Abstract
Background & Aims
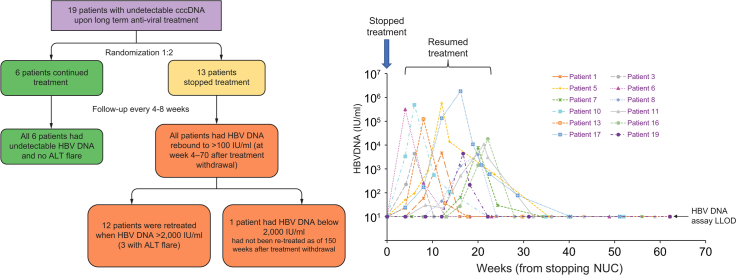
Nucleos(t)ide analogues (NUCs) effectively suppress serum HBV DNA. Previously, we have identified 21 patients with undetectable covalently closed circular DNA (cccDNA) upon long-term NUC therapy. This study investigated the effect of NUC withdrawal in patients with undetectable cccDNA.
Methods
Nineteen patients on long term NUCs (median 13.4 years) were recruited: 13 were randomized to discontinue NUCs; 6 to continue taking NUCs. All had undetectable cccDNA at the time of last liver biopsy (median time 2.9 years prior to randomization). Serum HBV DNA, hepatitis B surface antigen (HBsAg), hepatitis B core-related antigen (HBcrAg), liver biochemistry, and serum HBV RNA were monitored.
Results
At the time of randomization, all patients had undetectable serum HBV DNA and HBV RNA. Twelve of the 13 patients had HBV DNA rebound to 100 IU/ml within 20 weeks of NUC discontinuation. The thirteenth patient had HBV DNA rebound at week 70. Three patients experienced biochemical flares after re-treatment which subsequently resolved. There was no significant association between the time of HBV DNA rebound and baseline HBsAg, HBcrAg and alanine aminotransferase, duration of treatment, and age at which treatment was stopped (all p >0.05). At the time of HBV DNA rebound, HBV DNA levels correlated with HBcrAg levels (p = 0.003), but not with HBsAg levels (p = 0.262).
Conclusions
In patients with undetectable intrahepatic cccDNA, virologic rebound still occurred after NUC cessation. At the rebound of HBV DNA, the kinetics of HBsAg production were independent of those of viral DNA replication. Additional studies are required to determine the factors that may predict virologic rebound and when NUCs can be discontinued in HBsAg-positive patients with chronic hepatitis B.
Lay summary
It has been shown that following long-term nucleos(t)ide analogue treatment for chronic hepatitis B, some patients have undetectable levels of viral DNA in their livers. We tested the results of withdrawing nucleos(t)ide analogue treatment in these patients and found that viral relapse could occur in patients with undetectable viral DNA. Further research is required to determine whether nucleos(t)ide analogue treatment can be discontinued in specific patients with chronic hepatitis B.
Keywords: Chronic hepatitis B, hepatitis B virus DNA rebound, antiviral therapy, stopping therapy
Abbreviations: ALT, alanine aminotransferase; anti-HBe, antibody to HBeAg; cccDNA, covalently closed circular DNA; CHB, chronic hepatitis B; ETV, entecavir; HBcrAg, hepatitis B core-related antigen; HBeAg, hepatitis B e antigen; HBsAg, hepatitis B surface antigen; LdT, telbivudine; NUCs, nucleos(t)ide analogues; TDF, tenofovir disoproxil fumarate; ULN, upper limit of normal
Graphical abstract
Highlights
-
•
Patients on long-term nucleos(t)ide analogue treatment with undetectable HBV DNA may have undetectable cccDNA.
-
•
Stopping treatment for patients with undetectable cccDNA resulted in rebound of serum HBV DNA, mostly within 20 weeks.
-
•
There is no association between time of HBV DNA rebound and other viral markers, including HBsAg titers and HBcrAg.
-
•
Even in patients with undetectable cccDNA in liver biopsies, virologic relapse can still occur.
Introduction
Chronic hepatitis B (CHB) affects approximately 257 million people globally.1 Following hepatocyte entry, the HBV relaxed circular DNA genome enters the nuclei, associates with cellular histones and other proteins and forms a covalently closed circular DNA (cccDNA) minichromosome. cccDNA is the template for HBV transcription and can only be measured in liver biopsies. Since nucleos(t)ide analogues (NUCs) for the treatment of CHB have no direct action on cccDNA, the persistence of cccDNA is thought to be the main obstacle to achieving a “cure” for CHB.2 However, it has been postulated that with prolonged NUC treatment, cccDNA may become depleted through natural cell division and cell death of the hepatocytes.3,4 In a previous study from our center of 43 patients on long-term NUCs (median treatment duration 10.5 years) with 3 liver biopsies, cccDNA was reduced by 99.89%, with 21 patients (49%) having undetectable cccDNA, as measured by a real-time PCR assay.3 It is not known whether a reduction of cccDNA to very low or undetectable levels is a sufficient endpoint for cessation of NUC treatment. The current study was designed to investigate the effect of withdrawal of NUCs in patients with undetectable cccDNA levels.
