Abstract
The ovarian tumor domain (OTU) deubiquitinylating cysteine proteases OTUB1 and OTUB2 (OTU ubiquitin aldehyde binding 1 and 2) are representative members of the OTU subfamily of deubiquitinylases. Deubiquitinylation critically regulates a multitude of important cellular processes, such as apoptosis, cell signaling, and growth. Moreover, elevated OTUB expression has been observed in various cancers, including glioma, endometrial cancer, ovarian cancer, and breast cancer. Here, using molecular dynamics simulation approaches, we found that both OTUB1 and OTUB2 display a catalytic triad characteristic of proteases but differ in their configuration and protonation states. The OTUB1 protein had a prearranged catalytic site, with strong electrostatic interactions between the active-site residues His265 and Asp267. In OTUB2, however, the arrangement of the catalytic triad was different. In the absence of ubiquitin, the neutral states of the catalytic-site residues in OTUB2 were more stable, resulting in larger distances between these residues. Only upon ubiquitin binding did the catalytic triad in OTUB2 rearrange and bring the active site into a catalytically feasible state. An analysis of water access channels revealed only a few diffusion trajectories for the catalytically active form of OTUB1, whereas in OTUB2 the catalytic site was solvent-accessible, and a larger number of water molecules reached and left the binding pocket. Interestingly, in OTUB2, the catalytic residues His224 and Asn226 formed a stable hydrogen bond. We propose that the observed differences in activation kinetics, protonation states, water channels, and active-site accessibility between OTUB1 and OTUB2 may be relevant for the selective design of OTU inhibitors.
Keywords: ubiquitin thioesterase (OTUB1), deubiquitylation (deubiquitination), ovarian cancer, allosteric regulation, ubiquitin-dependent protease, cysteine protease, catalytic triad, computational analysis, molecular dynamics, OTU deubiquitinase ubiquitin aldehyde binding
Introduction
Ubiquitinylation is a post-translational modification and involves one or more covalent additions to the lysine residues of the target protein that is mediated by sequential action of ubiquitin ligases E1 (ubiquitin-activating enzyme), E2 (ubiquitin-conjugating enzyme), and E3 (ubiquitin ligase). It can be reversed by removal of ubiquitin (deubiquitinylation) catalyzed by ubiquitin hydrolases or deubiquitinylating enzymes (DUBs)2 (1). DUBs play an essential role in the regulation of cellular processes like apoptosis, putative cell growth, cell cycle control, and DNA repair (2, 3). 102 DUBs were identified and can be grouped into five subfamilies (4, 5). The 18 OTUs identified in human can further be grouped into four categories: OTUBs, OTUDs, A20s, and OTULINs (6, 7). Cysteine proteases, such as ovarian tumor deubitquitinylases Otubain-1 (OTUB1) and Otubain-2 (OTUB2) belong to the ovarian tumor protease (OTU) subfamily of DUB proteins (8) and were the first two OTU proteins to show DUB activity (8, 9).
Elevated OTUB1 expression is associated with glioma (10), esophageal squamous cell carcinoma metastasis (11), endometrial cancer (12), hepatocellular carcinoma (HCC) (13), ovarian cancer (14), colorectal cancer (15, 16), breast cancer (17), liver cancer (13), and gastric cancer (14). A recent study by Li et al. (18) have identified the role of OTUB2 and its overexpression in non–small cell lung cancer tissues. Several studies have described the therapeutic potential of DUBs for cancer treatment.
OTUB1 and OTUB2 belong to the cysteine protease class with a sequence identity and similarity of 48 and 70%, respectively. A 37-residue-long N-terminal region in OTUB1 is not present in OTUB2. Despite the considerable sequence conservation and structural overlap, there is perceivable substrate specificity. OTUB1 has a slower cleavage kinetics and favor Lys48-linked polyubiquitin (19), whereas OTUB2 cleaves differently linked polyubiquitin chains.
Cysteine proteases have a Cys-His-Asn/Asp catalytic triad as the active site. When the cysteine sulfur atom is deprotonated, the histidine residue acts a base and becomes positively charged. The thiolate's high nucleophilicity leads to an attack of the carbon of the substrate peptide bond to produce a tetrahedral intermediate; then the active-site histidine acts as a proton donor to release the N-terminal fragment of the substrate (20). The intermediate is further stabilized by hydrogen bonding between the substrate oxyanion and a conserved glutamine residue (21). However, this catalytic triad is performing the proteolytic activity, and the protonation states (neutral or zwitterionic form) of cysteine and histidine are highly debated (22–25). The third residue (Asp/Asn) of the catalytic triad is required for a correct alignment and polarization of catalytic histidine (5). The Asp in the catalytic triad plays an role in the charge relay systems and is involved in the stabilization of the imidazolinium intermediate and the positioning of the catalytic site histidine (26). The role of Asn in the catalytic triad is to orient the side chain of the catalytic histidine to its optimum position for various steps of the catalytic mechanism (27).
OTUB1 in the absence of ubiquitin (crystal structure PDB code 2ZFY) is catalytically incompetent because of long distances between the catalytic triad residues histidine (His265) and cysteine (Cys91) and aspartic acid (Asp267) of 5.5 and 4.6 Å, respectively (19). Access to the active site of OTUB1 is blocked by Glu214 of the α9-α10 loop and forms a backbone hydrogen bond with the catalytic Cys91. The surface charge property of the ubiquitin-binding site (P′ side) of OTUB1 is different from OTUB2 located within the close proximity of Cys91, which sterically restricts active-site access. Conformational changes and specificity binding studies (19) show that OTUB1 and OTUB2 may adopt two conformational states and display different lysine linkage specificities, probably because of structural differences in their catalytic centers. Although OTUB1 and OTUB2 are close homologs (8), OTUB1 is highly specific for Lys48 cleavage, whereas OTUB2 has a broad cleavage profile (28). OTUB1 is autoinhibited in the ubiquitin-free (apo) state and is activated upon ubiquitin binding, whereas the OTUB2 is catalytically active even in the absence of ubiquitin (29). Structural analysis of OTUB1 shows the preference of ubiquitin (Ub)-charged E2 acting as an N-terminal extension of OTU (30, 31) and also binding to free ubiquitin, both mimicking the configuration of the canonical product of its DUB activity, the Lys48-linked di-Ub chain (32).
A long N-terminal helix of OTUB1 is critical for E2-ubiquitin recognition and inhibition; OTUB2 thus fails to recognize and inhibit E2 because it lacks N-terminal helix (32). The proximal ubiquitin binds with OTUB2 in a different manner because of the striking structural differences in the N-terminal region (absence of N-terminal helix in OTUB2), and several reports (7, 32, 33) show that the absence of N-terminal helix may be the reason for the nonselective deubitquitinylase activity of OTUB2. The short loop (residues 44–48) preceding the active-site helix different from all other cysteine proteases and in the crystal structure is shifted toward the catalytic residues to generate a spatially restricted active site and stabilize the oxyanion intermediate (29). Interestingly, in OTUB2 only subtle changes were observed in the catalytic residues, but larger conformational changes can be seen in the disordered region 197–204 (turned into ordered region upon ubiquitin binding), helices 1 and 2, and β-sheets 3 and 4 (28).
Deubiquitinylating enzymes have recently been identified as relevant cancer drug targets (34–36). Molecular simulations have become essential tools in identifying protein conformational dynamics and rationalizing drug selectivity. For USP7 (37) two druggable binding sites were identified of which one was essential for the inactive to active state conformational transition. MD studies elucidated the open to close conformation transition in viral DUB (turnip yellow mosaic virus) (38) in WT and mutants. The drug selectivity of zinc-dependent human DUBs of the JAMM family Rpn11 and CSN5 toward capzimin and CSN5i-3 could be explained because of formation of heterodimeric protein–protein complexes (39).
