Abstract
Nucleotide analog inhibitors, including broad-spectrum remdesivir and favipiravir, have shown promise in in vitro assays and some clinical studies for COVID-19 treatment, this despite an incomplete mechanistic understanding of the viral RNA-dependent RNA polymerase nsp12 drug interactions. Here, we examine the molecular basis of SARS-CoV-2 RNA replication by determining the cryo-EM structures of the stalled pre- and post- translocated polymerase complexes. Compared with the apo complex, the structures show notable structural rearrangements happening to nsp12 and its co-factors nsp7 and nsp8 to accommodate the nucleic acid, whereas there are highly conserved residues in nsp12, positioning the template and primer for an in-line attack on the incoming nucleotide. Furthermore, we investigate the inhibition mechanism of the triphosphate metabolite of remdesivir through structural and kinetic analyses. A transition model from the nsp7-nsp8 hexadecameric primase complex to the nsp12-nsp7-nsp8 polymerase complex is also proposed to provide clues for the understanding of the coronavirus transcription and replication machinery.
Keywords: 2019-nCoV, SARS-CoV-2, COVID-19, remdesivir, favipiravir, polymerase, RdRP, virus, nsp12, nsp8
Graphical Abstract
Highlights
-
•
Structures of SARS-CoV-2 RNA polymerase in complexes with RNA revealed
-
•
Conformational changes in nsp8 and its interaction with the exiting RNA are observed
-
•
Incorporation and delayed-chain-termination mechanism of remdesivir is elucidated
-
•
Transition model from primase complex to polymerase complex is proposed
Cryo-EM structures of the SARS-CoV-2 RNA polymerase in complexes with RNA, before and after RNA translocation, reveals structural rearrangements that the RNA-dependent RNA polymerase (RdRp) nsp12 and its co-factors (nsp7 and nsp8) undergo to accommodate nucleic acid binding. Further insights into how the complex is inhibited by remdesivir, and into the primase to polymerase transition, are also presented.
Introduction
The current pandemic of coronavirus disease 2019 (COVID-19), a respiratory disease that has led to more than three million cases and a pandemic is caused by a novel virus strain, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), an enveloped, positive-sense, single-stranded RNA betacoronavirus of the family Coronaviridae (Zhu et al., 2020). Previously known coronaviruses (CoVs) that infect humans, e.g., hCoV-OC43, HKU1, and 229E, usually cause mild to moderate upper respiratory tract illnesses (Corman et al., 2018). However, over the past two decades, highly pathogenic human coronaviruses have emerged (de Wit et al., 2016), including SARS-CoV-1, Middle East respiratory syndrome coronavirus (MERS-CoV), and the current SARS-CoV-2. Infection with these highly pathogenic coronaviruses can result in acute respiratory distress syndrome (ARDS), which might lead to a long-term reduction in lung function, arrhythmia, and death (Graham et al., 2013, Huang et al., 2020). Distinct from MERS-CoV or SARS-CoV-1, SARS-CoV-2 appears to spread more efficiently (Petrosillo et al., 2020). Unfortunately, there are currently no highly effective drugs targeting SARS-CoV-2, SARS-CoV-1, or MERS-CoV.
One of the most promising druggable targets for coronaviruses is the RNA-dependent RNA polymerase (RdRP), which is a crucial enzyme in the life cycle of RNA viruses. In coronaviruses, it supports the transcription and replication of their ∼30,000-nucleotide large RNA genomes. RdRP is targeted in different RNA viruses, including hepatitis C virus (HCV), Zika virus (ZIKV), and CoVs (Mercorelli et al., 2018). Compared to several other different positive-sense RNA viruses, SARS-CoV-2 polymerase displays structural similarity with several key amino acid residues conserved in the active site (Gao et al., 2020). This similarity makes drug repurposing an effective strategy that could shorten the drug development time compared to that of de novo drug discovery.
Nucleotide and nucleoside analogs that inhibit polymerases are an important group of antiviral agents (Debing et al., 2015). Clinical trials are in progress for the treatment of COVID-19 by using the RdRP nucleotide inhibitors, such as remdesivir (RDV) and favipiravir. A substantial obstacle for the discovery, optimization, and comprehensive evaluation of effective nucleotide-based drugs for the SARS-CoV-2 is the lack of molecular detail concerning substrate recognition during replication.
To gain insight into the mechanism of SARS-CoV-2 RNA replication and its inhibition by nucleotide analog inhibitors, we have determined near-atomic-resolution structures of SARS-CoV-2 polymerase-RNA complex in its catalytic states, including a remdesivir monophosphate (RDV-MP) incorporated pre-translocated complex and a stalled post-translocated complex.
Results
Assembly and purification of RNA polymerase catalytic complexes
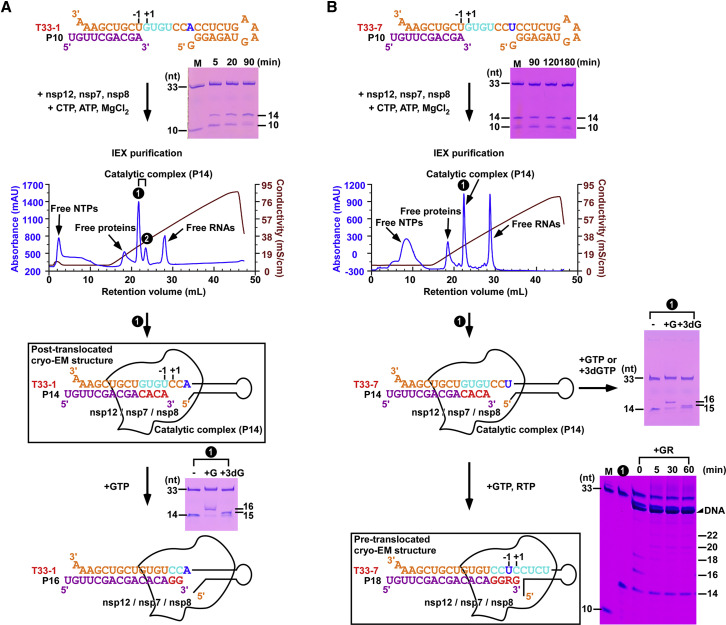
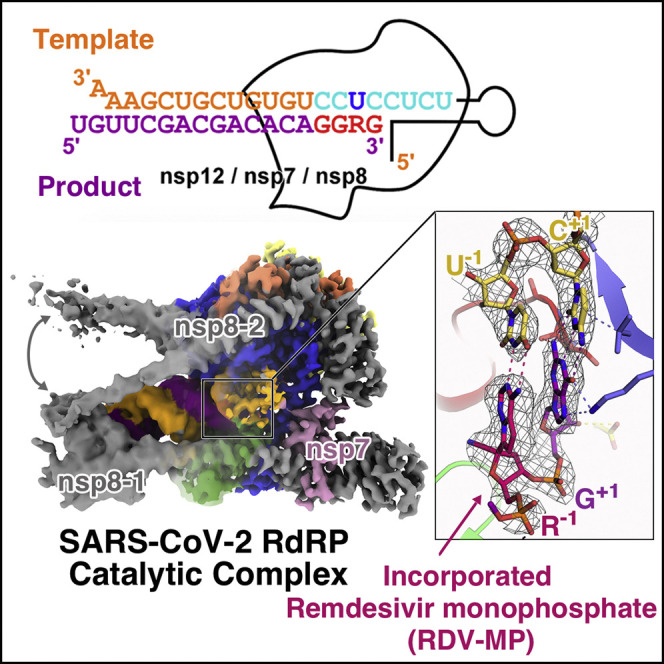
The structure of the RdRP from SARS-CoV-2 bound to non-structural protein 7 (nsp7) and nsp8 co-factors was previously determined in the absence of RNA, providing a view of the overall architecture of the RdRP complex assembly (Gao et al., 2020). However, the mechanism as to how RdRP recognizes and synthesizes RNA remains unsolved. To address this key issue, we have screened more than ten nucleic acid substrates (Table S1) for their polymerase activity in a primer-dependent RdRP assay (Figures 1 and S1 C). On the basis of the assessment of primer utilization efficiency and product homogeneity, we chose an RNA construct with a 33-mer template (T33-1) and a 10-mer primer (P10) for catalytic complex assembly and subsequent structural study. To obtain the catalytic complex, we mixed purified nsp12, nsp7, and nsp8 (Figures S1A and S1B) with the T33-1:P10 construct (Figure 1A). Cytidine triphosphate (CTP) and adenosine triphosphate (ATP) were supplied as the only nucleoside triphosphate (NTP) substrates to allow the incorporation of a C-A-C-A tetranucleotide, resulting in an RdRP catalytic complex containing a 14-mer product (P14) (Figure 1A). This complex represents a post-translocated state after four NMP incorporation events, therefore representing the SARS-CoV-2 RdRP-RNA catalytic complex in action.
Figure 1.
Assembly of SARS-CoV-2 RNA polymerase catalytic complexes
A schematic diagram of the catalytic complex assembly, purification, and reactivity assays of a stalled post-translocated complex (A) and an RDV-MP incorporated pre-translocated complex (B). Abbreviation is as follows: IEX: ion-exchange chromatography.
(A) The post-translocated complex assembly was performed by using a T33-1:P10 construct, the three nsp proteins, and CTP/ATP as the only NTP substrates to allow the synthesis of a 14-mer product (P14). The majority of the P10 was converted to P14 after a 90-min incubation (“M” indicates a marker with a mixture of 33-mer and 10-mer RNAs). The P14-containing catalytic complex was purified by anion exchange chromatography, and the purified complex can further react with GTP (G) or 3¢-deoxy-GTP (3dG) to yield the 16-mer (P16) or 15-mer containing complexes, respectively. The structure of this P14-containing complex was solved in cryo-EM trials at the post-translocated state.
(B) The pre-translocated complex was obtained through a two-step process. In the first step, a T33-7:P10 construct, the three nsp proteins, and CTP/ATP were mixed to allow the synthesis of a 14-mer product (P14). The majority of the P10 are converted to P14 after a 180-min incubation. The P14-containing catalytic complex was purified by anion exchange chromatography, and the purified complex can further react with G or 3dG to yield the 16-mer (P16) or 15-mer containing complexes, respectively. In the second step, the purified P14-containing complex was mixed with GTP and RDV-TP, and a pausing event corresponding to the synthesis of an 18-mer product (P18) was observed. The structure of this P18-containing complex was solved in cryo-EM trials at the pre-translocated state.
See also Figure S1.
