Abstract
Protein arginine methyltransferase 1 (PRMT1) is a key regulator of hepatic immune responses. Recently, we reported that PRMT1 regulates the tumor immune response in hepatocellular carcinoma (HCC). Here we found that PRMT1 expression in human HCC correlates with that of programmed cell death 1 ligand 1 (PD-L1), PD-L2, and other checkpoint genes. PRMT1 deletion in mice reduced PD-L1 and PD-L2 expression in tumors and reduced the efficiency of PD-1 antibody treatment in a diethylnitrosamine-induced HCC mouse model, suggesting that PRMT1 regulates the hepatic immune checkpoint. Mice had reduced PD-L1 and PD-L2 expression when PRMT1 was specifically deleted in tumor cells or macrophages, but PRMT1 deletion in dendritic cells did not alter PD-L1 and PD-L2 expression. rs975484 is a common polymorphism in the human PRMT1 gene promoter, and we found that it alters PRMT1 expression in blood monocytes and tumor-associated macrophages in human HCC. PRMT1 expression was higher in individuals with a GG genotype than in individuals with a CC genotype, and heterozygous carriers had intermediate expression. Luciferase reporter assays indicated that this differential expression is due to an extra C/EBPβ-binding site in the PRMT1 promoter of individuals carrying the minor G allele. The rs975484 genotype also correlated with PRMT1 target expression in HCC. Individuals with the GG genotype had significantly higher levels of the PRMT1 targets PD-L1, PD-L2, and VISTA than those with the CC genotype. We conclude that PRMT1 critically controls immune checkpoints in mice and humans and that the PRMT1 polymorphism rs975484 affects checkpoint gene expression in HCC.
Keywords: inflammation, CCAAT-enhancer-binding protein (C/EBP), macrophage, checkpoint control, tumor immunology, PD-L2, programmed cell death 1 ligand 1 (PD-L1), protein arginine methyltransferase 1 (PRMT1), rs975484, SNP
Introduction
Protein arginine methylation is a common posttranslational modification that plays a role in multiple pathways, including cell cycle control, RNA processing, and DNA replication. The protein arginine methyltransferase PRMT1 is responsible for about 85% of total cellular arginine methylation (1) and catalyzes arginine mono- and dimethylation using SAM as a methyl donor. PRMT1 methylates histone H4 at arginine 3, generating H4R3me2a, a transcriptional activation mark (1–5), contributing to the histone code. As a transcriptional coactivator, PRMT1 is recruited to promoters by a number of different transcription factors (3, 6, 7). Abnormal function of PRMT1 is closely associated with several types of cancer and cardiovascular diseases. Arginine methylation impacts gene transcription and splicing as well as upstream signal transduction (4). Recently, arginine methylation has emerged as an important regulator of the immune response (4, 7, 8). PRMT1 is induced during monocyte-to-macrophage differentiation. This induction results in deposition of the PRMT1-dependent histone mark H4R3me2a on several promoters involved in M2-like differentiation, including the PPARγ promoter. Data from PRMT1 knockout mice show that PRMT1 deficiency results in diminished PPARγ expression and lack of PPARγ-dependent M2 differentiation (9). In human livers, PRMT1 is involved in macrophage polarization and tumor-associated macrophage phenotype and function (10).
Hepatocellular carcinoma (HCC)2 is the third most common cause of cancer-related death worldwide (11, 12). Most HCC patients have a history of chronic liver disease and cirrhosis (13). Early-stage HCCs, are often treated with surgical resection, transplantation, and ablation and have a 5-year survival of 50%–80%. Despite that, metastases and de novo carcinogenesis occur even after potentially curative therapies, and the 5-year recurrence rate reaches up to 70% (14, 15). Patients with advanced HCC have an overall 5-year survival rate of less than 16%. Until recently, sorafenib, an oral multitargeted tyrosine kinase inhibitor, was the sole pharmacological treatment for advanced HCC. Recently, regorafenib (an angiogenesis inhibitor) and nivolumab (an immune checkpoint inhibitor) were approved as second-line HCC treatments for patients who fail prior sorafenib treatment.
Immune checkpoint inhibitors exert antitumor effects via checkpoint-mediated inhibition of programmed cell death 1/programmed cell death ligand 1 (PD-1/PD-L1) or cytotoxic T lymphocyte-associated protein 4 (CTLA-4) (14, 16). The PD-1/PD-L1 axis is a vital immune checkpoint signaling pathway that can down-regulate antitumor cell–mediated immune responses and prevent immune control of tumor progression (14, 17). However, the overall response rate of PD-1/PD-L1 inhibitors in patients is unsatisfactory, which limits application in clinical practice. Therefore, biomarkers that could effectively predict the efficacy of PD-1/PD-L1 inhibitors are crucial for patient selection. Biomarkers reflecting tumor immune microenvironment and tumor cell intrinsic features, such as PD-L1 expression, density of tumor infiltrating lymphocytes, tumor mutational burden, and mismatch repair deficiency, have been shown to be associated with treatment effect of anti-PD-1/anti-PD-L1 therapy in several tumor types. Furthermore, gut microbiota, circulating biomarkers, and a patient's previous history have been found to be valuable predictors as well (reviewed in Ref. 17).
In this work, we show that PRMT1 is an important regulator of immune checkpoint pathways in HCC. PRMT1 regulates PD-L1 and PD-L2 gene expression in mice and humans. The PRMT1 gene polymorphism rs975484 correlates with PRMT1 expression in immune cells and PD-L1 and PD-L2 expression in human HCC. Our findings suggest that rs975484 can be useful for predicting checkpoint gene expression in patients with HCC.