In this work, we focus on OTUB1 and OTUB2 DUBs to understand their differences in structural stability in the apo and holo forms and the role of water molecules in stabilizing the catalytic triad conformations using molecular dynamics simulation. Analyzing the conformational stability of the catalytic triad residues and the access and passage and pathways of solvent molecules helps us to understand the role of water molecules in stabilizing the active site. Our findings suggest that OTUB1 achieves its productive conformation in its charged state only upon ubiquitin binding, whereas in OTUB2 the productive conformation already prevails in its holo form conformation in the neutral protonation state. A very low number of water trajectories toward the catalytic state from the surface were observed in the productive conformation of OTUB1 in contrast to the very large number observed in the productive conformation of OTUB2. A number of conserved water molecules are located close to His265 and Asp267 in OTUB1 and in vicinity of Cys51 in OTUB2.
Results and discussion
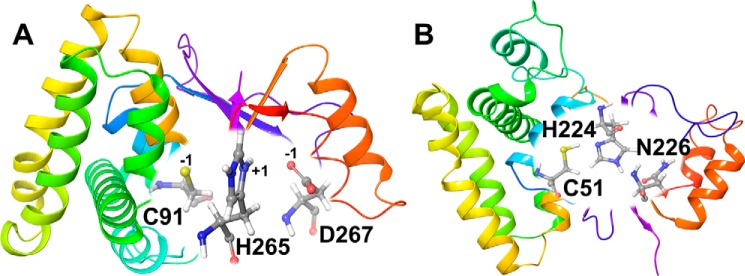
The comparison of OTUB1 and OTUB2 sequences shows 48% identity and 70% similarity (Fig. S1). Larger structural differences are apparent in the catalytic site and in the ubiquitin-binding pockets (19). A long N-terminal α1-helix is present in OTUB1 but missing in OTUB2. In our nomenclature, we refer to the apo OTUB1 structure in charged and neutral states as O1U0C and O1U0N, respectively; likewise the holo OTUB1 structures in charged and neutral states are O1U1C and O1U1N. Similarly, apo and holo OTUB2 charged and neutral states will be referenced as O2U0C/O2U0N and O2U1C/O2U1N. For representational purpose, we show a zwitterionic catalytic triad (in O1U1C) and a neutral charge catalytic site (in O2U0N) in Fig. 1.
Figure 1.
Structures of OTUB1 (O1U1C) in the ubiquitin-bound protein conformation (A) and OTUB2 (O2U0N) in the ubiquitin-free protein conformation (B). Residues of the catalytic triad are labeled, and their charged states are given in A.
Absence of Ub: apo OTUB1 (O1U0C, O1U0N) and OTUB2 (O2U0C, O2U0N)
Molecular dynamics simulations of OTUB1 (charged and neutral) and OTUB2 (charged and neutral) were replicated three times. The RMSD (Cα) plots of all apo OTUB1 and OTUB2 simulations were analyzed and show that the structures are stable throughout the simulations (Figs. S2 and S3). In addition, the global protein RMSFs reveal no large fluctuations. Only the loop region connecting sheets 2 and 3 in OTUB2 (O2U0C and O2U0N) shows a high degree of flexibility (Fig. S4 and S5).
Structural characteristics of the catalytic triad in ubiquitin-free OTUB1 and OTUB2
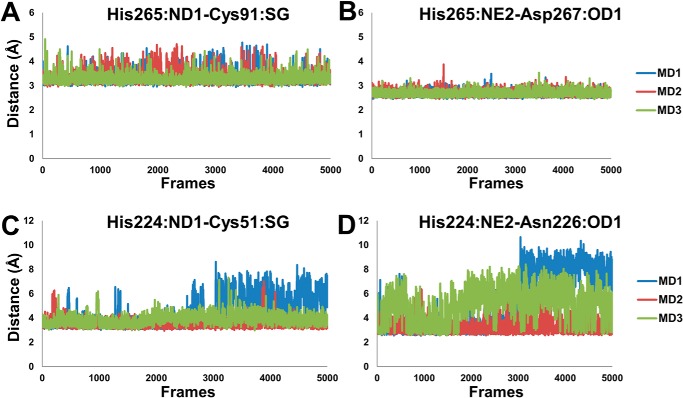
Catalytic protease activity requires an accurate positioning of the Cys/His residues to perform their catalytic function. The interatomic distances of the catalytic residues of both OTUB1 and OTUB2 were monitored. The average distances and their standard deviations are given in Table 1: in the charged state of OTUB1, the distances are significantly smaller than in the neutral state (His265ND1 … Cys91 sulfur atom were 5.8 ± 1.8 Å compared with 6.4 ± 0.9 Å; His265NE2 … Asp267OD1 4.8 ± 1.4 Å and 7.0 ± 1.2 Å). This is in good agreement with the catalytic geometry of the experimental structure and shows that the charged state is dominant in the X-ray structures.
Table 1.
Comparison of characteristic catalytic inter-residue distances from available X-ray structures and calculated averages ± S.D. from MD simulations of OTUB1 and OTUB2 in Å
| Protein | Protein conformation | Catalytic site protonation state | His-Cys | Asp-His | His (N-CA-CB-CG) dihedral angle |
|---|---|---|---|---|---|
| Å | Å | ° | |||
| OTUB1 | Ub-bound | 4DDG | 3.5 | 3.3 | 165 |
| O1U1C | Charged | 3.3 ± 0.2 | 2.7 ± 0.1 | 173 ± 05/−175 ± 04 | |
| O1U1N | Neutral | 4.7 ± 1.5 | 4.6 ± 2.1 | 170 ± 17/−147 ± 45 | |
| Ub-free | 2ZFY | 5.5 | 6.7 | −65 | |
| O1U0C | Charged | 5.8 ± 1.8 | 4.8 ± 1.4 | 084 ± 52/−156 ± 41 | |
| O1U0N | Neutral | 6.4 ± 0.9 | 7.0 ± 1.2 | 145 ± 45/-067 ± 19 | |
| OTUB2 | Ub-bound | 4FJV | 4.0 | 2.9 | −179 |
| O2U1C | Charged | 5.9 ± 0.7 | 7.5 ± 1.8 | 074 ± 44/−059 ± 23 | |
| O2U1N | Neutral | 5.4 ± 1.2 | 6.5 ± 1.7 | 126 ± 62/−079 ± 46 | |
| Ub-free | 1TFF | 4.0 | 2.9 | −176 | |
| O2U0C | Charged | 5.5 ± 0.9 | 4.7 ± 1.9 | 149 ± 44/−138 ± 52 | |
| O2U0N | Neutral | 3.9 ± 0.8 | 4.5 ± 1.8 | 174 ± 05/−150 ± 44 |
For the zwitterionic state O2U0C average contact distances for His224ND1 … Cys91SG of 5.5 ± 0.9 Å and His224NE2 … Asn226OD1 4.7 ± 1.9 Å were obtained. Unexpectedly, the neutral O2U0N state shows shorter interatomic distances for His224ND1…. Cys91SG of 3.9 ± 0.8 Å and His224NE2 … Asn226OD1 of 4.5 ± 1.8 Å (Figs. 2B and 3, C and D). The His224 dihedral angle Χ1(N-CA-CB-CG) is 174 ± 5°/−154 ± 44° degree in O2U0N, but in O2U0C, it is 149 ± 44°/−138 ± 52° and in O1U0C, O1U0N it is 84 ± 52°/−156 ± 41° and 145 ± 45°/−67 ± 19°, respectively (Table 1).
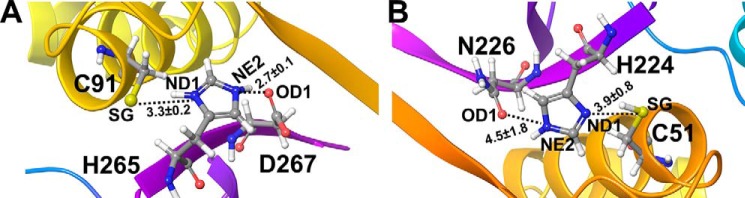
Figure 2.
Interatomic distances with standard deviations (in Å) of the key atoms of the catalytic residues monitored throughout simulations. A, OTUB1 (O1U1C). B, OTUB2 (O2U0N).
Figure 3.
