were tested for RNA synthesis on short model primer/templates mimicking a random elongation complex. Incorporation of a radiolabeled nucleotide allows gel-based detection of reaction products. Whereas WT SARS-CoV and SARS-CoV-2 RdRp complexes were able to synthesize RNA from a 4-mer primer and a 14-mer template, the corresponding active-site mutants showed no template-base–specific nucleotide incorporation (Fig. 1B). These data confirm that RNA synthesis activity is mediated by nsp12.
Figure 1.
Expression, purification, and characterization of the SARS-CoV and SARS-CoV-2 RdRp complexes. A, SDS-PAGE migration pattern of the purified enzyme preparations stained with Coomassie Brilliant Blue G-250 dye. Bands migrating at ∼100 kDa and ∼25 kDa contain nsp12 and nsp8, respectively. B, RNA synthesis on a short model primer/template substrate. Template and primer were both phosphorylated (p) at their 5′-ends. A radiolabeled 4-mer primer serves as a marker (m). G indicates incorporation of the radiolabeled nucleotide opposite template position 5. RNA synthesis was monitored with the purified RdRp complexes representing WT (wt, motif C = SDD) and the active-site mutant (motif C = SNN).
Selectivity measurements of RDV-TP with related and distant RdRp enzymes
The ratio of Michaelis–Menten steady-state kinetic parameters Vmax/Km for a single incorporation of a natural nucleotide over a nucleotide analogue defines the selectivity. With the limitations of a steady-state approach, a selectivity value lower than 1 suggests that the analogue is incorporated more efficiently than the natural NTP. Conversely, a selectivity value higher than 1 suggests that the analogue is incorporated less efficiently than the natural NTP. This approach enables comparisons of data with different enzymes and different nucleotide analogues. This approach does not provide distinct information on inhibitor binding, catalysis, or enzyme dissociation from its nucleic acid substrate. To measure selectivity for RDV-TP incorporation, we determined the steady-state kinetic parameters for single nucleotide incorporations compared with ATP (Fig. S1 and Table 1). Previously, we reported a selectivity value of 0.35 for RDV-TP incorporation with MERS-CoV RdRp (17). SARS-CoV and SARS-CoV-2 also showed low values in a similar range (0.32 and 0.26, respectively). For EBOV, RSV, and LASV enzymes, we measured higher values (4.0, 2.7, and 23, respectively). Because the RNA template used to measure selectivity in this study differs from the sequence previously used to study inhibition of EBOV and RSV RdRp (9), we repeated these experiments with EBOV and the current RNA template. The observed selectivity value of 4 is in good agreement with our previous measurement of 3.8 (9). LASV RdRp showed the highest selectivity value for RDV-TP of 23-fold, which provides evidence for target specificity. The combined results suggest that the ability of RDV-TP to compete with ATP is most pronounced with the coronavirus RdRp complexes.
Table 1.
Selectivity values for Remdesivir (RDV-TP) with related and distant RdRp enzymes
| ATP |
RDV-TP |
Selectivityc | |||||
|---|---|---|---|---|---|---|---|
| Vmaxa (product fraction) | Kmb | Vmax/Km | Vmax (product fraction) | Km | Vmax/Km | ||
| μm | μm | -fold | |||||
| MERS-CoVd | n = 7 | n = 6 | 0.35 | ||||
| 0.47 | 0.017 | 28 | 0.50 | 0.0063 | 79 | ||
| ±e | 0.011 | 0.0019 | 0.012 | 0.0006 | |||
| % errorf | 2 | 11 | 2 | 11 | |||
| SARS-CoV | n = 4g | n = 3 | 0.32 | ||||
| 0.73 | 0.03 | 25 | 0.70 | 0.010 | 68 | ||
| ± | 0.017 | 0.003 | 6.3 | 0.015 | 0.0008 | 5.4 | 0.026 |
| % error | 6 | 23 | 25 | 11 | 3 | 8 | 8 |
| SARS-CoV-2 | n = 8 | n = 3 | 0.28 | ||||
| 0.75 | 0.03 | 23 | 0.74 | 0.0089 | 84 | ||
| ± | 0.019 | 0.003 | 4.4 | 0.023 | 0.0010 | 14.3 | 0.045 |
| % error | 10 | 22 | 20 | 4 | 18 | 17 | 16 |
| EBOV | n = 3 | n = 3 | 4.0 | ||||
| 0.80 | 0.72 | 1.1 | 0.70 | 2.5 | 0.28 | ||
| ± | 0.048 | 0.21 | 0.065 | 0.047 | 0.76 | 0.048 | 0.49 |
| % error | 4 | 6 | 6 | 2 | 16 | 17 | 12 |
| RSVh | n = 3 | n = 3 | 2.7 | ||||
| 0.76 | 0.17 | 4.5 | 0.82 | 0.50 | 1.6 | ||
| ± | 0.022 | 0.023 | 0.027 | 0.089 | |||
| % error | 3 | 14 | 3 | 18 | |||
| LASV | n = 3 | n = 3 | 20 | ||||
| 0.57 | 0.11 | 5.6 | 0.35 | 1.3 | 0.29 | ||
| ± | 0.032 | 0.020 | 0.96 | 0.016 | 0.18 | 0.059 | 4.7 |
| % error | 2 | 18 | 17 | 40 | 53 | 20 | 24 |
a Vmax is a Michaelis–Menten parameter reflecting the maximal velocity of nucleotide incorporation.
b Km is a Michaelis–Menten parameter reflecting the concentration of the nucleotide substrate at which the velocity of nucleotide incorporation is half of Vmax.
c Selectivity of a viral RNA polymerase for a nucleotide substrate analogue is calculated as the ratio of the Vmax/Km values for NTP and NTP analogue, respectively.
d Data from Gordon et al. (17).
e S.D. of the average.
f Percent error.
g All reported values have been calculated on the basis of a 9-data point experiment repeated the indicated number of times (n).
h Data from Tchesnokov et al. (9).
Selectivity of other nucleotide analogues against SARS-CoV-2 RdRp
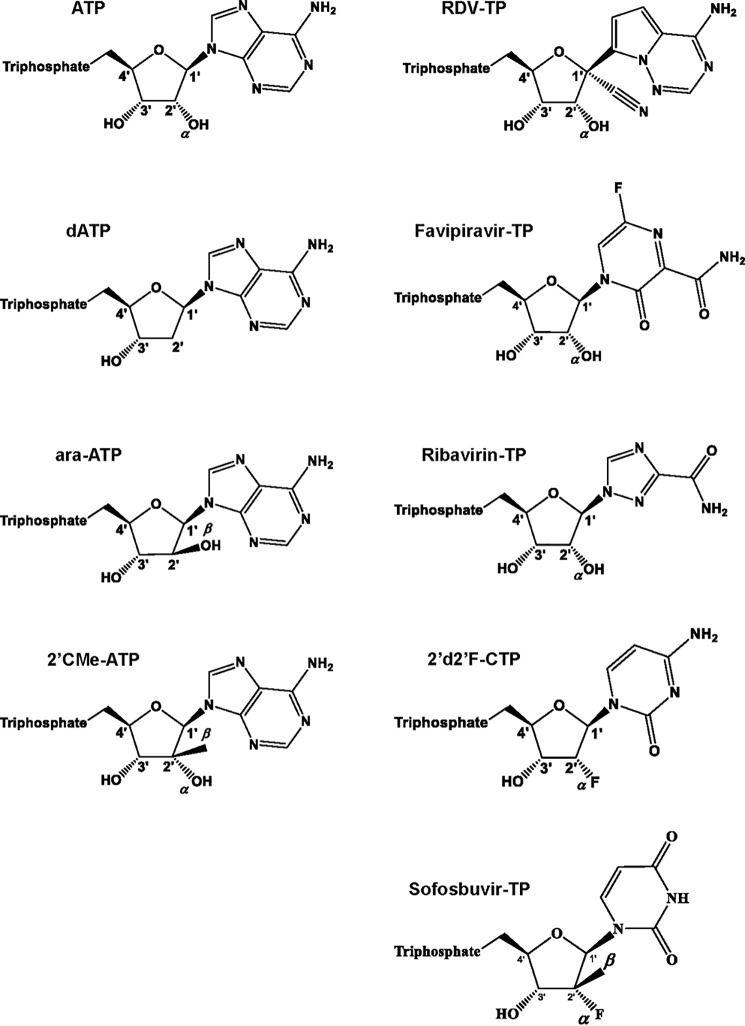
We determined selectivity values for various other nucleotide analogue inhibitors to provide a limited structure-activity relationship analysis with focus on SARS-CoV-2 RdRp (Fig. 2). Selectivity for dATP, which lacks the 2′α-hydroxyl group, 2′-C-methylated compounds, and the broad-spectrum antivirals favipiravir and ribavirin were included in these studies (Table 2). A high selectivity value of ∼950-fold was measured for dATP, which shows that the enzyme efficiently discriminates against dNTPs as one would expect for an RNA polymerase. Similarly, the 2′β-hydroxyl in ara-ATP also translates in a high selectivity value (>1000-fold). 2′-CMe-ATP contains a 2′β-methyl group, which shows a value of ∼170. 2-Deoxy-2′fluoro-CTP has been shown to exhibit antiviral activity against several RNA viruses (20–23). The selectivity value of 25 is relatively low compared with the aforementioned compounds and suggests that the 2′α-fluoro is largely tolerated by the enzyme. Sofosbuvir (SOF) is a uridine analogue that is approved for the treatment of hepatitis C virus (HCV) infection. The drug inhibits HCV RdRp (24). It contains a fluoro group at the 2′α-position and a methyl group at the 2′β-position. Here we measured a high selectivity value (∼1000-fold), which suggests that the 2′β-position may be the primary constraint for nucleotide incorporation by SARS-CoV-2 RdRp. The active triphosphate forms of favipiravir and ribavirin serve as substrates for several RdRp enzymes and mimic ATP and GTP (25–30). To compare efficiency of incorporation with RDV-TP, we measured selectivity values for incorporation opposite template uridine. High values of ∼500 and ≫1000, respectively, indicate that effective competition of these compounds with ATP is unlikely.
