Abstracts
This work used quantum chemical method via DFT to calculate molecular descriptors for the development of QSAR model to predict bioactivity (IC50- 50% inhibition concentration) of the selected 1, 2, 3-triazole-pyrimidine derivatives against receptor (human gastric cancer cell line, MGC-803). The selected molecular parameters were obtained by B3LYP/6-31G∗∗. QSAR model linked the molecular parameters of the studied compounds to their cytotoxicity and reproduced their observed bioactivities against MGC-803. The calculated IC50 tailored the observed IC50 and greater than standard compound, 5-fluorouracil, suggesting that the developed QSAR model reproduced the observed bioactivity. Statistical analyses (including R2, CV. R2 and gave 0.950, 0.970 and 0.844 respectively) revealed a very good fitness. Molecular docking studies revealed the hydrogen bonding with the amino acid residues in the binding site, as well as ligand conformations which are essential feature for ligand-receptor interactions. Therefore, the methods used in this study are veritable tools that can be employed in pharmacological and medicinal chemistry researches in designing better drugs with improve potency.
Keywords: Theoretical chemistry; 1, 2, 3-triazole-pyrimidine derivatives; Human gastric cancer cells (MGC-803); Anticancer; Molecular descriptors; Ligand conformations
Organic chemistry; Pharmaceutical chemistry; Theoretical chemistry; 1, 2, 3-triazole-pyrimidine derivatives, Human gastric cancer cells (MGC-803), Anticancer, Molecular descriptors, Ligand conformations.
1. Introduction
Globally, cancer kills within few months of diagnosis and over a million cases of death nowadays were resulted from cancer, thus posing serious burdens to the public (Song et al., 2017; Perez-Castillo et al., 2018; Ismail et al., 2019). Gastric cancer (GC) remains a common global malignancy with the second highest incidence and mortality rate of cancer diseases (Perez-Castillo et al., 2018; Ismail et al., 2019). Stomach cancer has been ranked 5th most common cancer globally, with diagnosed cases of 952,000 in 2012 and 723,000 deaths (WHO, 2014; Mustafa et al., 2017). Mostly, GC are gastric carcinoma and gastric antrum cancer, while the occurrence of gastroesophageal junction carcinoma is on the gradual increase. Analysis of the onset ages revealed that the occurrence of GC increases gradually among the youths (Li et al., 2015; Sun et al., 2015; Lee et al., 2017) and occurs twice as common in males than females (Mustafa et al., 2017), because estrogen probably protects women against the development of this form (Chandannos and Lagergren, 2008; Mustafa et al., 2017). Generally, GC patients are known to exhibit three high and three low characteristic features, this makes the incidence, metastasis and mortality rate to be significantly high (Ismail et al., 2019). To worsen the case, the early diagnosis and radical resection rates with five years survival rate are considerably low (Wu et al., 2015). Though, the exact causes of GC remain uncleared, its pathogenesis is the same as that of other malignant tumors such as multistep and multifactorial comprehensive diseases (Song et al., 2017). Universally, GC cases are often categorized into early- and advanced-stages (Song et al., 2017; Sitarz et al., 2018). Early-stage GC are limited to the mucosa or sub-mucosa, irrespective of the lesion size and the presence of lymph node metastasis. While the cancer that extends beyond the sub-mucosa stage to invade the gastric muscular layer is regarded as middle GC, whereas, when tumors infiltrate beyond or into the subserosa to the nearby organs or metastasizes, they are defined as the advanced GC Song et al. (2017). This stage includes intermediate and advanced tumors. The treatment strategies and effectiveness are determined by the stages of the tumor. If for instance, the early GC patient undergoes radical surgery followed by chemotherapy, and the postoperative 5-years, then the rate of survival is 90%; thus, making the therapeutic effects of early GC acceptable (Song et al., 2017).
Realizing that successful treatment of cancer patients remains serious concerns and/or challenges for the researchers worldwide. Thus, theoretical calculations of molecular descriptors as well as simulations of drug-like molecules with protein receptors are essentials for the prediction of the bioactivity and potency/efficacy of such molecules (Chill et al., 2003; Pogorzelska et al., 2015; Elshakre et al., 2020). Also, knowing the stable conformation, binding free energy and nonbonding interactions of the drug-like molecule in the active gouge of the protein receptor will help in understanding the phymacokinetic of the drug interactions in the system (Śledź and Caflisch, 2018; Marquina et al., 2019; Melge et al., 2019). Consequently, the role of computational chemist is not left out in the development of QSAR model that correlates molecular descriptors of the drug/drug like molecule to its bio-activity as well as simulating the drug-receptor interactions (docking) which govern kinetic mechanisms (Prachayasittikul et al., 2015; Gupta et al., 2018; Perez-Castillo et al., 2018).
Computational tools have gained tremendous significance in drug discoveries and design processes (Baldi, 2010; Ismail et al., 2019; Elshakre et al., 2020). Computational chemistry approach is often employed routinely for the study of drug-receptor complexes in atomic details and in calculating the properties of small-molecular drug candidates (Adedirin et al., 2018; Bello et al., 2018; Elshakre et al., 2020). Tool from information sciences and statistics are progressively important for organizing and managing the huge biological and chemical activity databases which are now possess by most pharmaceutical companies, so as to make the optimum use of these databases (Perez-Castillo et al., 2018). The most fundamental goal in using computer aided drug design is the prediction of various properties of drug, e.g. binding affinity to a certain target is necessary for biological activity (Ma et al., 2010; Vasaikar et al., 2016; Cruz-Monteagudo et al., 2017; Perez-Castillo et al., 2018). The principal benefit of this technique is that those properties can be predicted even before the candidates' drug molecules are synthesized (Alam and Khan, 2017; Elshakre et al., 2020).
1, 2, 3-triazole has been a profitable basis of motivation for medicinal chemists over the past years. Owing to its synthetic convenience by click chemistry as well as the diverse inhibitory actions, including antibacterial, antifungal, antiallergic, anti-inflammatory e.t.c (Demaray et al., 2008; Giffin et al., 2008; Aher et al., 2009; Wang et al., 2010; De-Simone et al., 2011; Patpi et al., 2012; Yu et al., 2013; Lebeau et al., 2017; Ahmed et al., 2017; Salah et al., 2018; Zaki et al., 2018; Schröder et al., 2019; Romad and Miranda, 2019). Pyrimidine containing sulfa drugs are classified on the basis of substitution and the classification with the respective example of drug is as follows. Monosubstituted and disubstituted sulfa drugs include sulfadimidine (sulfamethazine), sulfamerazine, sulfameythoxydiazine, methyldiazine and sulfadiazine. Trisubstituted sulfa drugs are sulfadoxine, sulfadimethoxine, sulfamethomidine, sulfisomidine, sulfamethoxine, and sulfacytine. Sulfadoxine which has a half-life of 7–9 days was used for malarial prophylaxis (White, 1996). Pyrimidinones scaffolds represent the class of a heterocyclic compounds that possess noteworthy pharmacological efficiencies (Sharma et al., 2014; Fumagalli et al., 2017; El-Shahawi and El-Ziaty, 2017; Rogerio et al., 2018) For example: antiviral (Choi et al., 2000; Parveen et al., 2010), anti-HIV (McGuigan et al., 2000; Pontikis et al., 2000), anti-bacterial (Palanki et al., 2000) and anticancer (Martin et al., 2000; Curtin et al., 2004; Rostom et al., 2009; Conejo-García et al., 2011; Lopez-Cara et al., 2011; Jin et al., 2011; Zhu et al., 2013; Morales et al., 2014). Abdel-Mohsen et al. (2010) reported the potent antitumor agent and cytotoxic activity of benzimidazole pyrimidine conjugate.
The study of 1, 2, 3-triazole moiety combined with pyrimidine derivatives is a new hybrid system that remains an unexplored research field in the area of theoretical/computation chemistry. Despite a notable success of the 1, 2, 3-triazole and pyrimidine as good anti-cancer activity, its theoretical investigations still remain unexplored and emergent area. Also, developing a new drug still remains major challenge, time consuming and cost intensive processes. Owing to the enormous expenses or failure of most candidate drugs found in the market, their developments presently face liability such as possible side effects, in addition to their therapeutic properties. This makes the computational approach a hot topic and novel method due to its ability to speed up and assist drug design processes (Kumari et al., 2017). Therefore, this research aimed at calculating the molecular descriptors for anticancer activities of 1, 2, 3-triazole-pyrimidine hybrids using quantum chemical method and calculations of binding free energy via molecular docking. This was limited to the use of DFT and to develop QSAR model that correlates molecular descriptor with bio-activities of the 20 molecules from 1, 2, 3-triazole-pyrimidine hybrids reported by Ma et al. (2014) and finally, to calculate binding free energies of the stable conformation for the ligand-receptor complexes.
2. Materials and methods
The major materials used for this work were software (Spartan 14 for quantum chemical calculations, Gretl for QSAR modeling, Discovery studio 4.1 for preparation of both ligand and receptor, Autodock (autodock tool 1.5.6 and autodock vina 1.1.2) for docking and bonding energy evaluation and pymol 1.7.4.4 for molecular visualization).
2.1. Quantum chemical software (Spartan)
Spartan 14 is a molecular modeling environment that is well-known for its equity, flexibility, ease of use and a tool for exploring organic, bio-organic, inorganic and organic chemistry through molecular mechanics and quantum chemical calculations, together with an array of graphical model for conveying the results of those calculations. It is also employed for molecular orbital calculations as well as wide ranges of graphical models with a full range of molecular mechanism, semi-empirical method, DFT and a section of wave function based on the important post-hartree fork model (Desai et al., 2001).
