Abstract
Angiogenesis is a hallmark of tumorigenesis, and hepatocellular carcinoma (HCC) is hypervascular and therefore very dependent on angiogenesis for tumor development and progression. Findings from previous studies suggest that in HCC cells, hypoxia-induced factor 1α (HIF1A) and zinc finger homeobox 3 (ZFHX3) transcription factors functionally interact in the regulation of genes in HCC cells. Here, we report that hypoxia increases the transcription of the ZFHX3 gene and enhances the binding of HIF1A to the ZFHX3 promoter in the HCC cell lines HepG2 and Huh-7. Moreover, ZFHX3, in turn, physically associated with and was functionally indispensable for HIF1A to exert its angiogenic activity, as indicated by in vitro migration and tube formation assays of human umbilical vein endothelial cells (HUVECs) and microvessel formation in xenograft tumors of HCC cells. Mechanistically, ZFHX3 was required for HIF1A to transcriptionally activate the vascular endothelial growth factor A (VEGFA) gene by binding to its promoter. Functionally, down-regulation of ZFHX3 in HCC cells slowed their tumor growth, and addition of VEGFA to conditioned medium from ZFHX3-silenced HCC cells partially rescued the inhibitory effect of this medium on HUVEC tube formation. In human HCC, ZFHX3 expression was up-regulated, and this up-regulation correlated with both HIF1A up-regulation and worse patient survival, confirming a functional association between ZFHX3 and HIF1A in human HCC. We conclude that ZFHX3 is an angiogenic transcription factor that is integral to the HIF1A/VEGFA signaling axis in HCC cells.
Keywords: gene regulation, gene silencing, hypoxia, hypoxia-inducible factor (HIF), liver cancer, transcription factor, transcription regulation, angiogenesis, vascular endothelial growth factor (VEGF), Hepatocellular carcinoma cells, ZFHX3
Introduction
Hypoxia is a common feature of hepatocellular carcinoma (HCC)2, and hypoxia-inducible factors (HIFs) are master regulators that activate diverse pathways under hypoxia, including angiogenesis, cellular metabolism, proliferation, and migration (1, 2). HIFs are composed of an oxygen-sensitive α-subunit and a constitutively expressed β-subunit, and hypoxia-induced factor 1α (HIF1A) is perhaps the most potent known HIF that promotes tumorigenesis by enhancing angiogenesis (3). In normal cells, HIF1A is maintained at a relatively low level by protein degradation; but in a hypoxic environment, HIF1A is stabilized and dimerizes with HIF1B to induce the transcription of VEGFA and other genes to promote tumor angiogenesis (4). Hypervascularization has thus been considered a prominent therapeutic target in HCC (5). For example, sorafenib, a kinase inhibitor that targets VEGF receptor, platelet-derived growth factor receptor, and multiple members of the MAPK pathway, has been approved by the Food and Drug Administration for the treatment of HCC and renal carcinoma (6). As a common form of liver tumor, HCC is ranked worldwide as the sixth most common cancer and the third most common cause of cancer deaths, with over 780,000 new cases and over 740,000 deaths annually (7). Compared with many other solid tumors, HCC is more hypervascular and thus more dependent on angiogenesis for development and progression. Understanding the molecular mechanisms of HCC pathogenesis, including HCC angiogenesis, is thus important for improving its detection and treatment.
Zinc finger homeobox 3 (ZFHX3), a large transcription factor containing 23 zinc finger domains, four homeodomains, and multiple other motifs, was originally identified as ATBF1 for the AT motif–binding factor 1 that represses the transcription of α-fetoprotein (AFP) by binding to its promoter (8–11). Interestingly, the transcription of AFP in HCC cells is down-regulated under hypoxic conditions via the binding of HIF1A to the AFP promoter (12), which suggests that ZFHX3 could functionally associate with HIF1A in gene regulation tumor angiogenesis.
ZFHX3 is frequently mutated in advanced prostate cancer (13, 14), and deletion of Zfhx3 in mouse prostates induces intraepithelial neoplasia and promotes tumorigenesis induced by the loss of Pten (15), indicating a tumor suppressor activity of ZFHX3 in prostate cancer. In HCC, ZFHX3 is infrequently altered (16), whereas its mRNA expression has been inconsistently reported in published studies (17, 18).
In this study, we examined whether ZFHX3 and HIF1A functionally interact with each other using in vitro and in vivo models of HCC angiogenesis. We found that the expression of ZFHX3 was significantly increased by hypoxia via the binding of HIF1A to ZFHX3's promoter in HCC cells, and ZFHX3 then became necessary for the angiogenic activity of HIF1A via transcriptional activation of the VEGFA angiogenic effector. ZFHX3 silencing attenuated HCC angiogenesis and inhibited tumor growth in nude mice. In human HCCs, higher levels of ZFHX3 expression correlated with higher HIF1A expression and worse disease-free survival. These findings indicate that ZFHX3 is integral to HIF1A function in HCC angiogenesis.
Results
Hypoxia increases the expression of ZFHX3 at both mRNA and protein levels
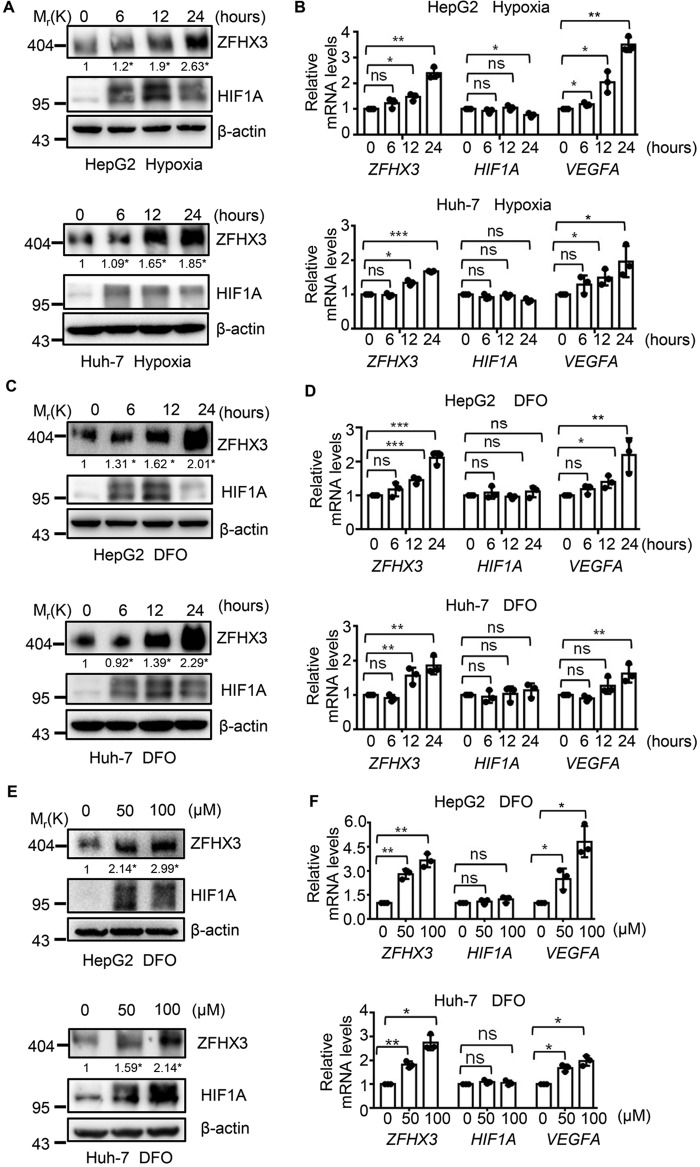
To explore whether ZFHX3 is functionally associated with HIF1A, we first determined whether ZFHX3 expression is affected by hypoxia, which induces the accumulation of HIF1A. The HCC cell lines HepG2 and Huh-7 were exposed to hypoxia (1% O2) for different times, and expression of ZFHX3 and HIF1A was analyzed. Consistent with previous studies (19), the HIF1A protein level was elevated after 6 h of hypoxia treatment, reached peak at 12 h, and then dropped at 24 h (Fig. 1A). Interestingly, the ZFHX3 protein level also increased after 6 h of hypoxia treatment and continued to increase at both 12 and 24 h of treatment (Fig. 1A and Fig. S1K). At the mRNA level, HIF1A was not increased by hypoxia, as hypoxia stabilizes the HIF1A protein mainly by post-translational modification (20, 21). ZFHX3 mRNA levels, however, were increased after 6 and 24 h of hypoxia treatment (Fig. 1B), which is consistent with changes in ZFHX3 protein level and suggests that hypoxia induces the up-regulation of ZFHX3 mRNA. As expected, VEGFA, a canonical downstream effector of HIF1A, was also up-regulated at the mRNA level by hypoxia (Fig. 1B).
Figure 1.
Hypoxia up-regulates ZFHX3 expression in HCC cells. A and B, HCC cell lines HepG2 and Huh-7 were cultured under hypoxia for the indicated times, and the expression of ZFHX3 and two regulators of hypoxia-induced angiogenesis, HIF1A and VEGFA, was detected by Western blotting and real-time PCR for protein (A) and mRNA (B), respectively. ZFHX3 band intensities were quantified and normalized to β-actin, and the results are shown below the ZFHX3 bands in A. C–F, HepG2 and Huh-7 cells were treated with the hypoxia-mimetic agent DFO for the indicated times at 50 μm (C and D) or the indicated concentrations for 24 h (E and F), and expression of the same set of molecules was analyzed as in A and B. Data are shown as mean ± S.D. Band intensity ratios below each lane of Western blottings in A, C, and E were the average from three independent experiments, and their scatter plots and statistical details are shown in Fig. S1 (S1K–S1M). *, p < 0.05. The statistical analysis of real-time PCR was based on three independent experiments (i.e. n = 3), and the value for each group in an experiment was the average of triplicates. *, p < 0.05; **, p < 0.01; ***, p < 0.001; ns, not significant.
HCC cell lines HepG2 and Huh-7 were also treated with deferoxamine (DFO), a chemical that has hypoxia-mimetic effects, and the same patterns of expression were detected for ZFHX3, HIF1A, and VEGFA in both a time- and dose-dependent manner (Fig. 1, C–F, and Fig. S1, L and M). Similar results were also obtained in the BEL-7402 cell line, which was originally reported to originate from a 53-year-old male patient with HCC but later was confirmed to be a HeLa derivative (22), and obtained in HeLa cells (Fig. S1, A–G). Therefore, hypoxia increases both the protein and mRNA levels of ZFHX3 in HCC cells.