Figure 2.
Chemical structures of ATP and ATP, CTP, and UTP nucleotide analogues used in this study.
Table 2.
Selectivity values for ATP, CTP, and UTP analogues against SARS-CoV-2 RdRp
| Vmaxa (product fraction) | Kmb | Vmax/Km | Selectivityc | |
|---|---|---|---|---|
| μm | -fold | |||
| ATPd (n = 8e) | 0.75 | 0.03 | 23 | Reference |
| ±f | 0.019 | 0.004 | 4.4 | |
| % errorg | 10 | 22 | 20 | |
| 2′-CMe-ATP (n = 3) | 0.84f | 6.4 | 0.13 | 173 |
| ± | 0.013 | 0.37 | 0.007 | 8.9 |
| % error | 1 | 5 | 5 | 5 |
| dATP (n = 3) | 0.63 | 27 | 0.02 | 975 |
| ± | 0.021 | 2.41 | 0.04 | 169 |
| % error | 11 | 28 | 16 | 17 |
| ara-ATP (n = 3) | 0.56 | 33 | 0.02 | 1329 |
| ± | 0.031 | 4.06 | 0.01 | 371 |
| % error | 15 | 39 | 30 | 28 |
| ATPh (n = 3) | 0.63 | 0.04 | 14 | Reference |
| ± | 0.019 | 0.006 | 1.3 | |
| % error | 3 | 12 | 9 | |
| Favipiravir-TP (n = 3) (as ATP analogue) | 0.52 | 21 | 0.03 | 570 |
| ± | 0.031 | 3.9 | 0.02 | 230 |
| % error | 7 | 41 | 52 | 40 |
| Ribavirin (n = 3) (as ATP analogue) | NAi | ≫g100 | NA | ≫1000 |
| ± | ||||
| % error | ||||
| CTP (n = 3) | 0.72 | 0.001 | 1022 | Reference |
| ± | 0.023 | 8.8 × 10−5 | 177 | |
| % error | 1 | 18 | 17 | |
| 2′d 2′ fluoro-CTP (n = 3) | 0.60 | 0.02 | 37 | 29 |
| ± | 0.036 | 0.003 | 8.5 | 7.6 |
| % error | 17 | 36 | 23 | 26 |
| UTP (n = 3) | 0.75 | 0.02 | 39 | Reference |
| ± | 0.021 | 0.003 | 9.3 | |
| % error | 4 | 22 | 24 | |
| SOF-TP (n = 3) | 0.77 | 21 | 0.04 | 1056 |
| ± | 0.030 | 2.3 | 0.01 | 212 |
| % error | 6 | 22 | 22 | 20 |
a Vmax is a Michaelis–Menten parameter reflecting the maximal velocity of nucleotide incorporation.
b Km is a Michaelis–Menten parameter reflecting the concentration of the nucleotide substrate at which the velocity of nucleotide incorporation is half of Vmax.
c Selectivity of a viral RNA polymerase for a nucleotide substrate analogue is calculated as the ratio of the Vmax/Km values for NTP and NTP analogue, respectively.
d These experiments were conducted on RNA template compatible with [α-32P]GTP incorporation at position 5.
e All reported values have been calculated on the basis of a 9-data point experiment repeated the indicated number of times (n).
f S.D. of the average.
g Percent error.
h These experiments were conducted on RNA template compatible with [α-32P]CTP incorporation at position 5.
i NA, not available.
Structural model of nucleotide binding by SARS-CoV-2 RdRp
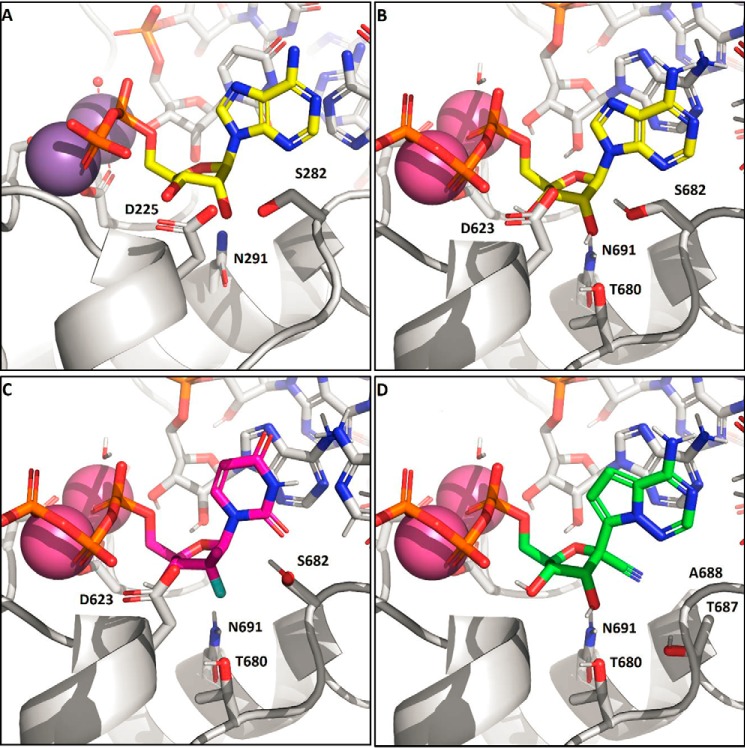
To provide plausible explanations for our experimental measurements, we generated a model of an elongating SARS-CoV-2 RdRp complex (Fig. 3). This model is based on the cryo-EM structure of the apo SARS-Co RdRp complex composed of nsp7, nsp8, and nsp12 (19). The active site is similar to other enzymes, for which ternary structures have been determined by X-ray crystallography, including HCV RdRp (Fig. 3A), norovirus, and poliovirus RdRp (24, 31, 32). The catalytic metal ions in SARS-CoV-2 nsp12 are coordinated by a trio of aspartates, Asp-618, Asp-760, and Asp-761, and the substrate β-phosphate is stabilized by Arg-555. Converting Asp-760 and Asp-761 into Asn-760 and Asn-761 rendered the enzyme inactive (Fig. 1). Of particular note, the residues Asp-623, Ser-682, and Asn-691 involved in 2′-OH recognition of the incoming nucleotide are conserved. However, whereas HCV, norovirus, and poliovirus RdRps use these serine and asparagine residues to coordinate the 2′-OH during incorporation, nsp12 appears to rely on an additional threonine residue (Thr-680) not present in the others enzymes (Fig. 3B). This interaction has the effect of pulling the substrate deeper into the pocket. The lower positioning of the NTP in the active site is also seen in the interaction of the 3′-OH with the protein. In HCV RdRp, 3′-OH forms a hydrogen bond to the Asp-225 backbone NH, whereas in the coronaviruses, the model instead suggests that the β-phosphate coordinates to the analogous Asp-623 NH. As a consequence of this repositioning of the substrate, the activity of various inhibitors across the coronaviruses is expected to diverge from that seen with other polymerases. The preference for 2′-OH > 2′d2′F ≫ 2′deoxy is evident from the nature of the polar residues coordinating that position and is similar to the other enzymes. In contrast, SOF-TP and 2′-CMe-ATP have increased clashes between the 2′β-Me and Asp-623 and Ser-682 (Fig. 3C). This is partially relieved by a conformational change in Ser-682, but overall, the poorer incorporation efficiencies of the 2′β-Me–substituted inhibitors is likely due to these putative steric clashes. In contrast, RDV-TP (Fig. 3D) is recognized at the 2′-OH in a manner similar to ATP, and the 1′-CN modification is well-positioned in a pocket formed between Thr-687 and Ala-688. The high efficiency of incorporation reported here is consistent with this model.