Results
PRMT1 gene polymorphisms regulate its expression in human blood monocytes
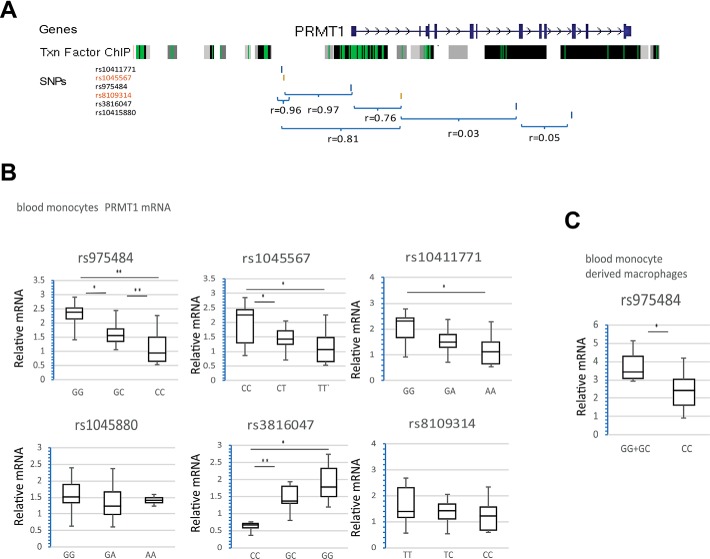
In humans, PRMT1 gene expression varies considerably between individuals. We hypothesized that genetic variations can account for some of these expression differences. Common genetic polymorphisms associated with the PRMT1 gene are presented in Fig. 1A; SNPs associated with phenotype in GWAS studies are highlighted in red.
Figure 1.
PRMT1 gene polymorphisms regulate its expression in human blood monocytes. A, map of the genomic region containing the PRMT1 gene. Shown are transcription factor ChIP sequencing clusters (161 factors) from ENCODE, with transcription (Txn) factor binding sites shown in green. Also shown are the location and linkage disequilibrium for the indicated SNPs. B, PRMT1 mRNA expression in blood monocytes from individuals with the indicated genotypes. n = 67. *, p < 0.05; **, p < 0.01. C, PRMT1 mRNA expression in blood monocyte–derived macrophages from individuals with the indicated genotypes. n = 26. *, p < 0.05.
We analyzed SNP genotype and PRMT1 mRNA expression in human monocytes from individuals who presented to the University of Kansas Liver Treatment Center during the period from November 2014 to June 2015 (Table S1). We found that the rs975484, rs1045567, rs10411771, and rs3816047 genotypes correlate with PRMT1 mRNA expression in human blood monocytes (Fig. 1B) and that rs975484 correlates with PRMT1 expression in human blood monocyte–derived macrophages (Fig. 1C). The genotype–mRNA correlation was independent of the severity of liver disease (Fig. S1).
It is important to note the linkage disequilibrium between rs975484, rs1045567, rs10411771, and rs8109314 (Fig. 1A). For example, the linkage disequilibrium between rs975484 and rs1045567 is 0.97; thus, the majority (in our case, 137 of 141) of individuals that have GG, GC, or CC for rs975484 also have CC, CT, or TT, respectively, for rs1045567. Because rs975484 showed the strongest correlation with PRMT1 expression and is located in the promoter region, where it is more likely to affect transcription factor binding (Fig. 1A), we focused further studies on rs975484.
The PRMT1 gene polymorphism rs975484 regulates its expression through C/EBPβ binding
First we confirmed that C-to-G substitution in the position of rs975484 can affect PRMT1 expression. We created PRMT1 promoter–driven luciferase constructs with C or G in the position of rs975484 (Fig. 2A) and expressed them in THP-1 cells. We found that C-to-G substitution results in an approximately 2-fold increase in luciferase mRNA expression (Fig. 2B), similar to the PRMT1 mRNA expression difference between GG and CC genotypes in human monocytes (Fig. 1B). Next we performed bioinformatic analysis of transcription factor binding to this promoter region (TfSiteScan, RRID:SCR_010667). We identified an extra C/EBPβ binding motif in the G but not in the C variant (Fig. 2C).
Figure 2.
PRMT1 gene polymorphism rs975484 regulate its expression through C/EBPβ binding. A, PRMT1 promoter–driven luciferase construct with C or G in the position of rs975484. B, relative luciferase mRNA expression in THP-1 cells transfected with C- or G-containing constructs or a control plasmid. n = 4 independent experiments; expression was normalized to C construct expression. *, p < 0.05, paired t test on the raw expression values. C, PRMT1 promoter sequence aligned with the CEBPB binding motif MA0466.2 (JASPAR database) and CEBPB binding sites in IL-6 and CCR5 gene promoters. D, relative luciferase mRNA expression in THP-1 cells transfected with C- or G-containing constructs or a control plasmid in the presence of a shRNA control plasmid or CEBPB-specific shRNAs. n = 3 independent experiments; expression was normalized to C (blue columns) or G (black columns) construct expression. *, p < 0.05, paired t test on the raw expression values. E, ChIP using anti-C/EBPβ antibody or IgG as a negative control in THP-1 cells expressing control vector or the C or G PRMT1 promoter construct. Data are presented as mean enrichment (over IgG) ± S.D. n = 3.
To evaluate the role of C/EBPβ in PRMT1 expression, we coexpressed PRMT1 promoter–driven luciferase in the presence of control shRNA or shRNAs specific to CEBPB (Fig. 2D and S2A). We found that C/EBPβ knockdown significantly reduced G construct expression, suggesting that C/EBPβ binding to the PRMT1 promoter promotes its expression.
Next we confirmed C/EBPβ binding to the PRMT1 promoter by ChIP in THP-1 cells expressing a C or G promoter construct. We found that C-to-G substitution results in a 3-fold increase in C/EBPβ binding to the PRMT1 promoter immediately upstream of the transcription start site (P1_I) (Fig. 2E). No binding was observed to the region downstream of the transcription start site (P1_II).
Interestingly, we also detected C/EBPβ binding to the C construct promoter, in agreement with bioinformatic analysis prediction. However, CEBPB knockdown did not decrease C construct expression. These data suggest that either the other C/EBPβ binding sites are nonfunctional or, alternatively, that loss of C/EBPβ binding results in an increase in binding of another transcription factor, such as NF-κB. Fig. S2B shows data to support the second hypothesis. C/EBPβ knockdown results in a 7-fold increase in PRMT1 expression in response to lipopolysaccharide. Similarly, in blood monocytes, GG monocytes (more C/EBPβ binding) showed lower PRMT1 mRNA expression in response to lipopolysaccharide compared with CC monocytes (less C/EBPβ binding) (Fig. S2C).