Up-regulation of ZFHX3 by hypoxia depends on HIF1A
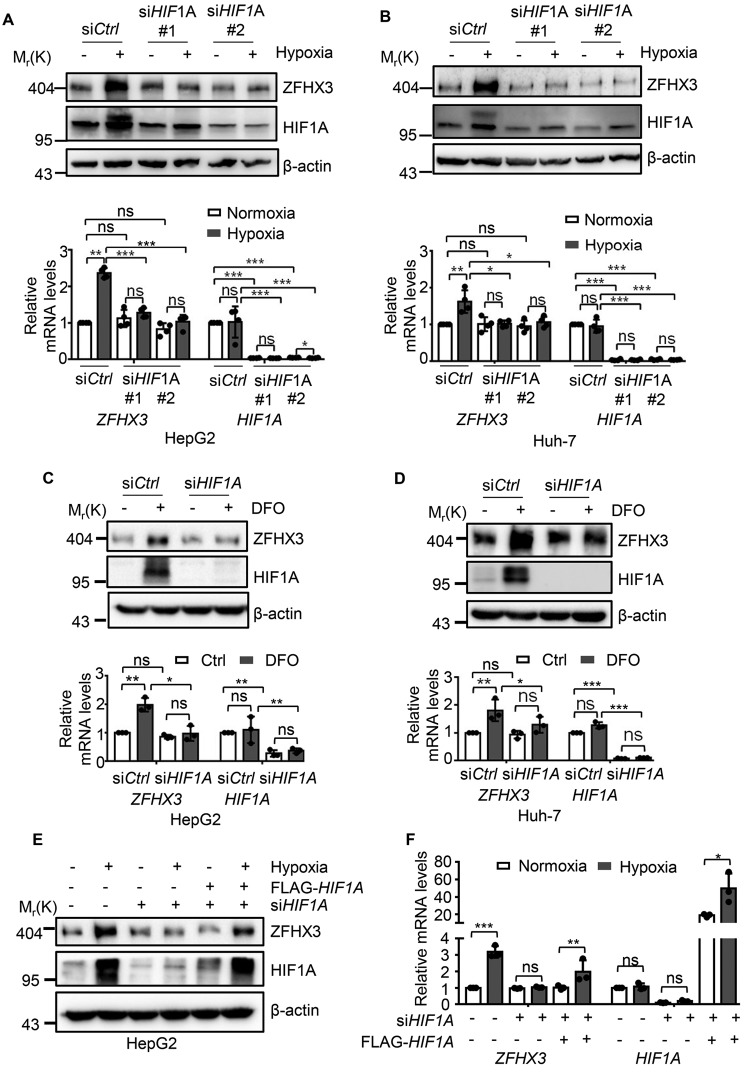
Hypoxia-induced factor-1A (HIF1A) is the key transcription factor that is stabilized by hypoxia to regulate the expression of hypoxia-responsive genes (3). We thus examined whether the up-regulation of ZFHX3 by hypoxia involves HIF1A. We silenced HIF1A by transfecting two siRNAs against HIF1A in the HCC cell lines HepG2 and Huh-7. Interestingly, the up-regulation of ZFHX3 protein and mRNA expression by hypoxia and DFO was dramatically inhibited after HIF1A silencing (Fig. 2, A–D). Considering that HIF2A has significant overlapping functions with HIF1A, we also knocked down HIF2A in HepG2 and Huh-7 cells and analyzed whether HIF2A is involved in ZFHX3 expression. Unlike HIF1A, silencing, HIF2A did not prevent the induction of ZFHX3 by hypoxia (Fig. S2, A–D). In addition, when HepG2 cells with HIF1A knockdown were transfected with HIF1A plasmid to restore the HIF1A protein level, hypoxia-induced ZFHX3 expression was partly restored (Fig. 2, E and F).
Figure 2.
HIF1A mediates hypoxia-induced ZFHX3 transcription in HCC cells. A–D, knockdown of HIF1A by RNAi in HepG2 (A and C) and Huh-7 (B and D) cells indicates that HIF1A is responsible for hypoxia-induced (A and B) or DFO-induced (C and D) ZFHX3 up-regulation, as measured for the expression of both protein (A–D, upper) and mRNA (A–D, lower) by Western blotting and real-time PCR, respectively. siHIF1A #1 and siHIF1A #2, siRNAs against HIF1A. E and F, ectopic expression of HIF1A by plasmid transfection in HepG2 cells with HIF1A silencing partially restored hypoxia-induced ZFHX3 expression, as measured by Western blotting (E) and real-time PCR (F) in HepG2 cells. Data are shown as means ± S.D. The statistical analysis for real-time PCR was based on three independent experiments (i.e. n = 3), and the value for each group in an experiment was the average of triplicates. *, p < 0.05; **, p < 0.01; ***, p < 0.001; ns, not significant.
Binding of HIF1A to ZFHX3 promoter is required for hypoxia to induce ZFHX3 transcription
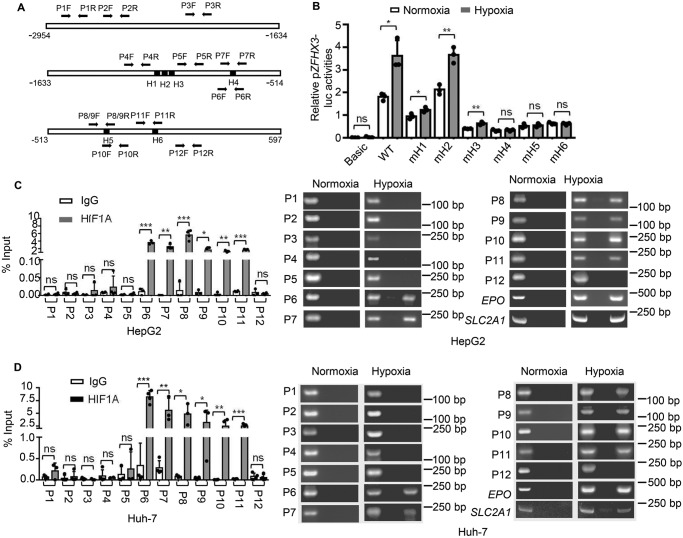
As a transcription factor, HIF1A binds to hypoxia-response elements (HREs) in the promoters of hypoxia-responsive genes to induce their transcription (23), and the core HRE sequence is RCGTG (R = A/G) (24). To determine whether ZFHX3 is a direct transcriptional target gene of HIF1A, we first analyzed ZFHX3's promoter and found six putative HREs (Fig. 3A). We then cloned the ZFHX3 promoter into the pGL3 luciferase reporter plasmid and analyzed ZFHX3's promoter activities. As expected, the ZFHX3 promoter displayed significant activity, and the activity was significantly increased by hypoxia (Fig. 3B, WT). Each of the six HREs in the ZFHX3 promoter was mutated, and the effect of promoter mutations on luciferase activity was analyzed. Mutation of HRE 2 (H2) did not affect promoter activity at all, suggesting that HRE 2 is not involved in ZFHX3 transactivation (Fig. 3B). Mutations in HREs 1 and 3 (H1 and H3) significantly reduced ZFHX3 promoter activity, but they did not eliminate the promoter's response to hypoxia (Fig. 3B). Mutations in HREs H4–H6 not only reduced ZFHX3 promoter activities but also eliminated the promoter's response to hypoxia (Fig. 3B). It is thus likely that, whereas HREs 1 and 3–6 are all involved in the maintenance of ZFHX3 transcription in HCC cells, only HREs 4–6 are responsible for the effect of hypoxia on ZFHX3 transcription.
Figure 3.
HIF1A binds to ZFHX3 promoter to mediate its induction by hypoxia. A, schematic of the ZFHX3 promoter from nucleotides −2954 to +597 relative to the transcription initiation site. Locations of six consensus HREs of pGL3-ZFHX3-Luc promoter are shown, which have the following sequences: H1, CCCCGTGC; H2, TCACGTGT; H3, TGACGTGG; H4, CCCGTGCT; H5, AGAGTGCA; and H6, GCCGTGCT. Location of PCR primers for ChIP–PCR are indicated by arrows (P1F to P9R). B, mutation of the 4th, 5th, or 6th HRE (mH4, mH5, and mH6, respectively) in the ZFHX3 promoter abolished its transactivation activity induced by hypoxia, as indicated by the promoter-luciferase reporter assay in HepG2 cells. C and D, HIF1A bound to the ZFHX3 promoter in HCC cells under hypoxia, as detected by ChIP–PCR in different cell lines (HepG2, C; Huh-7, D). EPO and SLC2A1, two known transcriptional targets of HIF1A, were used as positive controls. Data are shown as means ± S.D. The statistical analysis for luciferase assay was based on three independent experiments (n = 3) and that for real-time PCR was based on four independent experiments (n = 4). The value for each group in an experiment was the average of triplicates. *, p < 0.05; **, p < 0.01; ***, p < 0.001; ns, not significant.
ChIP–PCR assay was performed in HCC cell lines HepG2 and Huh-7 cells with HIF1A antibody to evaluate whether HIF1A physically binds to the promoter of ZFHX3 (Fig. 3, C and D). According to the six consensus HREs, 12 pairs of PCR primers were designed to amplify fragments that span different regions of the ZFHX3 promoter. Although no binding was detected under normoxia conditions, binding of HIF1A occurred in the proximal region (−367 to −168) of the ZFHX3 promoter under hypoxia, which contained HREs 5 and 6 (Fig. 3A). Binding was detectable for the fragment flanked by P6/7F and P6/7R, which spans HRE 4 (Fig. 3A). No binding to other fragments was detectable. Binding of HIF1A to the promoter of ZFHX3 was also detected by ChIP–PCR in the BEL-7402 cell line, a HeLa derivative (Fig. S3). Collectively, these findings suggest that under hypoxia, HIF1A directly binds to proximal HREs of the ZFHX3 promoter in HCC cells.