Figure 3.
A, X-ray structure of HCV RdRp with an incoming nonhydrolyzable ADP substrate (PDB entry 4WTD). The 2′-OH of the substrate is recognized by the trio of residues, Asp-225, Ser-282, and Asn-291, with hydrogen bonds formed to Ser-282 and Asn-291 in this preincorporation state. B, model of SARS-CoV-2 nsp12 with incoming ATP. In addition to the analogous Asp/Ser/Asn residues, Thr-680 is positioned to alter the hydrogen-bonding network and effectively pull the substrate lower into the pocket relative to NS5B. C, model of SARS-CoV-2 with SOF-TP. The greater occlusion of the 2′ position due to Asp-623 and Ser-682 makes 2′β-methyl substitution less effective than with NS5B. D, model of SARS-CoV-2 with remdesivir-TP. The remdesivir 1′CN sits in a pocket formed by residues Thr-687 and Ala-688. Residues Asp-623 and Ser-682 (not shown) adopt the same conformations as with ATP.
Patterns of inhibition of RNA synthesis by RDV-TP
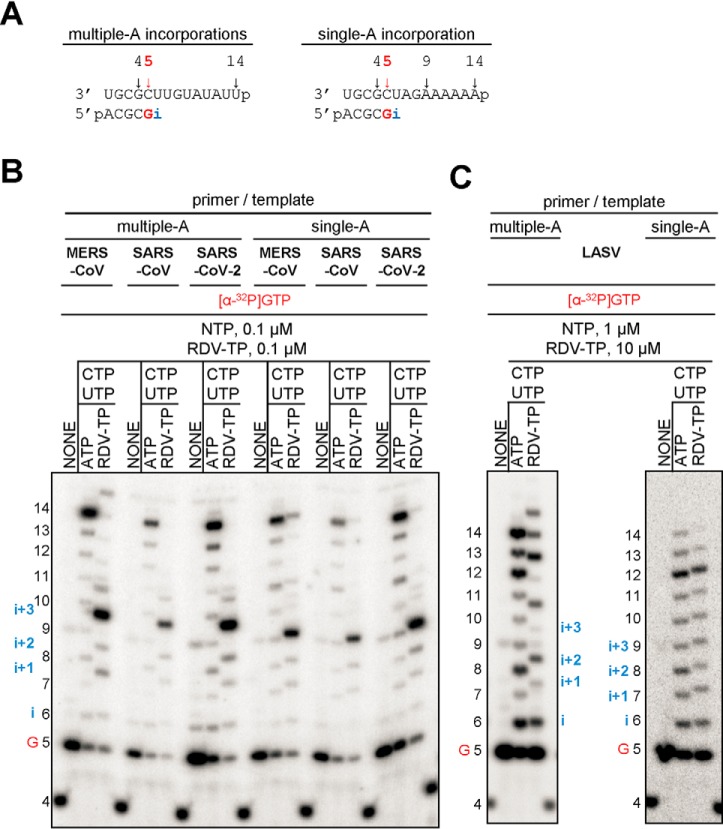
The incorporation of a nucleotide analogue into the growing RNA chain does not necessarily translate into inhibition. To determine the patterns of inhibition of RNA synthesis, we devised two RNA templates that contain single or multiple sites of incorporation (template uridines) (Fig. 4A). These sequences were used to compare the inhibitory effects of RDV-TP against MERS-CoV, SARS-CoV, SARS-CoV-2, and LASV RdRp. The latter enzyme showed a significantly higher selectivity value for RDV-TP (Table 1), and RDV does not show antiviral activity against LASV (10). For CoV RdRp complexes with templates allowing multiple incorporations, we observe termination of RNA synthesis at positions i+3 and i+4. A faint band representing the full-length product suggests low levels of read-through (Fig. 4B). On a template that allows only a single RDV-TP incorporation, termination is seen solely at position i+3. Full-length product formation is generally more pronounced under these conditions, following the order MERS-CoV RdRp > SARS-CoV RdRp > SARS-CoV-2 RdRp. For LASV RdRp, RNA synthesis patterns in the presence or absence of the nucleotide analogue are very similar. Chain termination or delayed chain termination is not evident. To further investigate the possibility of long-range effect of RDV-TP incorporation, we used a 26-mer RNA template and did not observe any change in the pattern of RNA synthesis (Fig. S3). Hence, LASV L protein is likely not inhibited by RDV-TP.
Figure 4.
Patterns of inhibition of RNA synthesis with RDV-TP. A, RNA primer/template substrates used to test multiple (left) or single (right) incorporations of RDV-TP. G indicates incorporation of the radiolabeled nucleotide opposite template position 5. RDV-TP incorporation is indicated by i. RDV-TP incorporation was monitored with purified CoV RdRp complexes (B) and LASV L protein (C) in the presence of the indicated combinations of NTPs and RDV-TP.
Examination of the structural model suggests that the primer with the incorporated RDV can translocate without obstruction to position i+1 (Fig. 5, A and B). Similarly, no obstructions can be discerned at the i+2 (Fig. 5C) or i+3 (Fig. 5D). This would allow the incorporation of three subsequent nucleotides, in agreement with our experimental data. However, at position i+4 (Fig. 5, E and F), a steric clash is seen between the 1′-CN substituent of RDV and residue Ser-861. The serine O is only 1.7 Å from the 1′-CN N. The short distance is expected to lead to a significant distortion of the positioning of the RNA, hampering translocation to the i+4 position.
Figure 5.
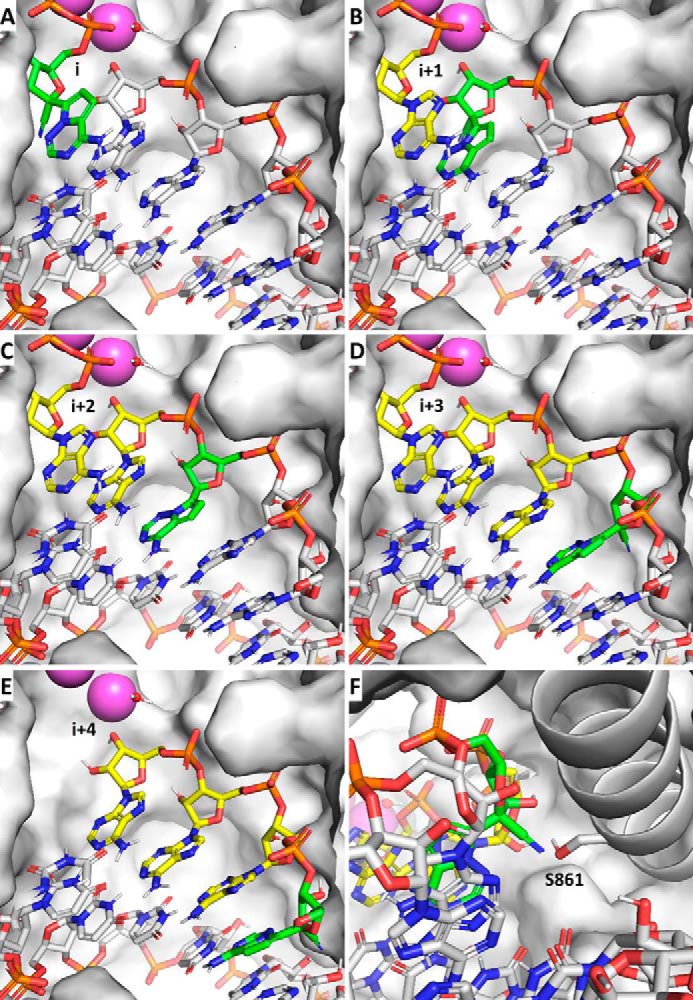
A steric clash between the incorporated RDV and Ser-861 prevents enzyme translocation at i+3. A–D, the primer with the incorporated RDV (green) translocates without obstruction from the substrate position i through i+3, allowing incorporation of three subsequent nucleotides (yellow). E, at i+4, the 1′-CN moiety of RDV encounters a steric clash with Ser-861 of nsp12. F, this clash likely prevents the enzyme from advancing into i+4.
