Abstract
Purpose
Mediator is a multiprotein complex that allows the transfer of genetic information from DNA binding proteins to the RNA polymerase II during transcription initiation. MED12L is a subunit of the kinase module, which is one of the four sub-complexes of the mediator complex. Other subunits of the kinase module have been already implicated in intellectual disability, namely MED12, MED13L, MED13 and CDK19.
Methods
We describe an international cohort of seven affected individuals harboring variants involving MED12L identified by array CGH, exome or genome sequencing.
Results
All affected individuals presented with intellectual disability and/or developmental delay, including speech impairment. Other features included autism spectrum disorder, aggressive behavior, corpus callosum abnormality and mild facial morphological features. Three individuals had a MED12L deletion or duplication. The other four individuals harbored single nucleotide variants (one nonsense, one frameshift and two splicing variants). Functional analysis confirmed a moderate and significant alteration of RNA synthesis in two individuals.
Conclusion
Overall data suggest that MED12L haploinsufficiency is responsible for intellectual disability and transcriptional defect. Our findings confirm that the integrity of this kinase module is a critical factor for neurological development.
Keywords: MED12L, intellectual disability, mediator complex, transcriptional defect, corpus callosum
INTRODUCTION
Mediator is a key regulator of gene expression involved in cell growth, homeostasis, development and differentiation 1. Mediator is a large multiprotein complex composed of four different modules (Kinase, Head, Middle and Tail), which conveys essential information from proteins bound at DNA response elements to the basal transcription machinery located around the transcription initiation site 2,3. Dysfunction of the transcription machinery components, including Mediator, has been shown to elicit a range of effects on cell states giving rise to diverse disorders including developmental delay and intellectual disability. Variants in Mediator subunits are associated with a wide range of genetic disorders, most of them exhibiting neurological disabilities 4. Genetic and biochemical studies have established the Mediator Kinase module as a major ingress of developmental and oncogenic signaling through the three other modules constituting the core component, and much of its function derives from its resident CDK8 kinase activity likely regulated by its association with MED13, MED12 and Cyclin C (CycC) subunits. Recent studies have shown that the kinase module can also encompass CDK19, MED12L, and MED13L, which are paralogs of CDK8, MED12, and MED13 respectively 5. Little is known about their biological roles, but each of these proteins appears to assemble in a mutually exclusive fashion with its paralog.
Germline variants of MED12 have already been found in several genetic disorders associated with X-linked intellectual disability, such as Opitz-Kaveggia syndrome also named FG syndrome 6–8, Lujan syndrome (p.N1007S) 9 and Ohdo syndrome 10,11. All of these MED12-related disorders exhibit defects in gene expression 12. Variants in CDK19, MED13 and MED13L have also been associated with neurodevelopmental disorders 13–15, likely due to defects in gene transcription.
Here, we report a series of individuals sharing variants involving MED12L (mediator complex subunit 12 like) [OMIM 611318] recruited through an international collaboration and identified by array Comparative Genomic Hybridization (aCGH), Exome or Genome Sequencing. Our attention focused on the effect of MED12L on gene expression, studying transcription machinery of two individuals’ fibroblasts. Whereas CDK19, MED12L, and MED13L are now all linked to neuronal and developmental disorders, their basic biological importance relative to CDK8, MED12, or MED13 remains unclear.
MATERIALS AND METHODS
Individual recruitment
The compilation of this case series resulted from an international collaborative effort among Centre Hospitalier Universitaire (CHU) Nantes, Strasbourg University, CHU Caen, Baylor Genetics Laboratories (BG), GeneDX, HudsonAlpha Institute for Biotechnology and University of Oklahoma School of Medicine. It was also partly facilitated by the web-based tools GeneMatcher and DECIPHER 16,17. All participants were clinically assessed by at least one clinical geneticist from one of the participating centers. The study was approved by the CHU de Nantes-ethics committee (number CCTIRS: 14.556). Consents for the publication of photographs (Figure 1) were obtained for individuals 1, 2 and 5.