Patients and methods
In a previous study of patients undergoing long-term NUC treatment, we had recruited patients with 3 liver biopsies: baseline (before NUC treatment), 1 year after treatment, and ≥6 years after treatment.3 cccDNA inside the liver tissues was measured by real-time PCR, as previously described.3,5 Briefly, total liver DNA extracted from liver tissues was subjected to digestion with plasmid-safe DNase (Epicenter, Wisconsin, MD), followed by “over-gap” real-time PCR amplification using primers spanning across the nicked regions of the HBV relaxed circular genome. Real-time PCR quantification was performed using the QuantiFast Probe PCR kit in a RotorGene Q real-time PCR system (Qiagen, GmbH, Hilden, Germany) with the “over-gap” primers and dual hybridization probes with fluorescence resonance energy transfer technology, sequences of which have been described previously.5 Serial diluted plasmids containing cloned HBV DNA were used as quantitation standards. Human genomic DNA content, which reflected the number of cells in the liver tissues, was measured by the Light-Cycler Control DNA kit (Roche Diagnostics, Branchburg, NJ). The lower limit of detection was 0.005 cccDNA copies/cell.3
In the previous study, 21 patients with undetectable cccDNA were identified. Of them, 19 were recruited in the present study. Of the 2 remaining patients, 1 defaulted; the other was positive for HBV DNA at screening. Two of the 19 recruited patients were positive for the hepatitis B e antigen (HBeAg); 14 were positive for antibody against HBeAg (anti-HBe); and 3 were negative for both HBeAg and anti-HBe.
Computer-assisted randomization was performed to randomize the 19 patients to either discontinue NUCs or continue NUCs at a ratio of 2:1. Thirteen patients were randomized to discontinue NUCs, and 6 to continue NUCs.
After the discontinuation of NUCs, all patients were seen every 4 to 8 weeks to monitor serum HBV DNA by the COBAS HBV Test (Roche Molecular Diagnostics, Pleasanton, CA, USA; lower limit of detection 1 log IU/ml), hepatitis B surface antigen (HBsAg) titers by the Elecsys HBsAg II Quant Assay (Roche Diagnostics, Indianapolis, IN, USA; lower limit of detection 0.05 IU/ml), hepatitis B core-related antigen (HBcrAg) titers by the Lumipulse HBcrAg Assay (Fujirebio Inc, Tokyo, Japan; lower limit of detection 1 kU/ml), and liver biochemistry. Serum HBV RNA levels were measured using a rapid amplification of cDNA ends (RACE)-based real-time PCR method, with a lower limit of detection of 800 copies/ml, as previously described.6 Rebound of HBV DNA was defined as having detectable serum HBV DNA of ≥100 IU/ml, which was equivalent to a ≥1 log increase from undetectable levels. NUCs were recommenced when HBV DNA levels further rose to ≥2,000 IU/ml (3.3 log IU/ml). After resumption of NUCs, patients were followed up every 8 to 16 weeks. The study was approved by the Institution Review Board of the University of Hong Kong and Hong Kong Hospital Authority West Cluster (Reference: UW 16-413) and was registered at The University of Hong Kong Clinical Trial Registry (Reference: HKUCTR-2110). Written consents were obtained from all patients recruited in this study.
Statistical analyses were performed by using IBM SPSS Statistics 25 (IBM, Armonk, NY, USA). Continuous variables were expressed as median and range. Related samples were tested with the Wilcoxon sign ranks test. The correlation between 2 variables was tested using the Pearson correlation analysis. Statistical significance was defined as a p value of less than 0.05.
Results
Nineteen patients (13 male and 6 female; median age 56 years [range 42–75]) with undetectable cccDNA were recruited in this study. Prior to randomization, of the 19 patients, 12 were taking entecavir (ETV), 4 were taking telbivudine (LdT), 3 were taking tenofovir disoproxil fumarate (TDF). At the time of randomization, the median duration of treatment was 13.4 years (range 8.7–14.9 years). The median duration between the last liver biopsy and randomization was 2.9 years (range 2.5–3.2 years), during which all 19 patients had at least 2 measurements of HBV DNA and HBV RNA performed. All 19 patients had persistently undetectable serum HBV DNA and HBV RNA levels prior to randomization.