Overcoming of the delayed chain termination by incorporated RDV
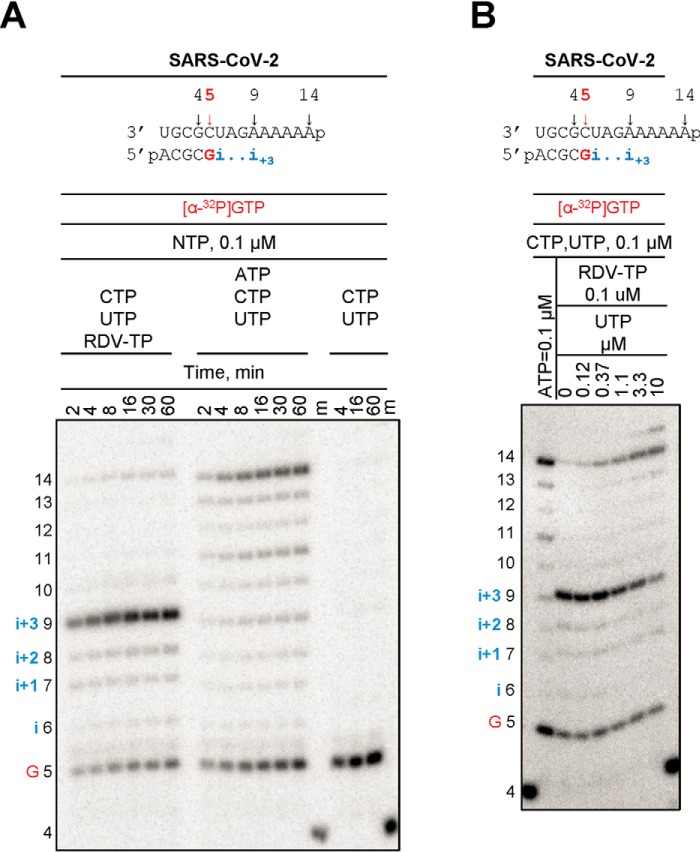
Read-through at a site of delayed chain termination may reduce the inhibitory effect of RDV-TP. We therefore tested whether termination of RNA synthesis could be overcome with time or with increasing concentrations of nucleotide pools (Fig. 6). In the presence of equivalent concentrations of RDV-TP and UTP (0.1 μm), which is the next nucleotide to be incorporated following the RDV-TP incorporation site and also following the site of delayed chain termination, the full-length product formation is negligible. The signal for delayed chain termination is not alleviated with time, which provides more evidence for a bona fide termination site and not enzyme pausing (Fig. 6A, left). In contrast, full-length product formation is time-dependent when ATP replaces RDV-TP in the reaction mixture (Fig. 6A, middle). As expected, the omission of both ATP and RDV-TP prevents RNA synthesis beyond product 5, thus controlling for C:U and U:U misincorporations under the present reaction conditions (Fig. 6A, right). However, delayed chain termination can be overcome with higher NTP concentrations. In the presence of increasing concentrations of UTP, the signal at i+3 decreases concomitantly with an increase in the full-length product. UTP concentrations that are ∼100-fold higher than RDV-TP can cause significant reductions in delayed chain termination (Fig. 6B).
Figure 6.
Overcoming of delayed chain termination. The RNA primer/template substrate used in this assay is shown above the gels. G indicates incorporation of the radiolabeled nucleotide opposite template position 5. Position i allows incorporation of ATP or RDV-TP. RNA synthesis was monitored with purified SARS-CoV-2 RdRp complex in the presence of indicated concentrations of NTP mixtures. A, time dependence of delayed chain termination. B, overcoming delayed chain termination with increasing concentrations of UTP.
Discussion
RDV is an investigational nucleotide analogue with a broad spectrum of antiviral activities against several RNA viruses, including filoviruses and coronaviruses (5, 8, 10, 12–14). Studies in mice and rhesus macaques have helped to assess the therapeutic potential of this drug against EBOV, SARS-CoV, and MERS-CoV (8, 13–15). Antiviral activity of RDV has also been demonstrated against SARS-CoV-2 in cell culture (33), whereas data from animal models and clinical trials are pending. Moreover, it remains to be seen whether the mechanism of inhibition described for EBOV RdRp and MERS RdRp is also relevant for SARS-CoV-2. Here, we expressed and purified active SARS-CoV-2 RdRp to study RNA synthesis and its inhibition by RDV-TP. Based on our biochemical data, we propose a unifying, refined mechanism of inhibition of SARS-CoV, MERS-CoV, and SARS-CoV-2 (Fig. 7).
Figure 7.
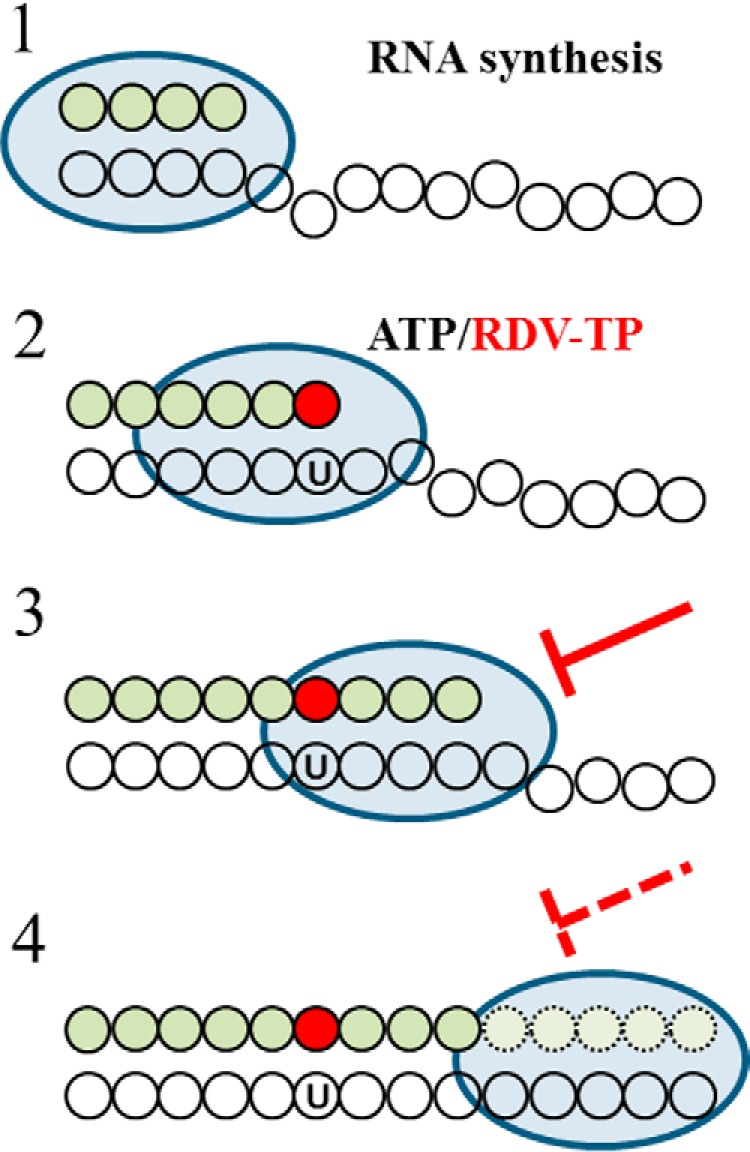
Mechanism of inhibition of CoV RdRp by RDV-TP. 1, the priming strand is shown with green circles, colorless circles represent residues of the template, and the blue oval represents the active CoV RdRp complex. This is a schematic representation of a random elongation complex. The footprint of RdRp on its primer/template is unknown. 2, competition of RDV-TP with its natural counterpart ATP opposite template uridine (U). The incorporated nucleotide analogue is illustrated by the red circle. 3, RNA synthesis is terminated after the addition of three more nucleotides, which is referred to as delayed chain termination. 4, delayed chain termination can be overcome by high ratios of NTP/RDV-TP.