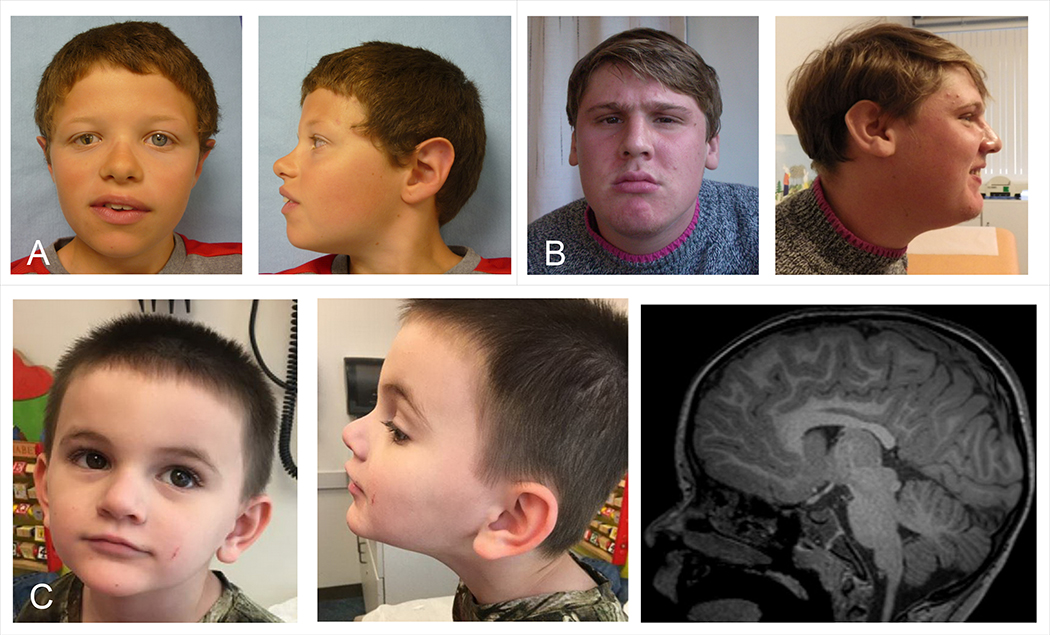
Figure 1: Photographs and brain MRI of individuals with variants in MED12L.
A, Individual 1: unilateral ptosis with iris coloboma, hypertelorism, sparse eyebrows, downslanted palpebral fissures, bulbous nasal tip. B, Individual 2: prominent nasal bridge, short philtrum, everted lower lip, small mouth. C, Individual 5: deep-set eyes, bulbous nasal tip, thin upper lip, triangular face. Brain MRI shows mildly hypoplastic corpus callosum, which is foreshortened with a small splenium.
Molecular analysis
Institutional review board-approved written informed consents were obtained for all subjects. DNA was extracted from leukocytes according to standard procedures.
Copy number variants (CNV) were found by microarray-based comparative genomic hybridization (array CGH). Different array platforms were used for genomic copy number analyses which were carried out according to manufacturers’ recommendations: Agilent CGH Microarray 60K or 180K (Agilent Technologies, Santa Clara, CA). Chromosomal rearrangements were confirmed by fluorescence in situ hybridization (FISH) with various specific probes on chromosome preparations from leukocyte cultures or by quantitative polymerase chain reaction (PCR) using standard protocols. Several different oligonucleotide probes were used to try to sequence the breakpoints of the CNV carried by individual 1. Parental testing was performed when DNA samples were available. Genomic positions are relative to human genome Build GRCh37/hg19. The CNVs were submitted to the DECIPHER database.
Single nucleotide variants (SNV) were all identified by exome or genome sequencing. The protocols used by each participating center have been detailed elsewhere 18–19. Reads were aligned to the human reference genome sequence (NCBI build 37.3, hg19). For SNV, segregation analysis was made by direct sequencing and then performed in the families to confirm inheritance when parental DNA samples were available. The variants were submitted to the ClinVar database (https://www.ncbi.nlm.nih.gov/clinvar/).
RRS assay
Recovery of RNA synthesis (RRS) was evaluated on primary fibroblast cultures using fluorescent non-radioactive assay as described previously 20. RRS evaluates the transcription-coupled repair pathway (TC-NER). Cells were plated on coverslips in 6-well plates at a confluence of 7×104 cells per well. After 2 days in culture, cells were washed with phosphate-buffered saline (PBS), followed by irradiation with a range of UV-C doses (6–12-20 J/m2). The non-irradiated plates acted as references. After UV-irradiation, cells were incubated for 23 h for RNA Synthesis recovery in DMEM (Dulbecco’s modified Eagle’s medium) supplemented with fetal bovine serum. Then, after washing with PBS, cells were labeled with 5-ethynyl-uridine (EU; Invitrogen) for 2 h. Cells were then washed again with PBS, followed by fixation and permeabilization. The last step involved an azide-coupling reaction and DAPI (6’-diaminido-2-phenylindole) staining (Click-iT RNA HCS Assay, Invitrogen). Finally coverslips were washed in PBS and mounted on glass slides with Ibidi Mounting Medium (Biovalley). Photographs of the cells were taken with a fluorescent microscope (Imager.Z2) equipped with a CCD (charge-coupled device) camera (AxioCam, Zeiss). The images were processed and analyzed with the ImageJ software. At least 50 cells were randomly selected, and the average nuclear fluorescent intensity was calculated.