Monitoring of interatomic distances of key catalytic residues (His-Cys-Asp/Asn) in MD simulations. A and B, His+ … Cys− (A) and His+ … Asp− (B) residues in OTUB1 (O1U1C). C and D, His0 … Cys0 (C) and His0 … Asn0 (D) residues of OTUB2 (O2U0N).
Apparently, there is no conformational change of the catalytic residues associated upon activation of OTUB2. The differences in structural parameters such as interatomic distances and dihedral angle are only minor and show that this is in agreement with experimental results that demonstrate the prevalence of the neutral state of apo OTUB2 to be catalytically active.
The Ub-bound protein conformation of OTUB1 and OTUB2
Simulations of the charged and neutral states of the catalytic triad residues of OTUB1 and OTUB2 in the ubiquitin-bound conformation were performed in triplicate. The RMSD (Cα) plots of all holo OTUB1 and OTUB2 show the stability of the charged and neutral states of OTUB2 but not for OTUB1. Whereas the OTU of OTUB1 is rather rigid with a RMSD of 2 Å, it is the flexible N terminus that is stabilized by interactions with the E2-linked ubiquitin binding and responsible for the very large RMSD of 6–8 Å in OTUB2 (Fig. S6). The RMSF analysis further confirms that the large fluctuations originate from the N-terminal helical region (first 21 amino acids of the protein), and the rest of the protein is less flexible (Fig. S7). The analysis of the RMSF further also shows local residue fluctuations in the loop connecting the helices 2 and 3 in both charge states. Less pronounced flexible regions are helices 8 and 9 in the charged state (O1U1C) and the loop connecting sheets 3 and 4 in the neutral state (O1U1N) (Fig. S7).
OTUB2, which does not possess the N-terminal helix, reveals a smaller RMSD in both charge states (O2U1C and O2U1N) during the entire simulations (Fig. S8). The analysis of the RMSFs of holo OTUB2 shows that the neutral state O2U1N is structurally more stable than the O2U1C except that the latter one shows sharp fluctuations in the short N-terminal helix and the small loop connecting sheet 5 and 6 (Fig. S9).
Analysis of catalytic triad conformation in ubiquitin-bound OTUB1 versus OTUB2
In O1U1C, the average contact distance in the MD trajectories between His265ND1 . . Cys91SG is 3.3 ± 0.2 Å, and the His265NE2 maintains a close contact distance of 2.7 ± 0.1 Å with Asp267OD1 (Figs. 2A and 3, A and B). The dihedral angle Χ1(N-CA-CB-CG) of His265 is very well-preserved throughout the simulation with the range of 173.7 ± 4.6° and −175.4 ± 3.6°. In O1U1N, the average distance of His265ND1 with Cys91SG in the simulations is significantly larger with 4.7 ± 1.5 Å, and the His265NE2 … Asp267OD1 distance of 4.6 ± 2.1 Å is likewise large and not arranged for catalysis. The dihedral of the His265 (Χ1) falls in the range of 170 ± 17°/−147 ± 45°. In the case of OTUB2, the different charge states O2U1C and O2U1N display average inter-residue distances between His224ND1 and Cys91SG of 5.9 ± 0.7 Å and 5.4 ± 1.2 Å, respectively. The average His224NE2 … Asn226OD1 distances are 7.5 ± 1.8 Å and 6.5 ± 1.7 Å, respectively. The histidine dihedral (Χ1) for O2U1C is 74 ± 44°/−59 ± 23°, and for O2U1N it is 126 ± 62°/−79 ± 46° (Table 1). This shows that structural parameters for the holo form in the charged state of OTUB1 (O1U1C) are in good agreement with the crystal structure. The other feasible charge states for holo conformations of OTUB1 and OTUB2 are significantly deviating from the experimental structural structures.
In OTUB1, throughout the total simulation time of the O1U1C state, the strong salt-bridge interaction between the catalytic site His265+ and Asp267− is persistent. Conformational restrictions of the active-site residues in OTUB1 are also due to Pro87 that is located in close proximity of Cys91, and also the small loop before the His265 has an effect on the Cys91 orientation. The role of α9-α10 loop to influence the conformational change on Cys91 and His265 upon the ubiquitin binding was only inferred from a static picture in the X-ray structure (19). A hydrogen-bonding interaction between the C-terminal end of α3 and the backbone of the catalytic Cys91 was speculated to inhibit substrate access to the active site and be the reason for an OTUB1 ligand-induced activation step.
Water access of active site in Ub-free in apo OTUB1 and OTUB2
Water molecules in the active site play an important role in regulating the activity, stability and selectivity of enzymes (40–42). The buried active sites in OTUB1 and OTUB2 are connected with the bulk solvents by water tunnels. The molecular properties of the amino acids of the water tunnels control the access of the water molecules. Studying the water tunnels is important, because it is the major route of the substrate delivery to the active-site center. The analysis of water molecules diffusing from the surface of the protein to the active site is a versatile tool to discriminate structurally similar proteins. Despite our special focus on dissecting the active state conformations O1U1C and O2U0N, we have also analyzed the complementary conformations of the OTUB1 and OTUB2 proteins in the different charge states.
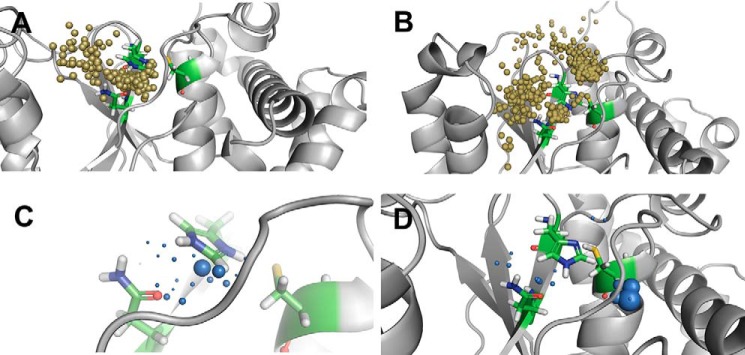
In O1U0C, the number of water inlets is 1125, of which 471 are incoming paths and 654 are outgoing paths. The localization of water molecules in the vicinity of the active-site residues His265 and Cys91 is shown in Fig. 4A. A high density of water molecules is also observed close to these residues with longer times of residence caused by strong hydrogen bonding and electrostatic interactions (Fig. 4C). In O1U0N, the number of inlets is smaller with 776, of which 387 are incoming paths and 389 are outgoing paths. The water inlets are mostly located close to the region of His265 and Asp267 (Fig. 4B). The local distribution of water molecules is high near the side chain oxygen of Asp267, low close to His265, and not present in the vicinity of Cys91 (Fig. 4D). This shows that the catalytic aspartate residue plays a vital role in trapping water molecules close to the active-site region of OTUB1.
Figure 4.
Analysis of water access in ubiquitin-free OTUB1 structures. Top panel, water inlets to the catalytic site for incoming and outgoing water molecules. Bottom panels, local distribution density of water molecules to identify stable water molecules. A and C, charged state of active-site O1U0C. B and D, neutral charge state of active-site O1U0N.
In O2U0C, 260 water inlets can be identified, and the numbers of incoming and outgoing channels are almost evenly distributed with 135 and 123, respectively. In this case, the number of water molecules that enter or reside close to the active-site His224 is high compared with two other active-site residues, cysteine and aspartate (Fig. 5A). The local distribution of water molecules is high near the His224 side chain compared with the other active-site residue, Asn226 (Fig. 5C). In O2U0N, on the other hand, 1499 inlets are observed, of which 833 are incoming paths and 666 are outgoing paths (Fig. 5B). O2U0N has numerous water trajectory paths with more incoming and outgoing paths compared with a small number of water molecules residing trapped in the active-site region (Fig. 5D). In O2U0N, the absence of the long N-terminal helix allows the opening of two large cavities located near the entrance of the catalytic triad, which act as the main route of access for the water molecules in this truncated structure. The water molecules are not densely located close to one catalytic residue but sparsely distributed around the whole catalytic triad. More specifically, analysis of the local water distribution density shows that the water molecules reside longer in the vicinity of the catalytic cysteine residue and can less frequently be found surrounding the catalytic histidine and asparagine residues.