Thirteen patients (8 on ETV, 3 on LdT, and 2 on TDF) were randomized to stop NUCs, of whom 1 was HBeAg-positive and 12 were HBeAg-negative. The median HBsAg and HBcrAg levels at the time of randomization were 414 IU/ml (range 70–2,780 IU/ml) and 2.6 kU/ml (range <1–36.5 kU/ml), respectively. Four of 13 patients had undetectable HBcrAg at the time of randomization. After stopping NUCs, all 13 patients had rebound of HBV DNA to >100 IU/ml, with a median time to rebound of 12 weeks (range 4–70 weeks). Three patients had an early HBV DNA rebound at their first follow up at week 4: 2 were receiving TDF and 1 LdT before stopping treatment. Three patients (1 was on ETV and 2 were on LdT) had HBV DNA rebound at week 8. The remaining 7 patients with HBV DNA rebound at weeks 12–70 (1 patient at week 12, 3 patients at week 16, 2 patients at week 20, and 1 at week 70) were all receiving ETV before stopping treatment. There was no significant association between the time of HBV DNA rebound and baseline parameters such as baseline HBsAg and HBcrAg levels, baseline alanine aminotransferase (ALT), duration of NUC treatment, and age at which treatment was stopped (all p >0.05).
The virologic and biochemical parameters at baseline and at the time of HBV DNA rebound of the 13 patients who stopped therapy were compared (Table 1). The median HBV DNA and HBcrAg level at the time of HBV DNA rebound was 3.16 log IU/ml and 5.80 kU/ml, respectively, both of which were significantly higher than at the time of stopping NUCs (p = 0.001 and 0.005, respectively). Compared with baseline, there was no significant increase in HBsAg levels at the time of HBV DNA rebound. All 13 patients were asymptomatic, and their bilirubin and prothrombin time remained normal throughout the study. The 6 patients who continued to receive NUCs had undetectable HBV DNA levels and normal ALT levels at last follow-up.
Table 1.
Comparison of virologic and biochemical parameters at baseline and at the time of HBV DNA rebound using the Wilcoxon signed ranks test.
| Parameter∗ | At baseline | At time of HBV DNA rebound (4–70 weeks) | p Value |
|---|---|---|---|
| HBV DNA, log IU/ml | <1 | 3.16 (2.16–5.76) | 0.001 |
| HBsAg, log IU/ml | 2.62 (1.84–3.44) | 2.48 (1.95–3.26) | 0.507 |
| HBcrAg, kU/ml | 2.60 (<1–36.5) | 5.80 (<1–143.3) | 0.005 |
| ALT levels, U/L | 27 (18–62) | 29 (16–54) | 0.726 |
ALT, alanine aminotransferase; HBcrAg, hepatitis B core-related antigen; HBsAg, hepatitis B surface antigen.
Values expressed as median (range).
The HBV DNA levels of the 12 patients with rebound before week 20 rose to >2,000 IU/ml. They were treated either with the NUCs they had been previously taking (for TDF and ETV patients) or with TDF (for LdT patients). The kinetics of virologic parameters and ALT levels after stopping NUC therapy (and resumption of NUC therapy) are shown in Fig. 1, Fig. 2, Fig. 3.
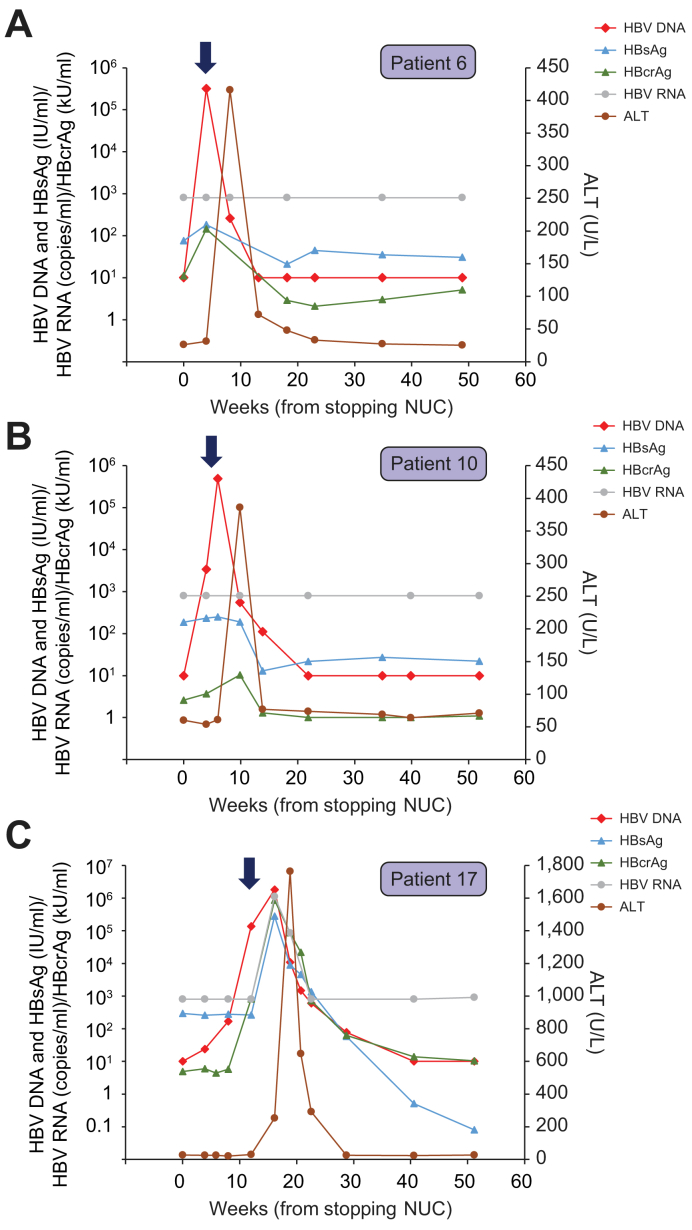
Fig. 1.