2.2. Minimization and optimization
In this research, 1, 2, 3-triazole-pyrimidine hybrids (Figure 1) were used to perform quantum calculations for molecular descriptors, QSAR model and docking simulation. The compounds (Figure 1) consist of twenty (20) derivatives (take from Ma et al. (2014) as presented in Table 1. The standard drug used was 5-fluorouracil which is a well-known anticancer drug (Zheng et al., 2013) employed in the experimental work by Ma et al. (2014).
Figure 1.
Schematic structure 1, 2, 3-triazole-pyrimidine.
Table 1.
Compound names of studied 1, 2, 3-triazole-pyrimidine hybrids system obtained from Figure 1.
| Compound | R1 | R2 | R3 | Compound Name |
|---|---|---|---|---|
| L1 | p-OCH3 | o-Cl | H | 2-((1-(2-chlorobenzyl)-1h-1, 2, 3-triazol-4-yl)methylthio)-4-((4-methoxyphenyl)amino)-6-phenylpyrimidine-5-carbonitrile. |
| L2 | m-CF3 | o-Cl | H | 2-((1-(2-Chlorobenzyl) -1H-1, 2, 3-triazol-4-yl) methylthio) -4- phenyl-6-((3-(trifluoromethyl) phenyl) amino) pyrimidine-5- carbonitrile. |
| L3 | o-Cl | o-Cl | H | 2-((1-(2-Chlorobenzyl) -1H-1, 2, 3-triazol-4-yl) methylthio) -4-((2-chlorophenyl) amino) -6-phenylpyrimidine-5-carbonitrile. |
| L4 | p-Cl | o-Cl | H | 2-((1-(2-Chlorobenzyl) -1H-1, 2, 3-triazol-4-yl) methylthio) -4- ((4-chlorophenyl) amino) -6-phenylpyrimidine-5-carbonitrile. |
| L5 | m-Cl | o-Cl | H | 2-((1-(2-Chlorobenzyl) -1H-1, 2, 3-triazol-4-yl) methylthio) -4-((3-chlorophenyl) amino) -6-phenylpyrimidine-5-carbonitrile. |
| L6 | o-OCH3 | o-Cl | H | 2-((1-(2-Chlorobenzyl) -1H-1, 2, 3-triazol-4-yl) methylthio) -4-((2-methoxyphenyl) amino) -6-phenylpyrimidine-5-carbonitrile. |
| L7 | m- CH3 | o-Cl | H | 2-((1-(2-Chlorobenzyl) -1H-1, 2, 3-triazol-4-yl) methylthio) -4-phenyl-6-(m-tolylamino) pyrimidine-5-carbonitrile. |
| L8 | m-NO2 | o-Cl | H | 2-((1-(2-Chlorobenzyl) -1H-1, 2, 3-triazol-4-yl) methylthio) -4-((3-nitrophenyl) amino) -6-phenylpyrimidine-5-carbonitrile. |
| L9 | o-F | o-Cl | H | 2-((1-(2-Chlorobenzyl) -1H-1, 2, 3-triazol-4-yl) methylthio) -4-((2-fluorophenyl) amino) -6-phenylpyrimidine-5Jcarbonitrile. |
| L10 | p-F | o-Cl | H | 2-((1-(2-Chlorobenzyl) -1H-1, 2, 3-triazol-4-yl) methylthio) -4-((4-fluorophenyl) amino) -6-phenylpyrimidine-5-carbonitrile. |
| L11 | o- CH3 | o-Cl | H | 2-((1-(2-Chlorobenzyl) -1H-1, 2, 3-triazol-4-yl) methylthio) -4-phenyl-6-(o-tolylamino) pyrimidine-5-carbonitrile. |
| L12 | o-F | p- CH3 | H | 4-((2-Fluorophenyl) amino) -2-((1-(4-methylbenzyl) -1H- 1, 2, 3-triazol-4-yl) methylthio) -6-phenylpyrimidine-5-carbonitrile. |
| L13 | p- CH3 | p- CH3 | H | 2-((1-(4-Methylbenzyl) -1H-1, 2, 3-triazol-4-yl) methylthio) -4-phenyl-6-(p-tolylamino) pyrimidine-5-carbonitrile. |
| L14 | o-Cl | p- CH3 | H | 4-((2-Fluorophenyl) amino) -2-((1-(4-methylbenzyl) -1H-1, 2, 3-triazol-4-yl) methylthio) -6-phenylpyrimidine-5-carbonitrile. |
| L15 | p- CH3 | o-Cl | H | 2-((1-(2-Chlorobenzyl) -1H-1, 2, 3-triazol-4-yl) methylthio) -4-phenyl-6-(p-tolylamino) pyrimidine-5-carbonitrile. |
| L16 | p- CH3 | o-Cl | p-CH(CH3)2 | 2-((1-(2-Chlorobenzyl) -1H-1, 2, 3-triazol-4-yl) methylthio) -4-(4-isopropylphenyl) -6-(p-tolylamino) pyrimidine-5-carbonitrile. |
| L17 | o-OCH3 | o-Cl | p-CH(CH3)2 | 2-((1-(2-Chlorobenzyl) -1H-1, 2, 3-triazol-4-yl) methylthio) -4-(4-isopropylphenyl) -6-(p-tolylamino) pyrimidine-5-carbonitrile. |
| 18 | p-CH3 | o-Cl | p-CH3 | 2-((1-(2-Chlorobenzyl) -1H-1, 2, 3-triazol-4-yl) methylthio) -4-(p-tolyl) -6-(p-tolylamino) pyrimidine-5-carbonitrile. |
| L19 | p- CH3 | o-Cl | m, p, m-triOCH3 | 2-((1-(2-Chlorobenzyl) -1H-1, 2, 3-triazol-4-yl) methylthio) -4-(p-tolylamino) -6-(3, 4, 5-trimethoxyphenyl) pyrimidine-5-carbonitrile. |
| L20 | p- CH3 | o-Cl | p-Cl | 2-((1-(2-Chlorobenzyl) -1H-1, 2, 3-triazol-4-yl) methylthio) -4-(4-chlorophenyl) -6-(p-tolylamino) pyrimidine-5-carbonitrile. |
All these compounds were minimized and optimized. Minimization is the process of refining-built molecules so as to get good structure that is suitable for optimization process. Optimization involves the geometry searching for the equilibrium or minimum energy of conformation. During this process, atoms, bond length and bond angle of the molecules displayed iteratively until new equilibrium geometry is reached which is called convergence.
2.3. Quantum chemical method
The equilibrium geometries for the 1, 2, 3-triazole-pyrimidine hybrids (Table 1) used in this study were optimized using DFT with the standard 6-31G∗∗ (d, p) basis set. The DFT method used consists of the three parameters density functional which includes Becke's gradient, exchange correlation (Becke, 1993) and the Lee, Yang, Parr correlation functional (i.e. B3LYP) (Yang et al., 2005). The choice of the selected functional and basis sets was attributed to the accuracy of DFT calculations. The sufficiency of polarized split-valence 6-31G∗∗ (d, p) basis sets have been demonstrated for the calculation of the excited properties of ligands (Jacquemin et al., 2008); this (i.e. 6-31G∗∗ (d, p) basis set) was therefore employed in this study. Frequency calculations were carried out at the same levels of the theory in order to characterize the stationary points as local minima and none of the optimized molecules has imaginary frequency. The molecules under study were designed to generate molecular descriptors that described the bioactivity (IC50) and binding affinity upon docking of the molecules with receptors (Table 1).
DFT is very useful in providing chemical descriptors such as chemical hardness (η), electronegativity (χ), softness (S), electrophilicity index (ω) and local Fukui function indices. Zhou and Navangul (1990) reported the principle of maximum hardness (absolute hardness) η, for an N-electron system with total energy E and η are defined as:
| (1) |
where IE is the vertical ionization energy which is approximated as -EHOMO and EA for the vertical electron affinity, denoted as -ELUMO (Koopmans, 1934). The global softness is the inverse of chemical hardness. The electron affinity can also be used in combination with ionization energy to give electronic chemical potential, μ as shown in Eq. (2). The negative of electron affinity (-χ) was defined by Parr and Pearson (Zhou and Navangul, 1990), as the characteristic of electronegativity of molecules:
| (2) |
The global electrophilicity index, ω, was introduced by Parr et al. (1999); this can be calculated using the electronic chemical potential, μ, and chemical hardness, η, as
| (3) |
As shown in the definition, this index measured the propensity of a species to accept electrons. Domingo (2002) proposed that high nucleophilicity and electrophilicity of heterocycles corresponded to the opposite extreme of the scale of global reactivity indexes. A good and more reactive nucleophile is characterized by a lower value of μ, ω, and in the opposite, a good electrophile was characterized by a high value of μ, ω.
2.4. Molecular descriptors
The molecular descriptors selected for this research were based on electronic properties of the studied compounds. These descriptors were the highest occupied molecular orbital (HOMO), the lowest unoccupied molecular orbital (LUMO), HOMO-LUMO band gap, softness, chemical hardness, chemical potential, diploe moment, solvation energy, global nucleophilicity, log P, Ovality, area volume, polar surface area (PSA) and polarizability.