ZFHX3 activates the transactivation of VEGFA in coordination with HIF1A under hypoxia
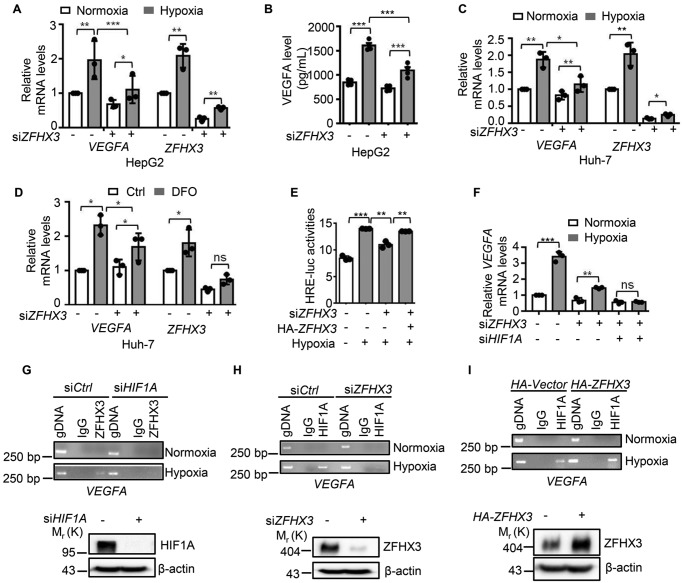
Considering that ZFHX3 is a transcription factor and that VEGFA is the most potent functional effector and a direct transcriptional target gene of HIF1A in hypoxia-induced tumor angiogenesis, it is likely that up-regulation of ZFHX3 by hypoxia also plays a role in the transcription of VEGFA. To test this notion, we knocked down ZFHX3 and detected VEGFA expression under hypoxia by real-time PCR and ELISA in HepG2 cells (Fig. 4, A and B), and we found that ZFHX3 knockdown greatly reduced hypoxia-induced VEGFA expression. Similar effects of ZFHX3 on VEGFA expression were also observed in Huh-7 cells under hypoxia and DFO treatment (Fig. 4, C and D). To further determine whether ZFHX3 plays a role in VEGFA transcription, we performed a promoter luciferase reporter assay using the HRE-luciferase reporter under different ZFHX3 conditions. As expected, RNAi-mediated ZFHX3 silencing significantly reduced hypoxia-induced HRE promoter activity, whereas ectopic expression of ZFHX3 partially rescued the effect (Fig. 4E). The functional necessity of ZFHX3 for VEGFA expression under hypoxia was also confirmed in the HeLa-derived BEL-7402 cells (Fig. S4, A and B). Although ZFHX3 was required for hypoxia to induce VEGFA expression (Fig. 4, A and B), knockdown of ZFHX3 did not completely eliminate the induction, as detected by real-time PCR (Fig. 4F). However, knockdown of both ZFHX3 and HIF1A completely eliminated the effect (Fig. 4F).
Figure 4.
ZFHX3 is required for transactivation of the hypoxia-responsive VEGFA in HepG2 cells. A and B, knockdown of ZFHX3 reduced hypoxia-induced VEGFA expression, as analyzed by both real-time PCR (A) and ELISA (B), and experiments were performed in duplicate for each group. C and D, similarly, in Huh-7 cells, knockdown of ZFHX3 reduced hypoxia-induced (C) or DFO-induced (D) VEGFA expression, as analyzed by real-time PCR. E, hypoxia-induced HRE promoter luciferase activity was reduced by the knockdown of ZFHX3 and increased by ectopic expression of ZFHX3 in HepG2 cells. F, ZFHX3 was required for hypoxia to induce VEGFA expression in HepG2 cells under hypoxia, as measured by real-time PCR. G, knockdown of HIF1A dramatically reduced the binding of ZFHX3 to VEGFA promoter in HepG2 cells under hypoxia. Western blotting (lower panel) confirmed the knockdown effect. H and I, knockdown of ZFHX3 dramatically reduced and ectopic expression of ZFHX3 increased the binding of HIF1A to the promoter of VEGFA in HepG2 cells under hypoxia. IgG was used as the isotype control. Western blotting (lower panel) confirmed the knockdown effect. Data are shown as means ± S.D. The statistical analysis for both luciferase assay and real-time PCR was based on three independent experiments (n = 3), and the value for each group in an experiment was the average of triplicates. *, p < 0.05; **, p < 0.01; ***, p < 0.001; ns, not significant.
To further test the effect of ZFHX3 on VEGFA transcription and whether it is related to the effect of HIF1A, we performed ChIP–PCR with ZFHX3 antibody in HepG2 cells under hypoxia, and we found that ZFHX3 bound to the promoter of VEGFA (Fig. 4G, left). Interestingly, knockdown of HIF1A decreased the binding of ZFHX3 to the VEGFA promoter, as no ZFHX3-bound VEGFA promoter DNA was detectable in the ChIP–PCR assay after HIF1A knockdown (Fig. 4G, right). Conversely, although HIF1A bound to the promoter of VEGFA under hypoxia as expected (Fig. 4, H and I), knockdown of ZFHX3 decreased (Fig. 4H) while ectopic expression of ZFHX3 increased (Fig. 4I) the amount of HIF1A-bound VEGFA promoter DNA. Therefore, ZFHX3 is also required for hypoxia to up-regulate VEGFA, and ZFHX3 appears to coordinate VEGFA's transcriptional activation under hypoxia.
ZFHX3 physically associates with HIF1A
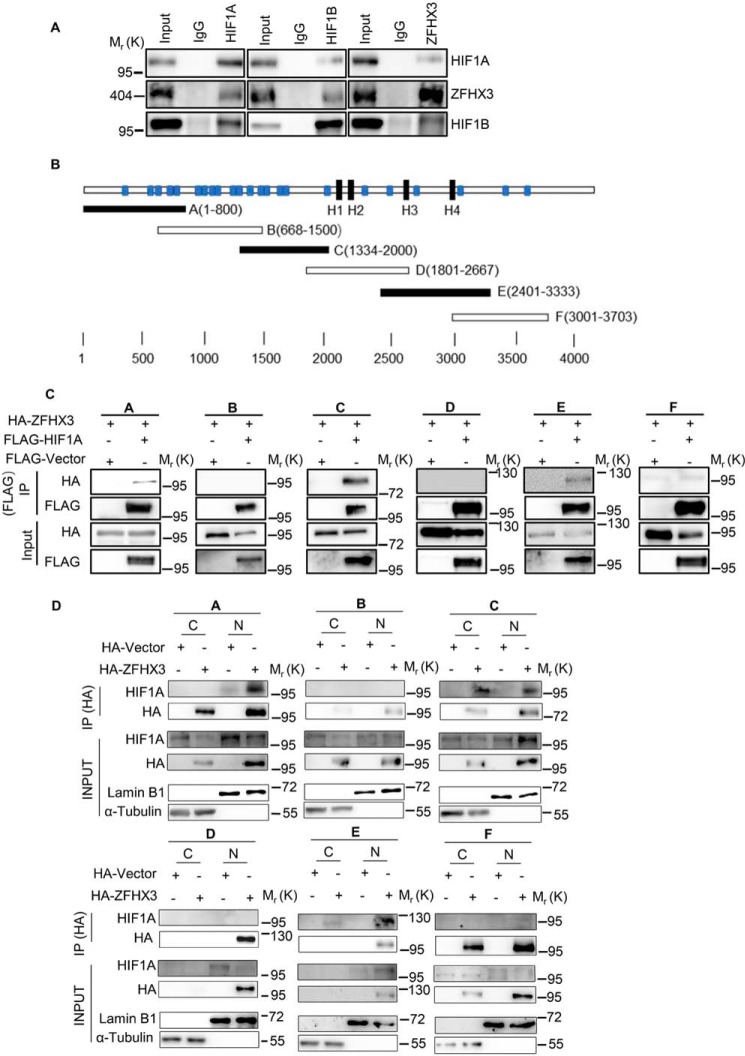
The findings that the ZFHX3 protein level affected the binding of HIF1A to VEGFA promoter (Fig. 4, H and I) and that HIF1A is required for ZFHX3 to bind the VEGFA promoter (Fig. 4G) suggest that ZFHX3 and HIF1A associate with each other in VEGFA transcription. To test this notion, we performed co-IP and Western blotting in HepG2 cells treated with hypoxia to detect the association between the two. HIF1A dimerizes with HIF1B to drive gene transcription, so both HIF1A and HIF1B were analyzed. Interestingly, both ZFHX3 and HIF1B were detected in the HIF1A protein precipitates (Fig. 5A, left); HIF1A and ZFHX3 were detected in the HIF1B protein precipitates (Fig. 5A, middle), and HIF1A and HIF1B were detected in the ZFHX3 protein precipitates (Fig. 5A, right), indicating an interaction between ZFHX3 and the HIF1A/HIF1B complex under hypoxia.
Figure 5.
ZFHX3 physically interacts with the HIF1A complex. A, detection of protein association between HIF1A, ZFHX3, and HIF1B in HepG2 cells by co-IP with HIF1A (left), HIF1B (middle), or ZFHX3 antibody (right) and Western blotting with the indicated antibodies. Input (1/20 of whole-cell lysate) indicates cell lysate not subjected to IP. B, schematic of full-length ZFHX3 (3703 residues, horizontal bar) with four homeodomains (black rectangle) and 23 zinc fingers (blue rectangle). The six overlapping fragments of ZFHX3 were named A–F. C, six HA-tagged overlapping ZFHX3 fragments and FLAG-tagged HIF1A were ectopically expressed in HepG2 cells under hypoxia, and IP and Western blotting were applied to test ZFHX3–HIF1A interactions. D, association of HIF1A with different fragments of ZFHX3 in HeLa cells ectopically-expressed HA-tagged ZFHX3 fragments under hypoxia. Nuclear and cytoplasmic fractions were separated, and IP and Western blotting were applied to each fraction. Input (1/50 of cytoplasmic or nuclear lysate) indicates the lysate not subjected to IP. C, cytoplasm; N, nucleus.
We also expressed six HA-tagged overlapping fragments of ZFHX3 (Fig. 5B) and FLAG-tagged HIF1A (FLAG-HIF1A) in 293T cells and performed IP and IB with FLAG antibody. Fragments A, C, and E of the six interacted with HIF1A (Fig. 5C). Different fragments of ZFHX3 may have different cellular localizations, which could affect their interactions with HIF1A, so we separated the nucleus and the cytosol from hypoxia-treated cells and performed IP and IB. Although all fragments were primarily located in the nucleus, fragments A–C and F were also detectable at varying levels in the cytoplasm, whereas fragments D and E were not (Fig. 5D). Again, HIF1A was detected in the precipitates of fragments A, C, and E from the nucleus but not in those of B, D, and E (Fig. 5D). A weak but detectable signal was also present in the cytoplasmic fraction for C and E (Fig. 5D). Therefore, ZFHX3 and HIF1A physically interact with each other in the nucleus involving multiple regions in fragments A, C, and E of ZFHX3.