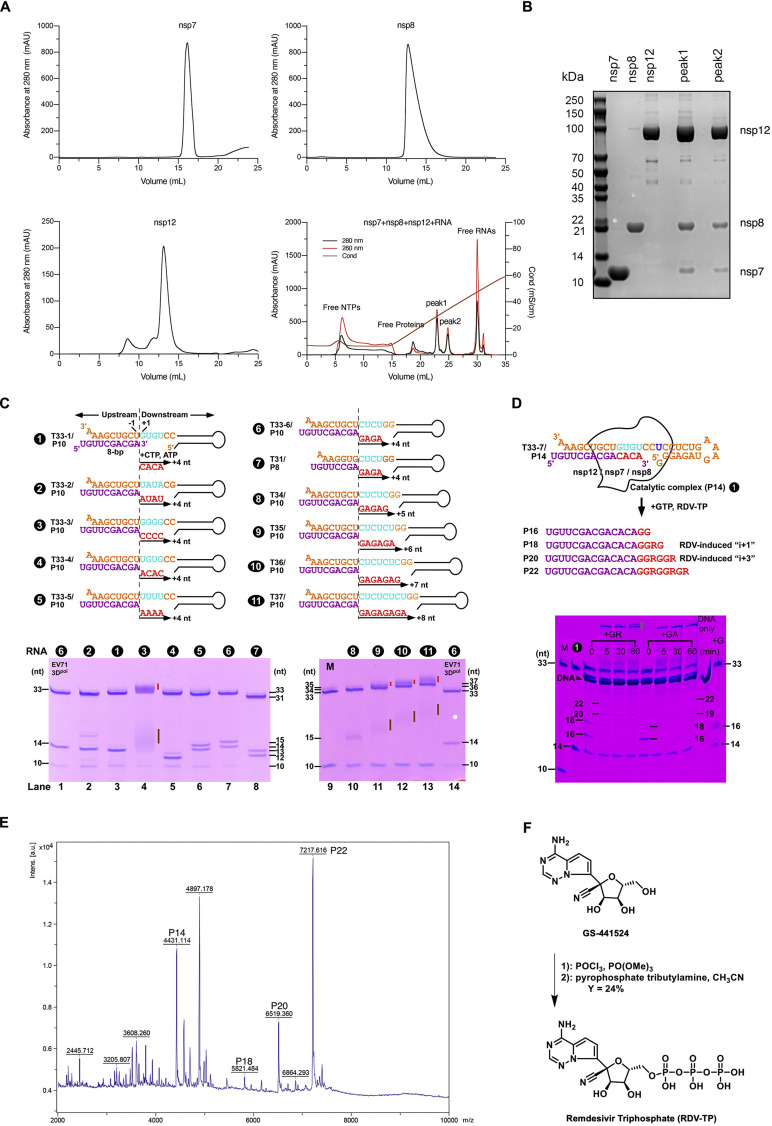
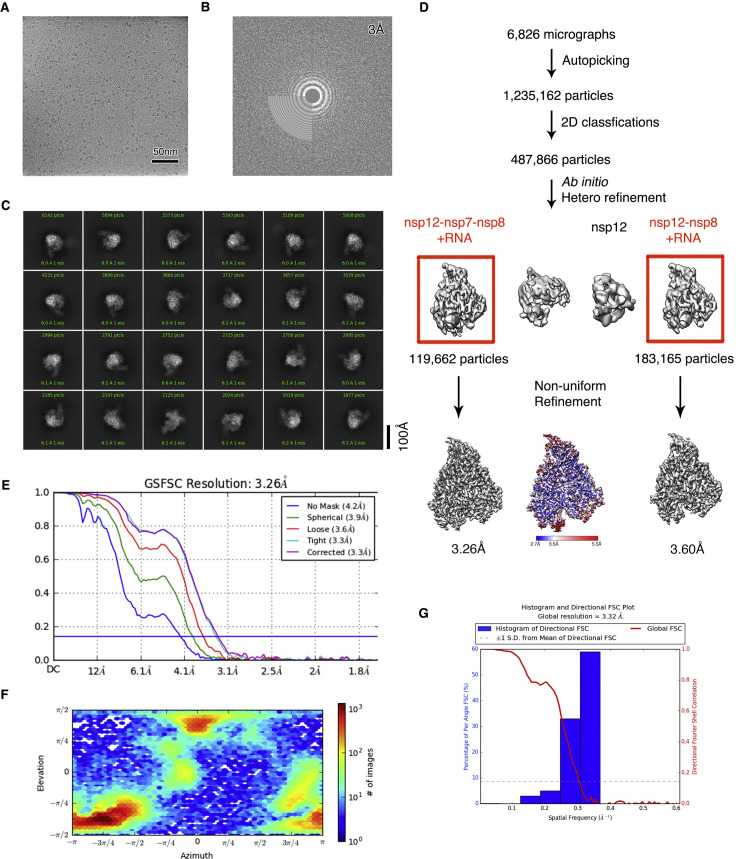
Figure S1.
SARS-CoV-2 polymerase catalytic complex purification and characterization, related to Figures 1, 2, 3, 5, and 6
(A) Gel filtration chromatography analysis of nsp7, nsp8, and nsp12. Ion exchange chromatography analysis of the RdRP-RNA complex (lower right).
(B) SDS-PAGE analysis of the chromatography peaks.
(C) Eleven RNA constructs tested in screening. The coloring scheme is the same as in Figure 1A and the downstream hairpin sequences are identical for all constructs. Besides the construct name (e.g., T33-1:P10), a number is assigned to each construct. RNA profile on the primer-extension reactions is tested on a gel (lower panels). The EV71 RdRP (3Dpol) capable of producing a homogeneous P14 product from the T33-6:P10 construct (number 6) was used as a reference (lanes 1 and 14). Different extent of primer utilization (compare lanes 2-8 and 10-13), misincorporation (lanes 2/6/7), product migration shift (lanes 4/11/12/13, brown vertical bars), and formation of product template-product-containing higher-order RNA structures (lanes 4/11/12/13, red vertical bars) were observed. One construct did not yield the product of the expected length (lane 5). The T33-1:P10 construct (number 1) was chosen for the catalytic complex assembly and structural study.
(D) The accumulation of the T33-7:P10-derived P18 product is related to RDV-MP incorporation. Purified T33-7/P14 complex was incubated with GTP/RDV-TP (+GR) or GTP/ATP (+GA) at 4°C for various periods (“0 min” corresponds to immediate reaction quench after manual mixing to initiate the reaction). For the +GR reaction, the P18 product is most prominent at “0 min” and diminishes over time, suggesting an RDV-induced pausing mechanism. The level of the P20 product is consistent at 5, 30, and 60 min time points, consistent with the RDV-induced “i+3” premature termination mechanism. In the +GA reaction, both the P18 and P20 products are not as prominent. Due to the formation of higher-order product-containing RNA structures (indicated by the green vertical bar), a 33-mer DNA complementary to the RNA template was supplied to help release the RNA products. Note that, in most cases, the release was still incomplete.
(E) Mass spectrometry analysis of the mixture sample for cryo-EM imaging of the pre-translocation complex. Purified T33-7/P14 complex was incubated with GTP/RDV-TP (+GR) at 4°C for 30 min. Proteins in the mixture were removed after a heat denature process.
(F) Synthesis of remdesivir triphosphate (RDV-TP).
To investigate the mechanism of RDV-TP (remdesivir triphosphate) inhibition, we designed another 33-mer template (T33-7) and successfully obtained an RDV-MP-incorporated catalytic complex in the pre-translocated state (Figure 1B). Briefly, the identical C-A-C-A tetranucleotide was incorporated by the RdRP to form the P14-containing complex. After purification by anion exchange chromatography, the P14-containing complex sample was incubated with guanosine-5'-triphosphate (GTP) and RDV-TP at 4°C for 30 min before being frozen for cryogenic electron microscopy (cryo-EM) data collection. The final structure showed a G-G-R-G tetranucleotide was further incorporated into the product RNA chain (corresponding to an 18-mer product or P18) (Figures 1B, S1D, and S1E). The cryo-EM map signal on the 1′-cyano (1′-CN) group of the −1 nucleotide, the guanine base of the +1 nucleotide in the product strand are unambiguous to ensure the assignment of this pre-translocated P18-containing catalytic complex in which the incorporated RDV-MP (R) has been translocated to the −1 position, whereas the 3′-guanosine occupies the +1 position (see below for details).
Structure architecture of the catalytic complex
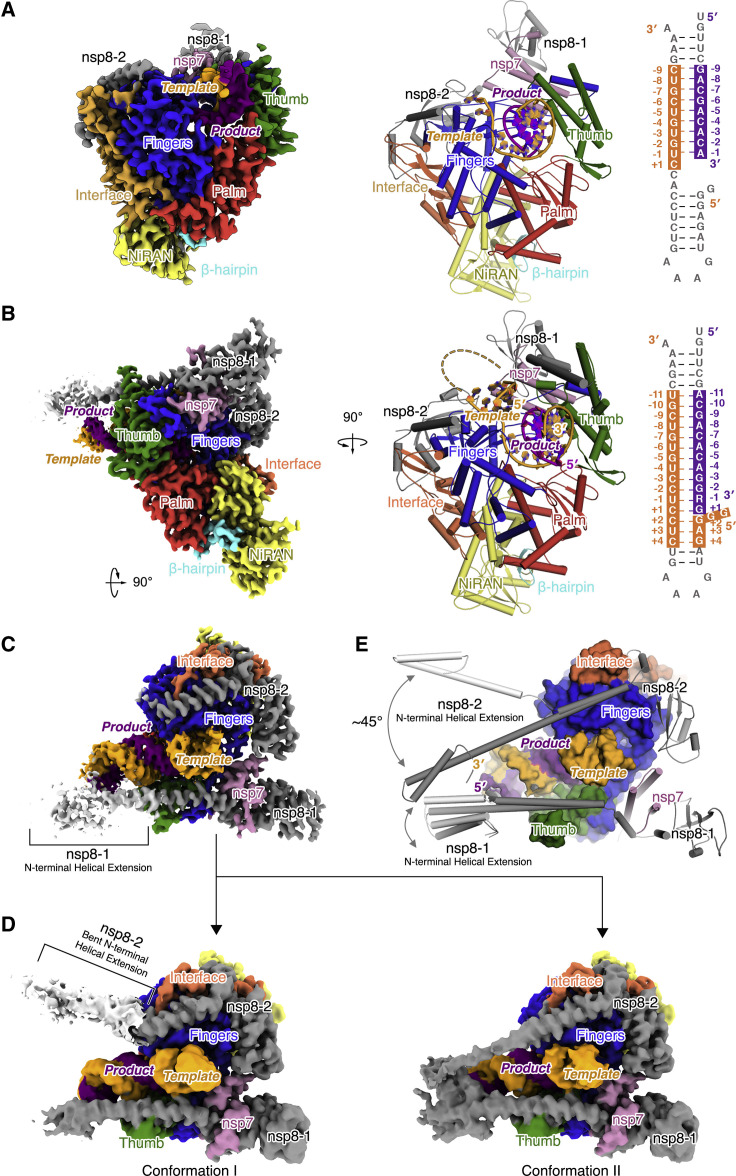
Cryo-EM single-particle analysis results in two near-atomic-resolution structures of the polymerase catalytic complexes, the complex in a pre-translocated state at 2.93 Å (Figure S2 ) and that in a post-translocated state at 3.26 Å (Figure S3 ). The complexes in the two states share similar structure architecture. They both comprise the viral proteins nsp12, nsp8, and nsp7, and RNA template-product duplex that lies in the active site cleft of nsp12 (Figures 2A and 2B). The most notable difference is that, in the pre-translocated complex with a longer P18 product, the existing duplex RNA meets the N-terminal helical extension of nsp8-1 not far from the exit (Figure 2C). Analysis of the two bound nsp8 promoters resolved two distinct conformations in the pre-translocated complex (Figure 2D). The major difference between these two conformations is at the N-terminal helical extension. Unlike the subtle difference at the most N-terminal end of nsp8-1, the N-terminal extension of nsp8-2 shows an orientation change of ∼45 degrees (Figure 2D). In conformation II, which shows more structural rigidity, the two copies of nsp8 bind from two opposite sides of the RNA duplex as it exits (Figure 2D, right). Long helical extensions in nsp8 protrude along with the exiting RNA, forming a stable platform that might enable processive replication of the large coronavirus genome (Figure 2E). Meanwhile, in the pre-translocated complex, the downstream RNA is partially resolved and interacts with the fingers domain and the product strand, whereas the +2 template nucleotide is still base-paired (Figure 3 A). The establishment of these interactions between the downstream RNA and other parts of the complex and the requirement for strand displacement at the immediate downstream might play a role in stabilizing or trapping of this complex, allowing a successful cryo-EM reconstruction.