PRMT1 gene polymorphisms regulate its expression in tumor-associated macrophages
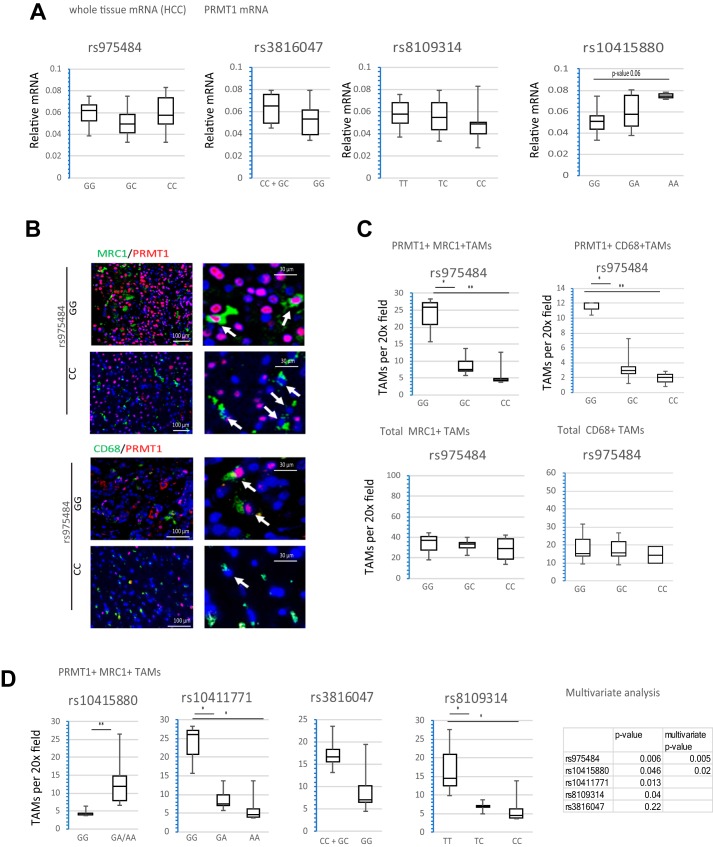
We did not find a significant correlation between PRMT1 polymorphisms and PRMT1 mRNA in total human HCC mRNA from individual specimens (Fig. 3A). In contrast, we found that PRMT1 expression in tumor-associated macrophages, measured as number of PRMT1-positive TAMs per field, was significantly different between individuals with the GG>GC>CC rs975484 genotype (Fig. 3, B and C). The number of total TAMs per field was not significantly different between genotypes (Fig. 3C). Similar results were observed for the rs10411771 and rs8109314 genotypes. The rs10415880 genotype also correlated with the number of PRMT1-positive TAMs, and this correlation was independent from rs975484 in a multivariate analysis, whereas rs1045567, rs10411771, and rs8109314 correlation with PRMT1 was not significant in the multivariate analysis. Interestingly, rs3816047 minor allele individuals had higher expression of PRMT1 than individuals with a major allele, in contrast to the results for blood monocytes (Fig. 3D).
Figure 3.
PRMT1 gene polymorphisms regulate its expression in tumor-associated macrophages. A, PRMT1 mRNA expression in whole-tissue mRNA of HCC specimens from individuals with the indicated genotypes. n = 36. B, representative images of immunohistochemistry staining of HCC sections from individuals with the GG and CC genotypes using anti-MRC1 or anti-CD68 to detect TAMs and anti-PRMT1 antibodies. Arrows indicate TAMs. C, number of PRMT1-positive TAMs counted by a blinded person unaware of the genotype of the samples in HCC sections from individuals with the GG (n = 3), GC (n = 6), or CC (n = 3) genotypes. MRC1-positive or CD68-positive TAMs double-positive for PRMT1 were counted as PRMT1+ TAMs. *, p < 0.05; **, p < 0.01. D, Left, number of PRMT1-positive TAMs (MRC1-positive cells) in HCC specimens of individuals with the indicated genotypes. n = 12. *, p < 0.05; **, p < 0.01. Right, multivariate analysis of PRMT1-positive TAMs correlation with indicated SNPs.
PRMT1 regulates immune checkpoint gene expression in mice and humans
Our previous work has demonstrated that PRMT1 is an important regulator of immune signaling in multiple cell types, including monocytes, liver macrophages, hepatocytes, liver tumor–associated macrophages, and hepatoma cells (8–10, 18). In human HCC, PRMT1 overexpression is associated with poor prognosis (Fig. S3), suggesting that PRMT1 might promote tumor growth. One of the PRMT1 targets we found in the liver is PD-L2 (10).
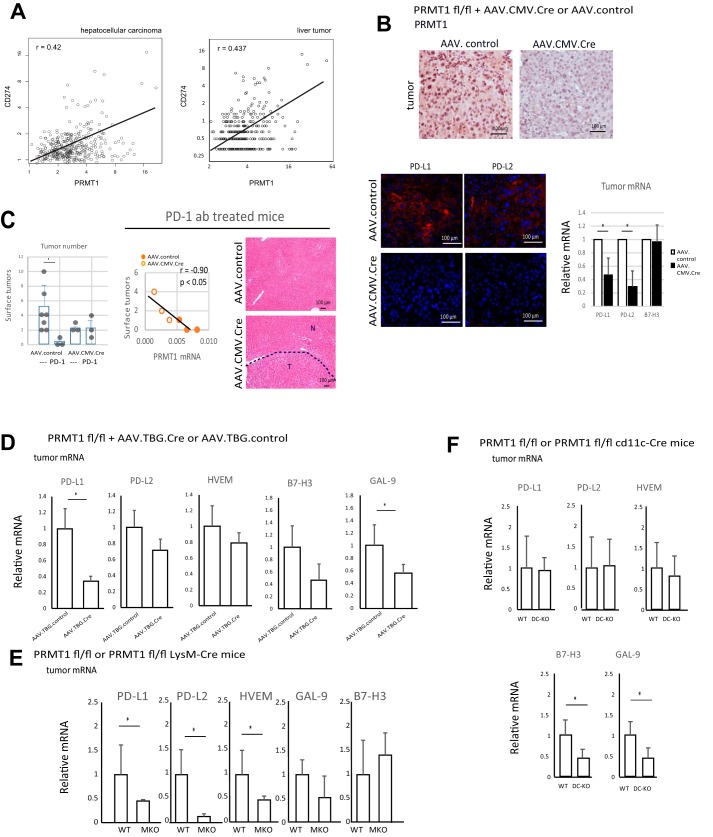
To test the role of PRMT1 in immune checkpoint regulation in the liver, we examined the gene expression of PRMT1 and immune checkpoint ligands in human HCC. We found significant correlation between PRMT1 expression and expression of genes coding for PD-L1 (CD274), PD-L2 (PDCD1L2), and others (VSIR or C10orf54 coding for VISTA, CD276 coding for B7-H3, TNFRSF14 coding for HVEM, and LGALS9 coding for GAL-9) (Fig. 4A and Fig. S4) in two different data sets: GEPIA (Gene Expression Profiling Analysis) (19) and Human Protein Atlas (RRID:SCR_006710). We found similar results in HCC specimens from the University of Kansas Medical Center liver bank (Fig. S5). Interestingly, we did not find any correlation in other types of tumors (Fig. S6).