Virological and biochemical parameters of the 3 patients who experienced ALT flare of ≥2× ULN after stopping treatment.
(A) Profile of patient 6; (B) Profile of patient 10; and (C) profile of patient 17. Thick arrows denote the time of resumption of NUCs. ALT, alanine aminotransferase; NUCs, nucleos(t)ide analogues; ULN, upper limit of normal.
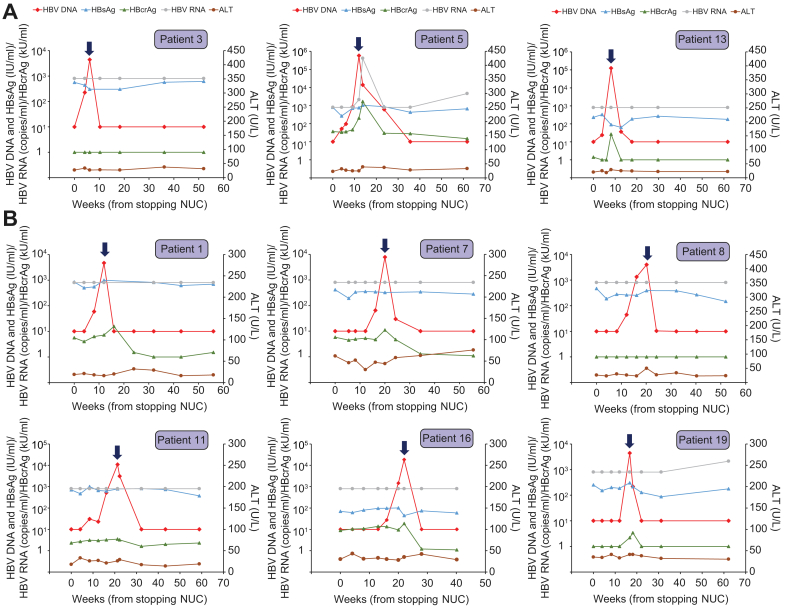
Fig. 2.
Virological and biochemical parameters of the 9 patients who did not have ALT flare.
Thick arrows denote the time of resumption of NUCs. (A) Patients with early HBV DNA rebound on or before week 8. (B) Patients with late HBV DNA rebound between weeks 12–20. ALT, alanine aminotransferase; NUCs, nucleos(t)ide analogues.
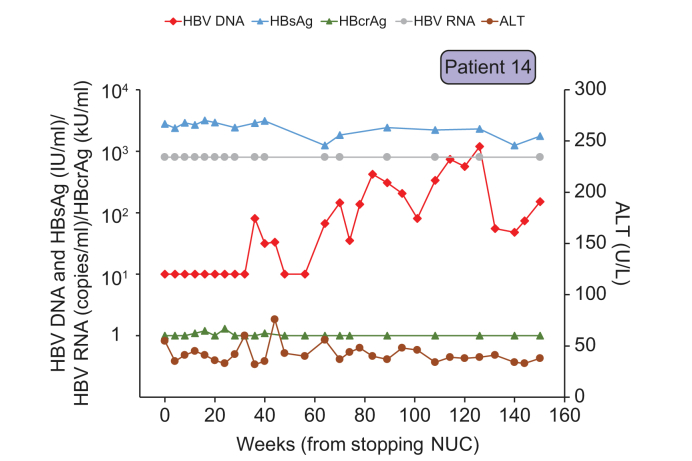
Fig. 3.
Virological and biochemical parameters of the patient whose HBV DNA remained below 2,000 IU/ml and hence did not resume NUC therapy.
NUCs, nucleos(t)ide analogues.