Figure 5.
Analysis of water access to the active site in ubiquitin-free OTUB2 structures. Top panels, water inlets to the catalytic site for incoming and outgoing water molecules. Bottom panels, local distribution density of water molecules to identify stable water molecules. A and C, charged states of catalytic triad residues O2U0C. B and D, neutral charge state of catalytic triad residues O2U0N.
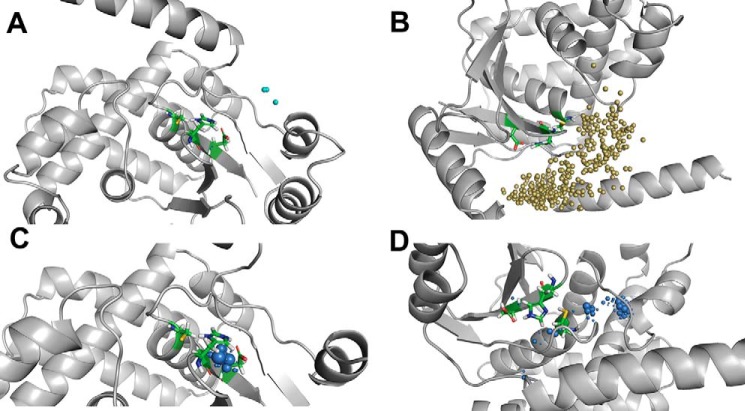
Water access in the Ub-bound conformation of OTUB1 and OTUB2
In the catalytically active state of OTUB1 (O1U1C), the water access to the active site is limited with a total of just 19 inlets comprising of 10 outgoing and 9 incoming paths (Fig. 6A). In O1U1C, the highly fluctuating extended N-terminal helix may acts as a swinging gate and not enable many water molecules to diffuse from the surface to the active-site region and vice versa. In O1U1C, very few molecules enter and leave the active site; compared with the number of incoming and outgoing water molecules' trajectories, more water molecules seem to be trapped inside the catalytic triad. This can be attributed to the strong salt bridge and stable interactions observed in the charged catalytic site state and also the presence of the long N-terminal helix. The local distribution density of water molecules enables the identification of essential residues capable of trapping water molecules, and it can also be used for conserved water identification or regulation of ligand access (43). In O1U1C, a very low water distribution density is found. In O1U1C, the water molecules are localized around the catalytic residues His265 and Asp267, especially close to His265 (Fig. 6C). In contrast to the charge state of OTUB1, in the neutral state (O1U1N), the total number of inlets is larger with 361, of which almost equal halves of them are incoming and outgoing paths (Fig. 6B). Analysis of the water residence time distribution shows that the water molecules are trapped far from the catalytic triad and remain in that pocket during the entire simulation. Although O1U1N has a larger number of water sites compared with O1U1C, they are not located in the vicinity of the catalytic sites, and hence the access to the catalytic site is restricted. The water distribution density also shows that during the simulations the water molecules are found distant from the catalytic sites, and only very few approach the catalytic cysteine residue (Fig. 6D).
Figure 6.
Water trajectory analysis of ubiquitin-bound OTUB1 structures. Top panels, water inlets to the catalytic site for incoming and outgoing water molecules. Bottom panels, local distribution density of water molecules to identify stable water molecules. A and C, charged state of catalytic residues O1U1C. B and D, neutral state of active-site O1U1N.
In the charged state of the Ub-bound conformation of OTUB2 (O2U1C), 586 water inlets were identified, of which 307 are incoming and 279 are outgoing (Fig. 7A). The local distribution density of water molecules is higher for the catalytic residue histidine (His224) than for the other two residues (Fig. 7C). In the neutral state of OTUB2 (O2U1N), of 896 water inlets identified, the numbers of incoming and outgoing paths are equally distributed with 450 and 446, respectively (Fig. 7B). The local water distribution density is not high close to the catalytic residues but at a certain distance from the active-site histidine and asparagine residues and remote from the active-site cysteine (see Fig. 7D).
Figure 7.
Water trajectory analysis of ubiquitin-bound OTUB2 structures. Top panels, water inlets to the catalytic site for incoming and outgoing water molecules. Bottom panels, local distribution density of water molecules to identify stable water molecules. A and C, O2U1C. B and D, O2U1N.
Comparative analysis of water trajectories of OTUB1 in Ub-free versus Ub-bound protein conformations
The comparison of the OTUB1 zwitterionic state in the ubiquitin-free (apo) and ubiquitin-bound (holo) protein conformations shows that there are thousands of water inlets, and the outgoing paths are larger in number in the free protein. The holo state shows fewer water inlets with just ∼20, which are equally distributed among the incoming and outgoing. If you compare the local water distribution density, both apo and holo conformations show a preferred water distribution density close to the catalytic histidine and cysteine 91 in the apo form and aspartate 265 in the holo form.
As an aside, the speculative neutral protonation states of OTUB1 in the apo and holo protein conformations show very different results. In the apo state, ∼780 inlets are equally distributed between incoming and outgoing paths. In the holo state, there is a larger number of water inlets compared with the charged state; there are ∼360, which are equally distributed as in the apo state. The major site of localization is remote from the catalytic region and not significant for the stabilization of the catalytic triad residues.
Comparative analysis of water trajectories of OTUB2 in Ub-free versus Ub-bound protein conformations
The comparison of charged states of apo and holo structures of OTUB2 shows a significant difference between Ub-free and Ub-bound protein conformations. In the apo conformation, only half of the water inlets are present compared with the holo conformation. In the apo state (O2U0C), 260 water inlets are almost equally distributed among incoming and outgoing channels (only slightly less outgoing paths), and the large number of 586 water inlets in the holo form (O2U1C) are also almost equally distributed among the incoming and outgoing (slightly less in outgoing). In both cases, the local water distribution density is high in the catalytic site.
The same observation can also be made for the neutral states. In the OTUB2 apo conformation (O2U0N), of the total 1500 water inlets, there are more (833 inlets) incoming and outgoing (666 inlets) identified. However, in the holo conformation (O2U1N), ∼900 inlets are almost equally distributed between incoming and outgoing. Interestingly in the Ub-free conformation, the local water distribution density is high in the vicinity of the side-chain atoms of all three catalytic residues, whereas as in the holo conformation state, it is remote from the active site.
Catalytically active forms OTUB1 (O1U1C) versus OTUB2 (O2U0N)
OTUB1 is catalytically active only when ubiquitin is bound, which points to a substrate-induced activation process. The active site in OTUB1 is not competent for catalysis in its apo state because the short loop connecting β4-β5 and the loop between helices 9 and 10 have an influence on the catalytic cysteine orientation and mediate the conformational change upon ubiquitin binding. Based on the careful analysis of catalytic site geometry from our molecular dynamics simulations, we infer that the zwitterionic state of OTUB1 in the Ub-bound conformation (O1U1C) represents the catalytically active state.
In contrast, OTUB2 is already active in the Ub-free apo state, and the active-site residues are in a prearranged, catalytically active conformation. However, according to our results, we stress that the neutral charge state of OTUB2 (Cys-SH, His0, Asn0, Asp−) is in a catalytically active conformation (O2U0N), and only minor proton transfer events are required to initiate the protease activity.