RESULTS
Clinical description
We identified seven affected individuals from seven independent families, including four males and three females. The main clinical features of our cohort are summarized in Table 1 and Figure 1 and detailed in Supplementary data.
Table 1. Detailed clinical features of the individuals with nucleotide and copy number variants involving MED12L.
NA: not available, SD: standard deviation, OFC: occipital frontal circumference.
| Individual | Individual 1 (Decipher 284908) | Individual 2 (Decipher 280845) | Individual 3 (Decipher 277489) | Individual 4 SCV000611598 | Individual 5 SCV000853261 | Individual 6 SCV000853262 | Individual 7 SCV000853263 |
|---|---|---|---|---|---|---|---|
| Mutation in MED12L (according to NCBI reference sequence NM_053002.5, NC_000003.11) | g.151129252C>T, c.5992C>T, p.(Gln1998Ter) | g.150906260dup, c.1747dup, p.(Ser583PhefsTer8) | g.151097900G>A, c.4374–1G>A | g.151101870G>A, c.4686–1G>A | |||
| Size of CNV (Mb) | 460 Kb duplication | 291 Kb deletion | 147 Kb duplication | ||||
| Proximal breakpoint (Hg19) | 150,983,389 | 150,876,508 | 150,966,686 | ||||
| Distal breakpoint (Hg19) | 151,441,372 | 151,167,962 | 151,114,133 | ||||
| Inheritance | de novo | de novo | NA | NA | de novo | NA | de novo |
| Origin | France | France | France | Ukraine | USA | USA | Caucasian |
| Gender | Male | Male | Male | Female | Male | Female | Female |
| Birth term (WG) | At term | At term | 39 | NA | 37 | 32 | 39 |
| Pregnancy complications | − | Acute foetal distress at birth | − | NA | Suspected cardiac anomaly, maternal pre-eclampsia | prenatal drug exposures (cocaine, tobacco) | − |
| Birth weight (grams/SD) | 3160 (−1 SD) | 4000 (+1,5 SD) | 3630 (+1 SD) | NA | 3000 (0 SD) | 1729 (0 SD) | 2404 (−2 SD) |
| Birth length (cm/SD) | 47.5 (−2 SD) | 53 (+1.5 SD) | 50 (0 SD) | NA | 48.5 (0 SD) | 38.1 (−1.5 SD) | NA |
| OFC at birth (cm/SD) | 37 (+1.5 SD) | 35 (+1 SD) | 34 (−0.5 SD) | NA | 34 (+0.5 SD) | NA | NA |
| Age at assessment | 12 years | 22 years | 13 years 8 months | 11 years | 5 years | 8 years | 3 years 10 months |
| Weight (kg/SD) | 27 (−2 SD) | 98 (+6 SD) under neuroleptic | 47 (0 DS) | 34 (−1 SD) | 21.1 (+0.5 SD) | 22.6 (0 SD) | 13.3 (−1 SD) |
| Height (cm/SD) | 131.5 (−2.5 SD) | 185 (+2 SD) | 156 (0 DS) | 142 (−0.5 SD) | 109 (−0.5 SD) | 122.5 (0 SD) | 91.5 (−2 SD) |
| OFC (cm/SD) | 53 (−1 SD) | 57 (+1 SD) | 56 (+1.5 SD) | NA | 50.1 (−0.5 SD) | 49.5 (−1.5 SD) | NA |
| Neurological abnormalities | |||||||
| Intellectual disability | moderate | moderate | mild (IQ 74) | moderate | mild | mild | severe |
| Hypotonia | − | − | − | NA | + | − | + |
| Motor delay | + (walking at 19 months) | − (walking at 16 months) | − | NA | + (walking at 18 months) | + (walking at 20 months) | + |
| Speech impairment | + (mild, sentences at 4 yo) | + (severe, can associate words) | + (pronounciation) can make a conversation | + (speech delay) | + (pronounciation), but good vocabulary and can make a conversation | + (speech delay) | + (no language) |
| Abnormal behavior | + | ++ | + | − | + | + | + |
| Aggressive behavior | − | + | ++ | − | + | ++ | − |
| Autistic features | + | ++ | + | − | + | − | − |
| Anxiety | ++ | + | − | − | − | − | − |
| Attention deficit | − | + | − | − | + | + | − |
| Hyperactivity | − | − | − | − | + | + | − |
| Sleeping disorder | − | + | − | − | + | − | + |
| Seizures | − | − | − | − | − | staring spells | + |