ZFHX3 is crucial for hypoxia to promote tube formation and migration of endothelial cells via VEGFA
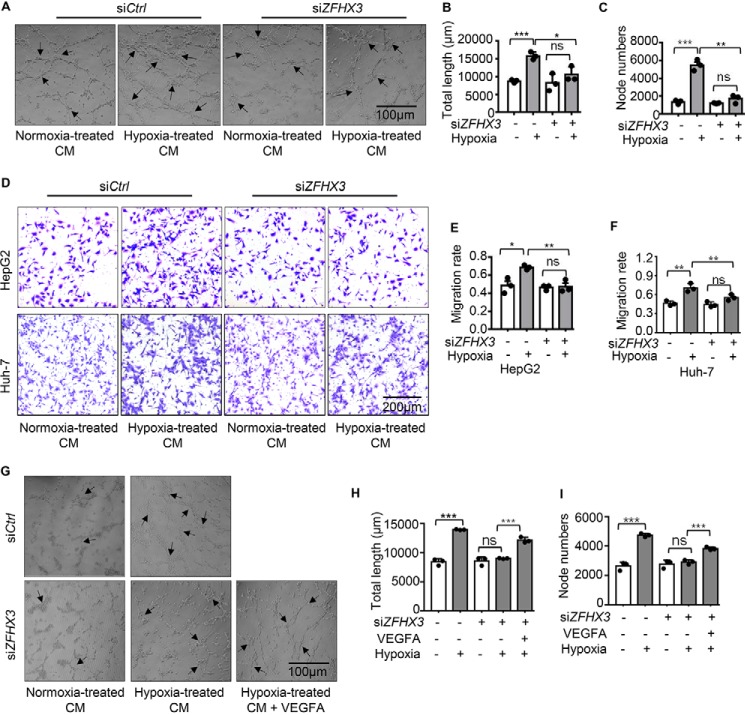
Hypoxia promotes angiogenesis by activating multiple pro-angiogenic pathways, particularly the HIF1A pathway, and VEGFA is an essential functional mediator of HIF1A. Based on the findings that ZFHX3 was necessary for VEGFA transcription (Fig. 4), ZFHX3 and HIF1A coordinated with each other in VEGFA transcription, and ZFHX3 was up-regulated by hypoxia via HIF1A, it is reasonable to propose a functional significance of ZFHX3 in angiogenesis. In testing this idea, we performed tube formation and migration assays using human umbilical vein endothelial cells (HUVECs), which are in vitro indicators of hypoxia-induced angiogenesis. Conditioned media (CM) from HepG2 cells with or without ZFHX3 knockdown and hypoxia treatment were used to treat HUVECs. Although CM of hypoxia-treated tumor cells significantly increased the total length of tubes and the number of tube nodes as expected, knockdown of ZFHX3 almost eliminated the increases (Fig. 6, A–C), indicating a crucial role of ZFHX3 in HUVEC tube formation. In a transwell assay, CM from hypoxia-treated HepG2 and Huh-7 cells promoted the migration of HUVECs as expected, but again hypoxia's promoting effect was eliminated by ZFHX3 knockdown (Fig. 6, D–F). Collectively, these findings indicate that attenuation of ZFHX3 up-regulation in HCC cells prevents hypoxia from promoting tube formation and migration of HUVECs, in vitro indicators of angiogenesis.
Figure 6.
ZFHX3 plays a necessary role in the migration and tube formation of HUVECs involving VEGFA. A–C, tube formation of HUVECs did not increase after incubation with CM from HepG2 cells with RNAi-mediated ZFHX3 silencing under hypoxia, as indicated by cell images (A), the total length of tubes (B), and the number of nodes (C). D–F, migration of HUVECs was not significantly affected by CM from HepG2 or Huh-7 cells treated with siZFHX3 as in A–C, as indicated by migrated cells (D) and their quantification in the transwell assay (E and F). G–I, addition of VEGFA to CM partially rescued the inhibitory effect of ZFHX3 knockdown on tube formation in HUVECs, as indicated by cell images (G), the total length of tubes (H), and the number of nodes (I). Statistical analysis was based on three independent experiments (n = 3), and the value for each group in an experiment was the average of triplicate (transwell) or five fields (tube formation). Scale bar for D is 200 μm, and scale bars for A and G are 100 μm. *, p < 0.05; **, p < 0.01; ***, p < 0.001; ns, not significant.
VEGFA was transcriptionally up-regulated by ZFHX3 and HIF1A under hypoxia, and ZFHX3 silencing down-regulated VEGFA, which suggests that down-regulation of VEGFA by ZFHX3 silencing has functional significance. We thus added VEGFA to the CM from cells with ZFHX3 knockdown rescued tube formation of HUVECs (Fig. 6, G–I). Therefore, ZFHX3 plays a crucial role in hypoxia-induced angiogenesis. Given that both HIF1A and ZFHX3 play important roles in hypoxia-induced angiogenesis, we also performed migration assays with CM from HepG2 and Huh-7 cells. The results show that ZFHX3 or HIF1A was sufficient to prevent the promotion of endothelial cell migration by hypoxia, and simultaneous knockdown of both ZFHX3 and HIF1A did not show an additive effect (Fig. S5).
ZFHX3 promotes xenograft tumor growth likely due to its angiogenic activity
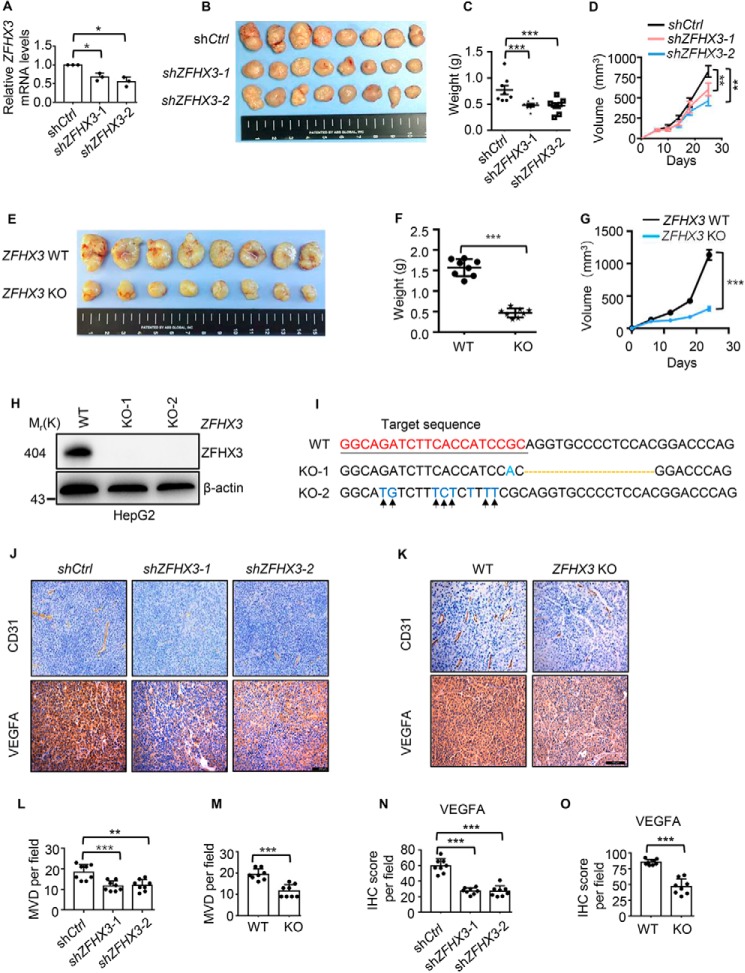
To test whether the necessity of ZFHX3 for hypoxia–HIF1A–VEGFA signaling to promote angiogenic activity affects tumor growth, we knocked down ZFHX3 using lentiviruses expressing ZFHX3 shRNAs or knocked out ZFHX3 using the CRISPR–Cas9 system in HepG2 cells, and we injected cells into nude mice subcutaneously for tumor growth analysis. Both knockdown and knockout of ZFHX3 significantly reduced tumor growth, as indicated by tumor images and weights at excision and tumor volume-based growth curve (Fig. 7, A–G). The inhibitory effect of knockout was more potent than that of knockdown as expected. Deletion of ZFHX3 in isolated clones of HepG2 cells was confirmed by Western blotting (Fig. 7H) and sequencing (Fig. 7I). Tumor tissue sections were immunohistochemically stained with anti-CD31 to detect microvessels and with VEGFA antibody to detect its expression (Fig. 7, J and K). Knockout or knockdown of ZFHX3 significantly decreased the number of microvessels in xenograft tumors, as indicated by IHC staining of CD31 (Fig. 7, J–M). Knockout or knockdown of ZFHX3 also reduced VEGFA protein expression (Fig. 7, J, K, N, and O), which is consistent with in vitro findings. IHC staining also detected the hypoxia marker CA9 in foci within xenograft tumors (Fig. S6), a typical pattern of CA9 expression in hypoxic areas. ZFHX3 thus has a promoting effect on both tumor growth and angiogenesis of HCC cells.
Figure 7.
Loss of ZFHX3 attenuates xenograft tumor growth of HCC cells likely via compromised angiogenesis. A–D, ZFHX3 was knocked down by shRNAs against ZFHX3 (shZFHX3-1 and -2) in HepG2 cells, as confirmed by real-time qPCR (A), and subcutaneous tumorigenesis assay was performed. Tumor growth is indicated by tumor images (B) and tumor weights (C) at excision and tumor growth curves (D). E–G, clone KO-1 was subjected to tumorigenesis assay as in B–D. H and I, two clones of HepG2 cells with CRISPR-Cas9–mediated ZFHX3 truncation, KO-1 and KO-2, were confirmed for lack of ZFHX3 protein by Western blotting (H) and mutations in ZFHX3 by DNA sequencing (I). I, CRISPR-Cas9 target sequence is underlined, and nucleotide deletion in clone KO-1 is indicated by a dotted line, and the mutations in clone KO-2 are marked by arrows. J and K, detection of microvessels by IHC staining of CD31 (upper) and VEGFA (lower) expression in xenograft tumors by IHC staining. L and M, microvessel densities (MVD) were quantified based on CD31 staining of endothelial cells. N and O, quantitative analyses of VEGFA expression based on IHC staining in J and K. The statistical analysis for microvessel densities (L and M) and VEGFA score of IHC staining was based on the average of three fields from all eight tumors (n = 8). Scale bars in J and K, 200 μm. *, p < 0.05; **, p < 0.01; ***, p < 0.001; ns, not significant.
Up-regulation of ZFHX3 and its correlation with both HIF1A up-regulation and worse patient survival in HCC
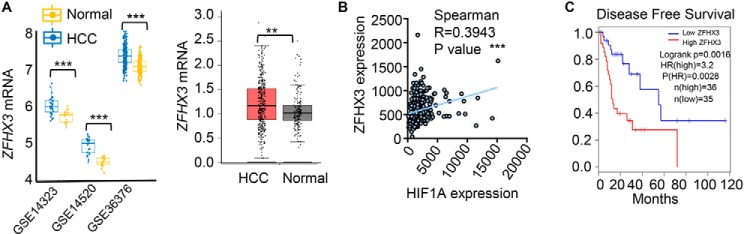
To determine the clinical relevance of ZFHX3-promoted angiogenesis in HCC, we collected HCC samples in the GEO and TCGA databases that had genome-wide expression data, and we analyzed ZFHX3 expression. The ZFHX3 mRNA level was clearly higher in HCC samples than in normal liver tissues based on the GEO and RNA-Seq data (Fig. 8A). We then tested whether ZFHX3 expression correlates with HIF1A expression in HCC specimens. Spearman correlation analysis demonstrated that expression levels of ZFHX3 and HIF1A positively correlated with each other in the TCGA database (Fig. 8B).
Figure 8.