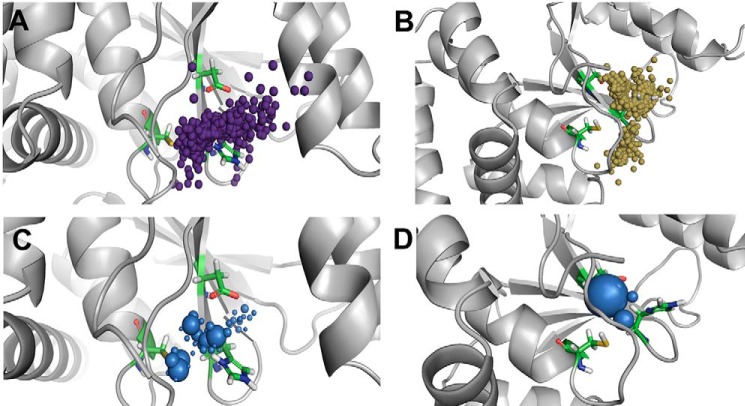
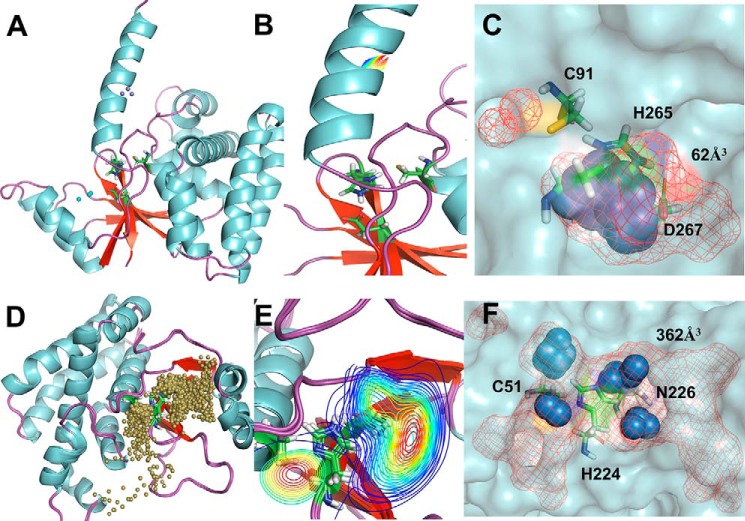
The comparison of the water trajectories between the catalytically active OTUB states, however, shows that there are significant differences between the OTUs. OTUB2 (O2U0N) possesses a huge number of water inlets compared with OTUB1 (O1U1C), and the clustering of water molecules is close to the catalytic site region in OTUB2 but remote from the catalytic site in OTUB1. The preferred sites of localization are all three catalytic residues in OTUB2 but only histidine and aspartate in OTUB1. The inner pocket volumes calculated based on the water molecule distribution densities reveal a smaller pocket of just 62 Å3 in OTUB1, whereas in OTUB2 it is nearly 6-fold larger with a volume of 362 Å3 (Fig. 8).
Figure 8.
Solvent accessibility of active sites. The top panels show O1U1C, and the bottom panels show O2U0N. A and D, localization of water molecules (represented as small balls) with passages to and from the active site of OTUB1 and OTUB2. B and E, water path cluster density was absent in O1U1C and spread across the entire catalytic triad in O2U0N. C and F, local water distribution densities represented as spheres, and inner pocket volume represented as mesh.
Water-mediated interactions in the catalytic triad residues
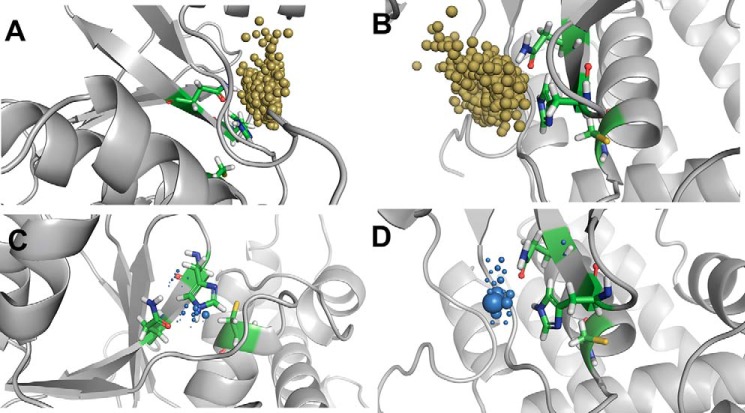
After the completion of pathway analysis of water molecules and its distribution in the active-site region, we also analyzed the hydrogen-bonding interactions of the conserved water molecules with the key catalytic residues. We only mention the water-mediated hydrogen bond interactions between the catalytic residues for the productive conformations of OTUB1 and OTUB2.
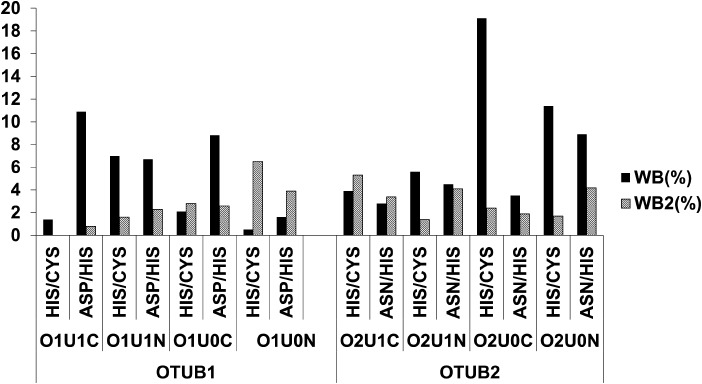
In O1U1C, in only 1.4% of the simulation time, direct water-mediated interactions between the ND1 of His265 and the sulfur atom of cysteine residue (Cys91) can be observed (Fig. 9). No extended water-mediated stabilizations of two or more water molecules are found. The NE2 of His265 and OD1 of Asp267 show water-mediated interactions for ∼11% of the simulation time. For only 1% of the simulation time, extended water-mediated contacts are observed. In O1U1C, the salt-bridge interaction between the positively charged His265 and the negatively charged Asp267 is stable and likewise a hydrogen bond interaction between the proton at the NE2 atom of His265 and OD1 atom of Asp267. The water molecule also makes a hydrogen bond interaction with OD2 atom of Asp267 and backbone oxygen atom of the His265 (Fig. 10A).
Figure 9.
Comparison of hydrogen-bonding interactions in different states and protein conformations of OTUB1 and OTUB2. Direct water-mediated interactions (WB) by one water molecule and extended water-mediated interactions by more than one water molecule (WB2).
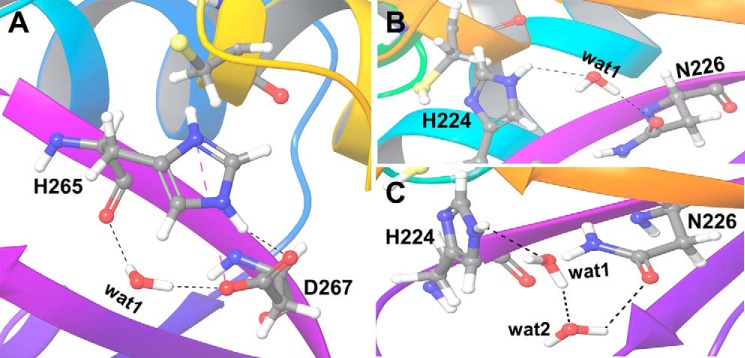
Figure 10.
A and B, water-mediated hydrogen bond (WB) interactions by one water molecule between the key catalytic residues in O1U1C (A) and O2U0N (B). C, extended water-mediated hydrogen bond (WB2) by two water molecules in O2U0N. Hydrogen-bonding interactions are shown as black dashed lines, and salt-bridge interactions are shown as blue dashed lines.
In the case of O2U0N, ND1 of His224 and the sulfur atom of Cys51 show direct water-mediated interactions for 11.4% of the simulation time and furthermore extended water-mediated contacts for nearly 2% of the simulation time. Similarly, the NE2 of His224 has direct and extended water-mediated interaction for nearly 9 and 4.2%, respectively, of the simulation time with OD1 of Asn226 (Fig. 9).
In case of O2U0N, the water molecule mediates the interaction between histidine and asparagine only when the distance between these residues increases sufficiently during the simulation. Then the water molecule bridges the OD1 atom of Asn226 and NE2 atom of His224 (Fig. 10B). If the distance further increases, extended water-mediated interactions can be observed in which more than one water molecule form an interaction bridging the OD1 atom of Asn226 and the NE2 atom of His224 (Fig. 10C). The results show that the persistent and strong electrostatic interactions between the zwitterionic catalytic residues in O1U1C and the presence of a long flexible N-terminal helix restrict and control the entry and egress of water molecules to the catalytic triad.