| Abnormal EEG | NA | − | NA | − | − | − | + |
| Abnormal brain magnetic resonance imaging | NA | NA | NA | Agenesis of the Corpus callosum, enlargement of the posterior aspect of the right and left lateral ventricle | Mildly hypoplastic corpus callosum | Normal | Cortical signal abnormality and volume loss of bilateral putamen and globus pallidus at 3 years |
| Extra-neurological abnormalities | |||||||
| Gastro-intestinal anomalies | Chronic constipation, neonatal occlusive syndrome, encopresia | Gastroesophageal reflux | unilateral inguinoscrotal hernia | − | Feeding difficulties in early infancy, moderate chronic constipation | − | Feeding difficulties (G-tube dependent), gastroesophageal reflux, intermittent constipation |
| Congenital malformations | Unilateral coloboma of iris and retina | − | − | − | Suspected VSD prenatally but normal echocardiogram at birth, Hypospadias, voiding dysfunction | − | − |
| Skeletal abnormalities | Thoracolumbar kyphosis, hyperlaxity | − | − | Very large knees, appears to have bony prominence medially | − | − | − |
| Hands and feet anomalies | Long appearing fingers, unilateral single palmar transverse crease | Long appearing fingers | Bilateral 5th finger brachyphalangy P1, pes planus | Fingers-fetal padding, 5th hypoplastic nails | − | − | − |
| Sensory abnormalities | Hypermetropia | − | Myopia | − | − | − | Hypermetropia, strabismus |
| Other findings | − | Dilated cardiomyopathy (toxic origin) | − | Hypopigmented macules (oval shapped on right shoulder blade) | recurrent respiratory infections | − | − |
| Dysmorphic features | |||||||
| High forehead | − | − | − | − | + | − | − |
| Downslanted palpebral fissures | + | − | − | − | − | − | + |
| Fullness of the upper eyelids | + | + | − | − | − | − | + |
| Prominent nasal bridge | − | + | − | + | − | − | − |
| Bulbous nasal tip | + | − | − | − | + | − | − |
| Open mouth | − | − | − | − | − | − | + |
| High, narrow palate | − | − | − | + | − | − | + |
| Other | Unilateral ptosis, hypertelorism, sparse eyebrows | Short philtrum, everted lower lip, small mouth | − | medial eyebrow flare, inverted lower eyelid, pointed chin, high cheek bones, down-turned corners of mouth, prominent ear crease-left ear | Deep-set eyes, thin upper lip, triangular face | − | Flat nasal bridge, upturned nose |
| Other genetics investigations | |||||||
| Karyotype | normal | normal | normal | NA | 46,XY,t(9;18)(p13;q12.2) | NA | normal |
| Chromosomal microarray | duplication 22q11.2 inherited from the healthy mother | − | − | normal | arr[hg19]4q34.3(178,557,799–179,142,775)x3 (small gain on chromosome 4 in a non-disease associated region) | arr[hg19]2p16.3(51,080,824–51,193,164)x1 (intronic deletion of NRXN1) | arr[hg19]10q11.21(43,555,634–43,626,143)x3 (contains the entire coding region of RET) |
| Gene testing | − | FMR1 negative | FMR1 negative | WGS identified VUS in TUBB2B: c.43G>A; p.(Gln15Lys) | normal Fragile X testing, WES identified the variant c.2380 C>T; p.(His794Tyr) in LZTR1 paternally inherited | NRXN1 sequencing negative | PTPN11, SMN1 deletion, DNA methylation for Prader-Willi/Angelman syndrome, neuromuscular multi-gene panel : negative |
Nomenclature HGVS V2.0 according to mRNA reference sequence NM_053002.5. Nucleotide numbering uses +1 as the A of the ATG translation initiation codon in the reference sequence, with the initiation codon as codon 1.