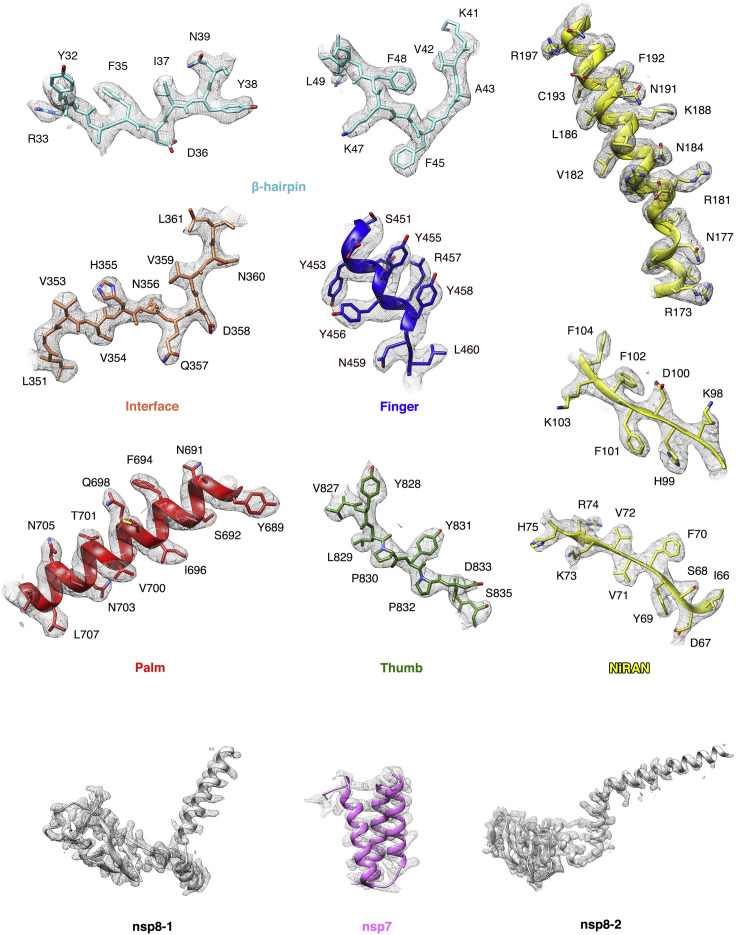
Figure S4.
Representative cryo-EM map, related to Figures 2 and 5
Representative regions of the protein model, colored as indicated, under cryo-EM map.
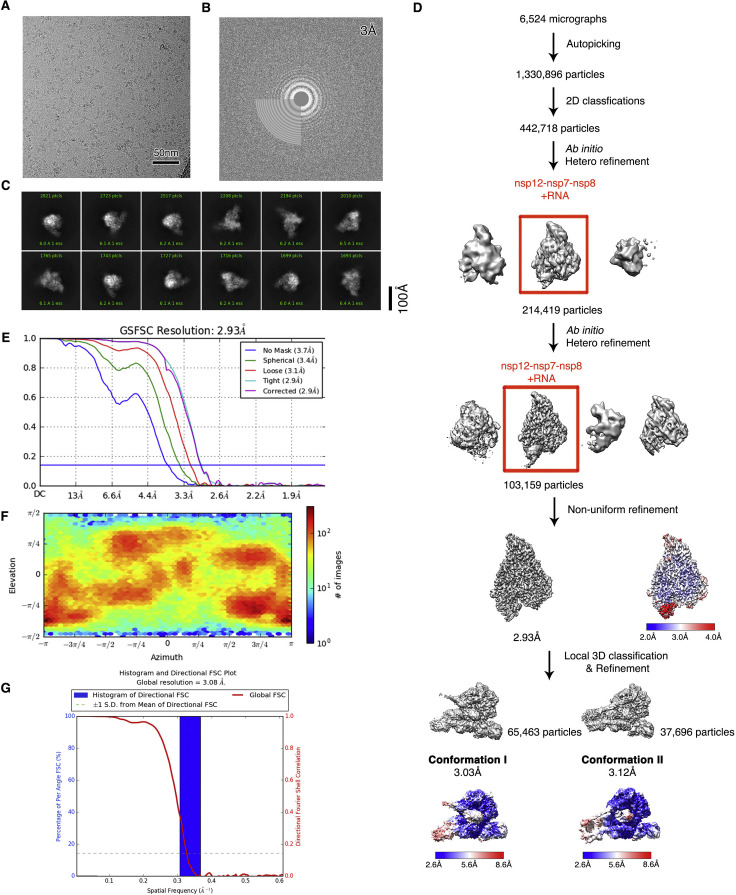
Figure S2.
Cryo-EM reconstruction of RdRP pre-translocated catalytic complex, related to Figures 2, 3, 4, 5, and 7
(A) Raw image of particles in vitreous ice recorded at defocus values of −1.0 to −2.0 μm. Scale bar, 50 nm.
(B) The power spectrum of the image shown in (A), with an indication of the spatial frequency corresponding to 3.0 Å resolution.
(C) Representative class averages. The edge of each square is 246 Å.
(D) The data processing scheme. Overview of nsp12-nsp7-nsp8-RNA pre-translocated catalytic complex reconstruction is shown in the bottom panel with the local resolution map.
(E) Fourier shell correlation (FSC) of the final 3D reconstruction following gold standard refinement. FSC curves are plotted before and after masking.
(F) Angular distribution heatmap of particles used for the refinement.
(G) 3DFSC result of the final map. Global FSC and histogram are shown.
Figure S3.
Cryo-EM reconstruction of RdRP post-translocated catalytic complex, Related to Figures 2, 3, 4, 5, and 7
(A) Raw image of particles in vitreous ice recorded at defocus values of −1.0 to −1.8 μm. Scale bar, 50 nm.
(B) The power spectrum of the image shown in (A), with an indication of the spatial frequency corresponding to 3.0 Å resolution.
(C) Representative class averages. The edge of each square is 246 Å.
(D) The data processing scheme. Overview of nsp12-nsp7-nsp8-RNA post-translocated catalytic complex reconstruction is shown in the bottom panel with the local resolution map.
(E) Fourier shell correlation (FSC) of the final 3D reconstruction following gold standard refinement. FSC curves are plotted before and after masking.
(F) Angular distribution heatmap of particles used for the refinement.
(G) 3DFSC result of the final map. Global FSC and histogram are shown.
Figure 2.
Overall structure of SARS-CoV-2 RNA polymerase catalytic complex
(A and B) The overall structure of the stalled post-translocated (A) and RDV-MP-incorporated pre-translocated (B) RNA polymerase catalytic complexes. Experimental cryo-EM maps are shown in the left graphics. In the right graphic, the protein is represented by ribbons and colored by subdomain (blue, fingers; red, palm; green, thumb; yellow, NiRAN; beta-hairpin, cyan; orange, interface). The RNA is depicted with the template strand in orange and the product strand in purple. Nucleotides resolved in the structures are indicated by color shading.
(C) Close-up view of the RNA in RDV-MP incorporated pre-translocated complex. Cryo-EM map is colored according to the same scheme in (B). The map threshold of the protein is 0.6, whereas that of the nucleic acid and nsp8 N-terminal helical extension (in white) is 0.3.
(D) Focus classification of the reconstruction in (C) results in two conformations of the complex with structural variations on the two nsp8 N-terminal helical extensions. The unsharpened maps (threshold 0.2) are colored according to the same scheme in (B). The threshold for the bent N-terminal helical extension of nsp8-2 (in white) in conformation I is 0.1.
(E) Structural variations between the two conformations in (D) are demonstrated in aligned complex structures depicted in the cartoon. N-terminal helical extensions of the nsp8 are docked with the corresponding portion in the SARS-CoV-1 nsp7-nsp8 hexadecameric primase complex (PDB: 2AHM). The gray two-way arrow indicates structural variations.
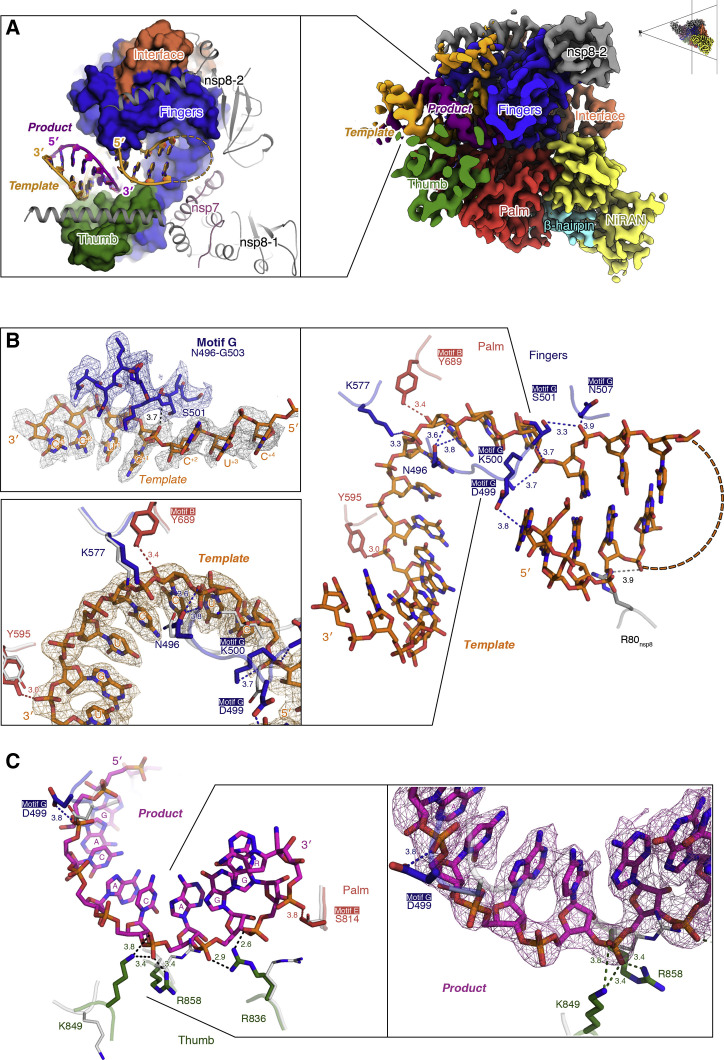
Figure 3.
Polymerase-nucleic acid interactions
(A) The interface between RNA template and product and the subdomains that coordinate them (thumb, palm, and fingers) in the pre-translocated complex. A map cross-section showing the nucleic acid signal near the center of the reaction is placed on the right graphic.
(B and C) The representative conserved amino acid residues and interactions that coordinate RNA. RNA bases are labeled according to standard polymerase numbering conventions. Protein-ligand hydrogen bonds are shown as dashed lines. The experimental cryo-EM map is superimposed on the RNA template (B) and product (C).
Polymerase-nucleic acid interactions
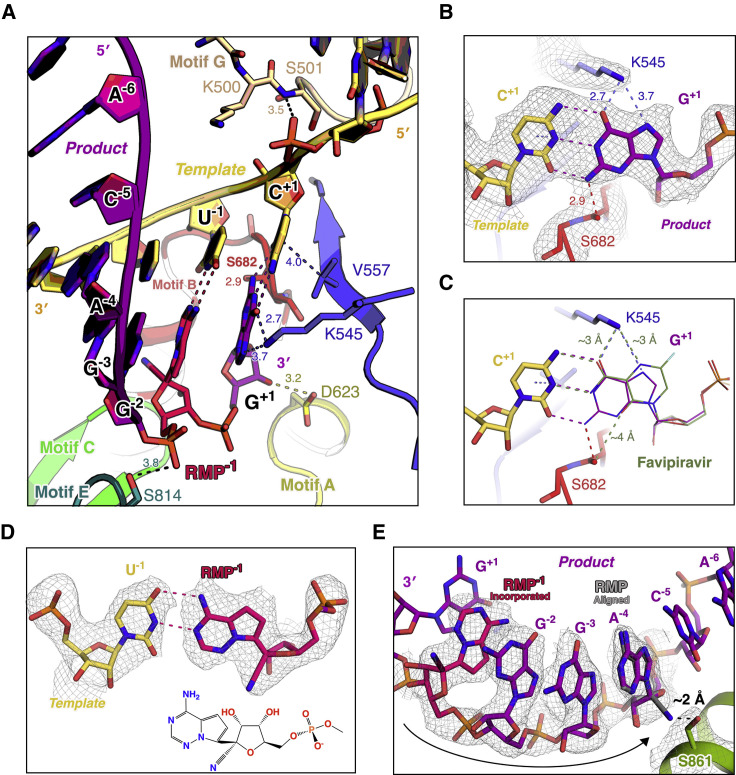
Fourteen base pairs in the pre-translocated complex and nine base pairs in the post-translocated complex double-stranded template-product RNA helix gripped by the fingers-palm-thumb domains, and three base pairs of the downstream template (corresponding to positions +2 to +4) in the pre-translocated complex, are resolved in the EM structure (Figure 3A). Non-sequence-specific protein-RNA interactions are observed between the template-product RNA and nsp12. In particular, in the pre-translocated complex, N496, K500, S501, N507, and K577 from the fingers domain and Y595 from the palm domain directly participate in stabilizing the phosphate backbone of the template (Figure 3B), whereas R836, K849, and R858, experience a significant rotation of their side chains to coordinate the phosphate backbone of the product (Figure 3C). Interestingly, D499 in motif G from the fingers domain interacts with both the 5′-terminal base of the template and the phosphate backbone of the product, whereas R80 from nsp8 interacts with the downstream base-paired template (Figures 3B and 3C). Meanwhile, Y689 of the fingers domain interacts with the ribose 2′-OH groups of nucleotides at the −2 position from the template strand (Figure 3B).