2.5. Multiple linear regressions
Multiple linear regression analysis (MLR) is used to examine the relationship between two or more independent variables and one dependent variable. MLR has been a veritable method used to investigate the correlation between biological activity and physicochemical properties of a set of bioactive compounds. It describes how a y-variable relates to two or more x-variables (or transformations of x-variables). The software used in this research work was Gretl which helped in generating equation which be expressed as:
| (4) |
where X is the regressor (also called the predictor or independent variable), Y is the response (also called the dependent variable), α and β are parameters that describe the relationship between X and Y, and the term ε represents the error model (the errors are also referred to as residuals) (Rob, 2014). Therefore, R2 was considered for the linearity and efficiency of the analysis,
Tolerance and variance inflation factor (V.I.F) were examined for the validity of the analysis. The highest correlation of independent variables with dependent variable was chosen for deriving the QSAR model. The statistical values, multiple correlation coefficients (r), standard error(s), cross validation R2 and standard error of prediction were used to evaluate the QSAR models. Several combinations of independent variables which obey the necessary rules for the validity of analysis were added in order to optimize the statistical values. The best model derived from the MLR analysis was used to predict the inhibitory activity of the 1, 2, 3-triazole-pyrimidine derivatives considered. To avoid self-correlation between the variables used for the derivation of QSAR model, the tolerance and V. I. F. rules were strictly adhered to. Figure 2 shows the various steps involved in QSAR modeling process.
Figure 2.
Schematic workflow of the present study.
2.6. Validation of QSAR model
The statistical equations were used to validate the QSAR model. The cross validation (R2), Adjusted R2, Chi-square, standard error, Root Mean Square Error (RMSE) and F-test were considered in this study. Cross-validation governed how reliable a QSAR model could be used for a particular set of data (Puzyn et al., 2010). It was also employed as an analytic instrument to estimate the prognostic control of an equation. Therefore, it was estimated using Eq. (5).
| (5) |
The R2 adjusted could be calculated using Eq. (6)
| (6) |
Thus, QSAR model is considered prognostic, if . R2 is a statistical measure of how close the data are to the fitted regression line. It is also known as the coefficient of determination, or the coefficient of multiple determinations for multiple regressions. The implication is that the closer the R2 value to 1 (unity), the better fits it is. Thus, the higher the R2 value, the better the model fits the data.
2.7. Molecular docking and binding energy evaluation
Molecular interactions are in several forms including protein-nucleic acid, enzyme substrate, protein-protein, drug protein and drug nucleic acid. They play imperative role in several essential biological processes, like cell regulations, gene expression controls, enzyme inhibitions, signal transduction, transport, antibody-antigen recognitions, and even the assemblies of multi-domain proteins. These types of interactions mostly bring about formations of stable protein–protein or protein-ligand complexes that are necessary to carry out their biological functions. Therefore, molecular docking helps in predicting the conformation of receptor-ligand complexes, the specific receptors of interest (usually proteins or nucleic acid molecules) was gotten from the protein data bank (www.proteindatabank.com) and the ligands are the molecules (Alejandra et al., 2013).
2.8. Discovery studio
Discovery studio is software for simulating molecules; it was developed and distributed by acceryls. It has a strong academic collaboration programmes that supports scientific researches (Young, 2001). Also, it is used in preparing (i.e. removal of water molecule and any other residues apart from the desired compound) the ligand and the receptor before subjecting them to docking using autodock tool software.
2.9. Autodock
Autodock tool was designed to simulate how small a molecule, such as substrate or drug candidate bind to receptor of known 3D structures. This helps to locate the active binding site (active gorge) in the receptor. In this work, Autodock vina was used, it is a novel generation of docking software from the molecular graphic laboratory (Trott and Olson, 2010). It achieves noteworthy improvement in the average accuracies of the binding mode predictions. It does not require the choose of atom type(s) and pre-calculated grid map (Morris et al., 2009). Instead, it calculates the grid internally; for the atom type that is needed, and does that virtually instantly (Eswar et al., 2006). The following commands are required to achieve the desired goals; vina --config conf.txt --log log.txt to do the calculation and vinasplit --input out.pdbqt to split the result in the order of scoring. Finally, Pymol, a post docking software was used to view the conformation and hydrophobic interactions of the ligands with the receptors.
3. Results and discussion
3.1. Calculated molecular properties of compounds L1-L20 used for testing anticancer property
In this work, several calculated molecular parameters including solvation energy, weight, hydrophobicity (LogP), volume (V), area, polar surface area (PSA), ovality, dipole moment (DM), heteroatoms (average of mulliken charges on all heteroatoms), HOMO, and LUMO energies were obtained as shown in Tables 2 and 3. The HOMO and LUMO are very important parameters which give convincing qualitative details about the excitation features of modeled compounds (Bouachrine et al., 2009, Yang et al., 2005 and Semire et al., 2012). As expected, the HOMO and LUMO energies along with the band gap energies of the compounds L1-L20 played essential role in binding the molecular compounds to the enzymes. Thus, the magnitude of these parameters determines the extent of nonbonding interactions such as hydrogen bonding and hydrophilic interactions between the receptor and ligand. The calculated HOMO and LUMO values for Compounds L1-L20 are presented in Table 2. High values of HOMO energy are indication of enhanced ability of the ligand to donate electron to the neighboring compounds (Oyebamiji and Semire, 2016a). Whereas, the lower values of LUMO energy imply that the studied molecular compounds have the ability to receive electrons from the neighboring compound which has the ability to donate electron (Oyebamiji and Semire, 2016a,b).
Table 2.
Selected molecular parameters obtained by B3LYP/6-31G∗∗ for anticancer.
| Comp | HOMO | LUMO | BG | SE | η | μ | MW | Ovality | Log P |
|---|---|---|---|---|---|---|---|---|---|
| L1 | -5.57 | -1.99 | 3.58 | -44.98 | 1.79 | -3.78 | 540.051 | 1.73 | 6.25 |
| L2 | -6.25 | -2.04 | 4.21 | -26.52 | 2.105 | -4.145 | 578.022 | 1.74 | 7.3 |
| L3 | -6.03 | -1.85 | 4.18 | -35.28 | 2.09 | -3.94 | 544.47 | 1.71 | 6.94 |
| L4 | -5.94 | -1.92 | 4.02 | -33.58 | 2.01 | -3.93 | 544.47 | 1.72 | 6.94 |
| L5 | -6.08 | -1.98 | 4.1 | -32.3 | 2.05 | -4.03 | 544.47 | 1.71 | 6.94 |
| L6 | -5.71 | -1.93 | 3.78 | -40.76 | 1.89 | -3.82 | 540.051 | 1.72 | 6.25 |
| L7 | -6.16 | -1.95 | 4.21 | -44.86 | 2.105 | -4.055 | 524.052 | 1.71 | 6.87 |
| L8 | -6.14 | -2.01 | 4.13 | -54.69 | 2.065 | -4.075 | 557.038 | 1.72 | 6.02 |
| L9 | -6.21 | -2.04 | 4.17 | -33.81 | 2.085 | -4.125 | 528.015 | 1.68 | 6.54 |
| L10 | -5.91 | -1.93 | 3.98 | -30.05 | 1.99 | -3.92 | 528.015 | 1.7 | 6.54 |
| L11 | -5.88 | -1.9 | 3.98 | -32.44 | 1.99 | -3.89 | 524.052 | 1.71 | 6.87 |
| L12 | -6.08 | -1.94 | 4.14 | -38.16 | 2.07 | -4.01 | 503.634 | 1.72 | 6.79 |
| L13 | -6.08 | -1.96 | 4.12 | -33.14 | 2.06 | -4.02 | 524.052 | 1.71 | 6.87 |
| L14 | -5.81 | -1.9 | 3.91 | -34.87 | 1.955 | -3.855 | 524.052 | 1.71 | 6.87 |
| L15 | -5.68 | -1.71 | 3.97 | -31.22 | 1.985 | -3.695 | 566.133 | 1.78 | 8.1 |
| L16 | -5.83 | -1.93 | 3.9 | -27.49 | 1.95 | -3.88 | 566.133 | 1.77 | 8.1 |
| L17 | -5.85 | -1.99 | 3.86 | -38.92 | 1.93 | -3.92 | 538.07 | 1.75 | 7.35 |
| L18 | -5.79 | -2.06 | 3.73 | -40.61 | 1.865 | -3.925 | 614.13 | 1.81 | 6.49 |
| L19 | -5.95 | -2.02 | 3.93 | -38.49 | 1.965 | -3.985 | 558.497 | 1.75 | 7.42 |
| L20 | -5.96 | -2.18 | 3.78 | -39.98 | 1.89 | -4.07 | 602.948 | 1.74 | 7.69 |
∗Comp: compound, HOMO: highest occupied molecular orbital, LUMO: lowest unoccupied molecular orbital, BG: band gap, SE: solvation energy, η: chemical hardness, μ: chemical potential, MW: molecular weight, LogP: hydrophobicity, DM: dipole moment.
Table 3.