Co-expression of SARS-CoV-2 nsp5, nsp7, nsp8, and nsp12 in insect cells yields an active RdRp complex composed of nsp8 and nsp12. The same data were obtained with MERS-CoV and SARS-CoV. RNA synthesis was monitored on short primer/templates that mimic the elongation stage (Fig. 7, step 1). We initially compared the efficiency of incorporation of RDV-TP with its natural counterpart ATP (Fig. 7, step 2). A steady-state kinetic approach was employed to translate our findings into quantitative terms and to facilitate comparisons among the various enzymes and compounds tested. Efficiency of incorporation of the natural nucleotide over the nucleotide analogue defines selectivity. For RDV-TP, we measured selectivity values of ∼0.3 with SARS-CoV, SARS-CoV-2, and previously also with MERS-CoV RdRp. SARS-CoV and SARS-CoV-2 both belong to the betacoronaviruses of the B lineage, and the nsp 12-amino acid sequences of the two viruses are 96% identical. In contrast, MERS-CoV belongs to the betacoronaviruses of the C lineage and is only 71% identical with SARS-CoV-2. Despite greater sequence variations, RdRp motifs that play important roles in substrate binding and catalysis are highly conserved among the three coronaviruses (Fig. S2). Hence, interactions with nucleotide analogue inhibitors are expected to be similar, and our selectivity data provide experimental evidence for this notion. In contrast, selectivity values for RDV-TP against EBOV and RSV RdRp are between 2 and 5, and LASV RdRp shows even higher selectivity values of ∼20. Whereas the active sites of EBOV and RSV enzymes still share a number of key residues within the classic polymerase motifs, LASV RdRp shows substantial differences when these motifs are compared with EBOV and RSV and also with SARS-CoV, SARS-CoV-2, and MERS-CoV (Fig. S2).
We have evaluated several other nucleotide analogues using the same protocol with a focus on SARS-CoV-2 RdRp (Fig. 2 and Table 2). The selectivity of ATP over dATP is ∼1000, which shows that the enzyme effectively discriminates against deoxyribonucleotides that are substrates for DNA polymerases. Similar results were obtained with the broad spectrum antivirals favipiravir and ribavirin (26, 27, 30). These compounds show high selectivity values of ∼500 and ∼10,000 in favor of ATP. Compounds with modifications at the 2′β-position are also associated with high selectivity values in favor of the natural nucleotide. SOF-TP and 2′-CMe-ATP show selectivity values of ∼1000 and ∼170 in favor of UTP and ATP, respectively. Selectivity of ATP over ara-ATP is likewise high (∼1000). A homology model of SARS-CoV-2 points to a putative steric clash of 2′β-modifications with residues Asp-623 and Ser-682. With the limitations of a steady-state approach, we are unable to ascribe differences in selectivity measurements solely to inhibitor binding; however, whereas the model is not based on structural data of SARS-CoV-2 RdRp, it provides a plausible explanation for our experimental observations.
RDV-TP is a nonobligate chain terminator, which contains a 3′-hydroxyl group that may still form a phosphodiester bond with the next incoming nucleotide. Indeed, as demonstrated for RdRp enzymes from RSV, EBOV, NiV, and MERS, delayed chain termination provides a likely mechanism of action (7–9, 17). For all three coronavirus RdRp complexes, we observe a specific termination site at position i+3 (Fig. 7, step 3). The structural reasons for the precise termination event remain to be elucidated; however, the underlying mechanism is likely to be common to all three coronaviruses if we consider the identical patterns of inhibition. At i+4, our model predicts a steric clash between the 1′-CN substituent of the incorporated RDV and residue Ser-861. This model is consistent with the observed termination at i+3 and the inability of the enzyme to translocate a single position further downstream to accommodate the next nucleotide. Sequences extracted from GenBankTM reveal that this serine residue is conserved across all alpha-, beta-, and deltacoronaviruses.
Termination of RNA synthesis can be overcome by higher concentrations of the natural nucleotide pools (Fig. 7, step 4). NTP can reach low millimolar concentrations (34, 35). The intracellular concentration of RDV-TP can vary between low and high micromolar levels in relevant cell cultures (8, 14), and high ratios of NTP/RDV-TP are likely detrimental to inhibition. However, the efficient incorporation of RDV-TP into the growing RNA chain may also provide a mechanism that counteracts reduced termination in the presence of high NTP concentrations. Our data confirm that consecutive and/or multiple sites of incorporation of RDV-TP increase termination and in turn inhibition; however, the efficiency of this effect remains to be determined (Fig. S2B). Another factor that can reduce the potency of nucleotide analogues is the 3′-5′ exonuclease activity of nsp14 (6, 36–38). This enzyme displays proofreading activity in conjunction with nsp10 (36, 37, 39). In this context, we have recently proposed that the additional three nucleotides that follow the incorporated RDV may provide protection from excision (17, 40). Future studies that take into account rates of nucleotide incorporation, rates of excision of multiple nucleotides, and the likelihood of RDV-TP reincorporation will be required to address this problem.
Several independent examples point to a significant correlation between the efficiency of selective incorporation of a given nucleotide analogue and the corresponding antiviral effect measured in cell culture. Half-maximal effective concentrations (EC50) of RDV against coronaviruses and filoviruses are in the submicromolar range, which is unusually low for a compound with a broad spectrum of antiviral activity (8, 10, 12, 14, 41). High potency in cell culture correlates with highly effective incorporation of RDV-TP with EBOV RdRp and, even more so, with the three coronavirus enzymes. Conversely, RDV shows a weak antiviral effect against LASV (10), and our biochemical data revealed low rates of incorporation by LASV RdRp. In this context, it is also important to note that our previous measurements with human mitochondrial RNA polymerase revealed high selectivity of ATP over RDV-TP (9), which is consistent with low levels of cytotoxicity of RDV (8, 14). Favipiravir-TP and ribavirin-TP are also less well-incorporated by SARS-CoV-2 RdRp. This is also evident with a template that offers multiple incorporation sites (Fig. S2B). In cell culture, these inhibitors often show EC50 values in the higher micromolar range depending on the nature of the RNA virus (26, 41, 42). High concentrations of favipiravir and ribavirin were also required to reduce infection with SARS-CoV-2 (EC50 = 109.50 μm and EC50 = 61.88 μm, respectively) (33). Moreover, ribavirin does not seem to provide clinical benefits in the context of SARS-CoV and MERS-CoV infection (41), 2′-CMe-ATP is not utilized as a substrate by EBOV RdRp, and 2′-C-methylated compounds show no significant antiviral effects in mini-genome replicons of EBOV (43). Equivalent studies are not yet available for coronaviruses; however, the high selectivity for the natural nucleotides over SOF-TP and 2′-CMe-ATP would not predict a potent antiviral effect.
In conclusion, the combined data provide evidence for a unifying mechanism of inhibition of RDV-TP against coronavirus RdRp. Favorable selectivity for the nucleotide analogue over its natural counterpart ATP and delayed chain termination at position i+3 are key elements of inhibition observed with SARS-CoV, MERS-CoV, and SARS-CoV-2 RdRp complexes. The availability of human safety data along with the antiviral studies in cell culture and in animal models as well as a clear mechanism of action provide a large body of evidence to justify the ongoing clinical trials with RDV for the treatment of COVID-19 (8, 10, 12–17, 33). The refined biochemical mechanism described in this study characterizes RDV as a direct-acting antiviral (DAA). This term was previously introduced to describe newer classes of HCV drugs that target a specific process in the viral life cycle (44), as opposed to older treatments with interferon and ribavirin that have been associated with multiple possible mechanisms (45). Compounds that are currently considered as potential treatments for COVID-19 include drugs approved for other conditions, repurposed drugs, and inhibitors with a broad spectrum of antiviral activities. Research into underlying mechanisms is needed to classify any of these compounds as a DAA.