Three patients (Patients 6, 10, and 17) experienced elevations of ALT levels (≥2× upper limit of normal), all of whom had early HBV DNA rebound on or before week 8 (Fig. 1). HBV DNA further increased to >2,000 IU/ml within 2–4 weeks of HBV DNA rebound, leading to early resumption of NUCs, after which HBV DNA returned to undetectable levels. In patients 6 and 10 (Fig. 1A,B), HBV DNA rose to >5 log IU/ml after stopping NUCs, followed by an increase in ALT to approximately 400 U/L. During HBV DNA/ALT flare, there was a slight increase in HBsAg and HBcrAg, which were then decreased back to a low level after resumption of NUCs. HBV RNA remained below the assay detection limit throughout the study in these 2 patients.
Patient 17 had a peak HBV DNA of 6.3 log IU/ml, followed by a substantial ALT flare to 1,764 U/L after the resumption of NUCs (Fig. 1C). This patient was positive for anti-HBe at baseline. At the time of HBV DNA flare, there was a flare of the other 3 markers measured (HBsAg, HBcrAg and HBV RNA), which were then decreased to a low/undetectable level after HBV DNA/ALT flare. An extended follow-up in this patient showed that this patient became HBsAg-negative at week 64 after stopping NUC therapy, which was equivalent to 48 weeks after re-treatment with TDF (data not shown).
Nine patients had HBV DNA rebound to >2,000 IU/ml but without ALT flare (Fig. 2). Three of them (Patients 3, 5 and 13; Fig. 2A) had early HBV DNA rebound on or before week 8, while 6 patients (Fig. 2B) had HBV DNA rebound between weeks 12-20. By week 22, all 9 patients resumed NUC treatment. In general, there was a slight (<1 log) increase of HBcrAg around the time of HBV DNA rebound, and HBsAg levels did not seem to correlate with HBV DNA flare. HBV RNA was mostly below the assay detection limit in all patients except Patient 5. In Patient 5, who was HBeAg-positive at the time of randomization, there was an approximately 2 log increase in HBcrAg and a more than 3 log increase in HBV RNA around the time of HBV DNA flare, at which point ALT peaked at only 39 U/L. HBcrAg and HBV RNA returned to levels comparable to baseline after resumption of NUCs.
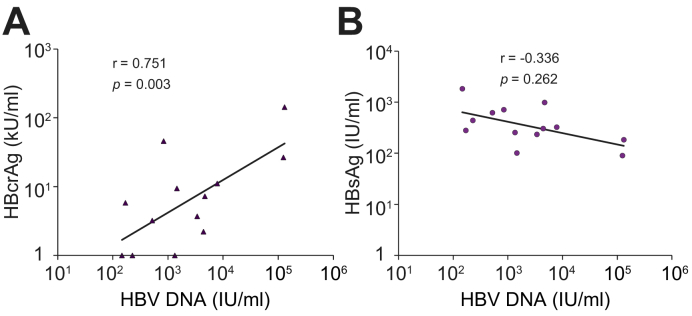
The thirteenth patient (Patient 14) had a late HBV DNA rebound to >100 IU/ml at week 70 (Fig. 3). HBV DNA levels remained below 2,000 IU/ml during all follow-up visits of 8 to 16 weeks apart, with the latest follow-up being 150 weeks. This patient had undetectable HBcrAg but with HBsAg of 2,780 IU/ml at baseline. HBV DNA peaked at 1,200 IU/ml at week 126. Thus, antiviral treatment had not been resumed in this patient. The peak ALT was 76 U/ml at week 44, and HBcrAg and HBV RNA levels were below or near the assay detection limit during follow-up. Interestingly, this patient had a high level of HBsAg (>3 log IU/ml) throughout the study. Fig. 4 shows the correlation between the levels of HBcrAg and HBsAg with HBV DNA levels at the time of HBV DNA rebound. HBcrAg showed good correlation with HBV DNA levels (p = 0.003) but there was no correlation between HBsAg and HBV DNA levels (p = 0.262).
Fig. 4.
Correlation between HBV DNA levels at the time of HBV DNA rebound and other viral markers.
Correlation between HBV DNA levels at the time of HBV DNA rebound and (A) HBcrAg; level of significance: p = 0.003 (Pearson correlation test); (B) HBsAg; level of significance: p = 0.262 (Pearson correlation test).
HBcrAg, hepatitis B core-related antigen; HBsAg, hepatitis B surface antigen.