Selected molecular parameters obtained by B3LYP/6-31G∗∗ for anticancer.
| Compd | DM | PSA | Area | HBD | HBA | POLAR | Volume | HET | NOR |
|---|---|---|---|---|---|---|---|---|---|
| L1 | 3.55 | 63.812 | 539.77 | 0 | 8 | 82.62 | 518.87 | 10 | 5 |
| L2 | 4.02 | 56.96 | 545.26 | 0 | 7 | 82.85 | 523.59 | 12 | 5 |
| L3 | 6.63 | 55.694 | 524.23 | 0 | 7 | 81.36 | 505.1 | 10 | 5 |
| L4 | 5.18 | 57.679 | 526.97 | 0 | 7 | 81.44 | 505.6 | 10 | 5 |
| L5 | 6.5 | 58.249 | 525.32 | 0 | 7 | 81.4 | 505.41 | 10 | 5 |
| L6 | 3.68 | 61.257 | 538.28 | 0 | 8 | 82.85 | 518.63 | 10 | 5 |
| L7 | 5.9 | 58.608 | 529.33 | 0 | 7 | 81.74 | 509.93 | 9 | 5 |
| L8 | 2.65 | 104.593 | 536.28 | 1 | 10 | 82.49 | 518.83 | 12 | 5 |
| L9 | 3.21 | 57.413 | 510.17 | 0 | 7 | 80.62 | 495.92 | 10 | 5 |
| L10 | 3.3 | 58.468 | 515.43 | 0 | 7 | 80.7 | 496.34 | 10 | 5 |
| L11 | 2.33 | 57.185 | 526.95 | 0 | 7 | 81.76 | 509.43 | 9 | 5 |
| L12 | 4.89 | 58.158 | 532.64 | 0 | 7 | 82.13 | 514.41 | 8 | 5 |
| L13 | 4.05 | 57.194 | 527.18 | 0 | 7 | 81.73 | 509.47 | 9 | 5 |
| L14 | 2.54 | 56.777 | 528.95 | 0 | 7 | 81.8 | 509.77 | 9 | 5 |
| L15 | 5.34 | 57.236 | 589.15 | 0 | 7 | 86.26 | 564.87 | 9 | 5 |
| L16 | 1.82 | 56.239 | 585.25 | 0 | 7 | 86.23 | 564.33 | 9 | 5 |
| L17 | 3.95 | 59.366 | 552.15 | 0 | 7 | 83.33 | 528.51 | 9 | 5 |
| L18 | 1.5 | 77.827 | 616.19 | 0 | 10 | 88.46 | 591.33 | 12 | 5 |
| L19 | 6.99 | 58.575 | 548.48 | 0 | 7 | 82.94 | 523.81 | 10 | 5 |
| L20 | 1.65 | 57.603 | 551.29 | 0 | 7 | 83.34 | 528.32 | 10 | 5 |
∗PSA: polar surface area, HBD: hydrogen bond donor, HBA: hydrogen bond acceptor, HET: heteroatoms, NOR: number of organic residues.
The calculated band gaps obtained by ground-state properties, from which the band gap is estimated from the energy difference between the LUMO and HOMO (Curioni et al., 1998; Hay, 2002) are presented in Table 2. The band gaps are essentially left-over energy ranges that are not concealed by any band as a result of the finite widths of the energy bands (Table 2) (Oyebamiji and Semire, 2016a,b). The order of the band gap is L1 < L18 < L6 = L20 < L17 < L16 < L14 < L19 < L15 < L10 = L11 < L4 < L5 < L13 < L8 < L12 < L9 < L3 < L2 = L7 (Table 2). The lower the band gap, the better the capacity of a compound to donate electron to the neighbouring molecules. Thus, based on aforementioned facts, band gap played an important role in protein–ligand interaction between bioactivity of the studied drug-like compounds.
Moreover, the calculated log P reveals the capacity of the drug-like molecule to melt in lipophilic (non-aqueous) solutions. Drug-like compounds need this to infuse through several biological membranes. Lipophilicity is classically measured as the distributions of the molecules between the aqueous and non-aqueous phases and it shows the cytotoxicity of ligands (Khaled et al., 2011). The calculated log P values are shown in Table 2 showing that the compounds L1-L20 were not effective in term of lipophilicity when considered the value of Meanwell (2011). The value obtained in this study are slightly higher than the log P value of 5 reported by Meanwell (2011). This suggest that the log P is not enough to validate effectiveness of the compounds under this study in term of lipophilicity.
Furthermore, the solvation energy (SE) was calculated using SM5.4 model based on semi-empirical (AM1) wave functions (Liu and Zou, 2006; Oyebamiji and Semire, 2016a,b). The SE consists of the summation of two terms which are: the required energy for creating a cavity in the solvent (water) and the energy of the electrostatic interactions between the solvent and the solute once the solute/molecule is placed in the cavity (Oyebamiji and Semire, 2016a). The equilibrium geometries as well as spectra and any property derived from the wave function are unaffected by SM5.4 model for SE calculation (Hehre, 2003). The SEs for compound L1-L20 are presented in Table 2. L8 was observed to be better in term of SE, since high value of SE adds to the drug resistance (Lowrey et al., 1997).
More so, DM which is the product of the charge at both ends of the dipole of the molecule and the distance between the charge of molecules were calculated as reported in Table 3. Moreover, the rare features of every single molecule studied was believed to be a function of larger value of DM (Debenedetti, 2003). The natures of non-bonded interactions are very critical in the relationship between ligand and the enzyme in which DM affects. The values established for DM of molecular compounds falls between 3 and 5 kJ/mol (David and Howard, 2002). This implies that almost all DM values obtained in this study fall within the accepted range except the value for L3, L4, L5, L7, L15 and L19 which are just slightly greater than 5 by only <2 suggesting their fairly acceptability as shown in Table 3. Therefore, all compounds having their values within the expected range should be stout in non-bonded interactions with the enzymes. Also, every molecular descriptor that was calculated was reviewed, in order to check if any relationship with bioactivity (IC50) could be observed.
3.2. QSAR model for human gastric cell line (MGC-803) using multiple linear regressions (MLR)
The use of QSAR brings about the proficient way of procuring a whole set of values, and this has nothing to do with the need of performing costly laboratory experiments. Also, in drug design and medicinal chemistry, QSAR is one of the treasured implements and most essential areas in chemometric which are comprehensively used (Hansch et al., 1990; Manly et al., 2001; Pourbasheer et al., 2010; Prachayasittikul et al., 2014, 2015). In this study, thirteen calculated descriptors (Eq. 7) were employed in the development of QSAR model which were used against human gastric cell line. The developed QSAR model related the activities of the studied compounds to their biological activities and this showed the effectiveness of the equation generated via the model as shown in Eq. (7).fd7
Moreover, the calculated IC50 tailored the observed IC50 as presented in Figure 3 which means that the developed QSAR model reproduced the observed bioactivity of the studied compounds. Thus, the combination of selected calculated descriptors as shown in Eq. (7) were observed to act well as anti-gastric cancer activity of the studied compounds. Furthermore, the QSAR model equation using thirteen descriptors for evaluation MGC-803 as shown in Eq. (7) was employed to determine the values CV.R2 and. The statistical analyses of R2, CV.R2 and) were 0.950, 0.970 and 0.844 respectively revealing a very good fitness. Table 4 presents the observed and predicted IC50 for MGC 803. By comparison, the observed and predicted are well correlated with only little negligible differences in some cases. However, while some predicted values were not perfectly correlated with the observed values, did not in any way imply their inactivity against MGC-803, rather, indicated their less activities, ranging from moderate to good or fair activity.
| (7) |
Figure 3.
The correlation between observed and predicted IC50 for MGC-803.
Table 4.
Experimental and predicted IC50 for MGC-803.
| Mol | Predicted | Observed |
|---|---|---|
| L1 | 11.59 | 7.56 |
| L2 | 24.44 | 22.83 |
| L3 | 5.48 | 4.64 |
| L4 | 3.30 | 5.85 |
| L5 | 6.72 | 15.82 |
| L6 | 5.90 | 5.80 |
| L7 | 24.34 | 16.15 |
| L8 | 43.07 | 43.07 |
| L9 | 9.18 | 7.58 |
| L10 | 4.75 | 8.74 |
| L11 | 13.05 | 7.28 |
| L12 | 56.60 | 64.00 |
| L13 | 22.74 | 17.28 |
| L14 | 1.03 | 7.19 |
| L15 | 66.59 | 64.00 |
| L16 | 60.19 | 64.00 |
| L17 | 27.66 | 24.44 |
| L18 | 29.38 | 30.75 |
| L19 | 19.00 | 18.54 |
| L20 | 16.14 | 15.63 |
3.3. Docking studies for human gastric cancer cell line (MGC-803)
Docking studies aimed at observing the interactions between ligand and the receptor to ascertain the precise configuration of the studied molecules in the gouge of receptor. In addition, it usually employs to predict the affinity of the ligand towards the protein (Harsha, 2011; Gupta et al., 2018). Therefore, in this present study, twenty compounds denoted as L1-L20 were docked against human gastric cell line; MGC-803 (PDB ID: 5ACM). The docking simulations performed on the compounds L1 to L20 are presented in Figures 4, 5, and 6. Table 5 is showing the interactions among residues of drugs and 5ACM obtained from the protein data bank. This showed that all compounds exhibited related binding energy (Table 5). The binding energies of compounds L1-L20 ranges from −3.2 to −10.3 kcal/mol and the distance of H-bond between amino acid and drug ranges from 1.0 to 3.3 Ǻ (Table 5). Whereas, the corresponding inhibition efficiency (IC50) of the compounds L1-L20 ranges from 4.64 μM to 64 μM. However, based on the existing fact that the compound(s) with low IC50 within ≤10 μM gives a better activity against the selected receptor (Walsky et al., 2006; Abdel-Mohsen et al., 2010; Zheng et al., 2013; Ma et al., 2014). This suggests that compounds L1, L3, L4, L6, L9-L11, L14 were perfectly aligned within the ≤10 μM range of the literature value while L12, L15 and L16 showed the highest IC50 and less activity. This, however, did not suggest a non-activity of the compounds which their IC50 were slightly higher or far greater than the 10 μM range.
Figure 4.
Transparent view of docked complexes showing ligand-receptor interactions in the binding pocket for the eight selected compounds against MGC-803 using Pymol.
Figure 5.