Conclusions
The detailed structural comparison of the zwitterionic and neutral states of Ub-free and Ub-bound protein conformations of OTUB1 and OTUB2 shows that the apo OTUB1 structure with a charged catalytic site (O1U1C) is in a stable but catalytically incompetent conformation. OTUB1 requires substrate-induced protease activation. A possible explanation is that OTUB1 is in inactive conformation in the absence of ubiquitin and a conformational change may induce a movement of His265 in closer proximity of Cys91 upon ubiquitin substrate binding. Also the DUBs USP7 (44) and USP12 crystallize in an autoinhibited state; USP12 requires an extra allosteric activation upon UAF1 and WDR20 binding (45). The allosteric binding to remote binding sites increases the catalytic rate, kcat, by a factor of 20, whereas the substrate affinity (Km) remains almost unchanged. OTUB1 is highly selective for Lys48-linked polyubiquitin chains, but not cleaving Lys63-, Lys29-, Lys6-, or Lys11-linked polyubiquitins (31). The selectivity could be rationalized by a bidentate ubiquitin-binding model with distinct sites for catalytic activity (Cys95) and a second distal binding site. Also, the role of the N-terminal extension of its core OTU domain is not fully resolved yet (32). According to our work, the long extended N-terminal helix resolved in the O1U1C is flexible throughout the simulation and acts as a gate of specificity regulation to control substrate and solvent access to the catalytic site.
In OTUB2, the catalytic triad is stabilized by unique hydrogen-bonding network configuration (29). The similarity in the side-chain orientations of Cys51 and His224 in OTUB2 to those of other cysteine protease structures implies that they are in a catalytically productive geometry even in the absence of ubiquitin binding. On the basis of sequence analysis of OTUs, the active-site catalytic triad is commonly annotated as Cys51/His224 and Asp48, with the latter stabilizing the orientation of the protonated histidine. In the apo and holo crystal structures and in our simulations, we can show that this aspartate is too far distant to directly interact with His224, but Asn226 forms hydrogen bonds with His224 and nearby other residues. This configuration is stable in both Ub-free and Ub-bound states and requires only a minor proton transfer event from Cys51 to His224 without any configurational reorientation to initiate the protease activity. The role of Asn226 in the isopeptidase reactions is corroborated by the absence of catalytic activity upon an N226A mutation. Based on the water trajectory analysis, OTUB2 is more prone to enable water diffusion and passage to and from the active site compared with OTUB1.
DUBs and OTUs have recently emerged and promising new drug targets for treating various types of cancer. The role of an asparagine residue in a stable configuration to replace an aspartate in cysteine proteases confirms that there are clearly distinct functional subclasses within the OTU family. The difference in protonation states of the residues of the catalytic triad (diad) plus selective ubiquitin-binding sites proximal and distal to the active site will allow the design of selective inhibitors.
Experimental procedures
Structure preparation
Crystal structures of OTUB1 and OTUB2 in the apo and co-crystallized ubiquitin-bound states are available from the PDB for OTUB1 structures in the absence of ubiquitin (code 2ZFY) and in the ubiquitin-bound state (code 4DDG); and for OTUB2 (ubiquitin-free structure, code 1TFF) and co-crystallized with the ubiquitin (code 4FJV) (Table 2). Downloaded structures were preprocessed to add missing loops, add amino acid side chains, and revert introduced mutations (in the crystal structure 4DDG-S1091C) using Prime module (Schrodinger) and then further processed to assign bond orders, add hydrogen atoms, and identify hydrogen bond interactions using the protein preparation wizard. In the resulting structures, only hydrogen atoms were minimized using the Epik module (Schrodinger). To carry out an unbiased study, the ubiquitin molecule, the E2 ligase and other structural units and solvent molecules were removed from both OTUB1 and OTUB2 structures. The cysteine and histidine residues in the catalytic triad were prepared in neutral (protonated Cys-SH/His0 Nϵ and charged (deprotonated Cys-S−/His+ imidazolium cation) using the 3D builder (Schrodinger).
Table 2.
Protein structures with PDB codes used in the study
| S No. | PDB code | Protein | Complex | Active site |
|---|---|---|---|---|
| 1 | 4DDG | OTUB1 | Ubiquitin | Ser1091-His1265-Asp1267a |
| 2 | 2ZFY | OTUB1 | NAb | Cys91-His265-Asp267 |
| 3 | 4FJV | OTUB2 | Ubiqutin | Cys51-His224-Asn226 |
| 4 | 1TFF | OTUB2 | NAb | Cys51-His224-Asn226 |
a Active site cysteine was mutated to serine.
b NA, not applicable.
Molecular dynamics simulation
Every simulation was performed in triplicate of all-atom 100-ns simulations using Gromacs 2018.3 (46) and the CHARMM36 force field (47). The TIP3P water model was used, and 0.15 m of salt (Na+Cl−) was added to neutralize the system. Energy minimization was carried out using the steepest descent algorithm for 5000 steps. Equilibrations were carried out in NVT and NPT ensembles for 1 and 2 ns, respectively. Temperature (310 K) and pressure (1 atm) were controlled by the velocity rescaling thermostat and a Parrinello–Rahman barostat (48), respectively. For the treatment of long-range electrostatic interactions, the particle-mesh Ewald summation was used. The three independent production runs for 100 ns each were carried out without any position restraints in an NPT ensemble. A time step of 2 fs was used, and the coordinates were saved every 2 ps.
Water pathway and analysis
The paths of the solvent diffusion to and from the active-site area (within a radius of 4 Å from the catalytic triad) to the surface of the protein were analyzed using AQUA-DUCT software (49). AQUA-DUCT is a tool to perform a detailed analysis of solvent access and channels in proteins. Each water molecule, which diffuses between a defined active-site sphere to the surface of the protein, is individually tracked. The raw paths connecting the surface of the protein and the active site are determined by looping over all frames of the simulation trajectories. A separate path for each water molecule that enters and leaves the surface of the protein and active-site sphere during the simulations is calculated. A clustering of the inlets (points where the traceable water molecules enters or leave the surface of the protein) is performed using different clustering algorithms. Finally, the results can be visualized using PyMOL (50). It also allows classification of pockets as “inner” or “outer” when analyzing the distribution of local water molecules on a grid spanning all paths with a default size of 1 Å. The maximal space explored by traced water molecules represents the outer pocket, whereas the inner pocket is the area, which is easily accessible by water molecules. Along the entire trajectory, the number of paths crossing the grid cells divided by number of frames gives the average density of traced residues. It can also detect high-density water points (also called “water hot spots”) from the distribution of local densities on the grid, traced water molecules trapped in hydrophobic cages or attracted by favorable interactions with nearby amino acids and long residence times. The results were visualized using PyMOL software (50).
Computation of direct and water-mediated interactions in the active site
The HBonds plugin in VMD was used to calculate the hydrogen bonds along the molecular dynamics simulations (51), and the geometric criteria of donor–acceptor distance should be <3.5 Å with a donor–hydrogen–acceptor angle of >100°. A water-mediated interaction between two residues is calculated when one water molecule is bridging a hydrogen-bond donor and acceptor. An extended water-mediated interaction is defined as a water-mediated hydrogen bond formed between two residues by two bridging water molecules. Although we have analyzed the entire structure, the focus in this work is on the water-mediated interaction between the key atoms of the catalytic triad residues.
Data availability
All prepared protein structural files in PDB format and topology files to reproduce the molecular dynamics simulations with GROMACS are freely available at Zednodo (3741678).
Author contributions
D. S. formal analysis; D. S., V. K., M. N., and M. S. investigation; D. S. and V. K. visualization; D. S. and V. K. methodology; D. S. and M. S. writing-original draft; D. S., M. N., and M. S. writing-review and editing; V. K. and M. S. software; M. N. and M. S. conceptualization; M. N. data curation; M. N. and M. S. supervision; M. N. and M. S. funding acquisition; M. S. resources; M. S. validation.
Supplementary Material
This work was supported by the Max Planck Society for the Advancement of Science. The work was also supported by Grant ZS/2016/04/78155 from the European Union Program European Regional Development Fund of the Ministry of Economy, Science and Digitalisation in Saxony Anhalt within the Center of Dynamic Systems. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S9.