Discussion
Recently there has been a debate as to whether NUC therapy can be stopped when there is substantial evidence of viral inactivity in patients with CHB.[7], [8], [9], [10], [11], [12] Several viral markers, such as HBsAg, HBcrAg, and HBV RNA, have been suggested as markers of viral inactivity.[13], [14], [15], [16], [17]
This is the first study to investigate the withdrawal of NUCs in patients with undetectable cccDNA in their hepatocytes. All 13 patients randomized to stop therapy had HBV DNA rebound. Six patients had rapid elevation of HBV DNA levels 4–8 weeks after cessation of therapy, 7 had HBV DNA rebound to >100 IU/ml at 12–20 weeks, and 1 had a late rebound at week 70. Compared with patients receiving ETV, patients receiving TDF or LdT seemed to have earlier HBV relapse. This is consistent with the findings of another study which shows that patients with TDF withdrawal have earlier HBV rebound.18 The biochemical flares were independent of the baseline HBeAg status, with the most severe flare of ALT (1,764 U/L) occurring in a patient who was anti-HBe positive at baseline. In the single HBeAg-positive patient who stopped treatment the highest ALT was 39 U/L only.
The rebound of HBV DNA after stopping NUCs suggests that even a small amount of cccDNA, not detectable by our assay, can still allow viral replication causing HBV DNA rebound. Nine out of 13 patients who stopped NUCs still had detectable HBcrAg, a marker of cccDNA transcription activity. This signifies that HBV replication still occurred in these “cccDNA-negative” patients. In fact, except for the few patients with very low or undetectable HBcrAg throughout the study (patients 3, 8 and 14), HBcrAg profile mirrored that of HBV DNA. In addition, HBcrAg showed good correlation with HBV DNA at the time of HBV DNA rebound. While serum HBV RNA may also be a useful marker of HBV transcription activity, the majority of patients had undetectable serum HBV RNA. It is possible that, in patients with long-term NUCs, the cccDNA pool may become very small, resulting in a serum HBV RNA level too low to be detected. Therefore, in patients with long term NUCs, HBcrAg may be useful to define HBV intrahepatic replicative activity, as demonstrated in previous studies,19,20 while the usefulness of serum HBV RNA requires further studies with more sensitive assays.
There was no correlation of HBV DNA rebound with HBsAg levels. This is probably due to the expression of HBsAg from integrated HBV DNA and the presence of excess free circulating HBsAg particles. HBsAg seroclearance occurs only in a small proportion of patients, some of whom may have hepatic flare prior to HBsAg seroclearance.[21], [22], [23] However, whether such beneficial hepatic flare is predictable and whether it outweighs the risk of uncontrolled flare are still controversial.7,8,[23], [24], [25] In the present study, we restarted treatment independent of the ALT levels. We therefore did not investigate the possibility of beneficial flares. The present study was designed to avoid severe reactivation. In the 3 patients who experienced severe ALT flares (400–1,760 U/L), the flares were observed after re-treatment. It was not known whether the flare would be more severe if the re-treatment was further delayed. The patient with an ALT flare of 1,760 U/L had HBsAg seroclearance 48 weeks after re-treatment with TDF, suggesting that beneficial flares leading to HBsAg seroclearance could still occur after resumption of NUCs. The timing of NUC resumption, as well as the identification of factors predicting HBsAg seroclearance at NUC withdrawal, require further investigation.
The limitation of this study is the small number of patients involved. However, this is unavoidable since repeated liver biopsies were required to document the levels of cccDNA in patients on prolonged NUC treatment. Our study is the only study to date to recruit patients with documented undetectable cccDNA levels. Another limitation concerns the sensitivity of the assays used for cccDNA and serum HBV RNA detection. To date, standardized assays for cccDNA and HBV RNA are not available. The early rebound of HBV DNA in this study might be due to small amounts of cccDNA not detectable by the current assay, or to liver biopsy sampling errors. Further studies should be performed with the development of more sensitive cccDNA and HBV RNA assays, such as digital droplet PCR-based assays for cccDNA.26
In conclusion, this study showed that absence of detectable cccDNA in liver biopsies does not preclude the occurrence of virologic rebound, and patients with undetectable cccDNA still harbor detectable virologic markers in the serum. Additional studies are required to determine whether the absence of cccDNA, in combination with measurement of other parameters, can help prevent uncontrolled hepatic flare and predict HBsAg seroclearance.
Financial support
The authors received no financial support to produce this manuscript.
Authors' contributions
CLL and DKHW contributed to the conception and design of the study. DKHW, and GTYW contributed to data collection. CLL, DKHW, WKS, and JF analyzed and interpreted the data. CLL and DKHW drafted the manuscript. WKS, JF and MFY critically revised the manuscript for important intellectual content. All authors approved the final version of the manuscript.
Conflicts of interest
CLL is an advisory board member and stockholder and received speaker's fees from Gilead Sciences. WKS received speaker's fees from Alfa-Wassermann, Astrazeneca, is an advisory board member of Celltrion, received speaker's fees and is an advisory board member of AbbVie, and received speaker's fees, research funding and is an advisory board member of Gilead Sciences. MFY is an advisory board member and received speaker's fees from AbbVie, Janssen, Biocartis NV, Bristol Myers Squibb, Fujirebio Incorporation, Gilead Sciences, Merck Sharp and Dohme, and Sysmex Corporation, and received research funding from Bristol Myers Squibb and Gilead Sciences. DKH Wong received travel support form Gilead Sciences. JF received travel support form Gilead Sciences and AbbVie. GTY Wong has no conflict of interest.