Experimental procedures
Nucleic acids and chemicals
All RNA primers and templates used for in this study were 5′-phosphorylated and purchased from Dharmacon (Lafayette, CO). 2′-CMe-ATP, RDV-TP, and SOF-TP were provided by Gilead Sciences (Foster City, CA). Ara-ATP, 2′deoxy-2′fluoro-CTP was purchased from TriLink (San Diego, CA). Ribavirin-TP was purchased from Jena Bioscience (Jena, Germany). Favipiravir-TP was purchased from Toronto Research Chemicals (North York, Ontario, Canada). NTPs and dATP were purchased from GE Healthcare. [α-32P]GTP was purchased from PerkinElmer Life Sciences.
Protein expression and purification
The pFastBac-1 (Invitrogen, Burlington, Ontario, Canada) plasmid with the codon-optimized synthetic DNA sequences (GenScript, Piscataway, NJ) coding for a portion of 1ab polyproteins of SARS-CoV (NCBI: AAP33696.1) and SARS-CoV-2 (NCBI: QHD43415.1) containing only nsp5, nsp7, nsp8, and nsp12 were used as a starting material for protein expression in insect cells (Sf9, Invitrogen). We employed the MultiBac (Geneva Biotech, Indianapolis, IN) system for protein expression in insect cells (Sf9, Invitrogen) according to published protocols (46, 47). SARS-CoV and SARS-CoV-2 protein complexes were purified using nickel-nitrilotriacetic acid affinity chromatography of the nsp8 N-terminal 8-histidine tag according to the manufacturer's specifications (Thermo Scientific). Expression and purification of EBOV RdRp complex were performed as described previously (48). LASV L protein full-length (NCBI: AIT17397.1) was also expressed in insect cells through the Baculovirus expression system and purified using Strep-Tactin affinity chromatography (IBA Lifesciences, Göttingen, Germany) of the N-terminal Strep-tag according to the manufacturer's specifications (IBA Lifesciences). The identities of the purified LASV, SARS-CoV, and SARS-CoV-2 proteins were confirmed by MS analysis (Dr. Jack Moore, Alberta Proteomics and Mass Spectrometry, Edmonton, Alberta, Canada).
Evaluation of RNA synthesis and inhibition studies
RNA synthesis assays, data acquisition, and quantification were done as previously reported by us (9, 17, 48). The following 5′-monophosphorylated RNA templates were used in this study (the portion of the template that is complementary to the 4-nt primer is underlined): 3′-UGCGCUAGAAAAAAp for measurements of the selectivity values of ATP analogues and determination of the patterns of inhibition with the incorporated RDV at a single position; 3′-UGCGCUAGAGAGAGAGAGAGAGAGAGp for determination of the pattern of inhibition of LASV RdRp with the incorporated RDV at a single position but on a 26-nt template; 3′-UGCGCUUGUAUAUUp for determination of the patterns of inhibition with the consecutively incorporated RDV; 3′-UGCGGUACUUUAUUp for measurements of the selectivity values of favipiravir-TP and ribavirin-TP; 3′-UGCGCGUAAAAAAAp for measurements of the selectivity values of 2′-d2′fluoro-CTP, and 3′-UCGCGAUCUUUAUU for measurements of the selectivity values of sofosbuvir-TP.
Briefly, RNA synthesis assay consisted of mixing (final concentrations) Tris-HCl (pH 8, 25 mm), RNA primer (200 μm), RNA template (2 μm), [α-32P]NTP (0.1 μm), various concentrations and combinations (as indicated) of NTP and NTP analogues, and MERS-, SARS- and SARS-CoV-2 RdRp complex (∼0.1 μm) on ice. Reactions with LASV L protein contained ∼0.02 μm enzyme. Reaction mixtures (10 μl) were incubated for 10 min at 30 °C followed by the addition of 5 μl of MgCl2 (5 mm). Reactions were stopped after 30 min by the addition of 15 μl of formamide/EDTA (50 mm) mixture and incubated at 95 °C for 10 min. Data were collected and analyzed using GraphPad Prism 7.0 (GraphPad Software, Inc., San Diego, CA) as reported previously (9, 17, 48). Our assay involves incorporation of [α-32P]GTP into the primer. With this approach, it is not possible to quantify the fraction of the extended primer; however, the reaction products are clearly defined by the correct incorporation of [α-32P]GTP.
Structural model of elongating SARS-CoV-2 nsp12
A model of the SARS-CoV-2 replication complex in its elongating state was constructed based on the recent cryo-EM structure of the apo-SARS-CoV nsp12/nsp7/nsp8 complex (PDB entry 6NUR) (1). With high sequence identity (nsp12 = 96%, nsp7 = 98%, nsp8 = 97%), a homology model was generated with Prime (49, 50). To this, an X-ray structure of HCV RdRp in its elongating state (PDB entry 4WTD) (3) was aligned to the SARS-CoV-2 model. Rather than doing a whole structure alignment, key residues that interact with the RNA in the RNA binding channel were aligned. These include Ala-97, Phe-193, Gly-283, and Ser-367 in NS5B and Ser-501, Gly-590, Gly-683, and Ser-814 in nsp12. The two Mn2+ ions and the ADP substrate from the HCV RdRp structure were changed to Mg2+ and ATP, and these, as well as the aligned RNA, were added to the SARS-CoV-2 model. Several active-site residues were manually adjusted to accommodate the incoming metals and substrate following the NS5B structure. These residues include Lys-545, Arg-555, Asp-618, Ser-682, Gly-683, Asp-760, and Asp-761. This was followed by a series of constrained optimizations in both Prime and Macromodel (Schrödinger, LLC, New York). The process was repeated several times using different types of constraints, and each time a similar model resulted. RDV-TP and other inhibitors were modeled into the nsp12 active site, allowing only limited protein flexibility. Homology models of other coronaviruses, including SARS and MERS, were generated from this model with Prime.
Data availability
All data are included within this article.
Author contributions
C. J. G., E. P. T., and M. G. data curation; C. J. G., E. P. T., J. K. P., and M. G. software; C. J. G., E. P. T., and M. G. formal analysis; C. J. G., E. P. T., and M. G. validation; C. J. G., E. P. T., E. W., J. K. P., and M. G. investigation; C. J. G., E. P.T., and M. G. visualization; C. J. G., E. P. T., E. W., J. K. P., and M. G. methodology; E. P. T. and M. G. conceptualization; E. P. T. and M. G. writing-original draft; E. P. T., J. K. P., J. Y. F., D. P. P., and M. G. writing-review and editing; M. G. resources; M. G. supervision; M. G. funding acquisition; M. G. project administration.
Supplementary Material
Acknowledgments
We thank Dr. Jack Moore (Alberta Proteomics and Mass Spectrometry Facility) for MS analysis. We also thank Dr. Ulrike Brockstedt for valuable suggestions for the design of Fig. 7.
This study was supported by Canadian Institutes of Health Research (CIHR) Grant 170343, Gilead Sciences, and the Alberta Ministry of Economic Development, Trade, and Tourism Major Innovation Fund Program for the AMR–One Health Consortium (to M. G.). M. G. has previously received funding from Gilead Sciences in support of the study of EBOV RdRp inhibition by RDV. This study is also sponsored in part by a grant from Gilead Sciences to M. G., J. K. P., J. Y. F., and D. P. P. are Gilead employees.
This article was selected as one of our Editors' Picks.
This article contains Figs. S1–S3.
- SARS
- severe acute respiratory syndrome
- COV
- coronavirus
- RdRp
- RNA-dependent RNA polymerase
- RDV
- remdesivir
- TP
- triphosphate
- 2′-CMe-ATP
- 2′C-methyl-ATP
- SOF
- sofosbuvir
- COVID-19
- coronavirus disease 2019
- MERS
- Middle East respiratory syndrome
- EBOV
- Ebola virus
- NiV
- Nipah virus
- RSV
- respiratory syncytial virus
- LASV
- Lassa virus
- HCV
- hepatitis C virus
- DAA
- direct-acting antiviral
- PDB
- Protein Data Bank
- nt
- nucleotide.