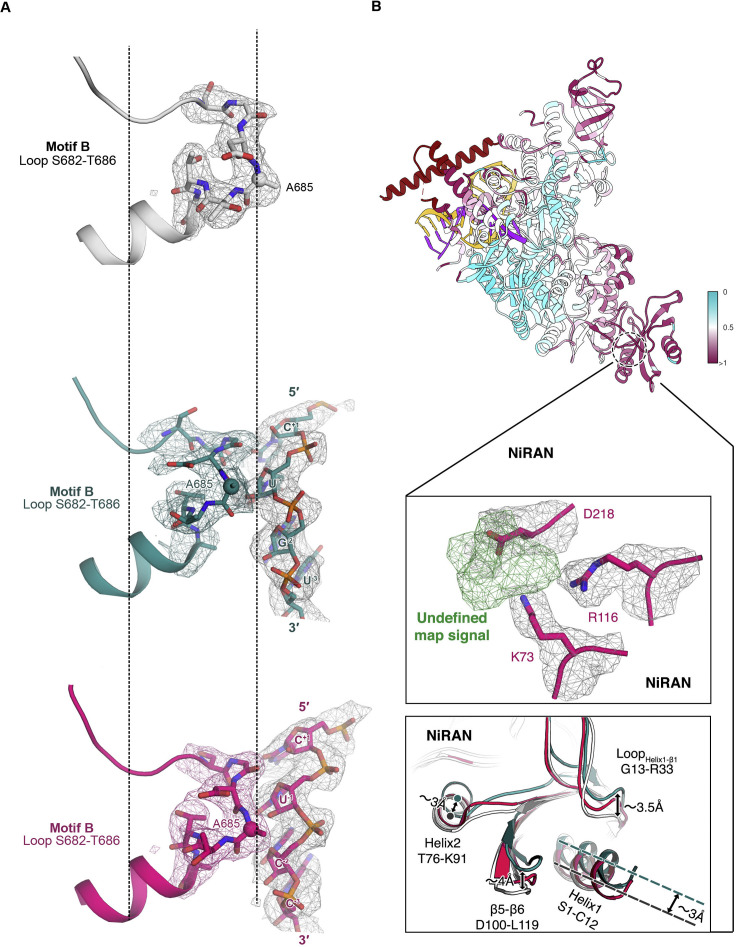
Besides the establishment of the non-sequence-specific RNA interactions, two motifs of the RdRP undergo conformational changes in response to accommodate the template-product RNA (Figure 4 A). A loop region (residues 682–686) within the RdRP catalytic motif B of the palm shows a dynamic interaction with RNA in both pre- and post-translocation states. It is “pushed” backward by the +1 to −2 template nucleotides with a maximum backbone shift of about 2.6 Å (residue A685) (Figures 4B and S5 A). This loop contains a glycine (G683) that is conserved in RNA-dependent polymerases (Gong and Peersen, 2010) and exhibits dynamic features in both positive- and negative-strand RNA virus RdRPs (Kouba et al., 2019, Sholders and Peersen, 2014). It participates in NTP ribose recognition and RdRP active site closure (Shu and Gong, 2016) and has been proposed to mediate RdRP translocation (Sholders and Peersen, 2014). At the same time, on the opposite side of the template strand, motif G (D499–L514) on the fingers also exhibits interaction with the RNA backbone (Figure 4C). Although the map signal for this region is relatively weak and undefined in the post-translocation structure, as mentioned earlier in the structure of the pre-translocation state, the conformation of motif G and its interaction with the +1 template nucleotide via S501 backbone nitrogen are well resolved (Figure 3B). In a very recent study characterizing the role of motif G in EV71 RdRP translocation, the motif G residues equivalent to K500 and S501 were shown to play a critical role in restricting the +1-template nucleotide (Wang et al., 2020). This restriction, on one hand, ensures precise positioning of the templating base for efficient NTP binding and catalysis, and on the other hand to control translocation by setting up a rate-limiting step (Wang et al., 2020).
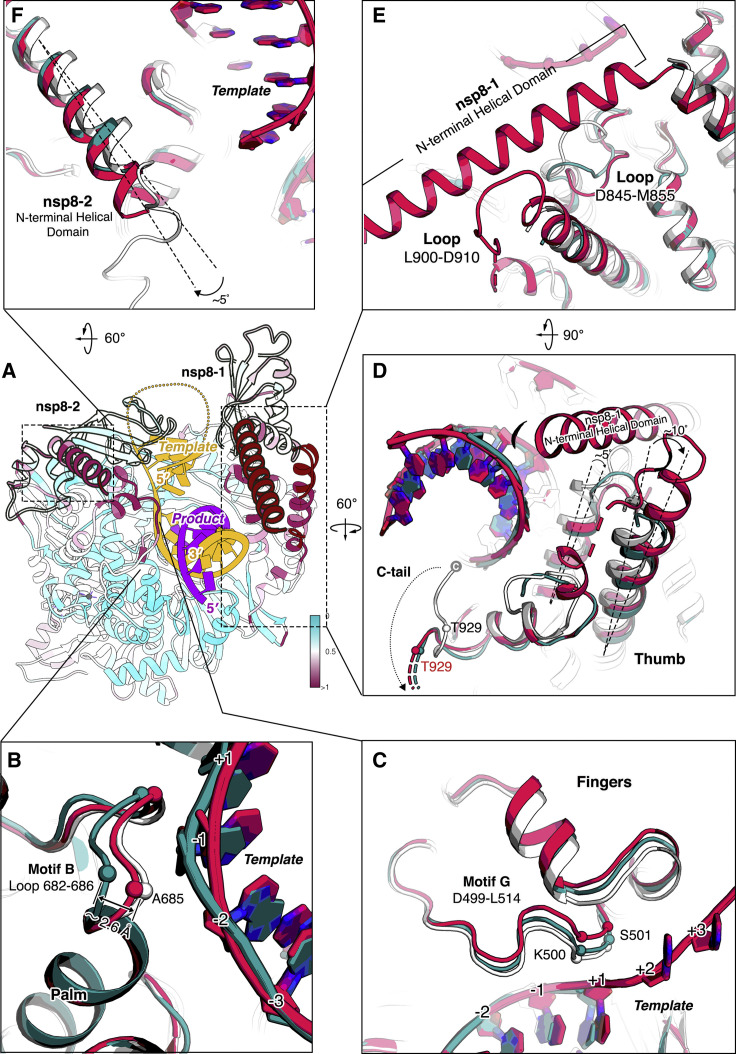
Figure 4.
Structural variations among apo and catalytic complexes
(A) The pre-translocated complex is colored according to the root mean square deviation (RMSD), which measures the average distance in Å between the C-alpha atoms of the three aligned complexes (pre-translocated complex in red; post-translocated complex in dark green; apo complex in white).
(B–F) Structural rearrangements of motif B on palm (B), motif G on fingers (C) and thumb (D), and two copies of nsp8 (E and F).
Figure S5.
Structural Variations and corresponding cryo-EM map in NiRAN and motif B among Apo and Catalytic Complexes, related to Figure 4
Pre-translocated complex in red; post-translocated complex in dark green; apo complex in white.
(A) The experimental cryo-EM map is superimposed by structural rearrangements of motif B on Palm. Three states are aligned by helix after loop S682 to T686 and residue A685 (shown as black dash lines).
(B) The pre-translocated complex is colored according to the root mean square deviation (RMSD) which measures the average distance in Å between the C-alpha atoms of the three aligned complexes(top). An extra undefined map signal (in green) in NiRAN is indicated under an unambiguous experimental cryo-EM map signal. Surrounding residues are shown in sticks and cartoons with the same threshold level (middle). Structural rearrangements of NiRAN are indicated (bottom).
Structural rearrangements of the co-factor nsp8
The interactions with RNA are manifested in broader conformational changes of the palm, fingers, and thumb domain of nsp12, which seem to be further propagated to the two copies of nsp8. Specifically, besides the C-terminal tail (residues 926–932) (C-tail) next to the RNA exit channel that points toward the active site in the apo state but flips out in the catalytic complexes, two α helices of the thumb domain are deflected 5–10 degrees to the outward of the complex in relation to the RNA (Figure 4D). What’s interesting is that, in the pre-translocation structure, the N-terminal helical domain of nsp8-1 lies on a notch formed at the top of these two helices and makes a conformational change in the 845–855 loop and stabilizes the 900–910 loop, which is missing in the apo and post-translocation structure (Figures 4D and 4E). Furthermore, the nsp8-2 N-terminal helix that lays on the fingers deflects by about 5 degrees (Figure 4F). In the pre-translocation structure, which has a longer RNA product, the most distal section of this helix appears to be able to switch between two particular conformations and interact with exiting RNA (Figures 2D and 2E).
Catalytic center, incorporation, and implications for inhibition by nucleotide analog inhibitors
The catalytic center of SARS-CoV-2 nsp12 is constructed by seven conserved motifs from A to G (Gao et al., 2020) (Figure 5 A). In the pre-translocation structure, the base pair of cytosine and guanosine at the +1 position have an unequivocal cryo-EM map signal (Figure 5B), which is tightly coordinated by the interactions from several conserved residues. Specifically, the backbone nitrogen atom of S501 and the sidechain of V557, respectively, position the phosphate and base moieties of cytosine on the template (Figure 5A). The backbone carbonyl oxygen atom of S682 and the nitrogen atom of the K545 side chain jointly stabilize the base group of guanosine from both sides, whereas D623 stabilizes its ribose group via an interaction with the 3′ hydroxyl group (Figure 5B). Given that favipiravir is a guanine analog, we aligned favipiravir monophosphate onto the guanosine to pair with the cytosine at +1 position of template, on the basis of a superposition on their phosphate and ribose groups. Interestingly, the distance between the side-chain nitrogen atom of K545 and the carboxamide oxygen atom and exocyclic amide of favipiravir are both about 3 Å (Figure 5B).
Figure 5.
Catalytic center, incorporation and implications on inhibition of nucleotide analog inhibitors
(A) Close view of the catalytic center in the RDV-MP incorporated pre-translocated complex. Interactions are shown as dashed lines and measured in Å.
(B) The base pair of cytosine and guanosine at +1 position and the coordinating residues under unambiguous experimental cryo-EM map signal.
(C) Favipiravir-DP is shown as the stick in olive, aligned on the guanosine at +1 position.
(D) The base pair of uracil and incorporated RDV-MP at −1 position under experimental cryo-EM map signal. The 2D chemical structure of the incorporated RDV-MP is provided.
(E) Structural demonstration of the delayed chain termination hypothesis of RDV. RDV (in gray) is aligned onto the product strand (in purple) to demonstrate their incorporated state at −4 position.