All individuals had neurological impairment. Intellectual disability/developmental delay was the main feature (7/7). Three individuals had a mild intellectual disability, three had a moderate intellectual disability with mild to severe speech impairment. One individual had a severe intellectual disability with seizures and brain abnormalities. Abnormal behavior was common (6/7) including mild to severe autism spectrum disorder (4/7) and aggressive behavior (4/7). Attention deficit with hyperactivity (3/7), sleep disorder (3/7) and hypotonia (1/7) were also noted. One individual had seizures and another one had staring spells with the EEG within normal limits. Brain MRI was performed in four individuals showing abnormalities for three of them: corpus callosum agenesis or hypoplasia (2/4), and secondary cortical signal abnormality with volume loss of bilateral putamen and globus pallidus (1/4).
Gastrointestinal issues were reported in 5/7 individuals, including chronic constipation (3/7), feeding difficulties (2/7, one individual required a gastrostomy tube placement), gastroesophageal reflux (2/7) and inguinal hernia (1/7). Various other miscellaneous extra-neurological features were reported such as congenital malformations (2/7, including iris coloboma and hypospadias), ophthalmologic features (3/7), skeletal abnormalities (2/7) and minor hand anomalies (4/7). Mild facial morphological features were reported in some individuals, but without a recognizable phenotype.
Genetic results
Genomic positions of copy number variants (CNVs) and single nucleotide variants (SNVs) identified in MED12L are mapped in Figure 2.
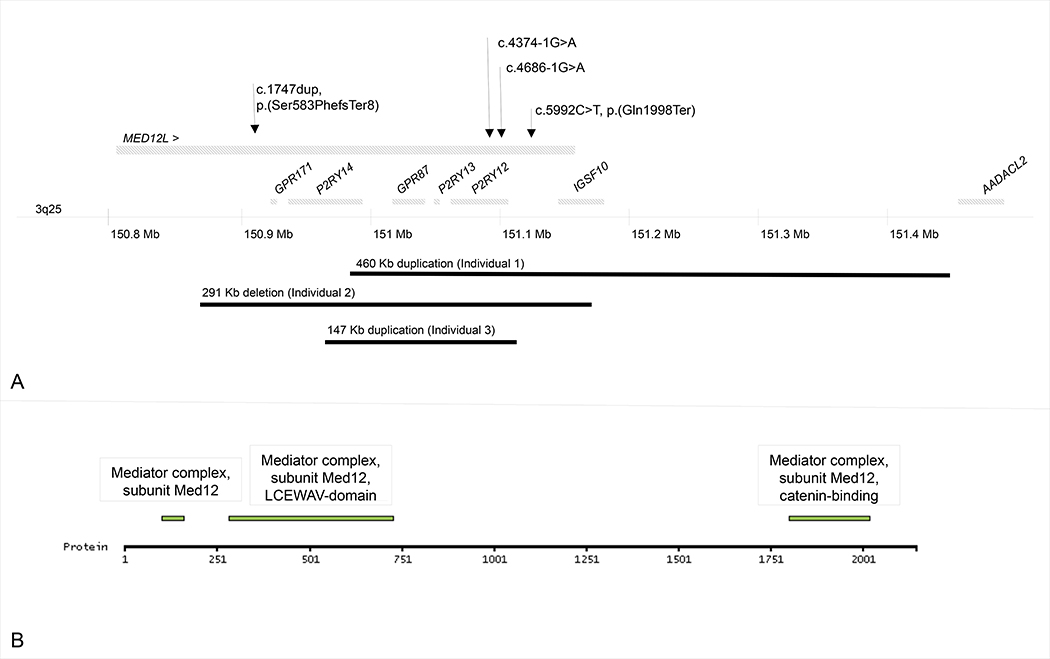
Figure 2:
A. Genomic position of MED12L variants identified in our series. Upper panel represents single nucleotide variants. Lower panel represents copy-number variants. B. Localization of predicted domains of MED12L protein.
Individuals 1, 2 and 3 harbored CNVs involving MED12L ranging in size from 147 Kb to 460 Kb (DECIPHER accession number: 284908, 280845 and 277489). Individual 1 carried a 460 Kb duplication involving the terminal part of MED12L. Attempts to sequence the breakpoints of the duplication by Sanger failed arguing for a complex rearrangement. Individual 2 carried a 291 Kb deletion involving the terminal part of MED12L. Individual 3 carried a 147 Kb intragenic MED12L duplication. Minimal and maximal coordinates of the CNVs are indicated in Table 1. CNVs were found to be de novo in individuals 1 and 2. Parents were not available for individual 3.