References
- 1. Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N., Bi Y., Ma X., Zhan F., Wang L., Hu T., et al. (2020) Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet 395, 565–574 10.1016/S0140-6736(20)30251-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., et al. (2020) A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 382, 727–733 10.1056/NEJMoa2001017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. World Health Organization (2020) Coronavirus Disease 2019 (COVID-19) Situation Report-50, March 10, World Health Organization, Geneva [Google Scholar]
- 4. Li H., Zhou Y., Zhang M., Wang H., Zhao Q., and Liu J. (2020) Updated approaches against SARS-CoV-2. Antimicrob. Agents Chemother. 10.1128/AAC.00483-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Siegel D., Hui H. C., Doerffler E., Clarke M. O., Chun K., Zhang L., Neville S., Carra E., Lew W., Ross B., Wang Q., Wolfe L., Jordan R., Soloveva V., Knox J., et al. (2017) Discovery and synthesis of a phosphoramidate prodrug of a pyrrolo[2,1-f][triazin-4-amino] adenine C-nucleoside (GS-5734) for the treatment of Ebola and emerging viruses. J. Med. Chem. 60, 1648–1661 10.1021/acs.jmedchem.6b01594 [DOI] [PubMed] [Google Scholar]
- 6. Agostini M. L., Andres E. L., Sims A. C., Graham R. L., Sheahan T. P., Lu X., Smith E. C., Case J. B., Feng J. Y., Jordan R., Ray A. S., Cihlar T., Siegel D., Mackman R. L., Clarke M. O., et al. (2018) Coronavirus susceptibility to the antiviral remdesivir (GS-5734) is mediated by the viral polymerase and the proofreading exoribonuclease. mBio 9, e00221–18 10.1128/mBio.00221-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jordan P. C., Liu C., Raynaud P., Lo M. K., Spiropoulou C. F., Symons J. A., Beigelman L., and Deval J. (2018) Initiation, extension, and termination of RNA synthesis by a paramyxovirus polymerase. PLoS Pathog. 14, e1006889 10.1371/journal.ppat.1006889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Warren T. K., Jordan R., Lo M. K., Ray A. S., Mackman R. L., Soloveva V., Siegel D., Perron M., Bannister R., Hui H. C., Larson N., Strickley R., Wells J., Stuthman K. S., Van Tongeren S. A., et al. (2016) Therapeutic efficacy of the small molecule GS-5734 against Ebola virus in rhesus monkeys. Nature 531, 381–385 10.1038/nature17180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tchesnokov E. P., Feng J. Y., Porter D. P., and Götte M. (2019) Mechanism of inhibition of Ebola virus RNA-dependent RNA polymerase by remdesivir. Viruses 11, E326 10.3390/v11040326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lo M. K., Jordan R., Arvey A., Sudhamsu J., Shrivastava-Ranjan P., Hotard A. L., Flint M., McMullan L. K., Siegel D., Clarke M. O., Mackman R. L., Hui H. C., Perron M., Ray A. S., Cihlar T., et al. (2017) GS-5734 and its parent nucleoside analog inhibit Filo-, Pneumo-, and Paramyxoviruses. Sci. Rep. 7, 43395 10.1038/srep43395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lo M. K., Feldmann F., Gary J. M., Jordan R., Bannister R., Cronin J., Patel N. R., Klena J. D., Nichol S. T., Cihlar T., Zaki S. R., Feldmann H., Spiropoulou C. F., and de Wit E. (2019) Remdesivir (GS-5734) protects African green monkeys from Nipah virus challenge. Sci. Transl. Med. 11, eaau9242 10.1126/scitranslmed.aau9242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Brown A. J., Won J. J., Graham R. L., Dinnon K. H. 3rd, Sims A. C., Feng J. Y., Cihlar T., Denison M. R., Baric R. S., and Sheahan T. P. (2019) Broad spectrum antiviral remdesivir inhibits human endemic and zoonotic deltacoronaviruses with a highly divergent RNA dependent RNA polymerase. Antiviral Res. 169, 104541 10.1016/j.antiviral.2019.104541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. de Wit E., Feldmann F., Cronin J., Jordan R., Okumura A., Thomas T., Scott D., Cihlar T., and Feldmann H. (2020) Prophylactic and therapeutic remdesivir (GS-5734) treatment in the rhesus macaque model of MERS-CoV infection. Proc. Natl. Acad. Sci. U.S.A. 117, 6771–6776 10.1073/pnas.1922083117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sheahan T. P., Sims A. C., Graham R. L., Menachery V. D., Gralinski L. E., Case J. B., Leist S. R., Pyrc K., Feng J. Y., Trantcheva I., Bannister R., Park Y., Babusis D., Clarke M. O., Mackman R. L., et al. (2017) Broad-spectrum antiviral GS-5734 inhibits both epidemic and zoonotic coronaviruses. Sci. Transl. Med. 9, eaal3653 10.1126/scitranslmed.aal3653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sheahan T. P., Sims A. C., Leist S. R., Schäfer A., Won J., Brown A. J., Montgomery S. A., Hogg A., Babusis D., Clarke M. O., Spahn J. E., Bauer L., Sellers S., Porter D., Feng J. Y., et al. (2020) Comparative therapeutic efficacy of remdesivir and combination lopinavir, ritonavir, and interferon β against MERS-CoV. Nat. Commun. 11, 222 10.1038/s41467-019-13940-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mulangu S., Dodd L. E., Davey R. T. Jr., Tshiani Mbaya O., Proschan M., Mukadi D., Lusakibanza Manzo M., Nzolo D., Tshomba Oloma A., Ibanda A., Ali R., Coulibaly S., Levine A. C., Grais R., Diaz J., et al. (2019) A Randomized, Controlled Trial of Ebola Virus Disease Therapeutics. The New England journal of medicine 381, 2293–2303 10.1056/NEJMoa1910993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gordon C. J., Tchesnokov E. P., Feng J. Y., Porter D. P., and Götte M. (2020) The antiviral compound remdesivir potently inhibits RNA-dependent RNA polymerase from Middle East respiratory syndrome coronavirus. J. Biol. Chem. 295, 4773–4779 10.1074/jbc.AC120.013056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Subissi L., Posthuma C. C., Collet A., Zevenhoven-Dobbe J. C., Gorbalenya A. E., Decroly E., Snijder E. J., Canard B., and Imbert I. (2014) One severe acute respiratory syndrome coronavirus protein complex integrates processive RNA polymerase and exonuclease activities. Proc. Natl. Acad. Sci. U.S.A. 111, E3900–E3909 10.1073/pnas.1323705111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kirchdoerfer R. N., and Ward A. B. (2019) Structure of the SARS-CoV nsp12 polymerase bound to nsp7 and nsp8 co-factors. Nat. Commun. 10, 2342 10.1038/s41467-019-10280-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Stuyver L. J., McBrayer T. R., Whitaker T., Tharnish P. M., Ramesh M., Lostia S., Cartee L., Shi J., Hobbs A., Schinazi R. F., Watanabe K. A., and Otto M. J. (2004) Inhibition of the subgenomic hepatitis C virus replicon in huh-7 cells by 2′-deoxy-2′-fluorocytidine. Antimicrob. Agents Chemother. 48, 651–654 10.1128/AAC.48.2.651-654.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Welch S. R., Scholte F. E. M., Flint M., Chatterjee P., Nichol S. T., Bergeron É., and Spiropoulou C. F. (2017) Identification of 2′-deoxy-2′-fluorocytidine as a potent inhibitor of Crimean-Congo hemorrhagic fever virus replication using a recombinant fluorescent reporter virus. Antiviral Res. 147, 91–99 10.1016/j.antiviral.2017.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kumaki Y., Day C. W., Smee D. F., Morrey J. D., and Barnard D. L. (2011) In vitro and in vivo efficacy of fluorodeoxycytidine analogs against highly pathogenic avian influenza H5N1, seasonal, and pandemic H1N1 virus infections. Antiviral Res. 92, 329–340 10.1016/j.antiviral.2011.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Welch S. R., Guerrero L. W., Chakrabarti A. K., McMullan L. K., Flint M., Bluemling G. R., Painter G. R., Nichol S. T., Spiropoulou C. F., and Albariño C. G. (2016) Lassa and Ebola virus inhibitors identified using minigenome and recombinant virus reporter systems. Antiviral Res. 136, 9–18 10.1016/j.antiviral.2016.10.007 [DOI] [PubMed] [Google Scholar]