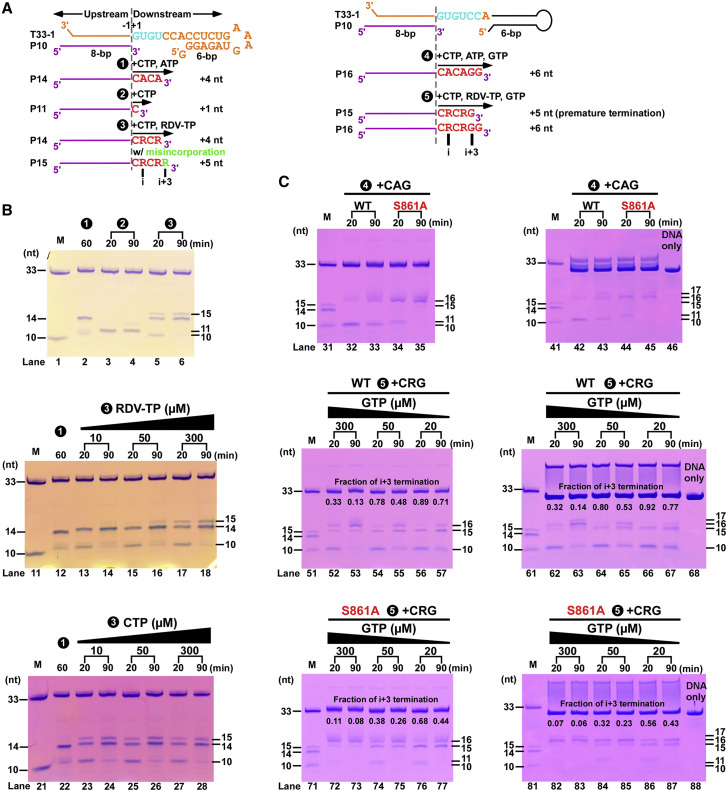
RDV is a phosphoramidate prodrug of a 1′-cyano-substituted adenosine nucleotide analog that shows broad-spectrum antiviral activity against several RNA viruses (Siegel et al., 2017). Its triphosphate form (RDV-TP) can be used as a substrate of several viral RdRP enzymes or complexes (Agostini et al., 2018, Tchesnokov et al., 2019). In contrast to classic chain terminators, inhibition of chain elongation is not observed immediately after the incorporated RDV monophosphate (RDV-MP) (Jordan et al., 2018, Tchesnokov et al., 2019). Its 3′-OH group allows the nucleophilic in-line attack on the incoming NTP. Indeed, in the pre-translocation complex, the 2.9 Å well-resolved map signal on the 1′-cyano group of the −1 product nucleotide ensures the assignment of RDV-MP, which had been incorporated and translocated to the −1 position, whereas the 3′-guanosine occupied the +1 position (Figures 5A and 5D). In a previous enzyme kinetics analysis (Gordon et al., 2020), favorable reactivity for the RDV-TP over ATP and delayed chain termination at position i+3 (where i corresponds to the position of the first incorporated RDV-MP) are observed to be the key features of the mechanism of inhibition with SARS-CoV-2 RdRP complexes. When RdRP synthesis proceeds to the i+3 position, the first incorporated RDV-MP will be at position −3 or −4 for the pre- or post-translocated state, respectively. According to the previous molecular modeling analysis, the 1′-CN substituent of incorporated RDV-MP could encounter a steric clash from the sidechain of S861 and probably lead to a distortion of the positioning of the RNA, hampering translocation to the −4 position (Gordon et al., 2020). On the basis of the current 2.9 Å catalytic pre-translocated complex structure, if one aligns the incorporated RDV-MP onto the adenosine at the −4 position, the serine hydroxyl oxygen would be as close as 2 Å from the 1′-cyano nitrogen (Figure 5E).
To further dissect the RDV-TP inhibition mechanism and to test this steric clash hypothesis, we compared the behaviors of RDV-TP with ATP in our T33-1:P10-based assays and the WT RdRP, and we also compared an S861A mutant regarding the i+3-related product profiles (Figure 6 ). We first compared the primer extension profiles between the CTP/ATP and CTP/RDV-TP combinations. It turned out that the latter combination led to both the regular 14-mer product (corresponding to the incorporation of a C-R-C-R tetranucleotide) and a misincorporation-derived 15-mer product. This misincorporation event likely arose from a C:R misincorporation given that this activity is greatly reduced when the RDV-TP concentration is lowered (Figure 6B, compare lanes 17–18 and 13–16) but not much affected by the variation of the CTP concentration (Figure 6B, compare lanes 27–28 and 23–26). We then compared the extension profiles between the CTP/ATP/GTP and CTP/RDV-TP/GTP combinations. For both the WT RdRP and the S861A mutant, the CTP/ATP/GTP combination led to the synthesis of the expected 16-mer product and a small amount of 17-mer product because of misincorporation, whereas the S861A mutant appeared to be more reactive (Figure 6C). As expected, the CTP/RDV-TP/GTP combination resulted in the production of a 15-mer product corresponding to the synthesis at the i+3 position, and the estimated fraction of termination (i.e., the fraction of 15-mer to 15-mer and longer products based on band intensities) values were 0.71–0.77 and 0.13–0.14 for low (20 μM) and high (300 μM) GTP concentrations at the 90-min reaction time point, respectively (Figure 6C, lane 57 versus 67 and 53 versus 63). This observation is consistent with the previous study showing the incoming NTP reduced the i+3 termination (Gordon et al., 2020). Compared with the WT RdRP, the S861A mutant had a smaller fraction of i+3 termination at all tested GTP concentrations, providing valid support of the steric clash hypothesis (Figure 6C, compare lanes 52–57 to 72–77 and 62–67 to 82–87).
Figure 6.
Behaviors of RDV-TP in primer extension assays
(A) The reaction flow charts for five different NTP combinations.
(B) The extension profiles are compared for three NTP combinations: (1) CTP+ATP; (2) CTP; and (3) CTP+RDV-TP. The R denotes incorporated RDV-MP. Shown on top, the CTP/RDV-TP reaction yields both 14-mer and misincorporation-related 15-mer products (lanes 5–6). Shown at the middle and bottom, this misincorporation is reduced when RDV-TP but not CTP concentration is lowered.
(C) The extension profiles are compared for two NTP combinations: (4) CTP+ATP+GTP (CAG) and (5) CTP+RDV-TP+GTP (CRG) in the absence (left) and presence (right) of a 33-mer DNA complementary to T33-1. Shown on top, both WT RdRP and the S861A mutant can synthesize the expected 16-mer product with the combination of C, A, and GTP. Shown in the middle, other than the 16-mer product, an RDV-related 15-mer product was made (corresponding to an RNA three nucleotides longer than that has the first incorporated RDV-MP or the “i+3” product) by the WT RdRP, indicating a premature termination event. Shown at the bottom, the S861A mutant also produced the 16-mer and the “i+3” products, albeit with a relatively lower amount of “i+3” termination. The addition of the complementary DNA helped visualize the 16- and 17-mer bands by trapping the RNA template. The same marker (M) was used as in Figure 1.
Discussion
We have obtained two cryo-EM structures of SARS-CoV-2 RdRP-RNA catalytic complex in both RDV-MP incorporated pre-translocation and stalled post-translocation states and provide a comprehensive view of the active site. This result is of significance for the design optimization of potential drug candidates, such as remdesivir and favipiravir. The N-terminalnidovirus RdRP-associated nucleotidyltransferase (NiRAN) domain of RdRP in apo and the two catalytic states shows dramatic conformational changes, and a strong undefined signal appears in the vicinity of K73, R116, and D218 in the pre-translocation complex (Figure S5B). Clarifying these features might require further investigation.
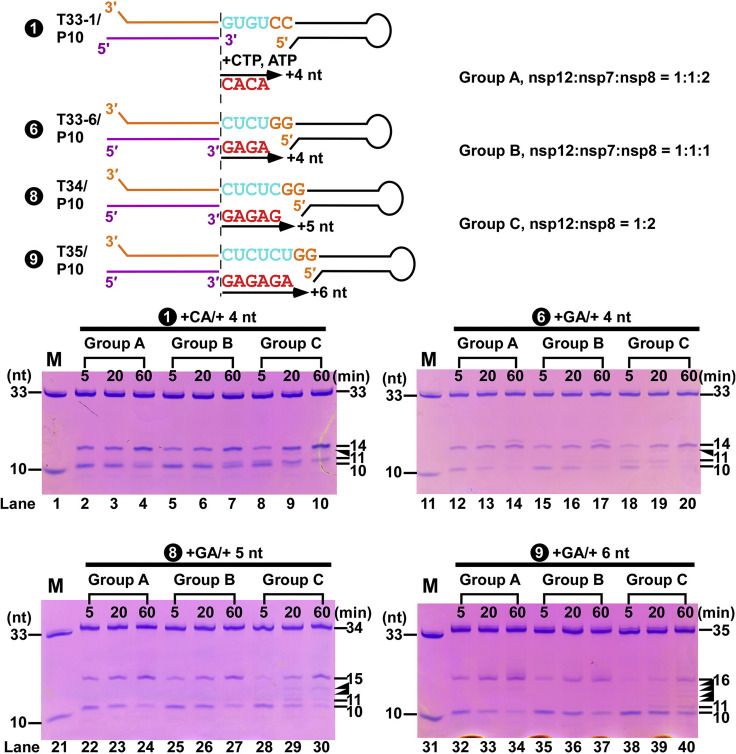
The coronavirus polymerase complex is composed of RdRP nsp12 and its co-factors nsp7 and nsp8. However, the existing structures of the complexes to date (Gao et al., 2020, Yin et al., 2020) have not revealed the complete nsp8. The functional role of the two co-factors has remained an enigma. Increased RNA synthesis efficiency associated with the simultaneous presence of nsp7, nsp8, and nsp12 has been characterized on SARS-CoV-1 (Subissi et al., 2014). A similar phenomenon on SARS-CoV-2 was also observed under our primer extension assay setup (Figure S6 ). In our work, we have revealed the overall structure of the two bound nsp8 molecules in different conformational states. The potential mechanism for promoting polymerase activity by stabilizing existing RNA through the interactions on their N-terminal helical extension is described. While this paper was under review, a study was posted as a pre-print reporting a cryo-EM structure of the SARS-CoV-2 RdRP complex with a 24-base-pair (bp) template-primer RNA (Hillen et al., 2020), which is formed by three repetitive RNA pairs connected through palindromic GUAC sticky ends. Although that structure likely represents a pre-catalytic or pre-initiation state that has not gone through catalysis, it seems to represent a similar global architecture to what is observed in the conformation II of our pre-translocation complex. The observation of such a conformation as the dominant form in the presence of their longer RNA duplex is informative. This observation led us to consider that, with respect to nsp8-upstream RNA interactions, conformation II might be the dominant state during RNA processive elongation, rather than conformation I. Conformation I in our structure is therefore likely to be a transition state before the RNA products getting long enough to establish the elongation-mode interactions with nsp8.
Figure S7.
Sequence alignment of RdRPs encoded by the virus mentioned, related to Figures 2, 3, and 5
Catalytic residues (in forest), product binding residues (in purple), template binding residues (in orange) and possible favipiravir binding residues (in sandy) are indicated by triangle. Motif A-G are indicated by rectangular.
Figure S6.
The role of nsp7 and nsp8-1, related to Figure 7
A comparison of primer-extension activities for three different nsp combinations on four RNA constructs. Top: the four RNA constructs differ in template sequences that direct the initial synthesis and the three combination modes (Groups A-C) of nsp12, nsp7, and nsp8 proteins. Bottom: primer-extension profiles. Solid triangles indicate the intermediate products that are more prominent in Group C combination (lanes 8-10, 18-20, 28-30, and 38-40). M: a marker with a mixture of 33-mer and 10-mer RNAs.