- DUB
- deubiquitinylating enzyme
- OTU
- ovarian tumor domain
- PDB
- Protein Data Bank
- Ub
- ubiquitin
- RMSD
- root-mean-square deviation
- RMSF
- root-mean-square fluctuations
- NVT
- constant particle number, volume, and temperature
- NPT
- constant particle number, pressure, and temperature.
References
- 1. Reyes-Turcu F. E., Ventii K. H., and Wilkinson K. D. (2009) Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu. Rev. Biochem. 78, 363–397 10.1146/annurev.biochem.78.082307.091526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zhang D., Zaugg K., Mak T. W., and Elledge S. J. (2006) A role for the deubiquitinating enzyme USP28 in control of the DNA-damage response. Cell 126, 529–542 10.1016/j.cell.2006.06.039 [DOI] [PubMed] [Google Scholar]
- 3. Adhikari A., Xu M., and Chen Z. J. (2007) Ubiquitin-mediated activation of TAK1 and IKK. Oncogene 26, 3214–3226 10.1038/sj.onc.1210413 [DOI] [PubMed] [Google Scholar]
- 4. Abdul Rehman S. A., Kristariyanto Y. A., Choi S. Y., Nkosi P. J., Weidlich S., Labib K., Hofmann K., and Kulathu Y. (2016) MINDY-1 is a member of an evolutionarily conserved and structurally distinct new family of deubiquitinating enzymes. Mol. Cell 63, 146–155 10.1016/j.molcel.2016.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Clague M. J., Barsukov I., Coulson J. M., Liu H., Rigden D. J., and Urbé S. (2013) Deubiquitylases from genes to organism. Physiol. Rev. 93, 1289–1315 10.1152/physrev.00002.2013 [DOI] [PubMed] [Google Scholar]
- 6. Sun J., Shi X., Mamun M. A. A., and Gao Y. (2020) The role of deubiquitinating enzymes in gastric cancer. Oncol. Lett. 19, 30–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mevissen T. E., Hospenthal M. K., Geurink P. P., Elliott P. R., Akutsu M., Arnaudo N., Ekkebus R., Kulathu Y., Wauer T., El Oualid F., Freund S. M., Ovaa H., and Komander D. (2013) OTU deubiquitinases reveal mechanisms of linkage specificity and enable ubiquitin chain restriction analysis. Cell 154, 169–184 10.1016/j.cell.2013.05.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Balakirev M. Y., Tcherniuk S. O., Jaquinod M., and Chroboczek J. (2003) Otubains: a new family of cysteine proteases in the ubiquitin pathway. EMBO Rep. 4, 517–522 10.1038/sj.embor.embor824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Borodovsky A., Ovaa H., Kolli N., Gan-Erdene T., Wilkinson K. D., Ploegh H. L., and Kessler B. M. (2002) Chemistry-based functional proteomics reveals novel members of the deubiquitinating enzyme family. Chem. Biol. 9, 1149–1159 10.1016/S1074-5521(02)00248-X [DOI] [PubMed] [Google Scholar]
- 10. Xu L., Li J., Bao Z., Xu P., Chang H., Wu J., Bei Y., Xia L., Wu P., Yan K., Lu B., and Cui G. (2017) Silencing of OTUB1 inhibits migration of human glioma cells in vitro. Neuropathology 37, 217–226 10.1111/neup.12366 [DOI] [PubMed] [Google Scholar]
- 11. Zhou H., Liu Y., Zhu R., Ding F., Cao X., Lin D., and Liu Z. (2018) OTUB1 promotes esophageal squamous cell carcinoma metastasis through modulating Snail stability. Oncogene 37, 3356–3368 10.1038/s41388-018-0224-1 [DOI] [PubMed] [Google Scholar]
- 12. Stanisić V., Malovannaya A., Qin J., Lonard D. M., and O'Malley B. W. (2009) OTU domain-containing ubiquitin aldehyde-binding protein 1 (OTUB1) deubiquitinates estrogen receptor (ER) α and affects ERα transcriptional activity. J. Biol. Chem. 284, 16135–16145 10.1074/jbc.M109.007484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ni Q., Chen J., Li X., Xu X., Zhang N., Zhou A., Zhou B., Lu Q., and Chen Z. (2017) Expression of OTUB1 in hepatocellular carcinoma and its effects on HCC cell migration and invasion. Acta Biochim. Biophys. Sin. (Shanghai) 49, 680–688 10.1093/abbs/gmx056 [DOI] [PubMed] [Google Scholar]
- 14. Weng W., Zhang Q., Xu M., Wu Y., Zhang M., Shen C., Chen X., Wang Y., and Sheng W. (2016) OTUB1 promotes tumor invasion and predicts a poor prognosis in gastric adenocarcinoma. Am. J. Transl. Res. 8, 2234–2244 [PMC free article] [PubMed] [Google Scholar]
- 15. Zhao L., Wang X., Yu Y., Deng L., Chen L., Peng X., Jiao C., Gao G., Tan X., Pan W., Ge X., and Wang P. (2018) OTUB1 protein suppresses mTOR complex 1 (mTORC1) activity by deubiquitinating the mTORC1 inhibitor DEPTOR. J. Biol. Chem. 293, 4883–4892 10.1074/jbc.M117.809533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Liu X., Jiang W. N., Wang J. G., and Chen H. (2014) Colon cancer bears overexpression of OTUB1. Pathol. Res. Pract. 210, 770–773 10.1016/j.prp.2014.05.008 [DOI] [PubMed] [Google Scholar]
- 17. Karunarathna U., Kongsema M., Zona S., Gong C., Cabrera E., Gomes A. R., Man E. P., Khongkow P., Tsang J. W., Khoo U. S., Medema R. H., Freire R., and Lam E. W. (2016) OTUB1 inhibits the ubiquitination and degradation of FOXM1 in breast cancer and epirubicin resistance. Oncogene 35, 1433–1444 10.1038/onc.2015.208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Li J., Cheng D., Zhu M., Yu H., Pan Z., Liu L., Geng Q., Pan H., Yan M., and Yao M. (2019) OTUB2 stabilizes U2AF2 to promote the Warburg effect and tumorigenesis via the AKT/mTOR signaling pathway in non-small cell lung cancer. Theranostics 9, 179–195 10.7150/thno.29545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Edelmann M. J., Iphöfer A., Akutsu M., Altun M., di Gleria K., Kramer H. B., Fiebiger E., Dhe-Paganon S., and Kessler B. M. (2009) Structural basis and specificity of human otubain 1-mediated deubiquitination. Biochem. J. 418, 379–390 10.1042/BJ20081318 [DOI] [PubMed] [Google Scholar]
- 20. Coulombe R., Grochulski P., Sivaraman J., Ménard R., Mort J. S., and Cygler M. (1996) Structure of human procathepsin L reveals the molecular basis of inhibition by the prosegment. EMBO J. 15, 5492–5503 10.1002/j.1460-2075.1996.tb00934.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Blow D. M., Birktoft J. J., and Hartley B. S. (1969) Role of a buried acid group in the mechanism of action of chymotrypsin. Nature 221, 337–340 10.1038/221337a0 [DOI] [PubMed] [Google Scholar]
- 22. Elsässer B., Zauner F. B., Messner J., Soh W. T., Dall E., and Brandstetter H. (2017) Distinct roles of catalytic cysteine and histidine in the protease and ligase mechanisms of human legumain as revealed by DFT-based QM/MM simulations. ACS Catalysis 7, 5585–5593 10.1021/acscatal.7b01505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wei D., Huang X., Liu J., Tang M., and Zhan C. G. (2013) Reaction pathway and free energy profile for papain-catalyzed hydrolysis of N-acetyl-Phe-Gly 4-nitroanilide. Biochemistry 52, 5145–5154 10.1021/bi400629r [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Shokhen M., Khazanov N., and Albeck A. (2011) The mechanism of papain inhibition by peptidyl aldehydes. Proteins 79, 975–985 10.1002/prot.22939 [DOI] [PubMed] [Google Scholar]
- 25. Shokhen M., Khazanov N., and Albeck A. (2009) Challenging a paradigm: theoretical calculations of the protonation state of the Cys25-His159 catalytic diad in free papain. Proteins 77, 916–926 10.1002/prot.22516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Craik C. S., Roczniak S., Largman C., and Rutter W. J. (1987) The catalytic role of the active site aspartic acid in serine proteases. Science 237, 909–913 10.1126/science.3303334 [DOI] [PubMed] [Google Scholar]