Figure 4.
PRMT1 regulates immune checkpoint gene expression in mice and humans. A, correlation between PRMT1 and CD274 (PD-L1) gene expression in HCC samples or liver tumor samples from the GEPIA database (A, The Cancer Genome Atlas (TCGA) samples, n = 369 HCC) or ProteinAtlas database (B, n = 365 liver tumors). p < 0.00001. B, PRMT1 floxed mice were injected with DEN (75 mg/kg) four times weekly starting at 4 weeks of age. At 6 months of age, mice were given AAV.CMV.CRE or AAV.control vectors at 1011 gc/mouse i.p. At 8 months of age, livers were analyzed. Liver sections containing tumor nodules were stained using anti-PRMT1 antibodies (top panel) or anti-PD-L1, anti-PD-L2 antibodies (bottom panel). Individual tumors were excised and analyzed for whole-tumor mRNA expression of genes coding for PD-L1 (Cd274), PD-L2 (Pdcd1lg2), and B7-H3 (Cd276). n = 3 mice. *, p < 0.05. C, PRMT1 mRNA expression in livers of mice correlates with tumor number. PRMT1 floxed mice were injected with DEN (75 mg/kg) four times weekly starting at 4 weeks of age. At 6 months of age, mice were given AAV.CMV.CRE or AAV.control vectors at 1011 gc/mouse i.p. Pairs of littermates, WT or KO, received four injections biweekly, starting at 6 months, of age of PD-1–specific antibody (12.5 mg/kg) or vehicle control. Left panel, surface tumor number was analyzed at 8 months of age. *, p < 0.05. Right panel, representative H&E staining of liver sections of WT and knockout mice). No tumor nodules were found in WT mice treated with PD-1 antibody. D, PRMT1 floxed mice were injected with DEN (10 mg/kg) at 2 weeks of age. At 6 months of age, mice were given AAV.TBG.CRE or AAV.control vectors at 1011 gc/mouse i.p. At 8 months of age, livers were analyzed. Individual tumors were excised and analyzed for whole-tumor mRNA expression. n = 3 mice. *, p < 0.05. E, relative mRNA in tumors of PRMT1 flox/flox LysM Cre mice and PRMT1 flox/flox WT littermates. Mice were injected with DEN (10 mg/kg) at 2 weeks of age. At 9 months of age, livers were analyzed. n = 3 mice. *, p < 0.05. F, PRMT1 flox/flox cd11c-Cre mice and PRMT1 flox/flox WT littermates were injected with DEN (10 mg/kg) at 2 weeks of age. At 9 months of age, livers were analyzed. Individual tumors were excised and analyzed for whole-tumor mRNA expression. n = 5–7 mice. *, p < 0.05.
PRMT1 is necessary for PD-1 antibody treatment efficiency
Next we determined whether PRMT1 regulates expression of these genes in a mouse model of HCC. PRMT1 floxed male mice were given DEN to induce HCC formation. At 6 months of age, mice were injected with AAV-control or AAV-Cre with Cre recombinase driven by a CMV, promoter and liver tumors were analyzed at 8 months of age (Fig. S7). AAV-CMV.Cre has broad cell type specificity, but the efficiency of knockout reached only 50%–80%. We confirmed that mice that received AAV-Cre lost expression of PRMT1 in tumor cells (Fig. 4B and Fig. S7). These tumors in PRMT1 knockout mice also lost immunohistochemically detectible expression of PD-L1 and PD-L2 proteins (Fig. 4B), which corresponded to significantly reduced mRNA expression of genes coding for PD-L1 and PD-L2 in whole-tumor mRNA (Fig. 4B).
WT and knockout littermates were treated every 2 weeks with four total injections of PD-1–specific antibodies at 12.5 mg/kg starting 1 week after AAV injection (Fig. 4C). PD-1 antibody treatment efficiently prevented tumor formation in WT mice. In liver sections of treated mice, we did not find tumor nodules. In contrast, PRMT1 knockout mice showed no effect of PD-1 treatment. We observed multiple tumors on the surface and within liver sections (Fig. 4C). The tumor number in anti-PD-1–treated mice correlated with PRMT1 expression in these mice, suggesting that PRMT1-dependent checkpoint regulation is a determinant of PD-1 treatment efficiency (Fig. 4C).
PRMT1 regulates PD-L1 and PD-L2 expression in tumor cells and macrophages in mice
The above results clearly show that PRMT1-dpendent regulation of the PD-1 pathway regulates tumor growth. To determine which cell(s) participate in this PRMT1 effect, we generated a series of cell-specific PRMT1 knockouts.
To knock out PRMT1 in tumor cells, we used AAV.TBG.Cre-mediated cre delivery to adult PRMT1 floxed mice at 6 months of age prior to tumor development. AAV injection results in hepatocyte-specific knockout and tumor cell–specific knockout in HCCs in these mice (20). Littermates were injected with AAV.TBG.cre or AAV.TBG.control vectors and developed tumors that were WT or tumor cell–specific PRMT1 knockout 2 months after AAV injection. We found that PRMT1 knockout tumors had significantly lower expression of genes coding for PD-L1 and GAL9 (Fig. 4D).
In isolated liver macrophages from PRMT1 WT or myeloid-specific PRMT1 knockout littermates (10), we found significant differences in expression of genes coding for PD-L1, PD-L2, HVEM, and B7-H3 (Fig. S8). We analyzed tumors from PRMT1 WT or myeloid-specific PRMT1 knockout littermates at 9 months of age. We found that tumors from myeloid-specific PRMT1 knockout mice had significantly lower expression of genes coding for PD-L1, PD-L2, and HVEM (Fig. 4E).