Up-regulation of ZFHX3 and its correlation with HIF1A up-regulation and worse patient survival in HCC. A, ZFHX3 mRNA levels were higher in HCC tissues than in normal liver tissues, as revealed by the analyses of microarray expression data from three GEO datasets (left) and RNA-Seq data (right) of HCC in the TCGA database. RNA sequence data for normal liver tissues was from the TCGA and GTEx database. B, ZFHX3 mRNA levels correlated with HIF1A levels in HCC, as revealed by Spearman correlation analysis using the TCGA database. C, HCC with higher ZFHX3 expression had poorer disease-free survival, as revealed by the survival analysis of HCC samples with both patient survival data and ZFHX3 expression information in the TCGA database. The Gene Expression Profiling Interactive Analysis (GEPIA; RRID:SCR_018294), an online tool, was used to test the relationship between ZFHX3 expression levels and disease-free survival. **, p < 0.01; ***, p < 0.001.
In the TCGA database, some HCC samples have both patient survival data and ZFHX3 expression information. Kaplan-Meier analysis of such cases demonstrated that patients with higher ZFHX3 expression levels had poorer disease-free survival (Fig. 8C), further implicating higher ZFHX3 expression in HCC development.
Discussion
ZFHX3 is an angiogenic transcription factor in HCC cells
Angiogenesis is crucial for the adaptation of tumor cells to hypoxic stress by providing oxygen and nutrients for the growth and progression of tumors. Angiogenesis occurs at a high level in HCC because the median O2 partial pressure in human HCC (6 mm Hg) is about one-fifth of that in normal liver tissue (30 mm Hg) (25), and thus HCC has greater vascularization and is more dependent on angiogenesis for growth (5). Angiogenesis is regulated by several signaling pathways via key transcription factors and downstream effectors. HIF1A is the most potent known transcription factor, and VEGFA is a cytokine that is induced by HIF1A to mediate HIF1A's function. HIF1A and VEGFA thus form a signaling axis that promotes tumor angiogenesis in various types of cancers, including HCC. In addition to hypoxia, various oncogenic signaling pathways promote tumor angiogenesis by activating the HIF1A/VEGF axis (26). For example, the small ubiquitin-like modifier E3 ligase Cbx4 enhances HIF1A sumoylation, which in turn increases its transcriptional activity and VEGF expression and subsequent angiogenesis and tumor growth in HCC (27). Activation of the PI3K or MAPK signaling also up-regulates VEGF (28). Accordingly, targeting the HIF1A/VEGF axis has become a meaningful approach for the treatment of HCC (29, 30).
Our findings in this study establish the ZFHX3 transcription factor as a novel angiogenic factor in HCC. This conclusion is supported by multiple lines of evidence, including tube formation and migration assays of HUVECs (Fig. 6 and Fig. S5) and the essential role of ZFHX3 in transcriptional induction of the VEGFA gene (Fig. 4, Fig. S4), which encodes a heparin-binding protein that induces proliferation and migration of vascular endothelial cells in both physiological and pathological angiogenesis (31). In addition, silencing ZFHX3 reduced tumor vascularization in the HepG2 xenograft model of HCC (Fig. 7, J–M). These functional studies indicate that, like HIF1A, ZFHX3 is also essential for hypoxia to induce angiogenesis in HCC cells. Nevertheless, whereas ZFHX3 clearly plays a role in angiogenesis, there is also a possibility that other mechanisms are also involved in ZFHX3-promoted tumor growth.
ZFHX3 is both a transcriptional target and a functional partner of HIF1A in HCC cells
HIF1A is the master regulatory transcription factor under hypoxia. It is composed of two subunits: HIF1A and HIF1B. Although HIF1B is constitutively expressed, HIF1A is maintained at a low protein level by the ubiquitin proteasome pathway under normoxia; only under hypoxia is HIF1A stabilized and translocated into the nucleus to promote angiogenesis (32). Although a large number of genes are regulated by HIF1A, only some are functional effectors of HIF1A. ZFHX3 is clearly a functional effector of HIF1A in HCC, as ZFHX3 is a transcriptional target gene of HIF1A (Fig. 3 and Fig. S3), and even in HCC specimens, ZFHX3 up-regulation significantly correlated with HIF1A up-regulation (Fig. 8). More importantly, up-regulated ZFHX3 is necessary for hypoxia to induce the expression of VEGFA (Fig. 4 and Fig. S4), a cytokine essential for tumor angiogenesis, as knockdown of ZFHX3 down-regulated VEGFA (Fig. 4 and Fig. S4) via direct binding to the promoter of VEGFA (Fig. 4 and Fig. S4). In addition, the angiogenic activity of ZFHX3, as indicated by tube formation and migration of HUVECs, clearly involved VEGFA (Fig. 6 and Fig. S5). Furthermore, ZFHX3 and HIF1A coordinated to induce VEGFA transcription, as the binding of ZFHX3 to VEGFA promoter depended on HIF1A (Fig. 4 and Fig. S4), and protein levels of ZFHX3 also affected the binding of HIF1A to the VEGFA promoter (Fig. 4 and Fig. S4). Interestingly, ZFHX3 and HIF1A proteins indeed associated with each other in hypoxia-treated HCC cells (Fig. 5), and the interaction involved more than one region of ZFHX3 protein (Fig. 5).
Therefore, while induced by hypoxia via HIF1A, ZFHX3 also functions as part of the HIF1A/VEGFA signaling axis in hypoxia-induced angiogenesis in HCC. Nevertheless, several important questions remain to be addressed regarding how ZFHX3 functions in the context of HIF1A/VEGFA signaling. For example, ZFHX3 is quite a large transcription factor with 23 zinc fingers, whereas HIF-1 functions in a heterodimer. Although certain cofactors of HIF1A have been identified to have a role in the binding of HIF1A to its target gene promoters, including the Tat-interactive protein Tip60 (33), whether ZFHX3 is a cofactor of HIF1A and how these two very different transcription factors coordinate to induce VEGFA transcription are interesting but unanswered questions. Furthermore, the SUMOylation status of HIF1A is important for its stability (34), and ZFHX3 SUMOylation could also modulate its function (35). Whether SUMOylation is involved in the ZFHX3–HIF1A interaction is also an interesting but unanswered question. Finally, whether genes other than VEGFA are transcriptionally regulated by ZFHX3 and HIF1A remains to be identified.
ZFHX3–HIF1A interaction has clinical implications in human HCC
The association between HIF1A and ZFHX3 also appeared to occur in human HCC, implicating ZFHX3 in HCC progression. HIF1A and VEGFA are overexpressed in HCC, and the HIF1A/VEGFA axis clearly plays an important role in the development and progression of human HCC (30). ZFHX3 not only promoted angiogenesis and tumor growth of HCC cells in a xenograft model (Figs. 6 and 7 and Fig. S5), it was also up-regulated in human HCC (Fig. 8), and its up-regulation correlated with HIF1A up-regulation as well as worse HCC patient survival (Fig. 8). It is thus likely that ZFHX3 also plays a role in human HCC via its function as part of the HIF1A/VEGFA axis. The role of ZFHX3 in HCC development and progression could have clinical implications. For example, as a therapeutic approach, inhibition of HIF1A activity can be achieved by selectively cutting off its functional dependence on its coactivator (33), and the HIF1A–ZFHX3 interaction could provide a similar opportunity for targeted therapy of HCC. In addition, reagents targeting ZFHX3 could be developed to constrain tumor angiogenesis and treat other hypoxia-related diseases.
Roles of ZFHX3 in tumorigenesis are tissue type–dependent
Interestingly, the role of ZFHX3 in human tumorigenesis appears to be tissue-dependent. As stated under the Introduction, ZFHX3 clearly plays a tumor suppressor role in prostate cancer because its inactivating deletions/mutations not only frequently occur in advanced human prostate cancers (13, 14) but also cause neoplastic lesions in mouse prostates (15). The findings in this study indicate an oncogenic role of ZFHX3 in HCC (Figs. 1–8 and Figs. S1–S6), even though genetic alterations of ZFHX3 are infrequent in HCC. A gene can clearly have opposing functions in tumor cells. For example, although the WT KLF5 transcription factor slows cell proliferation and tumorigenesis, its deacetylated mutant has opposing functions (36, 37). Our unpublished study suggests that ZFHX3 also plays an oncogenic role in breast cancer. Molecularly, although interaction with HIF1A to up-regulate VEGFA was established in this study as a mechanism by which ZFHX3 promotes HCC tumor growth, interaction of ZFHX3 with estrogen receptor beta (ERβ) to repress MYC and regulate other genes is an important mechanism underlying ZFHX3's tumor suppressor function in prostate cancer (38). Both mechanisms depend on ZFHX3's transcription factor function, yet the outcomes are opposing. Therefore, understanding the molecular mechanisms through which interactions of ZFHX3 with different transcription factors lead to different functions in different types of cancers is important not only to the field of gene regulation but also to the field of cancer biology. ZFHX3 provides a unique opportunity for addressing this question.
In this study, we examined whether and how ZFHX3 plays a role in the angiogenesis of human HCC cells. We found that ZFHX3 was dramatically up-regulated by hypoxia in HCC, and the up-regulation depended on the binding of HIF1A to the promoter of ZFHX3. Functionally, ZFHX3 was necessary for hypoxia to promote angiogenesis and tumor growth, and ZFHX3 exerted such functions by coordinating with HIF1A to induce the transcription of VEGFA in HCC cells. In human HCC, ZFHX3 was up-regulated, and the up-regulation correlated with HIF1A up-regulation and worse patient survival. ZFHX3 is thus a newly-identified angiogenic factor that could lead to novel therapeutic opportunities for the treatment of HCC.
Experimental procedures
Cell lines, plasmids, and transfection
HCC cell lines HepG2 and Huh-7, along with the BEL-7402/HeLa cell line and HUVECs, were purchased from the BeNa Culture Collection (Beijing, China). Human embryonic kidney 293T cells were purchased from the ATCC (Manassas, VA). These cell lines were cultured in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum (Gibco) in a humidified incubator (37 °C and 5% CO2). They were authenticated using short tandem repeats DNA profiling.
Mammalian expression plasmid for HIF1A pFLAG–CMV–HIF1A and that for HRE luciferase reporter pGL3–HRE–luciferase were kindly provided by Dr. Yushan Zhu of Nankai University (39). Expression plasmid for HA-tagged ZFHX3 pKXUa1–HA–ZFHX3 and those for the six fragments with HA tag, i.e. pcDNA3–HA–ZFHX3-A–F, were constructed in our previous study (40). The pZFHX3–Luc promoter luciferase reporter plasmid for ZFHX3, which was constructed and named as pATBF1-Luc1 in another previous study (41), was used as the template to generate mutants (CGTG to TACA) for the six HREs in ZFHX3 promoter using PCR-based cloning. These mutants were named pGL3–ZFHX3–Luc–mHRE1 to pGL3–ZFHX3–Luc–mHRE6, and primer sequences for PCR are shown in Table 1.
Table 1.