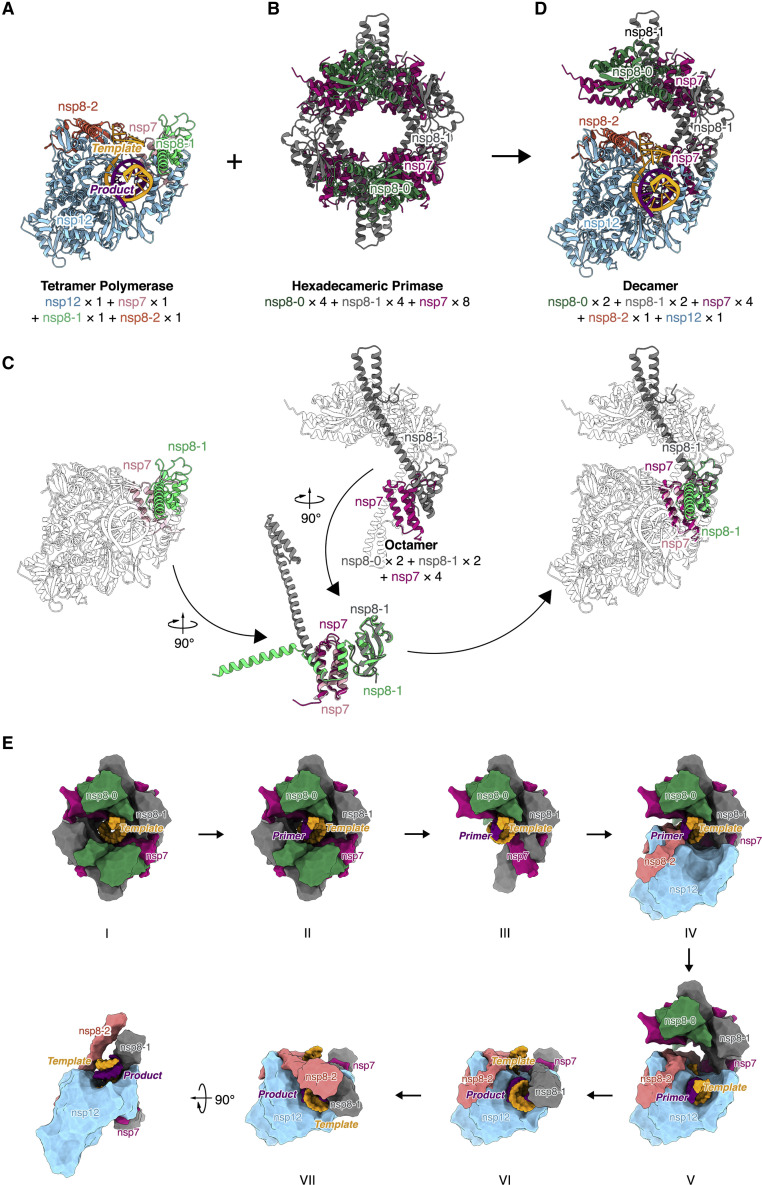
nsp7 and nsp8 can form a “non-canonical primase” to generate RNA primers (Imbert et al., 2006, te Velthuis et al., 2012, Xiao et al., 2012), although nsp8-associated primase or RdRP activities were not observed in some studies (Subissi et al., 2014, Tvarogová et al., 2019). After reviewing our previous work on the nsp7-nsp8 hexadecameric primase complex structure of SARS-CoV-1 (Zhai et al., 2005), we found that this pair of nsp7 and nsp8-1 have a similar relative position and conformation with one of the pairs in their hexadecameric complex (Figures 7 A to 7C). The ring-shaped hexadecameric complex is believed to dissociate into two octamers “half-rings” after primer synthesis (Li et al., 2010). If we superimpose the “half-ring” on to the nsp7-nsp8-nsp12 complex and assume that the nucleic acid passes through the central pore of the primase enzyme during primer synthesis (Zhai et al., 2005), we find that the substrate paths of the two are in a surprisingly close and parallel position (Figure 7D). Thus, there is the possibility of a fairly direct substrate channeling or “hand-over” between these two enzyme complexes. On the basis of this analysis, we propose a model for understanding the transition steps from the primase complex to the catalytic complex (Figure 7E).
Figure 7.
Transition model from the primase complex to the polymerase catalytic complex
(A and B) The SARS-CoV-2 nsp12-nsp7-nsp8 polymerase pre-translocation complex (A) and the SARS-CoV-1 nsp7-nsp8 hexadecameric primase complex (B) (PDB: 2AHM).
(C) Structure superimposition of the nsp12-nsp7-nsp8 polymerase catalytic complex and the nsp7-nsp8 octamer half-ring primase complex, on the basis of a common pair of nsp7 (SARS-CoV-1 in rose-red; SARS-CoV-2 in pink) and nsp8-1 (SARS-CoV-1 in dark gray; SARS-CoV-2 in light green). The rest of the subunits remain in a transparent outline.
(D) Proposed decamer assembly of nsp12-nsp7-nsp8 polymerase complex and half-ring primase complex.
(E) Proposed transition model from the primase complex to the polymerase complex. In state I, the RNA template (in orange) binds to the hexadecameric primase complex (nsp7 in rose-red; first nsp8 isoform, nsp8-0 in dark green; second nsp8 isoform, nsp8-1 in dark gray). In state II, the primase complex de novo synthesizes primer (in purple). In state III, half of the primase complex dissociates. In state IV, the remaining half of the primase complex recruits nsp12 (in blue) and the third isoform of nsp8 (nsp8-2 in pink) bound to it. In state V, the template and primer translocate into the active site of nsp12, the polymerase. RNA synthesis in nsp12-nsp7-nsp8 polymerase complex and the nsp8 N-terminal extension platform is forming (state VI, pre-translocation catalytic complex in conformation I; State VII, pre-translocation catalytic complex in conformation II).
Coronavirus replication and transcription are mediated by a replication-transcription complex (RTC) of which virus-encoded, nsps are the primary constituents. A broader spatiotemporal view of this complex viral replication-transcription mechanisms will help to develop antiviral drugs that interfere with this kernel process of viral replication. In addition to optimizing the inhibitor’s ability to target nsp12 for efficient incorporation into the RNA genome, one can also consider blocking the integrity of the polymerase complex, such as the interaction between nsp8 and nsp12, or the ability to evade or overcome the proofreading activity of the viral exoribonuclease (ExoN), such as β-d-N4-Hydroxycytidine (NHC) (Agostini et al., 2019).
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and Virus Strains | ||
| E. coli BL21(DE3) | ATCC | Cat#ATCCPTA-5073 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| ATP | Sigma | Cat#A2383 |
| GTP | Sigma | Cat#G8877 |
| CTP | Sigma | Cat#C1506 |
| UTP | Sigma | Cat#U6750 |
| DDM | Anatrace | Cat#D310 |
| Spermidine trihydrochloride | Sigma | Cat#S-2501 |
| D-Gluocoseamine-6-phosphate | Sigma | Cat#G-5509 |
| Acrylamide | Sangon Biotech | Cat#79-06-1 |
| DEPC-treated Water | ThermoFisher Scientific | Cat#4387937 |
| Boric acid | Sinopharm Chemical Reagent | Cat#10004818 |
| GS-441524 (CAS 1191237-69-0) | Topscience | Cat#T7222 |
| Deposited Data | ||
| Nsp12-nsp7-nsp8-RNA pre-translocated catalytic complex EM map | This paper | EMD-30275 |
| Nsp12-nsp7-nsp8-RNA pre-translocated catalytic complex EM map conformation I | This paper | EMD-30283 |
| Nsp12-nsp7-nsp8-RNA pre-translocated catalytic complex EM map conformation II | This paper | EMD-30284 |
| Nsp12-nsp7-nsp8-RNA post-translocated catalytic complex EM map | This paper | EMD-30252 |
| Nsp12-nsp7-nsp8-RNA pre-translocated catalytic complex structure | This paper | PDB: 7C2K |
| Nsp12-nsp7-nsp8-RNA post-translocated catalytic complex structure | This paper | PDB: 7BZF |
| Oligonucleotides | ||
| The oligonucleotides used in this study were listed in Table S1 | This paper | N/A |
| Recombinant DNA | ||
| pET22b-SARS-CoV-2-nsp12 | This paper | N/A |
| pET28a-SARS-CoV-2-nsp7 | This paper | N/A |
| pET28a-SARS-CoV-2-nsp8 | This paper | N/A |
| pRAV23 | Jeffrey Kieft laboratory, Colorado Denver School of Medicine. (Batey and Kieft, 2007) | N/A |
| Software and Algorithms | ||
| SerialEM | Mastronarde, 2003 | http://bio3d.colorado.edu/SerialEM/ |
| MotionCor2 | Zheng et al., 2017 | https://emcore.ucsf.edu/ucsf-motioncor2 |
| Gautomatch | Jack (Kai) Zhang | https://www.mrc-lmb.cam.ac.uk/kzhang/ |
| ImageJ | Schneider et al., 2012 | https://imagej.net/ |
| RELION 3.03 | Zivanov et al., 2018 | https://www2.mrc-lmb.cam.ac.uk/relion |
| cryoSPARC | Punjani et al., 2017 | https://cryosparc.com/ |
| UCSF Chimera | Pettersen et al., 2004 | https://www.cgl.ucsf.edu/chimera/ |
| COOT | Emsley et al., 2010 | https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/ |
| PHENIX | Afonine et al., 2012 | https://www.phenix-online.org/ |
| PyMOL | Schrodinger | https://pymol.org/2/ |
| UCSF ChimeraX | Goddard et al., 2018 | https://www.cgl.ucsf.edu/chimerax/ |
| Other | ||
| Quantifoil R0.6/1.0 200 mesh Cu holey carbon grids | Electron Microscopy Sciences | Cat#Q250CR-06 |
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Zihe Rao (raozh@tsinghua.edu.cn).
Materials Availability
All unique/stable reagents generated in this study are available from the Lead Contact without restriction.
Data and Code Availability
The cryo-EM maps and atomic models have been deposited at the Electron Microscopy Data Bank (EMD-30252, EMD-30275, EMD-30283 and EMD-30284) and the Protein Data Bank (PDB: 7BZF and PDB: 7C2K). A list of software used in this study can be found in the Key Resources Table.
Experimental Model and Subject Details
Proteins were obtained through recombinant expression in E. coli BL21(DE3).
Method Details
SARS-CoV-2 Polymerase Catalytic Complex Purification and Characterization
The SARS-CoV-2 nsp12 (GenBank: MN_908947) gene was inserted into a pET22b vector possessing a C-terminal 10 × His tag. Full-length SARS-CoV-2 nsp7 and nsp8 genes were respectively inserted into a pET28a vector with an N-terminal 6 × His tag. The plasmids were transformed into E. coli BL21(DE3). When bacterial cultures were grown in LB medium to an OD600 of 0.6-0.8 at 37°C, growth temperature was reduced to 16°C followed by the addition of a final concentration of 0.5 mM IPTG to induce the expression. After overnight induction, the harvested cell pellets were resuspended in lysis buffer (20 mM Tris-HCl, pH 8.0, 150 mM NaCl, 4 mM MgCl2, 10% glycerol) with benzonase (Yeasen) and lysed by passage through a high-pressure homogenizer at 4°C. The lysate was centrifugated at 18000 rpm for 30 min to obtain supernatant containing recombinant protein, which was purified by Ni-NTA affinity chromatography. The protein bound to the Ni-NTA column was eluted with lysis buffer supplemented with 300 mM imidazole and concentrated to load onto a Superdex 200 10/300 Increase column (GE Healthcare, USA) in gel filtration buffer (50 mM HEPES, pH 7.0, 100 mM NaCl, 4 mM MgCl2 and 4mM DTT). The collected protein fractions were concentrated to 5 mg/mL and stored at 4°C for further use.