- 27. Vernet T., Tessier D. C., Chatellier J., Plouffe C., Lee T. S., Thomas D. Y., Storer A. C., and Ménard R. (1995) Structural and functional roles of asparagine 175 in the cysteine protease papain. J. Biol. Chem. 270, 16645–16652 10.1074/jbc.270.28.16645 [DOI] [PubMed] [Google Scholar]
- 28. Altun M., Walter T. S., Kramer H. B., Herr P., Iphöfer A., Boström J., David Y., Komsany A., Ternette N., Navon A., Stuart D. I., Ren J., and Kessler B. M. (2015) The human otubain2-ubiquitin structure provides insights into the cleavage specificity of poly-ubiquitin-linkages. PLoS One 10, e0115344 10.1371/journal.pone.0115344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nanao M. H., Tcherniuk S. O., Chroboczek J., Dideberg O., Dessen A., and Balakirev M. Y. (2004) Crystal structure of human otubain 2. EMBO Rep. 5, 783–788 10.1038/sj.embor.7400201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nakada S., Tai I., Panier S., Al-Hakim A., Iemura S., Juang Y. C., O'Donnell L., Kumakubo A., Munro M., Sicheri F., Gingras A. C., Natsume T., Suda T., and Durocher D. (2010) Non-canonical inhibition of DNA damage-dependent ubiquitination by OTUB1. Nature 466, 941–946 10.1038/nature09297 [DOI] [PubMed] [Google Scholar]
- 31. Wang T., Yin L., Cooper E. M., Lai M. Y., Dickey S., Pickart C. M., Fushman D., Wilkinson K. D., Cohen R. E., and Wolberger C. (2009) Evidence for bidentate substrate binding as the basis for the K48 linkage specificity of otubain 1. J. Mol. Biol. 386, 1011–1023 10.1016/j.jmb.2008.12.085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Juang Y. C., Landry M. C., Sanches M., Vittal V., Leung C. C., Ceccarelli D. F., Mateo A. R., Pruneda J. N., Mao D. Y., Szilard R. K., Orlicky S., Munro M., Brzovic P. S., Klevit R. E., Sicheri F., et al. (2012) OTUB1 co-opts Lys48-linked ubiquitin recognition to suppress E2 enzyme function. Mol. Cell 45, 384–397 10.1016/j.molcel.2012.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wiener R., DiBello A. T., Lombardi P. M., Guzzo C. M., Zhang X., Matunis M. J., and Wolberger C. (2013) E2 ubiquitin-conjugating enzymes regulate the deubiquitinating activity of OTUB1. Nat. Struct. Mol. Biol. 20, 1033–1039 10.1038/nsmb.2655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. D'Arcy P., Wang X., and Linder S. (2015) Deubiquitinase inhibition as a cancer therapeutic strategy. Pharmacol. Ther. 147, 32–54 10.1016/j.pharmthera.2014.11.002 [DOI] [PubMed] [Google Scholar]
- 35. Fraile J. M., Quesada V., Rodríguez D., Freije J. M., and López-Otín C. (2012) Deubiquitinases in cancer: new functions and therapeutic options. Oncogene 31, 2373–2388 10.1038/onc.2011.443 [DOI] [PubMed] [Google Scholar]
- 36. Shen M., Schmitt S., Buac D., and Dou Q. P. (2013) Targeting the ubiquitin-proteasome system for cancer therapy. Expert Opin. Ther. Targets 17, 1091–1108 10.1517/14728222.2013.815728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Srivastava M., Suri C., Singh M., Mathur R., and Asthana S. (2018) Molecular dynamics simulation reveals the possible druggable hot-spots of USP7. Oncotarget 9, 34289–34305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Jupin I., Ayach M., Jomat L., Fieulaine S., and Bressanelli S. (2017) A mobile loop near the active site acts as a switch between the dual activities of a viral protease/deubiquitinase. PLoS Pathog. 13, e1006714 10.1371/journal.ppat.1006714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kumar V., Naumann M., and Stein M. (2018) Computational studies on the inhibitor selectivity of human JAMM deubiquitinylases Rpn11 and CSN5. Front. Chem. 6, 480 10.3389/fchem.2018.00480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Dyson P. J., and Jessop P. G. (2016) Solvent effects in catalysis: rational improvements of catalysts via manipulation of solvent interactions. Catalysis Sci. Technol. 6, 3302–3316 10.1039/C5CY02197A [DOI] [Google Scholar]
- 41. Stepankova V., Khabiri M., Brezovsky J., Pavelka A., Sykora J., Amaro M., Minofar B., Prokop Z., Hof M., Ettrich R., Chaloupkova R., and Damborsky J. (2013) Expansion of access tunnels and active-site cavities influence activity of haloalkane dehalogenases in organic cosolvents. ChemBioChem 14, 890–897 10.1002/cbic.201200733 [DOI] [PubMed] [Google Scholar]
- 42. Yang L., Dordick J. S., and Garde S. (2004) Hydration of enzyme in nonaqueous media is consistent with solvent dependence of its activity. Biophys. J. 87, 812–821 10.1529/biophysj.104.041269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Mitusińska K., Magdziarz T., Bzówka M., Stańczak A., and Gora A. (2018) Exploring Solanum tuberosum epoxide hydrolase internal architecture by water molecules tracking. Biomolecules 8, E143 10.3390/biom8040143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Özen A., Rougé L., Bashore C., Hearn B. R., Skelton N. J., and Dueber E. C. (2018) Selectively modulating conformational states of USP7 catalytic domain for activation. Structure 26, 72–84.e7 10.1016/j.str.2017.11.010 [DOI] [PubMed] [Google Scholar]
- 45. Li H., Lim K. S., Kim H., Hinds T. R., Jo U., Mao H., Weller C. E., Sun J., Chatterjee C., D'Andrea A. D., and Zheng N. (2016) Allosteric activation of ubiquitin-specific proteases by β-propeller proteins UAF1 and WDR20. Mol. Cell 63, 249–260 10.1016/j.molcel.2016.05.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kutzner C., Páll S., Fechner M., Esztermann A., de Groot B. L., and Grubmüller H. (2019) More bang for your buck: Improved use of GPU nodes for GROMACS 2018. J. Comput. Chem. 40, 2418–2431 10.1002/jcc.26011 [DOI] [PubMed] [Google Scholar]
- 47. Huang J., and MacKerell A. D. Jr. (2013) CHARMM36 all-atom additive protein force field: validation based on comparison to NMR data. J. Comput. Chem. 34, 2135–2145 10.1002/jcc.23354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Nosé S., and Klein M. L. (1983) Constant pressure molecular dynamics for molecular systems. Mol. Physics 50, 1055–1076 10.1080/00268978300102851 [DOI] [Google Scholar]
- 49. Magdziarz T., Mitusinska K., Bzowka M., Raczynska A., Stanczak A., Banas M., Bagrowska W., and Gora A. (2019) AQUA-DUCT 1.0: structural and functional analysis of macromolecules from an intramolecular voids perspective. Bioinformatics btz946 10.1093/bioinformatics/btz946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. DeLano W. L., and Lam J. W. (2017) PyMOL-The PyMOL Molecular Graphics System, Version 2.0, Schrödinger, LLC [Google Scholar]
- 51. Humphrey W., Dalke A., and Schulten K. (1996) VMD: visual molecular dynamics. J. Mol. Graph. 14, 33–38 10.1016/0263-7855(96)00018-5 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All prepared protein structural files in PDB format and topology files to reproduce the molecular dynamics simulations with GROMACS are freely available at Zednodo (3741678).