Primer sequences for cloning HRE mutants of pGL3-ZFHX3-Luc
| Primer name | Forward primer (5′–3′)/reverse primer (5′–3′) |
|---|---|
| mHRE1 | ACCCGGCCCCTACACTCTCACGTGTGACT/CACGTGAGAGTGTAGGGGCCGGGTGGGGG |
| mHRE2 | CGTGCTCTCATACATGACTGAATTGGGCT/AATTCAGTCATGTATGAGAGCACGGGGGC |
| mHRE3 | CTGGCTGTGATACAGACGGGGCCCAGGGA/GGGCCCCGTCTGTATCACAGCCAGGCCAC |
| mHRE4 | TGGGAAAGGGTGTAATCTTTGCTATCTCT/TAGCAAAGATTACACCCTTTCCCAGGGAG |
| mHRE5 | CCCGTGCTCTTGTATGTGACTGAATTGGG/TTCAGTCACATACAAGAGCACGGGGGCCG |
| mHRE6 | CTGCGCCCGGTGTAACTGTAGATGTCAGG/CATCTACAGTTACACCGGGCGCAGGCCGG |
Plasmids were transfected into cells using the Lipofectamine 2000 reagent (Invitrogen). Small interfering RNAs (siRNAs) were synthesized by RiboBio (Guangzhou, China) and transfected into cells using the Lipofectamine RNAiMax reagent (Invitrogen). Sequences of siRNAs used in this study are listed in Table 2. Hypoxic conditions were achieved by culturing cells in a hypoxia chamber (Billups Rothenberg, Del Mar, CA) with a mixed gas of 1% O2, 5% CO2, and 94% N2. Chemical deferoxamine mesylate (catalog no. ab120727, Abcam, Cambridge, MA) was also used to treat cells to mimic hypoxic conditions. All cell lines were authenticated by the short tandem repeats of DNA profiling.
Table 2.
List of sequences for siRNAs against different genes
| Gene name | siRNA sequences (5′–3′) |
|---|---|
| HIF1A-1 | GGACACAGAUUUAGACUUG |
| HIF1A-2 | GAUGGAAGCACUAGACAAA |
| HIF2A | GCAAAUGUACCCAAUGAUA |
| ZFHX3 | AGAAUAUCCUGCUAGUACA |
Antibodies and Western blotting
Cells were lysed using radioimmunoprecipitation assay (RIPA) buffer (150 mm NaCl, 50 mm Tris, pH 7.5, 1 mm EGTA, 1 mm EDTA, 1% Triton, 1% sodium deoxycholate, 0.1% SDS, and protease inhibitors (Roche Applied Science). Equal amounts of cellular protein were subjected to SDS-PAGE, and proteins were then transferred to polyvinylidene fluoride membranes. After incubation with 5% nonfat milk in TBST buffer (25 mm Tris, 150 mm NaCl, 0.1% Tween 20, pH 7.5) for 1 h at room temperature, the membrane was probed with primary antibodies overnight, and washed three times with TBST (each for 10 min). The membrane was incubated with HRP–conjugated anti-rabbit (catalog no. 7074S, Cell Signaling Technology, Danvers, MA) or anti-mouse (catalog no. 7076S, Cell Signaling Technology) antibodies at a 1:5000 dilution for 2 h. The blots were washed three times with TBST and incubated in Western Bright ECL substrate (Advansta, Menlo Park, CA), and images were captured using the ImageQuant LAS 4000 system (GE Healthcare). For the detection of ZFHX3 protein, 4% SDS-PAGE and the spectra multicolor high-range protein marker (catalog no. 26625; Thermo Fisher Scientific, Santa Clara, CA) were used. For other proteins, 8–12% SDS-PAGE and the PageRuler Plus Prestained protein marker (catalog no. 26616; Thermo Fisher Scientific) were used. The following antibodies were used in this study: anti-HIF1A (catalog no. NB100-479, 1:2000 dilution, Novus Biologicals, Littleton, CO), anti-HIF1B (catalog no. 5537, 1:1000 dilution, Cell Signaling Technology, Danvers, MA), β-actin (catalog no. A1978200UL, 1:10,000 dilution, Sigma), and ZFHX3 (homemade, 1:800 dilution).
Immunoprecipitation
Cells were collected and lysed in 1 ml of NP-40 lysis buffer (150 mm NaCl, 50 mm Tris-HCl, pH 7.5, 1% Nonidet P-40) plus protease inhibitors (Roche Applied Science) for 50 min on a rotor at 4 °C. After centrifugation at 12,000 × g for 10 min, the lysates were immunoprecipitated with specific antibodies overnight at 4 °C, and were then incubated with protein-G plus-Sepharose (GE Healthcare) for an additional 2 h. Thereafter, the precipitants were washed five times with the NP-40 lysis buffer, eluted by adding an equal volume of 2× loading buffer, boiled for 5 min, and analyzed by SDS-PAGE.
RNA isolation and real-time PCR
Total RNA was extracted from cultured cells with the TRIzol reagent (Invitrogen) according to the manufacturer's instructions. The first-strand cDNA was synthesized with oligo(dT) and random primers using the Moloney murine leukemia virus reverse transcriptase system (Promega, Madison, WI). The mRNA levels of indicated genes were quantified by qRT-PCR using the SYBR Green MasterMix reagent (Takara, Tokyo, Japan) on the Mastercycler ep Realplex thermal cycler system (Eppendorf, Shanghai, China). Expression levels were calculated using the 2−ΔΔCt method and normalized to that of β-actin. Primer sequences used for qRT-PCR are shown in Table 3.
Table 3.
Primer sequences for real-time qPCR analyses of different genes
| Gene name | Forward primer (5′–3′)/reverse primer (5′–3′) |
|---|---|
| ZFHX3 | TGTTCCAGATCGAGATGGGAAT/CTTTCCCAGATCCTCTGAGGTTT |
| VEGFA | GGCTGGCAACATAACAGAGAA/CCCCACATCTATACACACCTCC |
| HIF1A | CACCACAGGACAGTACAGGAT/CGTGCTGAATAATACCACTCACA |
| Actin | ATTGGCAATGAGCGGTTCC/GGTAGTTTCGTGGATGCCACA |
ChIP assay
The SimpleChIP® enzymatic chromatin IP kit (catalog no. 9003S, Cell Signaling Technology) was used for ChIP according to the manufacturer's protocol. Briefly, cultured cells were treated with 1% formaldehyde at room temperature for 10 min; 10× glycine was added to quench cross-linking, and cells were then washed, harvested, and lysed in the kit's lysis buffer. Cell lysates were digested with micrococcal nuclease, sonicated, and centrifuged to remove debris. ChIP was performed with the anti-HIF1A antibody (catalog no. NB100-479, Novus Biologicals, Littleton, CO), anti-ZFHX3 antibody, and IgG (negative control) overnight, and protein A–agarose beads were added. After a 2-h incubation, beads were washed sequentially with low-salt wash buffer, high-salt wash buffer, LiCl wash buffer, and Tris-EDTA buffer. The eluted immunocomplex was incubated with 5 m NaCl with proteinase K at 65 °C for 2 h, and DNA was purified and used as a template for PCR. Primer sequences for VEGF promoter (from −1216 to −883) are 5′-CACAGACCTTCACAGCCATC-3′ and 5′-CCCAGCGTAGACAGTTGAGT-3′; and those for ZFHX3 promoter are listed in Table 4.
Table 4.
Primer sequences for ChIP-PCR analysis of ZFHX3 promoter region
| Primer name | Forward primer (5′–3′)/reverse primer (5′–3′) |
|---|---|
| P1 | AAACCCGCTGTACTGTGA/ATTCTACCGAGCCAAACC |
| P2 | CAGCCAGCTCAGCGTTAG/CTCGCAGTTCTCCATACCC |
| P3 | TAGTCCCTCATTTCCATAA/TACTGCCACTGTCCCAAG |
| P4 | CACCTGTTCTTGGGCCTGAAGTCAG/GGGAATCATGTCCGATTA |
| P5 | ATACTGCTCTTCGCCTCAT/GTCAGAATCCCACCCTCA |
| P6 | GGGAGATAGAAGGCGCCC/ATAGCAAAGATCGTGCCC |
| P7 | CCGCTTTAAATCTTACCC/TTGAGGCCAGAGAAAGAG |
| P8 | AAAGCAGTTAATAGGATGGGTG/ATCGGGCGAGAAGAAAGG |
| P9 | AAAGCAGTTAATAGGATGGGTG/GAATCGGGCGAGAAGAAA |
| P10 | CACATTGGCTCCTGTCCC/CTCGCTCATCAAAGGTCA |
| P11 | ACCTTTGATGAGCGAGGGGTCA/CTGCGCTCGCTCCGCTTG |
| P12 | CTTCCGCTTTGTTGCTGT/TCACCCACGGGGCGCGGC |
ELISA
For the detection of secretory factors, conditioned culture media were collected from 100% confluent cells in 10-cm dishes, and a commercial ELISA kit against VEGF (catalog no. DVE00, R&D, Minneapolis, MN) was used to determine the expression level of VEGF following the manufacturer's instruction.
Luciferase reporter assay
ZFHX3 and HRE promoter activities were measured by the promoter luciferase assays using HepG2 cells, in which WT pZFHX3–Luc, mutants of pZFHX3–Luc with each of the HREs mutated, and the pGL3–HRE–luciferase reporter plasmid were transfected in combination with the pRT–TK Renilla luciferase plasmid (Promega) using the Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer's instructions. After 24 h of transfection, cells were incubated in normoxic or hypoxic conditions for 12 h and then collected. Luciferase activities were measured using the Dual-Luciferase® reporter assay system kit (Promega) according to the manufacturer's protocol. Each treatment was performed in triplicate, and mean ± S.D. was calculated for each group.
Transwell assay
For the migration assay, transwell inserts (pore size: 8 μm, BD Biosciences) in 24-well plates were used, with 1 × 105 HUVECs seeded into each upper chamber. HUVECs were treated with conditioned medium from normoxic or hypoxic conditions. After 24 h, the migrated cells were fixed with 4% paraformaldehyde; cells on the upper surface of the membrane were scraped with a cotton swab; cells on the lower side were stained with 0.1% crystal violet (Sigma) for 0.5 h and eluted in 250 μl of 10% acetic acid for 10 min, and the absorbance was measured at 570 nm. An equal number of seeded cells was also stained and measured for absorbance, and the reading was used to divide that of migrated cells. Each treatment was performed in triplicate, and each experiment was performed three times.
Tube formation assay in HUVECs
HCC cells were cultured in 10-cm dishes to 90% confluence after 48 h, washed with PBS three times, and cultured in 4 ml of medium with 1% serum under normal or hypoxic conditions for another 24 h to reach 100% density. Conditioned media were collected after centrifugation at 1500 rpm for 5 min. A total of 5 × 104 HUVECs were seeded into each well of a 24-well plate coated with growth factor–reduced Matrigel (BD Biosciences), and cultured for 6 h in conditioned medium. Images were captured using microscopy and analyzed for the extent of tube formation by measuring the tube length and counting the number of tube nodes using ImageJ software. At least 10 fields were examined for each group.