We analyzed tumors from PRMT1 WT or dendritic cell–specific PRMT1 knockout littermates. PRMT1 WT or dendritic cell–specific PRMT1 knockout littermates (PRMT1 flox/flox cd11c-Cre) were injected with DEN at 2 weeks of age, and livers were analyzed at 9 months of age. We found that tumors from PRMT1 DC-KO mice had significantly lower expression of genes coding for B7-H3 and GAL9 (Fig. 4F).
We confirmed the gene expression data by staining (Fig. 5). We found that tumors from PRMT1 hepatocyte-specific knockout mice had significantly less PD-L1 and PD-L2 compared with WT controls (Fig. 5A). Similarly, myeloid cell–specific knockout mice had significantly less PD-L1 and PD-L2 in tumor nodules compared with WT controls (Fig. 5B). In contrast, DC-KO mice and WT littermates had comparable PD-L1 and PD-L2 expression in the tumors (Fig. 5C). Taken together, these data suggest that PRMT1 regulates expression of multiple immune checkpoint genes in tumor cells as well as in immune cells such as macrophages.
Figure 5.
PRMT1 regulates PD-L1 and PD-L2 expression in tumor cells and macrophages. A, liver sections from WT or hepatocyte-specific PRMT1 knockout mice as in Fig. 4D were stained using anti-PD-L1 and anti-PD-L2 antibodies. Relative staining intensity is presented on the right. n = 3 mice. *, p < 0.05. B, liver sections from WT or myeloid-specific PRMT1 knockout littermates treated with DEN and analyzed at 6 months of age were stained using anti-PD-L1 and anti-PD-L2 antibodies. Relative staining intensity is presented on the right. n = 3 mice. *, p < 0.05. C, liver sections from WT or DC-specific PRMT1 knockout littermates treated with DEN and analyzed at 9 months of age were stained using anti-PD-L1 and anti-PD-L2 antibodies.
rs975484 defines expression of the PRMT1 targets PD-L1, PD-L2, and VISTA in human HCC
Finally we tested whether PRMT1 promoter polymorphisms correlate with PRMT1-dependent immune checkpoint gene expression in human HCC (Fig. 6). We found that the rs975484 genotype correlates best with immune checkpoint gene expression. Among other SNPs, rs10415880 showed a weak correlation with immune checkpoint expression (Fig. S9).
Figure 6.
The rs975484 genotype correlates with expression of the PRMT1 targets PD-L1, PD-L2, and VISTA in human HCC. A, mRNA expression in whole-tissue mRNA of HCC specimens from individuals with the indicated genotypes. n = 36. *, p < 0.05. B and C, liver sections containing HCC were stained for PD-L1, PD-L2, VISTA, B7-H3, and TIM3. B, relative staining intensity in individuals with the indicated genotypes. n = 3 (GG), 5 (GC), and 5 (CC). *, p < 0.05. C, representative images of staining in individuals with the GG and CC genotypes.
The rs975484 genotype correlates with mRNA expression of genes coding for PD-L2 and VISTA (Fig. 6A). Correlation with genes coding for PD-L1 showed a trend but did not reach statistical significance. We found no correlation with genes coding for B7-H3, GAL-9, or HVEM (Fig. 6A). We analyzed protein expression in these HCC specimens by staining and found that individuals with the GG genotype had significantly higher levels of PD-L1, PD-L2, and VISTA but not B7-H3 (Fig. 6, B and C). Additionally, we found that individuals with the GG genotype had significantly increased numbers of TIM3-positive exhausted T cells (Fig. 6, B and C).
Discussion
PRMT1 is an arginine methyltransferase that regulates many proteins directly and many more via histone arginine methylation. Recently, we and others found that PRMT1 is an immune regulator. Its targets include, but are not limited to, TRAF6, NF-κB, PPARγ, c-Myc, STAT3, FOXOs, and β-catenin, all of which are involved in immune cell regulation; specifically, immune cell polarization (7, 9, 10, 21). Thus, it is not surprising that PRMT1 is involved in regulation of the tumor immune environment. Specifically, we found that it regulates immune responses in hepatocellular carcinoma (10). In this work, we found that, in HCC, PRMT1 regulates expression of immune checkpoint genes in a cell type–specific manner. We examined the cell types that are known to express PD-L1 and PD-L2 and found that PRMT1 regulates different subsets of immune checkpoint genes in macrophages, DCs, and HCC tumor cells. PRMT1 promotes the expression of PD-L1 and PD-L2 in tumor cells and macrophages in DEN-induced tumors in mice. Interestingly, PRMT1 knockout mice did not show reduced tumor numbers despite a decrease in immune checkpoint ligands. This suggests that PD-L1/PD-L2–low tumors have alternative mechanisms for immune evasion. These data agree with studies published previously (17, 22). In humans, PRMT1 positively correlates with expression of these genes, suggesting that this regulation is relevant in human HCC as well.
Moreover, we found a SNP in the PRMT1 promoter, rs975484, that correlates with PRMT1 expression in immune cells in humans and correlates with PD-L1 and PD-L2 expression in human HCC. These data suggest that rs975484 might be a promising marker to predict PD-L1 and PD-L2 expression in tumors of patients with HCC and potentially predicts their response to PD-1/PD-L1–directed therapy. Further studies are necessary to confirm this hypothesis.
PRMT1 SNPs have been identified previously in several GWAS studies. rs1045567 is associated with general cognitive ability, and rs8109314 is associated with height, in agreement with PRMT1's role in development and brain function (GWAS catalog (23)). rs10415880 has been reported to associate with outcomes of arteriovenous fistulas for hemodialysis in Chinese men (24), possibly because of PRMT1-dependent asymmetric dimethyl arginine production and regulation of NO levels in these patients. These data and our findings here suggest that there are cell type–specific SNP–phenotype associations, possibly because of cell type–specific transcription factor binding to SNP-containing sites. In the case of rs975484, we found that it specifically regulates expression of PRMT1 in monocytes and macrophages through alteration of C/EBPβ-dependent PRMT1 expression. Individuals with the G allele have an extra C/EBPβ binding site in the PRMT1 promoter, which results in higher expression. The C/EBPβ transcription factor is highly expressed in immune cells compared with other liver cells, such as hepatocytes. C/EBPβ is involved in myeloid cell function, where it promotes tumor-induced tolerance and immune suppression (25–27). These data are in agreement with the specificity of rs975484-PRMT1 regulation for tumor-associated macrophages but not hepatoma cells.