- 24. Appleby T. C., Perry J. K., Murakami E., Barauskas O., Feng J., Cho A., Fox D. 3rd, Wetmore D. R., McGrath M. E., Ray A. S., Sofia M. J., Swaminathan S., and Edwards T. E. (2015) Viral replication: structural basis for RNA replication by the hepatitis C virus polymerase. Science 347, 771–775 10.1126/science.1259210 [DOI] [PubMed] [Google Scholar]
- 25. Crotty S., Maag D., Arnold J. J., Zhong W., Lau J. Y., Hong Z., Andino R., and Cameron C. E. (2000) The broad-spectrum antiviral ribonucleoside ribavirin is an RNA virus mutagen. Nat. Med. 6, 1375–1379 10.1038/82191 [DOI] [PubMed] [Google Scholar]
- 26. Furuta Y., Komeno T., and Nakamura T. (2017) Favipiravir (T-705), a broad spectrum inhibitor of viral RNA polymerase. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 93, 449–463 10.2183/pjab.93.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hawman D. W., Haddock E., Meade-White K., Williamson B., Hanley P. W., Rosenke K., Komeno T., Furuta Y., Gowen B. B., and Feldmann H. (2018) Favipiravir (T-705) but not ribavirin is effective against two distinct strains of Crimean-Congo hemorrhagic fever virus in mice. Antiviral Res. 157, 18–26 10.1016/j.antiviral.2018.06.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jin Z., Smith L. K., Rajwanshi V. K., Kim B., and Deval J. (2013) The ambiguous base-pairing and high substrate efficiency of T-705 (Favipiravir) Ribofuranosyl 5′-triphosphate towards influenza A virus polymerase. PLoS One 8, e68347 10.1371/journal.pone.0068347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Maag D., Castro C., Hong Z., and Cameron C. E. (2001) Hepatitis C virus RNA-dependent RNA polymerase (NS5B) as a mediator of the antiviral activity of ribavirin. J. Biol. Chem. 276, 46094–46098 10.1074/jbc.C100349200 [DOI] [PubMed] [Google Scholar]
- 30. Oestereich L., Rieger T., Neumann M., Bernreuther C., Lehmann M., Krasemann S., Wurr S., Emmerich P., de Lamballerie X., Ölschläger S., and Günther S. (2014) Evaluation of antiviral efficacy of ribavirin, arbidol, and T-705 (favipiravir) in a mouse model for Crimean-Congo hemorrhagic fever. PLoS Negl. Trop. Dis. 8, e2804 10.1371/journal.pntd.0002804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gong P., and Peersen O. B. (2010) Structural basis for active site closure by the poliovirus RNA-dependent RNA polymerase. Proc. Natl. Acad. Sci. U.S.A. 107, 22505–22510 10.1073/pnas.1007626107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zamyatkin D. F., Parra F., Alonso J. M., Harki D. A., Peterson B. R., Grochulski P., and Ng K. K. (2008) Structural insights into mechanisms of catalysis and inhibition in Norwalk virus polymerase. J. Biol. Chem. 283, 7705–7712 10.1074/jbc.M709563200 [DOI] [PubMed] [Google Scholar]
- 33. Wang M., Cao R., Zhang L., Yang X., Liu J., Xu M., Shi Z., Hu Z., Zhong W., and Xiao G. (2020) Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 30, 269–271 10.1038/s41422-020-0282-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Traut T. W. (1994) Physiological concentrations of purines and pyrimidines. Mol. Cell. Biochem. 140, 1–22 10.1007/BF00928361 [DOI] [PubMed] [Google Scholar]
- 35. Kennedy E. M., Gavegnano C., Nguyen L., Slater R., Lucas A., Fromentin E., Schinazi R. F., and Kim B. (2010) Ribonucleoside triphosphates as substrate of human immunodeficiency virus type 1 reverse transcriptase in human macrophages. J. Biol. Chem. 285, 39380–39391 10.1074/jbc.M110.178582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ferron F., Subissi L., Silveira De Morais A. T., Le N. T. T., Sevajol M., Gluais L., Decroly E., Vonrhein C., Bricogne G., Canard B., and Imbert I. (2018) Structural and molecular basis of mismatch correction and ribavirin excision from coronavirus RNA. Proc. Natl. Acad. Sci. U.S.A. 115, E162–E171 10.1073/pnas.1718806115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bouvet M., Imbert I., Subissi L., Gluais L., Canard B., and Decroly E. (2012) RNA 3′-end mismatch excision by the severe acute respiratory syndrome coronavirus nonstructural protein nsp10/nsp14 exoribonuclease complex. Proc. Natl. Acad. Sci. U.S.A. 109, 9372–9377 10.1073/pnas.1201130109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Posthuma C. C., Te Velthuis A. J. W., and Snijder E. J. (2017) Nidovirus RNA polymerases: complex enzymes handling exceptional RNA genomes. Virus Res. 234, 58–73 10.1016/j.virusres.2017.01.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Minskaia E., Hertzig T., Gorbalenya A. E., Campanacci V., Cambillau C., Canard B., and Ziebuhr J. (2006) Discovery of an RNA virus 3′ → 5′ exoribonuclease that is critically involved in coronavirus RNA synthesis. Proc. Natl. Acad. Sci. U.S.A. 103, 5108–5113 10.1073/pnas.0508200103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tchesnokov E. P., Obikhod A., Schinazi R. F., and Götte M. (2008) Delayed chain termination protects the anti-hepatitis B virus drug entecavir from excision by HIV-1 reverse transcriptase. J. Biol. Chem. 283, 34218–34228 10.1074/jbc.M806797200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pruijssers A. J., and Denison M. R. (2019) Nucleoside analogues for the treatment of coronavirus infections. Curr. Opin. Virol. 35, 57–62 10.1016/j.coviro.2019.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Delang L., Abdelnabi R., and Neyts J. (2018) Favipiravir as a potential countermeasure against neglected and emerging RNA viruses. Antiviral Res. 153, 85–94 10.1016/j.antiviral.2018.03.003 [DOI] [PubMed] [Google Scholar]
- 43. Uebelhoer L. S., Albariño C. G., McMullan L. K., Chakrabarti A. K., Vincent J. P., Nichol S. T., and Towner J. S. (2014) High-throughput, luciferase-based reverse genetics systems for identifying inhibitors of Marburg and Ebola viruses. Antiviral Res. 106, 86–94 10.1016/j.antiviral.2014.03.018 [DOI] [PubMed] [Google Scholar]
- 44. Schinazi R., Halfon P., Marcellin P., and Asselah T. (2014) HCV direct-acting antiviral agents: the best interferon-free combinations. Liver Int. 34, Suppl. 1, 69–78 10.1111/liv.12423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Feld J. J., and Hoofnagle J. H. (2005) Mechanism of action of interferon and ribavirin in treatment of hepatitis C. Nature 436, 967–972 10.1038/nature04082 [DOI] [PubMed] [Google Scholar]
- 46. Berger I., Fitzgerald D. J., and Richmond T. J. (2004) Baculovirus expression system for heterologous multiprotein complexes. Nat. Biotechnol. 22, 1583–1587 10.1038/nbt1036 [DOI] [PubMed] [Google Scholar]
- 47. Bieniossek C., Richmond T. J., and Berger I. (2008) MultiBac: multigene baculovirus-based eukaryotic protein complex production. Curr. Protoc. Protein Sci. Chapter 5, Unit 5.20 10.1002/0471140864.ps0520s51 [DOI] [PubMed] [Google Scholar]
- 48. Tchesnokov E. P., Raeisimakiani P., Ngure M., Marchant D., and Götte M. (2018) Recombinant RNA-dependent RNA polymerase complex of Ebola virus. Sci. Rep. 8, 3970 10.1038/s41598-018-22328-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Jacobson M. P., Friesner R. A., Xiang Z., and Honig B. (2002) On the role of the crystal environment in determining protein side-chain conformations. J. Mol. Biol. 320, 597–608 10.1016/S0022-2836(02)00470-9 [DOI] [PubMed] [Google Scholar]
- 50. Jacobson M. P., Pincus D. L., Rapp C. S., Day T. J., Honig B., Shaw D. E., and Friesner R. A. (2004) A hierarchical approach to all-atom protein loop prediction. Proteins 55, 351–367 10.1002/prot.10613 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are included within this article.