Deletion of ZFHX3 in HepG2 cells
The CRISPR–Cas9 system was used to introduce a deletion in ZFHX3 following the protocol from the Zhang laboratory (42). Briefly, sgRNA-encoding oligonucleotides for the ZFHX3 genome, 5′-CACCGGGCAGATCTTCACCATCCGC-3′ (forward) and 5′-AAACGCGGATGGTGAAGATCTGCCC-3′ (reverse), were synthesized and annealed; and annealed DNA was digested with BsmBI and cloned into the CRISPR–Cas9 lentiviral vector, which was kindly provided by Dr. Yushan Zhu of Nankai University. For the preparation of lentiviral particles, 293T cells in 6-cm dishes were transfected with 1 μg of CRISPR–Cas9 lentivirus–ZFHX3 plasmid, 750 ng of psPAX2 packaging plasmid, and 250 ng of pMD2.G envelope plasmid using the FuGENE 6 transfection reagent (Promega). HepG2 cells were seeded onto 6-well plates, grown to ∼80% confluency, infected with lentiviral supernatant containing 8 μg/ml of Polybrene, and selected in the medium containing puromycin (2 μg/ml) for 96 h. Single clones were then isolated, and deletion of ZFHX3 was confirmed by Western blotting and DNA sequencing after PCR amplification with primers 5′-TTTCCAGCCAGTAGCCCTTTGCA-3′ (forward) and 5′-GTTGGTGTAGTAGTCACAGGCGTTG-3′ (reverse).
Preparation of lentiviruses expressing ZFHX3 shRNAs
The two shRNAs for ZFHX3, which were validated in a previous study (43) with the following pairs of oligonucleotides: 5′-CCGGGCGATGCTCTTAGACTGTGATCTCGAGATCACAGTCTAAGAGCATCGCTTTTT-3′ and 5′-AATTCAAAAAGCGATGCTCTTAGACTGTGATCTCGAGATCACAGTCTAAGAGCATCGC-3′ for shZFHX3-1 and 5′-CCGGGCCAGGAAGAATTATGAGAATCTCGAGATTCTCATAATTCTTCCTGGCTTTTT-3′ and 5′-AATTCAAAAAGCCAGGAAGAATTATGAGAATCTCGAGATTCTCATAATTCTTCCTGGC-3′ for shZFHX3-2, were cloned into the pLKO.1 vector as described previously (43). Lentiviruses were produced in 293T cells by co-transfecting pLKO.1, pMD2.G, and psPAX2 plasmids as described in the previous paragraph.
Tumorigenesis assay
BALB/c nude mice aged 4–5 weeks were used for the tumorigenesis assay. For each mouse, a total of 2 × 106 HepG2 cells (control shRNA or ZFHX3 shRNAs) and a total of 5 × 106 HepG2 cells (WT or ZFHX3 knockout) were injected subcutaneously on both sides. Four mice were successfully injected for each group. Tumor volumes were measured twice a week for 4 weeks, and the size of a tumor was calculated using the following formula: tumor volume (cm3) = (length × width2)/2. At the end of the experiment, mice were euthanized, and tumors were surgically dissected and weighed. Use of mice was approved by the Institutional Animal Care and Use Committee at Nankai University, and all mice were maintained by facility technicians at the Center for Experimental Animals.
Immunohistochemistry
Tissue sections were rehydrated and boiled in a pressure cooker for 3 min in a citrate buffer (10 mm sodium citrate, pH 6.0) for antigen retrieval; treated with 0.3% H2O2 for 20 min to block endogenous peroxidase activity; incubated with 5% normal goat serum to block nonspecific antibody binding; incubated with primary antibodies at 4 °C overnight and then with the EnVision Polymer-HRP secondary antibodies (Dako, Glostrup, Denmark) at room temperature for 1 h; visualized with a DAB substrate kit (Maixin-Bio, Fuzhou, Fujian, China); stained with hematoxylin; dehydrated; and mounted. The following antibodies were used for immunohistochemistry: anti-CD31 (catalog no. ab28364, 1:500 dilution, Abcam), anti-VEGFA (catalog no. ab46154, 1:2000 dilution, Abcam), and anti-CA9 (catalog no. ab15086, 1:1000 dilution, Abcam).
Statistical and bioinformatic analyses
All experiments except for tumorigenesis were performed three times unless stated otherwise. Each treatment in an experiment was in triplicate. For all real-time qPCR, each biological sample was analyzed in triplicate. For quantification of Western blotting results, we used ImageJ software to measure the relative intensity of each band, and relative protein levels were normalized to that of the loading control. Data are presented as mean ± S.D. unless otherwise indicated. Details of statistics are provided in each figure legend.
Groups of means among different treatments were compared using the Student's t tests (two-tailed, unpaired), except that one-way analysis of variance was used for tumor growth curves (Fig. 7, D and G). The GraphPad Prism (Version 5.0, San Diego, CA) was used for analyses. p value smaller than 0.05 was considered statistically significant.
Three independent liver hepatocellular carcinoma (LIHC) gene expression profiles (GSE14323; GSE14520; and GSE36376) were downloaded from the Gene Expression Omnibus (GEO) database (RRID:SCR_005012) to investigate the mRNA expression distribution of ZFHX3 in hepatocellular carcinoma (LIHC) (44–46). Furthermore, the Gene Expression Profiling Interactive Analysis (GEPIA; RRID:SCR_018294) website, which is an on-line analysis tool based on the RNA-Seq expression data of 9736 tumors and 8587 normal samples from TCGA and the GTEx projects, was used to validate the mRNA expression of ZFHX3 between LIHC and adjacent normal tissues. Student's t tests were utilized for the comparison of two sample groups. Differences were considered as statistically significant when p < 0.05. Spearman correlation test from the GraphPad Prism was performed to determine the correlation between ZFHX3 and HIF1A mRNA levels in the LIHC of TCGA. The 369 patient samples from TCGA, in which both gene expression data and patient survival data are available, were used to test whether ZFHX3 expression levels correlate with disease-free survival using the Kaplan-Meier method and the log-rank test from the GEPIA.
Author contributions
C. F. and J.-T. D. conceptualization; C. F. and J. A. formal analysis; C. F., N. A., J. L., and M. L. investigation; C. F., N. A., and J.-T. D. visualization; C. F. writing-original draft; B. Z., M. L., Z. Z., and J.-T. D. writing-review and editing; Z. Z. and J.-T. D. supervision; L. F., X. T., and D. W. project administration; J.-T. D. funding acquisition.
Supplementary Material
Acknowledgments
We thank Dr. Yushan Zhu of Nankai University for kindly lending the hypoxia chamber and providing plasmids and Dr. Anthea Hammond of Emory University for editing the manuscript. We also thank Drs. Linbo Chen, Yang Yang, Dan Zhao, Biao Ma, Gui Ma, and Ang Gao of Nankai University and Lan Li of Southern University of Science and Technology for advice and help throughout the study.
This work was supported by National Natural Science Foundation of China (NSFC) Grants 81472464 and 31871466. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S6.
- HCC
- hepatocellular carcinoma
- HIF1A
- hypoxia-inducible factor-1α
- HIF1B
- hypoxia-inducible factor-1β
- DFO
- deferoxamine mesylate
- HRE
- hypoxia-response element
- ELISA
- enzyme-linked immunosorbent assay
- HRP
- horseradish peroxidase
- IP
- immunoprecipitation
- IB
- immunoblot
- HUVEC
- human umbilical vein endothelial cell
- CM
- conditioned medium
- VEGF
- vascular endothelial growth factor
- VEGFA
- vascular endothelial growth factor A
- IHC
- immunohistochemistry
- qPCR
- quantitative PCR
- LIHC
- liver hepatocellular carcinoma
- RNA-Seq
- RNA-sequencing
- GEPIA
- Gene Expression Profiling Interactive Analysis
- MAPK
- mitogen-activated protein kinase
- PI3K
- phosphatidylinositol 3-kinase.