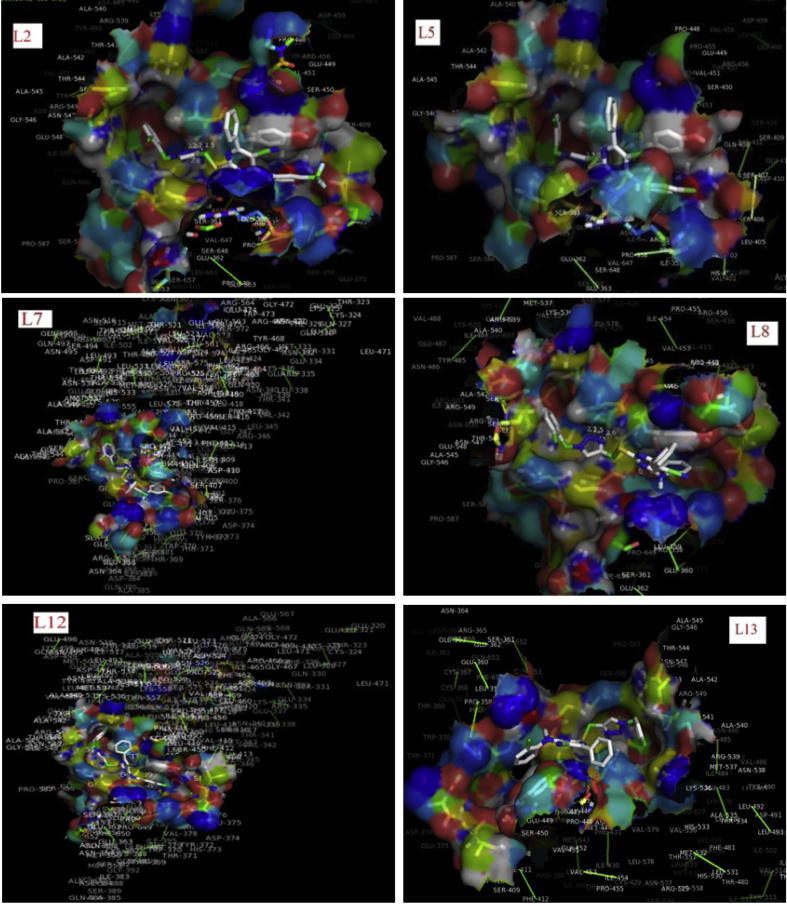
Transparent view of docked complexes showing ligand-receptor interactions in the binding pocket for the compounds L2, L5, L7, L8, L12-L13 against MGC-803 using Pymol.
Figure 6.
Transparent view of docked complexes showing ligand-receptor interactions in the binding pocket for the compounds L15-L20 against MGC-803 using Pymol.
Table 5.
Interaction among residues of drugs and 5ACM.
| Mol | Binding Energy (kcal/mol) | IC50 (μM) | H-Bond Between Amino Acid and Drug | Distance of H-Bond Between Amino Acid and Drug (Å) |
|---|---|---|---|---|
| L1 | -9.7 | 7.56 | THR-72, LIG:N | 2.7 |
| L2 | -9.9 | 22.83 | - | - |
| L3 | -10.0 | 4.64 | (i) GLU-52, LIG: N (ii) GLU-52, LIG:N (iii) GLU-52, LIG:N | ]2.8, 3.1, 2.8 |
| L4 | -10.2 | 5.85 | (i) THR-23, LIG:N (ii) SER-21 LIG:N | 1.9, 1.6 |
| L5 | -9.8 | 15.82 | - | - |
| L6 | -9.2 | 5.8 | - | - |
| L7 | -10.1 | 16.15 | (i) GLU-52, LIG:N (ii) GLU-52, LIG:N | 2.5, 2.8 |
| L8 | -10.2 | 43.07 | (i) TYR-38, LIG: H (ii) TYR-93, LIG:O (iii) TYR-34, LIG:O (iv) TYR-34, LIG:O (v) SER-91, LIG:O (vi) SER-91, LIG:N | 1.9, 2.7, 2.8, 3.1, 2.7, 2.2 |
| L9 | -9.6 | 7.58 | (i) SER-36, LIG:N (ii) TYR-38, LIG:H | 2.7, 2.1 |
| L10 | -9.7 | 8.74 | - | - |
| L11 | -10.3 | 7.28 | - | - |
| L12 | -9.0 | 64 | (i) TYR-34, LIG:N (ii) SER-91, LIG:N | 3.3, 3.0 |
| L13 | -8.7 | 17.28 | TYR-34, LIG: N | 2.2 |
| L14 | -10.0 | 7.19 | ASP-87 LIG:N | 3.0 |
| L15 | -9.4 | 64 | - | - |
| L16 | -9.3 | 64 | - | - |
| L17 | -10.3 | 24.44 | GLN—40, LIG: N | 3.5 |
| L18 | -8.0 | 30.75 | (i) THR-19, LIG: O (ii) SER-2.3, LIG: O (iii) SER-11, LIG:O | 1.5, 2.3, 2.3 |
| L19 | -3.2 | 18.54 | SER-91, LIG:N | 1.0 |
| L20 | -10.0 | 15.63 | (i) THR-72, LIG: N (ii) THR-19, LIG: N (ii) THR-19, LIG:N | 1.5, 2.0, 2.5 |
| 5-Fu | -4.2 | 7.69 |
Comparing the 8 compounds with better activity (Figure 4), the compound L3 with the lowest IC50 values (i. e. 4.64 μM) exhibited excellent activity against human gastric cancer cell line (MGC-803) under the study. This significant activity than the other compounds was ascribed to its chlorine at the ortho position (R1 = ortho-Cl). This further implies that compound L3 have more potency with about 14-fold higher than L12, L15 and L16 which both possess the same IC50 values of 64 μM. Whereas, in comparison with the standard (5-fluorouracil denoted as 5-Fu) a well-known anticancer drug (Zheng et al., 2013) which was used in the experimental work by Ma et al. (2014), L3 was about 2-fold potent than 5-Fu which have IC50 value of 7.69 μM (Table 5). This observation could be the influence of polarizability of chlorine atom on the ortho-position of L3. Although, majority of these compounds bearing a 1, 2, 3-triazole–pyrimidine hybrids exhibited moderate to good potency with IC50 values in the range of 4.64–64 μM. However, compounds L1-L11, L13, L17-L20 were more potent considering their IC50 values which are lower than L12, L15-L16, though they both possess IC50 values in the single- or double-digit micromolar (μM) ranges. Similar findings were reported by Walsky et al. (2006); Abdel-Mohsen et al., 2010; Zheng et al. (2013); Ma et al. (2014).
This study further examined the compounds with the best IC50 and binding energy based on the lowest IC50 of all the compounds presented in Table 5. Based on this, eight compounds including, L1, L3, L4, L6, L9-L11 and L14 were therefore selected as the best compounds (Figure 4) with the lowest IC50 in the single-digit micromolar (μM) range and their corresponding binding energies as presented Table 5. These were further compared by docking the standard compound (5-Fu) which is one of the anticancer drugs already in the market, in order to determine the potency of the eight (8) selected compounds. Here, it was observed that the compounds gave excellent inhibition properties against MGC-803 than the reference compound 5-Fu, due to their higher binding free energies than 5-Fu (with binding energy of -4.2 kcal/mol (Adejoro et al., 2016).
However, the correlation between binding energy and IC50 of the eight selected compounds is shown in Figure 7 which is in the order of increasing IC50 and decreasing binding energy. According to Table 5, comparing the binding energies with the standard, the best selected compounds were in the order of L3 > L6 > L14 > L11 > L1 >L9 > 10 > 5-Fu. The analysis of the docked complex (as shown in Figure 4) revealed the relationships between binding energy and IC50 (presented in Figure 7), implying the conformation of the ligand in the active gouge of the receptor. The binding interactions that occurred in-between the ligand and the receptor were displayed in Table 5. This suggests hydrogen bond and electrostatic relationship that further explained the hydrophobic interactions (as a result of non-polar residue interaction). The docked complexes of compounds L3 revealed a minimum free binding energy in terms of negativity as displayed in Table 5 and Figures 4 and 5 implying the binding pores and interactions of the complex and ligand. This result was in good agreement with the findings of Adejoro et al. (2016). The best conformation in each ligand-receptor complex with minimum free energy of interaction and inhibition efficiency was taken as shown in Table 5 and Figure 4. This was assumed to be correlated with IC50 and binding energy in each docking.
Figure 7.
Correlation between binding energy and IC50of the selected compounds against MGC-803.
4. Conclusion
The part played by triazole-pyrimidine hybrid in the clinical world as anti-cancer cannot be over emphasized. Quantum chemical method, quantitative structural activities relationship model and molecular docking approaches were used in the correlation of the activities of selected triazole-pyrimidine hybrid to their activities. In the QSAR evaluation, the statistical values, multiple correlation coefficients (r), standard error(s), cross validation R2 and standard error of prediction were used. Therefore, the best model derived from the analysis was employed in the prediction of the IC50 of the considered triazole-pyrimidine derivatives. Docking studies were performed to assess the effectiveness of triazole-pyrimidine hybrid attached to active residues of MGC-803.
The results obtained showed that quantum chemical calculations via DFT calculations of molecular parameters in building QSAR model linked the molecular parameters of the studied compounds to their cytotoxicity. The developed QSAR models exposed the responsibility taken by several calculated descriptors to link the electronic properties of the molecules to their bioactivities and the QSAR model reproduced the observed bioactivities of these compounds against MGC-803. Finally, the molecular docking studies help to know that hydrogen bonded with the amino acid residues in the binding site, and also, the essentiality of the ligand conformations as the significant features for ligand-receptor binding.
Declarations
Author contribution statement
Rhoda Oyeladun Oyewole: Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Abel Kolawole Oyebamiji: Contributed reagents, materials, analysis tools or data.
Banjo Semire: Conceived and designed the experiments; Contributed reagents, materials, analysis tools or data.
Funding statement
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Competing interest statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Acknowledgements
The authors acknowledge the Department of Pure and Applied Chemistry, Faculty of Pure and Applied Sciences, LAUTECH Ogbomoso, Oyo State, Nigeria for the platform to carry out this research and also appreciate Dr. Adegoke Adesina Kayode for the assistance in data interpretations and presentation.