Despite the remarkable clinical success of anti-PD-1/PD-L1 immunotherapy, a high proportion of patients fail to respond. Studies have suggested that the efficacy of PD inhibitors depends on different factors; for example, genomic features, including tumor mutation burden and oncogenic pathways, tumor microenvironment, systemic immunity status, microbiome, and others. Several biomarkers have been suggested to be associated with a positive response to PD pathway blockade, including high levels of PD-L1 expression in tumor cells or immune cells, mutations, and epithelial–mesenchymal transition/stem-like features, but none of these have a definitive predictive value (16). There are no GWAS studies to investigate the genetic factors involved in PD inhibitor response. Our results suggest that a genetic polymorphism in one of the upstream regulators, PRMT1, might be a candidate biomarker of PD treatment response because it is a good predictor of PD-L1 and PD-L2 expression. The advantage of using a genetic approach to make predictions regarding individual patient responses is that it avoids the need for biopsy and immunohistochemistry to detect of PD-L1 expression. There is a lot of disagreement regarding scoring of IHC results because different cutoff values and scoring systems are used in separate clinical trials, and different antibodies and IHC platforms lead to incomparability of results among trials (14, 17).
Another important consideration is that PD-L1 can be up-regulated by two main mechanisms. Immunity-dependent PD-L1 up-regulation is characterized by high T cell infiltration with a high number of exhausted TIM3-positive T cells, and it has been shown to be somewhat predictive of the efficiency of PD-directed therapy (17). The second mechanism for PD-L1 up-regulation is through intracellular oncogenic signaling pathways. This form of up-regulation during treatment has limited predictive value. In our work, we found that individuals with the G variant, who have higher PD-L1 and PD-L2 levels, also have a higher number of TIM3-positive T cells, suggesting that PD-L1/2 up-regulation, in this case, is immunity-dependent and potentially more relevant to PD-1 inhibitor therapy.
Experimental procedures
Mice
Prmt1 floxed mice have been described before (9). Prmt1 myeloid-specific knockout mice are described in Ref. 9, and hepatocyte-specific knockout mice are described in Ref. 20. To create dendritic cell–specific knockout Prmt1, floxed mice were bred with Itgax-Cre mice (cd11c-Cre from The Jackson Laboratory).
All mice were housed in a temperature-controlled, specific pathogen–free environment with 12-h light/dark cycles and fed regular mouse chow and water ad libitum. All animal handling procedures were approved by the Institutional Animal Care and Use Committees at the University of Kansas Medical Center (Kansas City, KS).
To induce HCC formation, mice were given DEN at 75 mg/kg four times weekly starting at 4 weeks of age. Alternatively, mice were given DEN at 10 mg/kg at 2 weeks of age.
Primary antibodies
Anti-PRMT1 (F339), anti-PD-L1(mouse-specific), anti-PD-L1 (human-specific), anti-PD-L2 (human-specific), anti-VISTA, and anti-TIM3 antibodies were from Cell Signaling Technology. Rat anti-PD-L2 antibodies were from Novus. Anti-Mrc1 and anti-C/EBPβ (sc-7962 X, for ChIP) antibodies were from Santa Cruz Biotechnology.
Cell culture
THP-1 cells were maintained in RPMI medium (Invitrogen) containing 10% FBS, 50 units ml−1 penicillin, and 50 mg ml−1 streptomycin. Cells were transfected using Lipofectamine 3000 transfection reagent (Invitrogen) according to the manufacturer's protocol.
Vectors
The PRMT1 promoter luciferase construct was from ActiveMotif (SwitchGear Genomics). The promoter point mutation was generated with the Q5 mutagenesis kit from New England Biolabs. AAV8.TBG.PI.Null, AAV8.TBG.PI.Cre, and AAV.CMV.CRE were from Vector Biolabs (Malvern, PA).
Human specimens
Deidentified human specimens were obtained from the Liver Center Tissue Bank at the University of Kansas Medical Center. All studies using human tissue samples were approved by the Human Subjects Committee of the University of Kansas Medical Center. All studies abided by the Declaration of Helsinki principles.
Peripheral blood mononuclear cells were isolated from the peripheral blood mononuclear cell fraction using magnetic beads (human CD14) (Miltenyi Biotec, 130-050-201) according to the manufacturer's instructions. To obtain blood monocyte–derived macrophages, cells were differentiated by treatment with 1 × 104 units/ml M-CSF for 7 days.
Real Time PCR
RNA was extracted from cultured cells using the RNeasy Mini Kit (Qiagen). Complementary DNA was generated using an RNA reverse transcription kit (Applied Biosystems, catalog no. 4368814). Quantitative real-time RT-PCR was performed in a CFX96 real-time system (Bio-Rad) using specific sense and antisense primers combined with iQ SYBR Green Supermix (Bio-Rad) for 40 amplification cycles: 5 s at 95 °C, 10 s at 57 °C, and 30 s at 72 °C.
Immunohistochemistry
Immunostaining on formalin-fixed sections was performed by deparaffinization and rehydration, followed by antigen retrieval by heating in a pressure cooker (121 °C) for 5 min in 10 mm sodium citrate (pH 6.0) as described previously (20). Peroxidase activity was blocked by incubation in 3% hydrogen peroxide for 10 min. Sections were rinsed three times in PBS/PBS-T (0.1% Tween 20) and incubated in Dako Protein Block (Dako) at room temperature for 1 h. After removal of blocking solution, slides were placed into a humidified chamber and incubated overnight with an antibody, diluted 1:300 in Dako Protein Block at 4 °C. Antigen was detected using SignalStain Boost IHC detection reagent (catalog no. 8114, Cell Signaling Technology, Beverly, MA), developed with diaminobenzidene (Dako, Carpinteria, CA), counterstained with hematoxylin (Sigma-Aldrich), and mounted. Signal intensity was analyzed by Aperio ImageScope 12.1.