Genome sequencing was performed in individual 4 and exome sequencing in individuals 5 to 7. One frameshift, one nonsense and two splice variants were identified in MED12L (NM_053002.5, exons are numbered like in NG_021244.1): c.1747dup, p.(Ser583PhefsTer8), c.5992C>T, p.(Gln1998Ter), c.4374–1G>A, c.4686–1G>A (ClinVar accessions: SCV000611598, SCV000853261, SCV000853262 and SCV000853263). Variants were absent from GnomAD, EVS and 1000 Genome databases. Nonsense variants likely result in the degradation of mRNA by nonsense-mediated decay. Segregation analysis showed that the variants occurred de novo when parents’ DNA was available (individuals 5 and 7).
Lastly, individual 4 harbored a variant of unknown significance in TUBB2B [OMIM 612850] that could not be tested for inheritance or excluded as contributing to the individual overall phenotype (see table 1).
Functional analysis
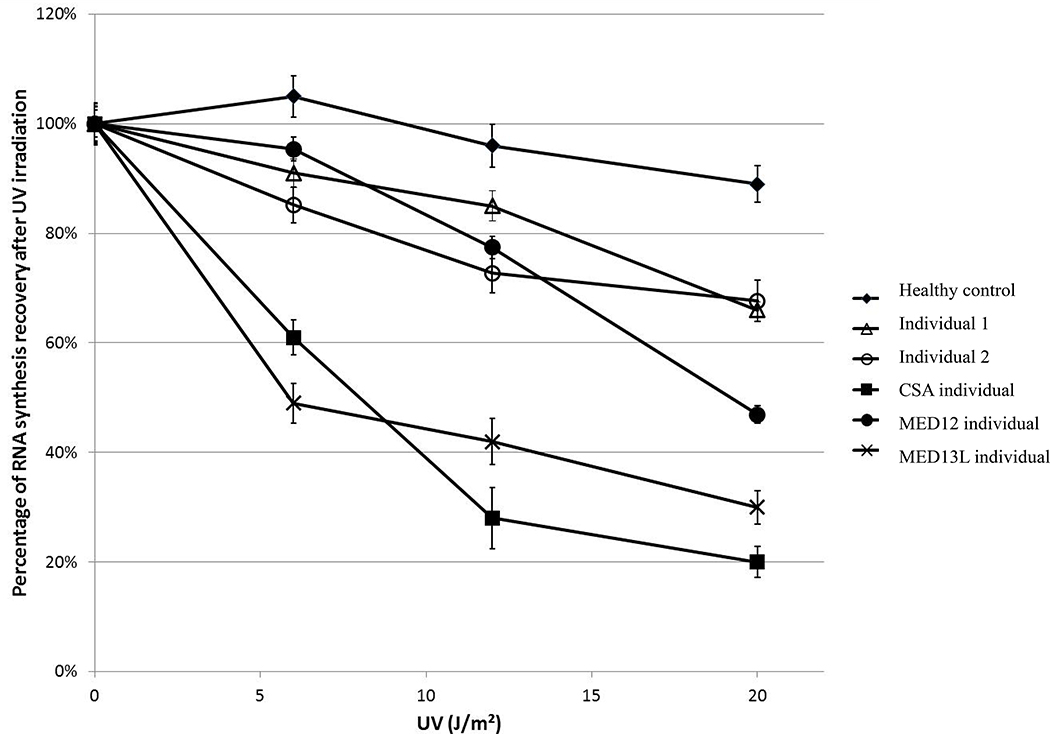
To test the functional consequences of the MED12L variants on transcriptional processes, we used the recovery of RNA synthesis (RRS) assay, which is the gold standard diagnostic tool in Cockayne syndrome (CS), another transcription-related human disease with syndromic intellectual disability 21. This assay reflects the global transcriptional activity by measuring the incorporation of fluorescent uridine 24 hours after UV irradiation. In wild type cells, UV irradiation temporarily halts the transcription of a large subset of genes, and 24 hours later they recover a normal RNA synthesis activity 22. Such an assay is used to partly synchronize cells and enable a standardized assessment of global transcriptional activity. Primary fibroblasts from two individuals with MED12L CNV (individuals 1 and 2) were available for RRS testing and were compared to fibroblasts from a healthy control, from a classical Cockayne individual (carrying pathogenic variants in the ERCC8/CSA gene) and from individuals carrying pathogenic variants in other mediator subunit genes, MED12 and MED13L. Both MED12L cell lines show a moderate but significant decreased RNA synthesis level 24 hours after UV irradiation, as compared to the healthy control, similar to the level observed in a control MED12 cell line (Figure 3). The control MED13L cell line showed a more severely reduced RNA synthesis level, closer to the classical and the severe transcriptional defect observed in the control Cockayne cell line 22.