References
- Abdel-Mohsen H.T., Ragab F.A.F., Ramla M.M., El-Diwani H.I. Novel benzimidazolee pyrimidine conjugates as potent antitumor agents. Eur. J. Med. Chem. 2010;45:2336–2344. doi: 10.1016/j.ejmech.2010.02.011. [DOI] [PubMed] [Google Scholar]
- Adedirin O., Uzairu A., Shallangwa G.A., Abechi S.E. QSAR studies on derivatives of quinazoline-4(3H)-ones with anticonvulsant activities. J. Eng. Exact Sci JCEC. 2018;4(2) [Google Scholar]
- Adejoro I.A., Waheed S.O., Adeboye O.O. Molecular docking studies of Lonchocarpus cyanescens triterpenoids as inhibitors for malaria. J. Phys. Chem. Biophys. 2016;6(213) [Google Scholar]
- Aher N.G., Pore V.S., Mishra N.N., Kumar A., Shukla P.K., Sharma A., Bhat M.K. Synthesis and antifungal activity of 1, 2, 3-triazole containing fluconazole analogues. Bioorg. Med. Chem. Lett. 2009;19:759–763. doi: 10.1016/j.bmcl.2008.12.026. [DOI] [PubMed] [Google Scholar]
- Ahmed M., Razaq H., Faisal M., Siyal A.N., Haider A. Metal-free and azide-free synthesis of 1,2,3-triazoles derivatives. Synth. Commun. 2017;47(13):1193–1200. [Google Scholar]
- Alam S., Khan F. 3D-QSAR studies on Maslinic acid analogs for Anticancer activity against Breast Cancer cell line MCF-7. Sci. Rep. 2017;7:6019. doi: 10.1038/s41598-017-06131-0. 1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alejandra H., Aldo Y., Victor A., Héctor V., Claudia M. Protein Engineering-Technology and Application. 2013. Protein-Protein and Protein-Ligand Docking Chapter 3; pp. 63–81. [Google Scholar]
- Baldi A. Computational approaches for drug design and discovery: an overview. Syst. Rev. Pharmaceut. 2010 [Google Scholar]
- Becke A. Density functional thermochemistry. III. The role of exact exchange. J. Phys. Chem. 1993;98:5648–5652. [Google Scholar]
- Bello A.S., Uzairu A., Shallangwa G.A., Ibrahim A. In-silico studies of some indole derivatives as anti-hepatitis drug. J. Eng. Exact Sci JCEC. 2018;4(2) [Google Scholar]
- Bouachrine M., Hamidi M., Bouzzine S.M., Taoufik H. Theoretical study on thestructure and electronic properties of new materials based on thiophene and oxadiazole. J. Chem. Res. 2009;10:29–37. [Google Scholar]
- Chandannos E., Lagergren J. Oestrogen and enigmatic male predominance of gastric cancer. Eur. J. Canc. 2008;44(16):2397. doi: 10.1016/j.ejca.2008.07.031. Chapter 1. 1. World Cancer Report (2014). World Health Organization. [DOI] [PubMed] [Google Scholar]
- Chill J.H., Quadt S.R., Levy R., Schreiber G., Anglister J. The human type I interferon receptor: NMR structure reveals the molecular basis of ligand binding. Structure. 2003;11(7):791–802. doi: 10.1016/s0969-2126(03)00120-5. [DOI] [PubMed] [Google Scholar]
- Choi Y., Li L., Grill S., Gullen E., Lee C.S., Gumina G., Tsujii E., Cheng Y.C., Chu C.K. Structure-activity relationships of (E)-5-(2-bromovinyl)uracil and related pyrimidine nucleosides as antiviral agents for Herpes viruses. J. Med. Chem. 2000;43:2538–2546. doi: 10.1021/jm990543n. [DOI] [PubMed] [Google Scholar]
- Conejo-García A., García-Rubino M.E., Marchal J.A., Núnez M.C., Ramírez A., Cimino S., García M.A., Aranega A., Gallo M.A., Campos J.M. Synthesis and anticancer activity of (RS)-9-(2, 3-dihydro-1, 4-benzoxaheteroin-2-ylmethyl)- 9H-purines. Eur. J. Med. Chem. 2011;46:3795–3801. doi: 10.1016/j.ejmech.2011.05.046. [DOI] [PubMed] [Google Scholar]
- Cruz-Monteagudo M., Schürer S., Tejera E., Pérez-Castillo Y., Medina-Franco J.L., Sánchez-Rodríguez A. Systemic QSAR and phenotypic virtual screening: chasing butterflies in drug discovery. Drug Discov. Today. 2017;22:994–1007. doi: 10.1016/j.drudis.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curioni A., Andreoni W., Treusch R., Himsel F.J., Haskal E., Seidler P., Terminello L.J. Atom-resolved electronic spectra for Alq3 from theory and experiment. Appl. Phys. Lett. 1998;72(13):1575–1577. [Google Scholar]
- Curtin N.J., Barlow H.C., Bowman K.J., Calvert A.H., Davison R., Golding B.T., Huang B., Loughlin P.J., Newell D.R., Smith P.G., Griffin R.J. Resistance-modifying agents. 11.1 pyrimido[5, 4-d]pyrimidine modulators of antitumor drug activity. Synthesis and structureeactivity relationships for nucleoside transport inhibition and binding to a1-acid glycoprotein. J. Med. Chem. 2004;47:4905–4922. doi: 10.1021/jm040772w. [DOI] [PubMed] [Google Scholar]
- David F.L.L., Howard B.B. Molecular binding interactions: their estimation and rationalization in QSARs in terms of theoretically derived parameters. Sci. World J. 2002;2:1776–1802. doi: 10.1100/tsw.2002.343. [DOI] [PubMed] [Google Scholar]
- Debenedetti P. Condensed matter. J. Phys. 2003;(15):1669–1670. [Google Scholar]
- Demaray J.A., Thuener J.E., Dawson M.N., Sucheck S.J. Synthesis of triazoleoxazolidinones via a one-pot reaction and evaluation of their antimicrobial activity. Bioorg. Med. Chem. Lett. 2008;18:4868–4871. doi: 10.1016/j.bmcl.2008.07.087. [DOI] [PubMed] [Google Scholar]
- Desai B., Sureja D., Naliapara Y., Shah A., Saxena A. Synthesis and QSAR studies of 4-substituted phenyl-2, 6-dimethyl-3, 5-bis-N-(substituted phenyl)carbamoyl-1, 4-dihydropyridines as potential antitubercular agents. Bioorg. Med. Chem. Lett. 2001;9(8):1993–1998. doi: 10.1016/s0968-0896(01)00141-9. [DOI] [PubMed] [Google Scholar]
- De-Simone R., Chini M.G., Bruno I., Riccio R., Mueller D., Werz O., Bifulco G. Structure-based discovery of inhibitors of microsomal prostaglandin E2 synthase-1, 5-lipoxygenase and 5-lipoxygenase-activating protein: promising hits for the development of new anti-inflammatory agents. J. Med. Chem. 2011;54:1565–1575. doi: 10.1021/jm101238d. [DOI] [PubMed] [Google Scholar]
- Domingo E. Quasispecies theory in virology. J. Virol. 2002;76:463–465. doi: 10.1128/JVI.76.1.463-465.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Shahawi M.M., El-Ziaty A.K. Enaminonitrile as building block in heterocyclic synthesis: synthesis of novel 4H-Furo[2,3-d][1,3]oxazin-4-one and furo[2,3-d]pyrimidin-4(3H)-one derivatives. J. Chem. 2017:1–6. [Google Scholar]
- Elshakre M.E., Noamaan M.A., Moustafa H., But H. Density functional theory, chemical reactivity, pharmacological potential and molecular docking of dihydrothiouracil-indenopyridopyrimidines with human-DNA topoisomerase II. Int. J. Mol. Sci. 2020;21:1253. doi: 10.3390/ijms21041253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eswar N., Marti-Renom M., Webb B., Madhusudhan M., Eramian D., Shen M., Sali A. Comparative protein structure Modeling using MODELLER. Curr. Protocol. Bioinformat. 2006;5(6):1–30. doi: 10.1002/0471250953.bi0506s15. Supplement, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fumagalli M., Lecca D., Abbracchio M.P., Ceruti S. Pathophysiological role of purines and pyrimidines in neurodevelopment: unveiling new pharmacological approaches to congenital brain diseases. Front. Pharmacol. 2017;8 doi: 10.3389/fphar.2017.00941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giffin M.J., Heaslet H., Brik A., Lin Y.C., Cauvi G., Wong C.H., McRee D.E., Elder J.H., Stout C.D., Torbett B.E. A copper(I)-catalyzed 1, 2, 3-triazole azideealkyne click compound is a potent inhibitor of a multidrug-resistant HIV-1 protease variant. J. Med. Chem. 2008;51:6263–6270. doi: 10.1021/jm800149m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta M., Sharma R., Kumar A. Docking techniques in pharmacology: how much promising? Comput. Biol. Chem. 2018;76:210–217. doi: 10.1016/j.compbiolchem.2018.06.005. [DOI] [PubMed] [Google Scholar]
- Hansch C., Sinclair J.F., Sinclair P.R. Induction of cytochromeP450 by barbiturates in chick embryo hepatocytes: aquantitative structure–activity analysis. Quant. Struct.-Activ. Relat. 1990;9:223–226. [Google Scholar]
- Harsha P. In silico drug designing and docking analysis for hypertension using Nifedipine as lead molecule. Int. J. Pharmaceut. Res. Dev. 2011;3(3):104–108. [Google Scholar]
- Hay P.J. Theoretical studies of the ground and excited electronic states in cyclometalated phenylpyridine Ir(III) complexes using density functional theory. J. Phys. Chem. 2002;106(8):1634–1641. [Google Scholar]