Immunofluorescence
Immunostaining on formalin-fixed sections was performed by deparaffinization and rehydration, followed by antigen retrieval by heating in a pressure cooker (121 °C) for 5 min in 10 mm sodium citrate (pH 6.0). Sections were rinsed three times in PBS/PBS-T (0.1% Tween 20) and incubated in Dako Protein Block (Dako) at room temperature for 1 h. After removal of blocking solution, slides were placed into a humidified chamber and incubated overnight with an antibody, diluted 1:300 in Dako Protein Block at 4 °C. After washing with PBS, coverslips were incubated with Alexa Fluor–conjugated secondary antibody (1:500) in 0.1 μg/ml DAPI for 1 h in the dark at room temperature. Coverslips were washed and mounted with FluorSave reagent (Calbiochem, La Jolla, CA). Slides were observed in a Nikon Eclipse 800 upright epifluorescence microscope (Nikon Instruments, Melville, NY). Images were acquired using a Nikon CoolSNAP camera.
Genotyping
Genomic DNA was isolated using a genomic DNA isolation kit (Promega). 80–200 ng of genomic DNA was used for real-time PCR in a CFX96 real-time system (Bio-Rad) using specific TaqMan probes for each SNP (ThermoFisher) combined with TaqMan Assay Mastermix (Applied Biosystems) for 40 amplification cycles.
ChIP assay
ChIP was performed as described previously (9, 28). THP-1 cells (1.5 × 107) were cross-linked by addition of 1% formaldehyde for 10 min. Cells were lysed with 10 mm Tris-HCl (pH 8.0), 10 mm NaCl, 3 mm MgCl2, and 0.5% NP-40. Nuclei were collected by centrifugation; resuspended in 1% SDS, 5 mmol/liter EDTA, and 50 mmol/liter Tris-HCl (pH 8.0); and sonicated to generate chromatin to an average length of ∼100–500 bp. Next, samples in 1% Triton X-100, 2 mm EDTA, 20 mm Tris-HCl (pH 8.1), and 150 mm NaCl were immunoprecipitated overnight at 4 °C with 4 μg of ChIP-grade antibody. 20 μl of magnetic beads (Dynabeads M-280, Invitrogen) was used to purify immunocomplexes. Following purification, cross-links were reverted by incubation at 65 °C for 6 h. Samples were purified with a Qiagen kit (catalog no. 27104).
Statistics
Results are expressed as mean ± S.D. Student t test, paired t test, Pearson's correlation, or one-way analysis of variance with Bonferroni post hoc test was used for statistical analyses. p < 0.05 was considered significant.
Data availability
All data described in the manuscript are contained within the manuscript. Additional data are available upon request.
Author contributions
M. S., J. Z., and I. T. data curation; M. S. and J. Z. investigation; M. S. methodology; A. K. and S. A. W. resources; S. A. W. and I. T. conceptualization; S. A. W. and I. T. formal analysis; S. A. W. and I. T. writing-review and editing; I. T. supervision; I. T. writing-original draft.
Supplementary Material
Acknowledgments
The human liver specimens used in this study were derived from samples provided by the University of Kansas Liver Center Tissue Bank. We thank Drs. Jameson Forster, Sean Kumer, Tim Schmitt, and Bashar Abdulkarim for assistance with obtaining these specimens.
This study was supported by grant AA027586 from the National Institute on Alcoholism and Alcohol Abuse and the 2016 Pinnacle Award from the American Association for the Study of Liver Diseases. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1–S9 and Table S1.
- HCC
- hepatocellular carcinoma
- IHC
- immunohistochemistry
- AAV
- adeno-associated virus
- DC
- dendritic cells
- DEN
- N-Nitrosodiethylamine
- FOXO
- forkhead box O
- GC
- genome copies
- GWAS
- genome-wide association study
- TAM
- tumor-associated macrophage
- CMV
- cytomegalovirus
- KO
- knockout.
References
- 1. Tang J., Frankel A., Cook R. J., Kim S., Paik W. K., Williams K. R., Clarke S., and Herschman H. R. (2000) PRMT1 is the predominant type I protein arginine methyltransferase in mammalian cells. J. Biol. Chem. 275, 7723–7730 10.1074/jbc.275.11.7723 [DOI] [PubMed] [Google Scholar]
- 2. Yang Y., McBride K. M., Hensley S., Lu Y., Chedin F., and Bedford M. T. (2014) Arginine methylation facilitates the recruitment of TOP3B to chromatin to prevent R loop accumulation. Mol. Cell 53, 484–497 10.1016/j.molcel.2014.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Barrero M. J., and Malik S. (2006) Two functional modes of a nuclear receptor-recruited arginine methyltransferase in transcriptional activation. Mol. Cell 24, 233–243 10.1016/j.molcel.2006.09.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bedford M. T., and Clarke S. G. (2009) Protein arginine methylation in mammals: who, what, and why. Mol. Cell 33, 1–13 10.1016/j.molcel.2008.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chittka A. (2010) Dynamic distribution of histone H4 arginine 3 methylation marks in the developing murine cortex. PLoS ONE 5, e13807 10.1371/journal.pone.0013807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. An W., Kim J., and Roeder R. G. (2004) Ordered cooperative functions of PRMT1, p300, and CARM1 in transcriptional activation by p53. Cell 117, 735–748 10.1016/j.cell.2004.05.009 [DOI] [PubMed] [Google Scholar]
- 7. Hassa P. O., Covic M., Bedford M. T., and Hottiger M. O. (2008) Protein arginine methyltransferase 1 coactivates NF-κB-dependent gene expression synergistically with CARM1 and PARP1. J. Mol. Biol. 377, 668–678 10.1016/j.jmb.2008.01.044 [DOI] [PubMed] [Google Scholar]