Eleven template-primer RNA constructs (Table S1) were initially screened to identify sequences that could generate stalled nsp12/nsp7/nsp8-RNA catalytic complexes stable enough to survive an anion exchange chromatography purification. A construct comprising a 33-mer template (T33-1) and a 10-mer primer (P10) was chosen in complex assembly to generate a 14-mer product (P14)-containing catalytic complex for structural studies. The RNA templates were prepared by in vitro T7 RNA polymerase-glmS ribozyme-based approaches described previously (Batey and Kieft, 2007, Gong and Peersen, 2010) (see below for more detail), and the P10 and an 8-mer primer (P8) was chemically synthesized (Integrated DNA Technologies). The T33-1:P10 construct was made with a 10% molar excess of P10 annealed to T33-1 via a 3-min incubation at 45°C followed slow cooling to room temperature (r.t.). For the P14-containing stalled catalytic complex assembly (post-translocation complex), a typical 1.5-mL reaction mixture containing 12 μM nsp12, 12 μM nsp7, 24 μM nsp8, 10 μM T33-1:P10, 300 μM CTP, and 300 μM ATP in a buffer of 50 mM HEPES (pH 7.0), 100 mM NaCl, 5 mM MgCl2, 4 mM dithiothreitol (DTT) was incubated at 25°C for 40-60 min. The reaction mixture was then loaded onto a MonoQ column (GE Healthcare) equilibrated in a Low-Salt buffer (50 mM HEPES (pH 7.0), 50 mM NaCl, 5 mM MgCl2, 4 mM DTT) and the catalytic complex was eluted with a linear gradient to 1 M NaCl. The pooled complex fractions were buffer exchanged to the Low-Salt buffer and concentrated to reach a concentration of 5 mg/mL for further cryo-EM experiments. To test the reactivity of the P14-containing complex, GTP or 3′-deoxy-GTP (3dGTP) was supplied at 300 μM to allow the synthesis of a 16-mer or 15-mer product, respectively. The RNA T33-7:P10 modified from T33-1:P10, a U substitution of A in the template, was designed for RDV-TP incorporation into the P14-containing complex (pre-translocation complex). The procedure for obtaining the P14-containing complex was the same as described above. The pooled complex fractions from the MonoQ column were buffer-exchanged to the Low-Salt buffer and concentrated to reach a concentration of 9 mg/mL. After the addition of 100 μM GTP and 100 μM RDV-TP, the complex was incubated at 4°C for 30min before conducting the cryo-EM experiment.
In the T33-1:P10-based primer-dependent polymerase assays for comparing the behaviors of RDV-TP and ATP, unless otherwise indicated, a typical 20 μL reaction mixture containing 6 μM WT nsp12 or its variant S861A, 6 μM nsp7, 12 μM nsp8, 4 μM T33-1:P10, and 300 μM each NTP, in a buffer of 50 mM HEPES (pH 7.0), 50 mM NaCl, 5 mM MgCl2, 4 mM DTT was incubated at 25°C for 20 and 90 min. The reaction mixture was quenched with an equal volume of stop solution (95% (vol./vol.) formamide, 20 mM EDTA (pH 8.0), 0.02% (wt./vol.) bromphenol blue, and/or 0.02% (wt./vol.) xylene cyanol). A 33-mer DNA complementary to T33-1 was added to the quenched samples at a 4:1 or 3:1 molar ratio (DNA:RNA) to help release the RNA products from the template. RNA species were resolved by 20% polyacrylamide/7 M urea gel electrophoresis and then visualized by staining with Stains-All (Sigma-Aldrich). Color images obtained by scanning the stained gels were converted to gray-scale images prior to quantitation by using ImageJ (version 1.50i, National Institute of Health, USA) (Schneider et al., 2012). The intensity-based fraction of 15-mer among 15- to 17-mer products was used to estimate the fraction of i+3 termination.
RNA Templates Preparation
RNA templates were obtained by in vitro T7 RNA polymerase transcription using a pRAV23 plasmid (kindly supplied by Jeffrey S. Kieft) that contains a T7 promoter and a glmS ribozyme sequence in the transcribed region. The pRAV23 plasmid and synthetic DNAs that contain sequences to generate the target RNAs were used in PCR reactions to generate T7-promoter containing DNA constructs, which in turn serve as the template in 8-16 mL transcription reactions in a buffer (60 mM HEPES (pH 7.8), 22 mM MgCl2, 2 mM spermidine, 0.01% (vol./vol.) Triton X-100, and 15 mM DTT) containing 4 mM each NTP and 25 μg/mL T7 RNA polymerase. After 3.5 h, the reaction mixtures were incubated at 65°C for 3 min and slowly cooled to room temperature to facilitate the folding of the glmS ribozyme. To generate the ribozyme-cleaved RNA products, gluocoseamine-6-phosphate and NaCl were added to the mixture at final concentrations of 5 mM and 100 mM, respectively. The cleavage reactions were carried out at 42°C for 0.5–1 h before being quenched by an equal volume of gel loading buffer (10 M Urea, 1 × TBE, 0.02% (wt./vol.) xylene cyanol, and 0.02% (wt./vol.) bromphenol blue). The target RNAs were purified by 12% polyacrylamide/7 M urea gel electrophoresis, excised from the gel, eluted using an Elu-Trap device (GE Healthcare), ethanol precipitated, dissolved in RNA annealing buffer (RAB) (50 mM NaCl, 5 mM Tris (pH 7.5), 5 mM MgCl2), and stored at −80°C after a self-annealing process (a 3-min incubation at 95°C followed by snap cooling to minimize inter-molecular annealing).
Synthesis of Remdesivir Triphosphate
The synthesis of remdesivir triphosphate (RDV-TP) was accomplished according to the established process (Warren et al., 2016). To a suspension of GS-441524 (300 mg, 1.03 mmol) (CAS 1191237-69-0, Catalog Number: T7222, Topscience, China) in PO(OMe)3 (5 mL) at 0°C was added POCl3 (236 mg, 1.54 mmol), the solution became slowly clear. The reaction mixture was stirred at 0°C for 2 h, and a suspension of pyrophosphate tributylamine salts (844 mg, 1.54 mmol) in CH3CN (3 mL) was added followed by the addition of Bu3N (762 mg, 4.12 mmol). The reaction mixture was stirred at 0°C for 2 h, the reaction was monitored by liquid chromatography/mass spectrometry (LC/MS), quenched by the addition of triethylammonium bicarbonate buffer (1 M, 12 mL). The resulting mixture was stirred at room temperature for 30 min, and then triethylamine (1 mL) was added. The mixture was stirred an additional 30 min and washed three times with EtOAc (20 mL × 3). The aqueous phase was purified by Pre-HPLC to afford the product RDV-TP salt (205 mg, 24%, 3.0 eq triethylamonium) as an off-white solid.
Cryo-EM Grid Preparation and Data Collection
In total, 3 μL of protein-RNA complex solution at 5 mg/mL (added with 0.025% DDM) was applied onto an H2/O2 glow-discharged, 200-mesh Quantifoil R0.6/1.0 grid (Quantifoil, Micro Tools GmbH, Germany). The grid was then blotted for 3.0 s with a blot force of 0 at 8°C and 100% humidity and plunge-frozen in liquid ethane using a Vitrobot (Thermo Fisher Scientific, USA). Cryo-EM data were collected with a 300 keV Titan Krios electron microscope (Thermo Fisher Scientific, USA) and a K2 Summit direct electron detector (Gatan, USA). Images were recorded at EFTEM with a 165000 × magnification and calibrated super-resolution pixel size 0.41 Å/pixel. The exposure time was set to 5 s with a total accumulated dose of 60 electrons per Å2. All images were automatically recorded using SerialEM (Mastronarde, 2005). For the sample of nsp12-nsp7-nsp8-RNA post-translocated catalytic complex, a total of 6,826 images were collected with a defocus range from 1.0 μm to 1.8 μm. For the sample of nsp12-nsp7-nsp8-RNA pre-translocated catalytic complex, a total of 6,524 images were collected with a defocus range from 1.0 μm to 2.0 μm. Statistics for data collection and refinement are in Table S2.
Cryo-EM Image Processing
All dose-fractioned images were motion-corrected and dose-weighted by MotionCorr2 software (Zheng et al., 2017) and their contrast transfer functions were estimated by cryoSPARC patch CTF estimation (Punjani et al., 2017). For the dataset of nsp12-nsp7-nsp8-RNA post-translocated catalytic complex, a total of 1,235,162 particles were auto-picked using blob picker and extracted with a box size of 300 pixels in cryoSPARC (Punjani et al., 2017). The following 2D, 3D classifications, and refinements were all performed in cryoSPARC. 487,866 particles were selected after two rounds of 2D classification. 100,000 particles were used to do Ab-Initio reconstruction in four classes, and then these four classes were used as 3D volume templates for heterogeneous refinement with all selected particles, with 119,662 particles converged into nsp12-nsp7-nsp8-RNA class and 183,165 particles converged into nsp12-nsp8-RNA class. Next, both particle sets were used to perform non-uniform refinement, yielding a resolution of 3.26 Å and 3.60 Å, respectively. For the dataset of nsp12-nsp7-nsp8-RNA pre-translocated catalytic complex, a total of 1,330,896 particles were auto-picked using the templates selected from the previous 2D classes and extracted with a box size of 320 pixels in cryoSPARC. 804,943 particles were selected after 2D classification. Two rounds of Ab-Initio reconstruction and heterogeneous refinement were performed, with 214,419 particles converged into nsp12-nsp7-nsp8-RNA class. After non-uniform refinement, the resolution reached 2.93 Å. Then, this particle set was imported into RELION-3.03 (Zivanov et al., 2018) to perform local 3D classification and two classes of different conformations could be distinguished. After non-uniform refinement in cryoSPARC, the final resolution of conformation I reached 3.03 Å (65463 particles) while that of conformation II reached 3.12 Å (37696 particles). Local resolution estimation was performed using Resmap (Kucukelbir et al., 2014).
Model Building and Refinement
To build the two structures of the SARS-CoV-2 nsp12-nsp7-nsp8-RNA complex, the structure of the SARS-CoV-2 nsp7-nsp8-nsp12 complex (PDB: 7BTF) was placed and rigid-body fitted into the cryo-EM map using UCSF Chimera (Pettersen et al., 2004). The RNA model was manually built in Coot 0.8 (Emsley et al., 2010) with the guidance of the cryo-EM map, and overall real-space refinement was performed using Phenix 1.9 (Afonine et al., 2012). The data validation statistics are shown in Table S2.
Creation of Figures
Figures of molecular structures were generated using PyMOL (The PyMOL Molecular Graphics System, Schrödinger, LLC.) (DeLano, 2010) and UCSF ChimeraX (Goddard et al., 2018).
Quantification and Statistical Analysis
Quantification and statistical analyses employed in this publication pertain to the analysis on electron microscopy data and the determination of structures by electron microscopy, which are integral parts of existing algorithms and software used. Resolution estimations of cryo-EM density maps are based on the 0.143 Fourier Shell Correlation (FSC) criterion (Chen et al., 2013, Rosenthal and Henderson, 2003). No new methods were used to determine whether the data met assumptions of the statistical approach.
Acknowledgments
We would like to pay an exceptional tribute to ShanghaiTech University and their administrative team, as well as the Bio-Electron Microscopy Facility for their great care and support to our research team to enable us to carry out this research in a safe and healthy environment. It would have been impossible for us to attain this achievement without their tremendous efforts in the last two months during the COVID-19 pandemic. We also must express our gratitude to the campus service team of ShanghaiTech University scientific research platform of Shanghai Institute for Advanced Immunochemical Studies (SIAIS) and National Center for Protein Science Shanghai (NCPSS), as well as all the manager and technician individuals who provided onsite or remote technical support. Their kind help and fearless support are pivotal to this work during the epidemic. We would like to thank the University of Queensland for their collaboration. We thank Meihua Wang for providing some of the RNA template samples. We are grateful to Lili Niu, Xiang Ding, and Fuquan Yang from the Laboratory of Proteomics, Core Facility of Protein Science, Institute of Biophysics, Chinese Academy of Sciences, for the RNA analysis with Mass spectrometry. This work was supported by the National Program on Key Research Project of China (2017YFC0840300 and 2020YFA0707500 to Z.R. and 2018YFA0507200 to P.G.), the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB08020200 to Z.R.), the National Natural Science Foundation of China (81520108019 and 813300237 to Z.R. and 32041007 to P.G.) and Science and Technology Commission of Shanghai Municipality, China (20431900200).