- Hehre W. 2003. A Guide to Molecular Mechanics and Quantum Chemical Calculations: Wave function, INC, Irvine. [Google Scholar]
- Ismail S.Y., Uzairu A.B., Sagag A. Insilico studies of sulfur-containing shikonin oxime derivatives as inhibitors of MGC 803 gastric cancer cell line. J. Eng. Exact Sci JCEC. 2019;5(1) [Google Scholar]
- Jacquemin D., Perpète E., Ciofini I., Adamo C. Accurate simulation of optical properties in dyes. Acc. Chem. Res. 2008;42:326–334. doi: 10.1021/ar800163d. [DOI] [PubMed] [Google Scholar]
- Jin C., Liang Y.J., He H., Fu L. Synthesis and antitumor activity of ureas containing pyrimidinyl group. Eur. J. Med. Chem. 2011;46:429–432. doi: 10.1016/j.ejmech.2010.11.026. [DOI] [PubMed] [Google Scholar]
- Khaled A., Petri R., Sampo M., Olavi P. Metabolism of α-Thujone in human hepatic preparations in vitro. Xenobiotica. 2011;41:101–111. doi: 10.3109/00498254.2010.528066. [DOI] [PubMed] [Google Scholar]
- Koopmans T. Ordering of wave functions and eigenvalues to the individual electrons of an atom. Physica. 1934;1:104–113. [Google Scholar]
- Kumari S., Chowdhury J., Sikka M., Verma P., Jha P., Mishra A.K.…Chopra M. Identification of potent cholecystokinin-B receptor antagonists: synthesis, molecular modeling and anti-cancer activity against pancreatic cancer cells. Med. Chem. Commun. 2017;8(7):1561–1574. doi: 10.1039/c7md00171a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebeau A., Abrioux C., Bénimèlis D., Benfodda Z., Meffre P. Synthesis of 1,4-disubstituted 1,2,3-triazole derivatives using click chemistry and their Src kinase activities. Med. Chem. 2017;13:40–48. doi: 10.2174/1573406412666160404125718. [DOI] [PubMed] [Google Scholar]
- Lee H.S., Kim W.H., Kwak Y. Molecular testing for gastrointestinal cancer. J. Pathol. Transl. Med. 2017;51:103–121. 3. doi: 10.4132/jptm.2017.01.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Liu H.Y., Guo S.H. Detection of microsatellite instability in gastric cancer and dysplasia tissues. Int. J. Clin. Exp. Med. 2015;8:21442–21447. [PMC free article] [PubMed] [Google Scholar]
- Liu H.Y., Zou X. Electrostatics of ligand binding: parametrization of the generalized Born model and comparison with the Poisson-Boltzmann approach. J. Phys. Chem. B. 2006;110:9304–9313. doi: 10.1021/jp060334w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Cara L.C., Conejo-García A., Marchal J.A., Macchione G., Cruz-Lopez O., Boulaiz H., García M.A., Rodríguez-Serrano F., Ramírez A., Cativiela C., Jimenez A.I., García-Ruiz J.M., Choquesillo-Lazarte D., Aranega A., Campos J.M. New (RS)-benzoxazepin-purines with antitumour activity: the chiral switch from (RS)-2, 6-dichloro-9-[1-(p-nitrobenzenesulfonyl)-1, 2, 3, 5-tetrahydro- 4, 1-benzoxazepin-3-yl]-9H-purine. Eur. J. Med. Chem. 2011;46:249–258. doi: 10.1016/j.ejmech.2010.11.011. [DOI] [PubMed] [Google Scholar]
- Lowrey A.H., Famini G.R., Loumbev V., Wilson L.Y., Tosk J.M. Modeling drug-melani interaction with theoretical linear solvation energy relationships. Pig. Cell Res. 1997;10(5):251–256. doi: 10.1111/j.1600-0749.1997.tb00684.x. [DOI] [PubMed] [Google Scholar]
- Ma L.Y., Pang L.P., Wang B., Zhang M., Hu B., Deng-Qi X., Kun-Peng S., Zhang B.L., Liu Y., Zhang E., Liu H.M. Design and synthesis of novel 1, 2, 3-triazole-pyrimidine hybrids as potential anticancer agents. Eur. J. Med. Chem. 2014;86:368–380. doi: 10.1016/j.ejmech.2014.08.010. [DOI] [PubMed] [Google Scholar]
- Ma X.H., Shi Z., Tan C., Jiang Y., Go M.L., Low B.C. In-silico approaches to multi-target drug discovery: computer aided multi-target drug design, multi-target virtual screening. Pharm. Res. (N. Y.) 2010;27:739–749. doi: 10.1007/s11095-010-0065-2. [DOI] [PubMed] [Google Scholar]
- Manly C.J., Louise M.S., Hammer J.D. The impact of informatics and computational chemistry on synthesis and screening. Drug Discov. Today. 2001;6:1101–1110. doi: 10.1016/s1359-6446(01)01990-0. [DOI] [PubMed] [Google Scholar]
- Marquina S., Santiago M.M., Carranza J.N.S., Mojica M.A., Maya L.G., Hernández R.S.R., Alvarez L. Design, synthesis and QSAR study of 2′-hydroxy-4′-alkoxy chalcone derivatives that exert cytotoxic activity by the mitochondrial apoptotic pathway. Bioorg. Med. Chem. 2019;27(1):43–54. doi: 10.1016/j.bmc.2018.10.045. [DOI] [PubMed] [Google Scholar]
- Martin D.S., Bertino J.R., Koutcher J.A. ATP depletion þ-pyrimidine depletion can markedly enhance cancer therapy: fresh insight for a new approach. Canc. Res. 2000;60:6776–6783. [PubMed] [Google Scholar]
- McGuigan C., Barucki H., Blewett S., Carangio A., Erichsen J.T., Andrei G., Snoeck R., De-Clercq E., Balzarini J. Highly potent and selective inhibition of Varicella-Zoster Virus by bicyclic furopyrimidine nucleosides bearing an aryl side chain. J. Med. Chem. 2000;43:4993–4997. doi: 10.1021/jm000210m. [DOI] [PubMed] [Google Scholar]
- Meanwell N.A. Synopsis of some recent tacticalapplication of biostereses in drug design. J. Med. Chem. 2011;54(8):2529–2591. doi: 10.1021/jm1013693. [DOI] [PubMed] [Google Scholar]
- Melge A.R., Kumar L.G., Pavithran K., Nair S.V., Manzoor K., Gopi Mohan C. Predictive models for designing potent tyrosine kinase inhibitors in chronic myeloid leukemia for understanding its molecular mechanism of resistance by molecular docking and dynamics simulations. J. Biomol. Struct. Dyn. 2019;37:4747–4766. doi: 10.1080/07391102.2018.1559765. [DOI] [PubMed] [Google Scholar]
- Morales F., Ramírez A., Conejo-García A., Morata C., Marchal J.A., Campos J.M. Anti-proliferative activity of 2, 6-dichloro-9- or 7-(ethoxycarbonylmethyl)- 9H- or 7H-purines against several human solid tumour cell lines. Eur. J. Med. Chem. 2014;76:118–124. doi: 10.1016/j.ejmech.2014.02.012. [DOI] [PubMed] [Google Scholar]
- Morris G.M., Huey R., Lindstrom W., Sanner M.F., Belew R.K., Goodsell D.S., Olson A.J. Autodock4 and AutoDockTools4: automated docking with selective receptor flexiblity. J. Comput. Chem. 2009;16:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustafa M., Menon J., Muniandy R.K., Illzam E.M., Nornazirah A., Nang M.K., Fairrul K., Sharifa A.M. Gastric cancer: risk factors, diagnosis and management. IOSR J. Dent. Med. Sci. 2017;16(3):69–74. [Google Scholar]
- Oyebamiji A.K., Semire B. DFT-QSAR model and docking studies of antiliver Cancer (Hepg-2). Activities of 1, 4-Diydropyridine based derivatives. Cancer Biol. 2016;6(2):69–78. [Google Scholar]
- Oyebamiji Kolawole Abel, Semire Banjo. Studies of 1, 4-Dihydropyridine derivatives for anti-breast cancer (MCF-7) activities: combinations of DFT-QSAR and docking methods. New York Sci. J. 2016;9(6):58–66. [Google Scholar]
- Palanki M.S.S., Erdman P.E., Gayo-Fung L.M., Shevlin G.I., Sullivan R.W., Goldman M.E., Ransone L.J., Bennett B.L., Manning A.M., Suto M.J. Inhibitors of NF-kB and AP-1 gene expression: SAR studies on the pyrimidine portion of 2- chloro-4-trifluoromethylpyrimidine-5-[N-(30, 50-bis(trifluoromethyl)phenyl). carboxamide] J. Med. Chem. 2000;43:3995–4004. doi: 10.1021/jm0001626. [DOI] [PubMed] [Google Scholar]
- Parr R.G., Szentpaly L., Liu S. Electrophilicity index. J. Am. Chem. Soc. 1999;121:1922–1924. [Google Scholar]
- Parveen H., Hayat F., Salahuddin A., Azam A. Synthesis, characterization and biological evaluation of novel 6-ferrocenyl-4-aryl-2-substituted pyrimidine derivatives. Eur. J. Med. Chem. 2010;45:3497–3503. doi: 10.1016/j.ejmech.2010.04.023. [DOI] [PubMed] [Google Scholar]
- Patpi S.R., Pulipati L., Yogeeswari P., Sriram D., Jain N., Sridhar B., Murthy R., Anjana Devi T., Kalivendi S.V., Kantevari S. Design, synthesis, and structureeactivity correlations of novel dibenzo[b, d]furan, dibenzo[b, d]thiophene, and N-methylcarbazole clubbed 1, 2, 3-triazoles as potent inhibitors of mycobacterium tuberculosis. J. Med. Chem. 2012;55:3911–3922. doi: 10.1021/jm300125e. [DOI] [PubMed] [Google Scholar]