Please refer to the accompanying ICMJE disclosure forms for further details.
Footnotes
Author names in bold designate shared co-first authorship
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jhepr.2020.100112.
Contributor Information
Ching-Lung Lai, Email: hrmelcl@hku.hk.
Danny Ka-Ho Wong, Email: dannykhwong@gmail.com.
Supplementary data
References
- 1.World Health Organization fact sheet. Hepatitis B. 2018. http://www.who.int/en/news-room/fact-sheets/detail/hepatitis-b [cited July 18, 2018]. Available at. [Google Scholar]
- 2.Nassal M. HBV cccDNA: viral persistence reservoir and key obstacle for a cure of chronic hepatitis B. Gut. 2015;64:1972–1984. doi: 10.1136/gutjnl-2015-309809. [DOI] [PubMed] [Google Scholar]
- 3.Lai C.L., Wong D., Ip P., Kopaniszen M., Seto W.K., Fung J. Reduction of covalently closed circular DNA with long-term nucleos(t)ide analogue treatment in chronic hepatitis B. J Hepatol. 2017;66:275–281. doi: 10.1016/j.jhep.2016.08.022. [DOI] [PubMed] [Google Scholar]
- 4.Mason W.S., Cullen J., Moraleda G., Saputelli J., Aldrich C.E., Miller D.S. Lamivudine therapy of WHV-infected woodchucks. Virology. 1998;245:18–32. doi: 10.1006/viro.1998.9150. [DOI] [PubMed] [Google Scholar]
- 5.Werle-Lapostolle B., Bowden S., Locarnini S., Wursthorn K., Petersen J., Lau G. Persistence of cccDNA during the natural history of chronic hepatitis B and decline during adefovir dipivoxil therapy. Gastroenterology. 2004;126:1750–1758. doi: 10.1053/j.gastro.2004.03.018. [DOI] [PubMed] [Google Scholar]
- 6.van Bommel F., Bartens A., Mysickova A., Hofmann J., Kruger D.H., Berg T. Serum hepatitis B virus RNA levels as an early predictor of hepatitis B envelope antigen seroconversion during treatment with polymerase inhibitors. Hepatology. 2015;61:66–76. doi: 10.1002/hep.27381. [DOI] [PubMed] [Google Scholar]
- 7.Marciano S., Gadano A. Why not to stop antiviral treatment in patients with chronic hepatitis B. Liver Int. 2018;38(Suppl 1):97–101. doi: 10.1111/liv.13627. [DOI] [PubMed] [Google Scholar]
- 8.Moreno-Cubero E., Del Arco R.T.S., Pena-Asensio J., de Villalobos E.S., Miquel J., Larrubia J.R. Is it possible to stop nucleos(t)ide analogue treatment in chronic hepatitis B patients? World J Gastroenterol. 2018;24:1825–1838. doi: 10.3748/wjg.v24.i17.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Papatheodoridis G., Vlachogiannakos I., Cholongitas E., Wursthorn K., Thomadakis C., Touloumi G. Discontinuation of oral antivirals in chronic hepatitis B: a systematic review. Hepatology. 2016;63:1481–1492. doi: 10.1002/hep.28438. [DOI] [PubMed] [Google Scholar]
- 10.Terrault N.A., Lok A.S.F., McMahon B.J., Chang K.M., Hwang J.P., Jonas M.M. Update on prevention, diagnosis, and treatment of chronic hepatitis B: AASLD 2018 hepatitis B guidance. Hepatology. 2018;67:1560–1599. doi: 10.1002/hep.29800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.European Association for the Study of the Liver EASL 2017 clinical practice guidelines on the management of hepatitis B virus infection. J Hepatol. 2017;67:370–398. doi: 10.1016/j.jhep.2017.03.021. [DOI] [PubMed] [Google Scholar]
- 12.Sarin S.K., Kumar M., Lau G.K., Abbas Z., Chan H.L., Chen C.J. Asian-Pacific clinical practice guidelines on the management of hepatitis B: a 2015 update. Hepatol Int. 2016;10:1–98. doi: 10.1007/s12072-015-9675-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen C.H., Lu S.N., Hung C.H., Wang J.H., Hu T.H., Changchien C.S. The role of hepatitis B surface antigen quantification in predicting HBsAg loss and HBV relapse after discontinuation of lamivudine treatment. J Hepatol. 2014;61:515–522. doi: 10.1016/j.jhep.2014.04.029. [DOI] [PubMed] [Google Scholar]