References
- 1. Blatchley M. R., Hall F., Wang S., Pruitt H. C., and Gerecht S. (2019) Hypoxia and matrix viscoelasticity sequentially regulate endothelial progenitor cluster-based vasculogenesis. Sci. Adv. 5, eaau7518 10.1126/sciadv.aau7518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Masoud G. N., and Li W. (2015) HIF-1α pathway: role, regulation and intervention for cancer therapy. Acta Pharm. Sin. B 5, 378–389 10.1016/j.apsb.2015.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Schito L., and Semenza G. L. (2016) Hypoxia-inducible factors: master regulators of cancer progression. Trends Cancer 2, 758–770 10.1016/j.trecan.2016.10.016 [DOI] [PubMed] [Google Scholar]
- 4. Choudhry H., and Harris A. L. (2018) Advances in hypoxia-inducible factor biology. Cell Metab. 27, 281–298 10.1016/j.cmet.2017.10.005 [DOI] [PubMed] [Google Scholar]
- 5. Zhu A. X., Duda D. G., Sahani D. V., and Jain R. K. (2011) HCC and angiogenesis: possible targets and future directions. Nat. Rev. Clin. Oncol. 8, 292–301 10.1038/nrclinonc.2011.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bodnar R. J. (2014) Anti-angiogenic drugs: involvement in cutaneous side effects and wound-healing complication. Adv. Wound Care 3, 635–646 10.1089/wound.2013.0496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Forner A., Reig M., and Bruix J. (2018) Hepatocellular carcinoma. Lancet 391, 1301–1314 10.1016/S0140-6736(18)30010-2 [DOI] [PubMed] [Google Scholar]
- 8. Morinaga T., Yasuda H., Hashimoto T., Higashio K., and Tamaoki T. (1991) A human α-fetoprotein enhancer-binding protein, ATBF1, contains four homeodomains and seventeen zinc fingers. Mol. Cell. Biol. 11, 6041–6049 10.1128/MCB.11.12.6041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ninomiya T., Mihara K., Fushimi K., Hayashi Y., Hashimoto-Tamaoki T., and Tamaoki T. (2002) Regulation of the α-fetoprotein gene by the isoforms of ATBF1 transcription factor in human hepatoma. Hepatology 35, 82–87 10.1053/jhep.2002.30420 [DOI] [PubMed] [Google Scholar]
- 10. Kataoka H., Miura Y., Joh T., Seno K., Tada T., Tamaoki T., Nakabayashi H., Kawaguchi M., Asai K., Kato T., and Itoh M. (2001) α-Fetoprotein producing gastric cancer lacks transcription factor ATBF1. Oncogene 20, 869–873 10.1038/sj.onc.1204160 [DOI] [PubMed] [Google Scholar]
- 11. Miura K., Okita K., Furukawa Y., Matsuno S., and Nakamura Y. (1995) Deletion mapping in squamous cell carcinomas of the esophagus defines a region containing a tumor suppressor gene within a 4-centimorgan interval of the distal long arm of chromosome 9. Cancer Res. 55, 1828–1830 [PubMed] [Google Scholar]
- 12. Mazure N. M., Chauvet C., Bois-Joyeux B., Bernard M. A., Nacer-Chérif H., and Danan J. L. (2002) Repression of α-fetoprotein gene expression under hypoxic conditions in human hepatoma cells: characterization of a negative hypoxia response element that mediates opposite effects of hypoxia inducible factor-1 and c-Myc. Cancer Res. 62, 1158–1165 [PubMed] [Google Scholar]
- 13. Sun X., Frierson H. F., Chen C., Li C., Ran Q., Otto K. B., Cantarel B. M., Vessella R. L., Gao A. C., Petros J., Miura Y., Simons J. W., and Dong J. T. (2005) Frequent somatic mutations of the transcription factor ATBF1 in human prostate cancer. Nat. Genet. 37, 407–412 10.1038/ng1528 [DOI] [PubMed] [Google Scholar]
- 14. Grasso C. S., Wu Y. M., Robinson D. R., Cao X., Dhanasekaran S. M., Khan A. P., Quist M. J., Jing X., Lonigro R. J., Brenner J. C., Asangani I. A., Ateeq B., Chun S. Y., Siddiqui J., Sam L., et al. (2012) The mutational landscape of lethal castration-resistant prostate cancer. Nature 487, 239–243 10.1038/nature11125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sun X., Fu X., Li J., Xing C., Frierson H. F., Wu H., Ding X., Ju T., Cummings R. D., and Dong J. T. (2014) Deletion of Atbf1/Zfhx3 in mouse prostate causes neoplastic lesions, likely by attenuation of membrane and secretory proteins and multiple signaling pathways. Neoplasia 16, 377–389 10.1016/j.neo.2014.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gao J., Aksoy B. A., Dogrusoz U., Dresdner G., Gross B., Sumer S. O., Sun Y., Jacobsen A., Sinha R., Larsson E., Cerami E., Sander C., and Schultz N. (2013) Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 6, pl1 10.1126/scisignal.2004088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kim C. J., Song J. H., Cho Y. G., Cao Z., Lee Y. S., Nam S. W., Lee J. Y., and Park W. S. (2008) Down-regulation of ATBF1 is a major inactivating mechanism in hepatocellular carcinoma. Histopathology 52, 552–559 10.1111/j.1365-2559.2008.02980.x [DOI] [PubMed] [Google Scholar]
- 18. Tang Z., Li C., Kang B., Gao G., Li C., and Zhang Z. (2017) GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 45, W98–W102 10.1093/nar/gkx247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chen D., Wu L., Liu L., Gong Q., Zheng J., Peng C., and Deng J. (2017) Comparison of HIF1AAS1 and HIF1AAS2 in regulating HIF1α and the osteogenic differentiation of PDLCs under hypoxia. Int. J. Mol. Med. 40, 1529–1536 10.3892/ijmm.2017.3138 [DOI] [PubMed] [Google Scholar]
- 20. Pugh C. W., and Ratcliffe P. J. (2003) Regulation of angiogenesis by hypoxia: role of the HIF system. Nat. Med. 9, 677–684 10.1038/nm0603-677 [DOI] [PubMed] [Google Scholar]
- 21. Hirota K., and Semenza G. L. (2006) Regulation of angiogenesis by hypoxia-inducible factor 1. Crit. Rev. Oncol. Hematol. 59, 15–26 10.1016/j.critrevonc.2005.12.003 [DOI] [PubMed] [Google Scholar]
- 22. Rebouissou S., Zucman-Rossi J., Moreau R., Qiu Z., and Hui L. (2017) Note of caution: contaminations of hepatocellular cell lines. J. Hepatol. 67, 896–897 10.1016/j.jhep.2017.08.002 [DOI] [PubMed] [Google Scholar]
- 23. Ameri K., Burke B., Lewis C. E., and Harris A. L. (2002) Regulation of a rat VL30 element in human breast cancer cells in hypoxia and anoxia: role of HIF-1. Br. J. Cancer 87, 1173–1181 10.1038/sj.bjc.6600576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wenger R. H., Stiehl D. P., and Camenisch G. (2005) Integration of oxygen signaling at the consensus HRE. Sci. STKE 2005, re12 10.1126/stke.3062005re12 [DOI] [PubMed] [Google Scholar]
- 25. Chiu D. K., Tse A. P., Xu I. M., Di Cui J., Lai R. K., Li L. L., Koh H. Y., Tsang F. H., Wei L. L., Wong C. M., Ng I. O., and Wong C. C. (2017) Hypoxia inducible factor HIF-1 promotes myeloid-derived suppressor cells accumulation through ENTPD2/CD39L1 in hepatocellular carcinoma. Nat. Commun. 8, 517 10.1038/s41467-017-00530-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mazure N. M., Brahimi-Horn M. C., and Pouysségur J. (2003) Protein kinases and the hypoxia-inducible factor-1, two switches in angiogenesis. Curr. Pharm. Des. 9, 531–541 10.2174/1381612033391469 [DOI] [PubMed] [Google Scholar]
- 27. Li J., Xu Y., Long X. D., Wang W., Jiao H. K., Mei Z., Yin Q. Q., Ma L. N., Zhou A. W., Wang L. S., Yao M., Xia Q., and Chen G. Q. (2014) Cbx4 governs HIF-1α to potentiate angiogenesis of hepatocellular carcinoma by its SUMO E3 ligase activity. Cancer Cell 25, 118–131 10.1016/j.ccr.2013.12.008 [DOI] [PubMed] [Google Scholar]
- 28. Berra E., Pagès G., and Pouysségur J. (2000) MAP kinases and hypoxia in the control of VEGF expression. Cancer Metastasis Rev. 19, 139–145 10.1023/A:1026506011458 [DOI] [PubMed] [Google Scholar]
- 29. Carbajo-Pescador S., Ordoñez R., Benet M., Jover R., García-Palomo A., Mauriz J. L., and González-Gallego J. (2013) Inhibition of VEGF expression through blockade of Hif1α and STAT3 signalling mediates the anti-angiogenic effect of melatonin in HepG2 liver cancer cells. Br. J. Cancer 109, 83–91 10.1038/bjc.2013.285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Liu L. P., Ho R. L., Chen G. G., and Lai P. B. (2012) Sorafenib inhibits hypoxia-inducible factor-1α synthesis: implications for antiangiogenic activity in hepatocellular carcinoma. Clin. Cancer Res. 18, 5662–5671 10.1158/1078-0432.CCR-12-0552 [DOI] [PubMed] [Google Scholar]
- 31. Takahashi H., and Shibuya M. (2005) The vascular endothelial growth factor (VEGF)/VEGF receptor system and its role under physiological and pathological conditions. Clin. Sci. 109, 227–241 10.1042/CS20040370 [DOI] [PubMed] [Google Scholar]
- 32. Pouysségur J., Dayan F., and Mazure N. M. (2006) Hypoxia signalling in cancer and approaches to enforce tumour regression. Nature 441, 437–443 10.1038/nature04871 [DOI] [PubMed] [Google Scholar]
- 33. Perez-Perri J. I., Dengler V. L., Audetat K. A., Pandey A., Bonner E. A., Urh M., Mendez J., Daniels D. L., Wappner P., Galbraith M. D., and Espinosa J. M. (2016) The TIP60 complex is a conserved coactivator of HIF1A. Cell Rep. 16, 37–47 10.1016/j.celrep.2016.05.082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cheng J., Kang X., Zhang S., and Yeh E. T. (2007) SUMO-specific protease 1 is essential for stabilization of HIF1α during hypoxia. Cell 131, 584–595 10.1016/j.cell.2007.08.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sun X., Li J., Dong F. N., and Dong J. T. (2014) Characterization of nuclear localization and SUMOylation of the ATBF1 transcription factor in epithelial cells. PLoS ONE 9, e92746 10.1371/journal.pone.0092746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Dong J. T., and Chen C. (2009) Essential role of KLF5 transcription factor in cell proliferation and differentiation and its implications for human diseases. Cell. Mol. Life Sci. 66, 2691–2706 10.1007/s00018-009-0045-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Diakiw S. M., D'Andrea R. J., and Brown A. L. (2013) The double life of KLF5: opposing roles in regulation of gene-expression, cellular function, and transformation. IUBMB Life 65, 999–1011 10.1002/iub.1233 [DOI] [PubMed] [Google Scholar]
- 38. Hu Q., Zhang B., Chen R., Fu C., A J., Fu X., Li J., Fu L., Zhang Z., and Dong J. T. (2019) ZFHX3 is indispensable for ERβ to inhibit cell proliferation via MYC downregulation in prostate cancer cells. Oncogenesis 8, 28 10.1038/s41389-019-0138-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ma B., Chen Y., Chen L., Cheng H., Mu C., Li J., Gao R., Zhou C., Cao L., Liu J., Zhu Y., Chen Q., and Wu S. (2015) Hypoxia regulates Hippo signalling through the SIAH2 ubiquitin E3 ligase. Nat. Cell Biol. 17, 95–103 10.1038/ncb3073 [DOI] [PubMed] [Google Scholar]
- 40. Dong X. Y., Sun X., Guo P., Li Q., Sasahara M., Ishii Y., and Dong J. T. (2010) ATBF1 inhibits estrogen receptor (ER) function by selectively competing with AIB1 for binding to the ER in ER-positive breast cancer cells. J. Biol. Chem. 285, 32801–32809 10.1074/jbc.M110.128330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Dong X. Y., Guo P., Sun X., Li Q., and Dong J. T. (2011) Estrogen up-regulates ATBF1 transcription but causes its protein degradation in estrogen receptor-α–positive breast cancer cells. J. Biol. Chem. 286, 13879–13890 10.1074/jbc.M110.187849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ran F. A., Hsu P. D., Wright J., Agarwala V., Scott D. A., and Zhang F. (2013) Genome engineering using the CRISPR–Cas9 system. Nat. Protoc. 8, 2281–2308 10.1038/nprot.2013.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zhao D., Ma G., Zhang X., He Y., Li M., Han X., Fu L., Dong X. Y., Nagy T., Zhao Q., Fu L., and Dong J. T. (2016) Zinc finger homeodomain factor Zfhx3 is essential for mammary lactogenic differentiation by maintaining prolactin signaling activity. J. Biol. Chem. 291, 12809–12820 10.1074/jbc.M116.719377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mas V. R., Maluf D. G., Archer K. J., Yanek K., Kong X., Kulik L., Freise C. E., Olthoff K. M., Ghobrial R. M., McIver P., and Fisher R. A. (2009) RMA expression data for liver samples from subjects with HCV, HCV-HCC, or normal liver. Gene Expression Omnibus. GSE14323
- 45. Wang X. W. (2010) Gene expression data of human hepatocellular carcinoma (HCC). Gene Expression Omnibus. GSE14520
- 46. Park C. K. (2012) Gene expression profiles of both tumor and adjacent non-tumor liver identify hepatocellular carcinoma patients at high risk of recurrence after curative hepatectomy. Gene Expression Omnibus. GSE36376
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.