Figure 3: Recovery of RNA synthesis (RRS) following UV irradiation.
Fibroblasts of individuals 1 (∆) and 2 (○) show a moderately decreased level of RRS as compared to the normal control cell line (♦), similar to a MED12 mutated cell line (●). A MED13L mutated cell line (▯) shows a more severely decreased RRS level, closer to the typical severely decreased level of RRS in a CSA defective cell line (■). Error bars represent standard errors of the mean.
DISCUSSION
Here we described a series of seven individuals presenting with variants (CNVs and SNVs) involving MED12L. Individual 1 carried a partial de novo duplication of MED12L. Despite not being proven by our study, we suspected a complex rearrangement. We also showed that the duplication was associated with a defect in transcriptional activity of fibroblasts. Individual 2 carried a partial deletion with a similar transcriptional defect as for individual 1. Individuals 3, 4 and 5 carried respectively an intragenic duplication, a nonsense variant (localized in exon 39/43), and a frameshift variant with a premature nonsense variant, all predicted to result in an altered mRNA likely eliminated by nonsense-mediated decay (NMD). Individuals 6 and 7 carried intronic variants predicted to alter splicing resulting in a premature nonsense codon. Most variants were de novo. Unfortunately, segregation was not possible for three patients either because they were adopted (individuals 4 and 6) or because parents’ DNA were not available (individual 3). Functional studies were performed only for two French patients. Fibroblasts were not available for the other individuals. All in all, despite these limitations, those data argue for a haploinsufficiency mechanism leading to transcriptional defect.
All individuals described here showed intellectual disability/developmental delay or speech impairment, sometimes associated with abnormal behavior, corpus callosum agenesis, and mild facial morphological features. No genotype-phenotype correlation could be identified in our series. To note that individual 1 also harbored a 22q11.2 duplication, which is considered as a risk factor for attention deficit hyperactivity disorder, autism and intellectual disability with a low penetrance estimated around 10% 23 . The healthy mother of individual 1 carried the same 22q11.2 duplication. Even if we could not exclude a partial role of this duplication, functional data provided and the severity of intellectual disability tended to suggest that MED12L CNV had a more important role in the phenotype of individual 1.
MED12L contains 43 exons and encodes a protein component of Mediator, which is involved in transcriptional coactivation of nearly all RNA polymerase II-dependent genes 22. MED12L is localized to the nucleus and mainly expressed in brain (www.proteinatlas.org). By homology, three domains are predicted to be common with MED12, including a LCEWAV-domain (AA 283–730), with a conserved sequence motif of unknown function, and a catenin-binding domain (AA 1802–2019), activating the canonical Wnt/beta-catenin pathway. The measure of probability of intolerance to loss of function (pLI) score, based on the difference in observed and expected loss-of-function variants in the gene, indicates MED12L is extremely intolerant to heterozygous loss of function (pLI score of 1) 24. MED12L overlaps with six genes, five of which P2RY12, P2RY13, P2RY14, GPR171 and GPR87 encode purinergic receptors participating in vascular and immune response to injury, and currently have no known link to neurodevelopment. MED12L also overlaps IGSF10 which encodes an immunoglobin involved in the control of early migration of neurons expressing gonadotropin-releasing hormone. A MED12L variant (rs1554120) was associated (p = 5.25E-06) with cortical thickness of right Heschl’s gyrus (HG) by genome-wide association study 25. HG is a core region of the auditory cortex whose morphology is highly variable across individuals. This variability has been linked to sound perception ability in both speech and music domains. MED12L interacts with NCAPD2 in human 26. NCAPD2 encodes a non-SMC (Structural maintenance of chromosomes) subunit of the condensin complex and causes autosomal recessive microcephaly 27.