- Perez-Castillo Y., Sanchez-Rodrıguez A., Tejera E., Cruz-Monteagudo M., Borges F., Cordeiro M.N.D.S. A desirability-based multi objective approach for the virtual screening discovery of broad-spectrum anti-gastric cancer agents. PloS One. 2018;13(2) doi: 10.1371/journal.pone.0192176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogorzelska A., Sławinski J., Brozewicz K., Ulenberg S., Baczek T. Novel 3-Amino-6-chloro-7-(azol-2 or 5-yl)-1,1-dioxo1,4,2-benzodithiazine derivatives with anticancer activity: synthesis and QSAR study. Molecules. 2015;20:21960–21970. doi: 10.3390/molecules201219821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontikis R., Dolle V., Guillaumel J., Dechaux E., Note R., Nguyen C.H., Legraverend M., Bisagni E., Aubertin A.M., Grierson D.S., Monneret C. Synthesis and evaluation of “AZT-HEPT”, “AZT-pyridinone”, and “ddC-HEPT” conjugates as inhibitors of HIV reverse transcriptase1. J. Med. Chem. 2000;43:1927–1939. doi: 10.1021/jm991125l. [DOI] [PubMed] [Google Scholar]
- Pourbasheer E., Riahi S., Ganjali M.R., Norouzi P. Quantitative structure–activity relationship (QSAR). study of interleukin-1 receptor associated kinase 4 (IRAK-4). inhibitor activity by thegenetic algorithm and multiple linear regression (GA–MLR)method. J. Enzym. Inhib. Med. Chem. 2010;25:844–853. doi: 10.3109/14756361003757893. [DOI] [PubMed] [Google Scholar]
- Prachayasittikul V., Pingaew R., Worachartcheewan A., Nantasenamat C., Prachayasittikul S., Ruchirawat S., Prachayasittikul V. Synthesis, anticancer activity and QSAR study of 1,4-naphthoquinone derivatives. Eur. J. Med. Chem. 2014;84:247–263. doi: 10.1016/j.ejmech.2014.07.024. [DOI] [PubMed] [Google Scholar]
- Prachayasittikul V., Pingew R., Anuwongcharoen N., Worachartcheewan A., Nantasenamat C., Prachayasittikul S.…Prachayasittikul V. Discovery of novel 1,2,3-triazole derivatives as anticancer agents using QSAR and in silico structural modification. Springer Plus. 2015;4(1) doi: 10.1186/s40064-015-1352-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puzyn T., Leszczyński J., Cronin M. Vol. 8. 2010. Recent Advances in QSAR Studies: Methods and Applications. (Challenges and Advances in Computational Chemistry and Physics). [Google Scholar]
- Rob N. 2014. Tutorial 6 : Linear Regression; pp. 1–14. [Google Scholar]
- Rogerio K.R., Carvalho L.J.M., Domingues L.H.P., Neves B.J., Moreira Filho J.T., Castro R.N., Graebin C.S. Synthesis and molecular modelling studies of pyrimidinones and pyrrolo[3,4-d]-pyrimidinodiones as new antiplasmodial compounds. Memórias Do Instituto Oswaldo Cruz. 2018;113(8) doi: 10.1590/0074-02760170452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romad S., Miranda L. Strategies towards the synthesis of N2-substituted 1,2,3-triazoles. An. Acad. Bras. Cienc. 2019;91 doi: 10.1590/0001-3765201820180751. [DOI] [PubMed] [Google Scholar]
- Rostom S.A.F., Ashour H.M.A., Abd-ElRazik H.A. Synthesis and biological evaluation of some novel polysubstituted pyrimidine derivatives as potential antimicrobial and anticancer agents. Archive Pharm. 2009;342:299–310. doi: 10.1002/ardp.200800223. [DOI] [PubMed] [Google Scholar]
- Salah B.H.K., Das S., Ruiz N., Andreu V., Martinez J., Wenger E. How are 1,2,3-triazoles accommodated in helical secondary structures? Org. Biomol. Chem. 2018;16:3576–3583. doi: 10.1039/c8ob00686e. [DOI] [PubMed] [Google Scholar]
- Schröder D.C., Kracker O., Fröhr T., Góra J., Jewginski M., Nieß A., Antes I., Latajka R., Marion A., Sewald N. 1,4-Disubstituted 1H-1,2,3-Triazole containing peptidotriazolamers: a new class of peptidomimetics with interesting foldamer properties. Front. Chem. 2019;7:155. doi: 10.3389/fchem.2019.00155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semire B., Oyebamiji A., Ahmad M. Theoretical study on structure and electronic properties of 2, 5-bis [4-N, N-diethylaminostyryl] thiophene and its furan and pyrrole derivatives using density functional theory (DFT) Pak. J. Chem. 2012;2(4):166–173. [Google Scholar]
- Sharma V., Chitranshi N., Agarwal A.K. Significance and biological importance of pyrimidine in the microbial world. Int. J. Med. Chem. 2014:1–31. doi: 10.1155/2014/202784. 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitarz R., Skierucha M., Mielko J., Offerhaus J., Maciejewski R., Polkowski W. Gastric cancer: epidemiology, prevention, classification, and treatment. Canc. Manag. Res. 2018;10:239–248. doi: 10.2147/CMAR.S149619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Śledź P., Caflisch A. Protein structure-based drug design: from docking to molecular dynamics. Curr. Opin. Struct. Biol. 2018;48:93–102. doi: 10.1016/j.sbi.2017.10.010. [DOI] [PubMed] [Google Scholar]
- Song Z., Wu Y., Yang J., Yang D., Fang X. Progress in the treatment of advanced gastric cancer. Tumor Biol. 2017:1–7. doi: 10.1177/1010428317714626. [DOI] [PubMed] [Google Scholar]
- Sun Z., Wang Q., Yu X. Risk factors associated with splenic hilar lymph node metastasis in patients with advanced gastric cancer in northwest China. Int. J. Clin. Exp. Med. 2015;8:21358–21364. [PMC free article] [PubMed] [Google Scholar]
- Trott O., Olson A. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization and multithreading. J. Comput. Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasaikar S., Bhatia P., Bhatia P.G., Chu Yaiw K. Complementary approaches to existing target based drug discovery for identifying novel drug targets. Biomedicines. 2016;4 doi: 10.3390/biomedicines4040027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsky R.L., Astuccio A.V., Obach R.S. Evaluation of 227 drugs for in vitro inhibition of cytochrome P450 2B6. J. Clin. Pharmacol. 2006;46 doi: 10.1177/0091270006293753. [DOI] [PubMed] [Google Scholar]
- Wang X.L., Wan K., Zhou C.H. Synthesis of novel sulfanilamide-derived 1, 2, 3-triazoles and their evaluation for antibacterial and antifungal activities. Eur. J. Med. Chem. 2010;45:4631–4639. doi: 10.1016/j.ejmech.2010.07.031. [DOI] [PubMed] [Google Scholar]
- White N.J. The treatment of malaria. N. Engl. J. Med. 1996;335(11):800–806. doi: 10.1056/NEJM199609123351107. http://www.scopus.com/scopus/inward/record.url?eid=2-s2.0-0029792910&partnerID=K84CvKBR&rel=3.0.0&md5=0134e84ce9f61c7db4e1b02510062ebd [DOI] [PubMed] [Google Scholar]
- World Health Organization . 2014. Chapter 1.1. World Cancer Report 2014. ISBN 9283204298. [Google Scholar]
- Wu H., Wang W., Tong S. Nucleostemin regulates proliferation and migration of gastric cancer and correlates with its malignancy. Int. J. Clin. Exp. Med. 2015;8:17634–17643. [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Yang L., Feng J., Ren A. Theoretical studies on the electronic and optical properties of two thiophene–fluorene based π-conjugated copolymers. Polymer. 2005;46:10970–10982. [Google Scholar]
- Young D. John Wiley & Sons; 2001. Introduction to Computational Chemistry, A Practical Guide for Applying Techniques to Real World Problems; p. 408. [Google Scholar]
- Yu S., Wang N., Chai X., Wang B., Cui H., Zhao Q., Zou Y., Sun Q., Meng Q., Wu Q. Synthesis and antifungal activity of the novel triazole derivatives containing 1, 2, 3-triazole fragment. Arch Pharm. Res. (Seoul) 2013;36:1215–1222. doi: 10.1007/s12272-013-0063-0. [DOI] [PubMed] [Google Scholar]
- Zaki M., Oukhrib A., El-Hakmaoui A., Hiebel M.A., Raboin S.B., Akssira M. Synthesis of novel 1,2,3-triazole-substituted tomentosins. Z. Naturforsch. B Chem. Sci. 2018;74(3):273–281. [Google Scholar]
- Zheng Y.C., Duan Y.C., Ma J.L., Xu R.M., Zi X., Lv W.L., Liu H.M. Triazole–dithiocarbamate based selective lysine specific demethylase 1 (LSD1) inactivators inhibit gastric cancer cell growth, invasion, and migration. J. Med. Chem. 2013;56(21):8543–8560. doi: 10.1021/jm401002r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z., Navangul H.V. Absolute hardness and aromatisity: MNDO study of benzenoid hydrocarbons. J. Phys. Org. Chem. 1990;3:784–788. [Google Scholar]
- Zhu S.L., Wu Y., Liu C.J., Wei C.Y., Tao J.C., Liu H.M. Design and stereoselective synthesis of novel isosteviol-fused pyrazolines and pyrazoles as potential anticancer agents. Eur. J. Med. Chem. 2013;65:70–82. doi: 10.1016/j.ejmech.2013.04.044. [DOI] [PubMed] [Google Scholar]