- 14.Wang C.C., Tseng K.C., Hsieh T.Y., Tseng T.C., Lin H.H., Kao J.H. Assessing the durability of entecavir-treated hepatitis B using quantitative HBsAg. Am J Gastroenterol. 2016;111:1286–1294. doi: 10.1038/ajg.2016.109. [DOI] [PubMed] [Google Scholar]
- 15.Tanaka E., Matsumoto A. Guidelines for avoiding risks resulting from discontinuation of nucleoside/nucleotide analogs in patients with chronic hepatitis B. Hepatol Res. 2014;44:1–8. doi: 10.1111/hepr.12108. [DOI] [PubMed] [Google Scholar]
- 16.Jung K.S., Park J.Y., Chon Y.E., Kim H.S., Kang W., Kim B.K. Clinical outcomes and predictors for relapse after cessation of oral antiviral treatment in chronic hepatitis B patients. J Gastroenterol. 2016;51:830–839. doi: 10.1007/s00535-015-1153-1. [DOI] [PubMed] [Google Scholar]
- 17.Wang J., Shen T., Huang X., Kumar G.R., Chen X., Zeng Z. Serum hepatitis B virus RNA is encapsidated pregenome RNA that may be associated with persistence of viral infection and rebound. J Hepatol. 2016;65:700–710. doi: 10.1016/j.jhep.2016.05.029. [DOI] [PubMed] [Google Scholar]
- 18.Kuo M.T., Hu T.H., Hung C.H., Wang J.H., Lu S.N., Tsai K.L. Hepatitis B virus relapse rates in chronic hepatitis B patients who discontinue either entecavir or tenofovir. Aliment Pharmacol Ther. 2019;49:218–228. doi: 10.1111/apt.15053. [DOI] [PubMed] [Google Scholar]
- 19.Wong D.K., Seto W.K., Cheung K.S., Chong C.K., Huang F.Y., Fung J. Hepatitis B virus core-related antigen as a surrogate marker for covalently closed circular DNA. Liver Int. 2017;37:995–1001. doi: 10.1111/liv.13346. [DOI] [PubMed] [Google Scholar]
- 20.Testoni B., Lebosse F., Scholtes C., Berby F., Miaglia C., Subic M. Serum hepatitis B core-related antigen (HBcrAg) correlates with covalently closed circular DNA transcriptional activity in chronic hepatitis B patients. J Hepatol. 2019;70:615–625. doi: 10.1016/j.jhep.2018.11.030. [DOI] [PubMed] [Google Scholar]
- 21.Chevaliez S., Hezode C., Bahrami S., Grare M., Pawlotsky J.M. Long-term hepatitis B surface antigen (HBsAg) kinetics during nucleoside/nucleotide analogue therapy: finite treatment duration unlikely. J Hepatol. 2013;58:676–683. doi: 10.1016/j.jhep.2012.11.039. [DOI] [PubMed] [Google Scholar]
- 22.Seto W.K., Wong D.K., Fung J., Huang F.Y., Lai C.L., Yuen M.F. Reduction of hepatitis B surface antigen levels and hepatitis B surface antigen seroclearance in chronic hepatitis B patients receiving 10 years of nucleoside analogue therapy. Hepatology. 2013;58:923–931. doi: 10.1002/hep.26376. [DOI] [PubMed] [Google Scholar]
- 23.Berg T., Simon K.G., Mauss S., Schott E., Heyne R., Klass D.M. Long-term response after stopping tenofovir disoproxil fumarate in non-cirrhotic HBeAg-negative patients - FINITE study. J Hepatol. 2017;67:918–924. doi: 10.1016/j.jhep.2017.07.012. [DOI] [PubMed] [Google Scholar]
- 24.Liem K.S., Fung S., Wong D.K., Yim C., Noureldin S., Chen J. Limited sustained response after stopping nucleos(t)ide analogues in patients with chronic hepatitis B: results from a randomised controlled trial (Toronto STOP study) Gut. 2019;68:2206–2213. doi: 10.1136/gutjnl-2019-318981. [DOI] [PubMed] [Google Scholar]
- 25.Cao J., Chi H., Yu T., Li Z., Hansen B.E., Zhang X. Off-treatment hepatitis B virus (HBV) DNA levels and the prediction of relapse after discontinuation of nucleos(t)ide analogue therapy in patients with chronic hepatitis B: a Prospective stop study. J Infect Dis. 2017;215:581–589. doi: 10.1093/infdis/jix025. [DOI] [PubMed] [Google Scholar]
- 26.Caviglia G.P., Abate M.L., Tandoi F., Ciancio A., Amoroso A., Salizzoni M. Quantitation of HBV cccDNA in anti-HBc-positive liver donors by droplet digital PCR: a new tool to detect occult infection. J Hepatol. 2018;69:301–307. doi: 10.1016/j.jhep.2018.03.021. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.