Author Contributions
Z.R., P.G., and Q.W. conceived, initiated, and coordinated the project. H.W., Y.Z., C.Z., L.Y., H.G., Y.H., and J.G. purified the protein; J.W., Q.L., and X.F. performed the RNA template preparation; J.W., Q.L., and H.W. assembled and purified the catalytic complex; J.W. and Q.L. carried out the enzymology experiments; P.G. supervised the RNA construct design and the catalytic complex assembly and purification; Xiaobao Yang synthesized RDV-TP; Y.G., F.L., W.J. and Q.S. collected the cryo-EM data; Y.G., A.M., W.J. and F.L. processed cryo-EM data; Q.W., A.M., W.J., Y.G. built and refined the structure model; and the manuscript was written by Q.W., P.G., Y.G., J.W., H.W., W.J., A.M., Xiaobao Yang, L.G., and Z.R. All authors discussed the experiments and results, read, and approved the manuscript.
Declaration of Interests
The authors declare no competing interests.
Published: May 22, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.cell.2020.05.034.
Supplemental Information
References
- Afonine P.V., Grosse-Kunstleve R.W., Echols N., Headd J.J., Moriarty N.W., Mustyakimov M., Terwilliger T.C., Urzhumtsev A., Zwart P.H., Adams P.D. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr. 2012;68:352–367. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agostini M.L., Andres E.L., Sims A.C., Graham R.L., Sheahan T.P., Lu X., Smith E.C., Case J.B., Feng J.Y., Jordan R. Coronavirus Susceptibility to the Antiviral Remdesivir (GS-5734) Is Mediated by the Viral Polymerase and the Proofreading Exoribonuclease. MBio. 2018 doi: 10.1128/mBio.00221-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agostini M.L., Pruijssers A.J., Chappell J.D., Gribble J., Lu X., Andres E.L., Bluemling G.R., Lockwood M.A., Sheahan T.P., Sims A.C. Small-Molecule Antiviral β-d-N4-Hydroxycytidine Inhibits a Proofreading-Intact Coronavirus with a High Genetic Barrier to Resistance. J. Virol. 2019 doi: 10.1128/JVI.01348-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batey R.T., Kieft J.S. Improved native affinity purification of RNA. RNA. 2007;13:1384–1389. doi: 10.1261/rna.528007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S., McMullan G., Faruqi A.R., Murshudov G.N., Short J.M., Scheres S.H., Henderson R. High-resolution noise substitution to measure overfitting and validate resolution in 3D structure determination by single particle electron cryomicroscopy. Ultramicroscopy. 2013;135:24–35. doi: 10.1016/j.ultramic.2013.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman V.M., Muth D., Niemeyer D., Drosten C. Hosts and Sources of Endemic Human Coronaviruses. Adv. Virus Res. 2018;100:163–188. doi: 10.1016/bs.aivir.2018.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit E., van Doremalen N., Falzarano D., Munster V.J. SARS and MERS: recent insights into emerging coronaviruses. Nat. Rev. Microbiol. 2016;14:523–534. doi: 10.1038/nrmicro.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debing Y., Neyts J., Delang L. The future of antivirals: broad-spectrum inhibitors. Curr. Opin. Infect. Dis. 2015;28:596–602. doi: 10.1097/QCO.0000000000000212. [DOI] [PubMed] [Google Scholar]
- DeLano W. Schrödinger, LLC; New York: 2010. The PyMOL molecular graphics system. [Google Scholar]
- Emsley P., Lohkamp B., Scott W.G., Cowtan K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y., Yan L., Huang Y., Liu F., Zhao Y., Cao L., Wang T., Sun Q., Ming Z., Zhang L. Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science. 2020;368:779–782. doi: 10.1126/science.abb7498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddard T.D., Huang C.C., Meng E.C., Pettersen E.F., Couch G.S., Morris J.H., Ferrin T.E. UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci. 2018;27:14–25. doi: 10.1002/pro.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong P., Peersen O.B. Structural basis for active site closure by the poliovirus RNA-dependent RNA polymerase. Proc. Natl. Acad. Sci. USA. 2010;107:22505–22510. doi: 10.1073/pnas.1007626107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon C.J., Tchesnokov E.P., Woolner E., Perry J.K., Feng J.Y., Porter D.P., Götte M. Remdesivir is a direct-acting antiviral that inhibits RNA-dependent RNA polymerase from severe acute respiratory syndrome coronavirus 2 with high potency. J. Biol. Chem. 2020;295:6785–6797. doi: 10.1074/jbc.RA120.013679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham R.L., Donaldson E.F., Baric R.S. A decade after SARS: strategies for controlling emerging coronaviruses. Nat. Rev. Microbiol. 2013;11:836–848. doi: 10.1038/nrmicro3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillen H.S., Kokic G., Farnung L., Dienemann C., Tegunov D., Cramer P. Structure of replicating SARS-CoV-2 polymerase. bioRxiv. 2020 doi: 10.1101/2020.04.27.063180. [DOI] [PubMed] [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imbert I., Guillemot J.C., Bourhis J.M., Bussetta C., Coutard B., Egloff M.P., Ferron F., Gorbalenya A.E., Canard B. A second, non-canonical RNA-dependent RNA polymerase in SARS coronavirus. EMBO J. 2006;25:4933–4942. doi: 10.1038/sj.emboj.7601368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan P.C., Liu C., Raynaud P., Lo M.K., Spiropoulou C.F., Symons J.A., Beigelman L., Deval J. Initiation, extension, and termination of RNA synthesis by a paramyxovirus polymerase. PLoS Pathog. 2018;14:e1006889. doi: 10.1371/journal.ppat.1006889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouba T., Drncová P., Cusack S. Structural snapshots of actively transcribing influenza polymerase. Nat. Struct. Mol. Biol. 2019;26:460–470. doi: 10.1038/s41594-019-0232-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucukelbir A., Sigworth F.J., Tagare H.D. Quantifying the local resolution of cryo-EM density maps. Nat. Methods. 2014;11:63–65. doi: 10.1038/nmeth.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Zhao Q., Zhang Y., Zhang Y., Bartlam M., Li X., Rao Z. New nsp8 isoform suggests mechanism for tuning viral RNA synthesis. Protein Cell. 2010;1:198–204. doi: 10.1007/s13238-010-0028-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastronarde D.N. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 2005;152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- Mercorelli B., Palù G., Loregian A. Drug Repurposing for Viral Infectious Diseases: How Far Are We? Trends Microbiol. 2018;26:865–876. doi: 10.1016/j.tim.2018.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrosillo N., Viceconte G., Ergonul O., Ippolito G., Petersen E. COVID-19, SARS and MERS: are they closely related? Clin Microbiol Infect. 2020 doi: 10.1016/j.cmi.2020.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Punjani A., Rubinstein J.L., Fleet D.J., Brubaker M.A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods. 2017;14:290–296. doi: 10.1038/nmeth.4169. [DOI] [PubMed] [Google Scholar]
- Rosenthal P.B., Henderson R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J. Mol. Biol. 2003;333:721–745. doi: 10.1016/j.jmb.2003.07.013. [DOI] [PubMed] [Google Scholar]
- Schneider Caroline, Rasband Wayne, Eliceiri Kevin. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sholders A.J., Peersen O.B. Distinct conformations of a putative translocation element in poliovirus polymerase. J. Mol. Biol. 2014;426:1407–1419. doi: 10.1016/j.jmb.2013.12.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu B., Gong P. Structural basis of viral RNA-dependent RNA polymerase catalysis and translocation. Proc. Natl. Acad. Sci. USA. 2016;113:E4005–E4014. doi: 10.1073/pnas.1602591113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel D., Hui H.C., Doerffler E., Clarke M.O., Chun K., Zhang L., Neville S., Carra E., Lew W., Ross B. Discovery and Synthesis of a Phosphoramidate Prodrug of a Pyrrolo[2,1-f][triazin-4-amino] Adenine C-Nucleoside (GS-5734) for the Treatment of Ebola and Emerging Viruses. J. Med. Chem. 2017;60:1648–1661. doi: 10.1021/acs.jmedchem.6b01594. [DOI] [PubMed] [Google Scholar]
- Subissi L., Posthuma C.C., Collet A., Zevenhoven-Dobbe J.C., Gorbalenya A.E., Decroly E., Snijder E.J., Canard B., Imbert I. One severe acute respiratory syndrome coronavirus protein complex integrates processive RNA polymerase and exonuclease activities. Proc. Natl. Acad. Sci. USA. 2014;111:E3900–E3909. doi: 10.1073/pnas.1323705111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tchesnokov E.P., Feng J.Y., Porter D.P., Götte M. Mechanism of Inhibition of Ebola Virus RNA-Dependent RNA Polymerase by Remdesivir. Viruses. 2019;11:326. doi: 10.3390/v11040326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- te Velthuis A.J., van den Worm S.H., Snijder E.J. The SARS-coronavirus nsp7+nsp8 complex is a unique multimeric RNA polymerase capable of both de novo initiation and primer extension. Nucleic Acids Res. 2012;40:1737–1747. doi: 10.1093/nar/gkr893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tvarogová J., Madhugiri R., Bylapudi G., Ferguson L.J., Karl N., Ziebuhr J. Identification and Characterization of a Human Coronavirus 229E Nonstructural Protein 8-Associated RNA 3¢-Terminal Adenylyltransferase Activity. J. Virol. 2019 doi: 10.1128/JVI.00291-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Li R., Shu B., Jing X., Ye H.Q., Gong P. Stringent control of the RNA-dependent RNA polymerase translocation revealed by multiple intermediate structures. Nat. Commun. 2020;11:2605. doi: 10.1038/s41467-020-16234-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren T.K., Jordan R., Lo M.K., Ray A.S., Mackman R.L., Soloveva V., Siegel D., Perron M., Bannister R., Hui H.C. Therapeutic efficacy of the small molecule GS-5734 against Ebola virus in rhesus monkeys. Nature. 2016;531:381–385. doi: 10.1038/nature17180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y., Ma Q., Restle T., Shang W., Svergun D.I., Ponnusamy R., Sczakiel G., Hilgenfeld R. Nonstructural proteins 7 and 8 of feline coronavirus form a 2:1 heterotrimer that exhibits primer-independent RNA polymerase activity. J. Virol. 2012;86:4444–4454. doi: 10.1128/JVI.06635-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin W., Mao C., Luan X., Shen D.D., Shen Q., Su H., Wang X., Zhou F., Zhao W., Gao M. Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir. Science. 2020:eabc1560. doi: 10.1126/science.abc1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai Y., Sun F., Li X., Pang H., Xu X., Bartlam M., Rao Z. Insights into SARS-CoV transcription and replication from the structure of the nsp7-nsp8 hexadecamer. Nat. Struct. Mol. Biol. 2005;12:980–986. doi: 10.1038/nsmb999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng S.Q., Palovcak E., Armache J.-P., Verba K.A., Cheng Y., Agard D.A. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods. 2017;14:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., China Novel Coronavirus Investigating and Research Team A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zivanov J., Nakane T., Forsberg B.O., Kimanius D., Hagen W.J., Lindahl E., Scheres S.H. New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife. 2018;7:e42166. doi: 10.7554/eLife.42166. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The cryo-EM maps and atomic models have been deposited at the Electron Microscopy Data Bank (EMD-30252, EMD-30275, EMD-30283 and EMD-30284) and the Protein Data Bank (PDB: 7BZF and PDB: 7C2K). A list of software used in this study can be found in the Key Resources Table.