- 8. Tikhanovich I., Kuravi S., Artigues A., Villar M. T., Dorko K., Nawabi A., Roberts B., and Weinman S. A. (2015) Dynamic arginine methylation of tumor necrosis factor (TNF) receptor-associated factor 6 regulates Toll-like receptor signaling. J. Biol. Chem. 290, 22236–22249 10.1074/jbc.M115.653543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tikhanovich I., Zhao J., Olson J., Adams A., Taylor R., Bridges B., Marshall L., Roberts B., and Weinman S. A. (2017) Protein arginine methyltransferase 1 modulates innate immune responses through regulation of peroxisome proliferator-activated receptor γ-dependent macrophage differentiation. J. Biol. Chem. 292, 6882–6894 10.1074/jbc.M117.778761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhao J., O'Neil M., Vittal A., Weinman S. A., and Tikhanovich I. (2019) PRMT1-dependent macrophage IL-6 production is required for alcohol-induced HCC progression. Gene Expr. 19, 137–150 10.3727/105221618X15372014086197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. El-Serag H. B. (2011) Hepatocellular carcinoma. N. Engl. J. Med. 365, 1118–1127 10.1056/NEJMra1001683 [DOI] [PubMed] [Google Scholar]
- 12. Soerjomataram I., Lortet-Tieulent J., Parkin D. M., Ferlay J., Mathers C., Forman D., and Bray F. (2012) Global burden of cancer in 2008: a systematic analysis of disability-adjusted life-years in 12 world regions. Lancet 380, 1840–1850 10.1016/S0140-6736(12)60919-2 [DOI] [PubMed] [Google Scholar]
- 13. Sanyal A. J., Yoon S. K., and Lencioni R. (2010) The etiology of hepatocellular carcinoma and consequences for treatment. Oncologist 15, 14–22 10.1634/theoncologist.2010-S4-14 [DOI] [PubMed] [Google Scholar]
- 14. Siu E. H., Chan A. W., Chong C. C., Chan S. L., Lo K. W., and Cheung S. T. (2018) Treatment of advanced hepatocellular carcinoma: immunotherapy from checkpoint blockade to potential of cellular treatment. Transl. Gastroenterol. Hepatol. 3, 89 10.21037/tgh.2018.10.16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Altekruse S. F., Henley S. J., Cucinelli J. E., and McGlynn K. A. (2014) Changing hepatocellular carcinoma incidence and liver cancer mortality rates in the United States. Am. J. Gastroenterol. 109, 542–553 10.1038/ajg.2014.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zongyi Y., and Xiaowu L. (2020) Immunotherapy for hepatocellular carcinoma. Cancer Lett. 470, 8–17 10.1016/j.canlet.2019.12.002 [DOI] [PubMed] [Google Scholar]
- 17. Yi M., Jiao D., Xu H., Liu Q., Zhao W., Han X., and Wu K. (2018) Biomarkers for predicting efficacy of PD-1/PD-L1 inhibitors. Mol. Cancer 17, 129 10.1186/s12943-018-0864-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tikhanovich I., Olson J. C., Taylor R., Bridges B., Dorko K., Roberts B. R., and Weinman S. A. (2015) Impaired TRAF6 methylation and TLR responses in liver tissue and circulating monocytes from patients with spontaneous bacterial peritonitis. Hepatology 62, 846a [Google Scholar]
- 19. Tang Z., Li C., Kang B., Gao G., Li C., and Zhang Z. (2017) GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 45, W98–W102 10.1093/nar/gkx247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhao J., Adams A., Roberts B., O'Neil M., Vittal A., Schmitt T., Kumer S., Cox J., Li Z., Weinman S. A., and Tikhanovich I. (2018) PRMT1 and JMJD6 dependent arginine methylation regulate HNF4 expression and hepatocyte proliferation. Hepatology 67, 1109–1126 10.1002/hep.29587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tikhanovich I., Zhao J., Bridges B., Kumer S., Roberts B., and Weinman S. A. (2017) Arginine methylation regulates c-Myc-dependent transcription by altering promoter recruitment of the acetyltransferase p300. J. Biol. Chem. 292, 13333–13344 10.1074/jbc.M117.797928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lin H., Wei S., Hurt E. M., Green M. D., Zhao L., Vatan L., Szeliga W., Herbst R., Harms P. W., Fecher L. A., Vats P., Chinnaiyan A. M., Lao C. D., Lawrence T. S., Wicha M., et al. (2018) Host expression of PD-L1 determines efficacy of PD-L1 pathway blockade-mediated tumor regression. J. Clin. Invest. 128, 1708 10.1172/JCI120803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Buniello A., MacArthur J. A. L., Cerezo M., Harris L. W., Hayhurst J., Malangone C., McMahon A., Morales J., Mountjoy E., Sollis E., Suveges D., Vrousgou O., Whetzel P. L., Amode R., Guillen J. A., et al. (2019) The NHGRI-EBI GWAS catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 47, D1005–D1012 10.1093/nar/gky1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lee K. H., Tsai W. J., Chen Y. W., Yang W. C., Lee C. Y., Ou S. M., Chen Y. T., Chien C. C., Lee P. C., Chung M. Y., and Lin C. C. (2016) Genotype polymorphisms of genes regulating nitric oxide synthesis determine long-term arteriovenous fistula patency in male hemodialysis patients. Renal Failure 38, 228–237 10.3109/0886022X.2015.1120096 [DOI] [PubMed] [Google Scholar]
- 25. He L., Ronis M. J., and Badger T. M. (2002) Ethanol induction of class I alcohol dehydrogenase expression in the rat occurs through alterations in CCAAT/enhancer binding proteins β and γ. J. Biol. Chem. 277, 43572–43577 10.1074/jbc.M204535200 [DOI] [PubMed] [Google Scholar]
- 26. Gao Y., Sun W., Shang W., Li Y., Zhang D., Wang T., Zhang X., Zhang S., Zhang Y., and Yang R. (2018) Lnc-C/EBPβ negatively regulates the suppressive function of myeloid-derived suppressor cells. Cancer Immunol. Res. 6, 1352–1363 10.1158/2326-6066.CIR-18-0108 [DOI] [PubMed] [Google Scholar]
- 27. McPeak M. B., Youssef D., Williams D. A., Pritchett C. L., Yao Z. Q., McCall C. E., and El Gazzar M. (2017) Frontline science: myeloid cell-specific deletion of Cebpb decreases sepsis-induced immunosuppression in mice. J. Leukoc. Biol. 102, 191–200 10.1189/jlb.4HI1216-537R [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Li Z., Zhao J., Tikhanovich I., Kuravi S., Helzberg J., Dorko K., Roberts B., Kumer S., and Weinman S. A. (2016) Serine 574 phosphorylation alters transcriptional programming of FOXO3 by selectively enhancing apoptotic gene expression. Cell Death Differ. 23, 583–595 10.1038/cdd.2015.125 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data described in the manuscript are contained within the manuscript. Additional data are available upon request.