Mediator is highly conserved across eukaryotes 22. It links gene-specific transcription activators with the basal transcription machinery. Mediator contains 30 subunits organized into four modules: MED12L is predicted to be a subunit of the kinase module with MED12, MED13, MED13L, CDK8, CDK19 and Cyclin C. The kinase module is reversibly associated with the Mediator core and is involved in transcriptional repression and activation. The function of this module could be mediated by its kinase activity when it is recruited to an upstream activation region or enhancers, or by the mutually exclusive interaction between the kinase module and RNA polymerase II with the Mediator core integrating the pre-initiation complex 28,29. The Mediator complex is also involved in chromatin regulation, and mRNA processing 3,30,31.
Other Mediator kinase module subunits have already been implicated in human disease, namely MED12, MED13, MED13L and CDK19. Variants in MED12 have been reported in Opitz-Kaveggia (FG) syndrome, Lujan-Fryns syndrome and Ohdo syndrome, Maat-Kievit-Brunner type 6,8–10. Those three X-linked intellectual disability syndromes are allelic and caused by missense MED12 variants and defects in gene expression. FG syndrome and Lujan-Fryns syndrome share some overlapping features such as dysgenesis of the corpus callosum, relative macrocephaly, a tall forehead and hypotonia. Chronic constipation or anal anomalies are a frequent finding in individuals with FG syndrome. Males with FG syndrome also have deficits in communication skills despite affability and excessive talkativeness, attention deficit hyperactivity disorder, maladaptive behavior with aggressive behavior and anxiety 32. Marfanoid habitus and hypernasal speech are related to Lujan-Fryns syndrome. Hyperactivity, aggressive behavior, shyness, and attention-seeking behavior are also common in individuals with Lujan-Fryns syndrome. Consistent facial morphological features include downslanted palpebral fissures, high narrow palate, dental crowding and microretrognathia. Ohdo syndrome is responsible for intellectual disability with hyperactivity and aggressive behavior, blepharophimosis, microretrognathia, constipation and feeding difficulties 30.
Recently, 13 individuals were described harboring de novo missense and nonsense MED13 [OMIM 603808] variants. All individuals had mild to moderate intellectual disability and speech disorder. Other features mainly included autism spectrum disorder, attention deficit hyperactivity, optic nerve abnormalities, Duane anomaly, hypotonia, mild congenital heart malformations, and dysmorphism 15. Missense and nonsense MED13L [OMIM 608771] variants are also responsible for moderate to severe intellectual disability and severe speech disorder with poor language. More than 60 individuals have been reported so far. Other features included hypotonia, autism and behavioral disorder, corpus callosum hypoplasia or agenesis, and miscellaneous malformations 14,33–35. Finally, disruption of CDK19 has been reported in an individual with intellectual disability, microcephaly and bilateral congenital retinal folds 13.
Therefore, all the subunits of the kinase module seem to be crucial for neurological development. Some features are common between the four conditions linked to MED12, MED13, MED12L and MED13L such as intellectual disability, behavior abnormalities and autism spectrum disorders. A CDK19 variant has been reported in only one individual with a different neurological phenotype. Table 2 summarizes common features between the four main MED-related conditions. Corpus callosum anomalies seem common with MED12 and MED12L variants and can be seen less frequently with MED13L variants. MED12 and MED13L variants appear to lead to a more severe condition, with poor speech and facial hypotonia, than MED13 and MED12L related disorders. Finally, the wide range of other organs affected is an inconsistent feature and can be related to the ubiquitous expression of those genes and their broad regulatory role. Mild facial morphological features are common for all those disorders, but does not help in pointing to a clinical diagnosis.
Table 2. Comparison of different phenotypes associated with MED12, MED13, MED13L and MED12L variants.
OSMKB: Ohdo, syndrome, Maat-Kievit-Brunner type. To note that intellectual disability was classified as mainly mild to moderate (+) or mainly moderate to severe (++). Others signs were considered as very frequent (+), occasional (+/−) or absent/rare (−).
| MED12 | MED13L | MED13 | MED12L | |||
|---|---|---|---|---|---|---|
| Lujan syndrome | FG syndrome | OSMKB | ||||
| Growth | ||||||
| Tall stature | + | − | − | − | − | − |
| Macrocephaly | + | + | − | − | − | − |
| Facies | ||||||
| Tall prominent forehead | + | + | + | − | − | − |
| Blepharophimosis | − | − | + | − | − | − |
| Downslanting palpebrae | + | + | + | − | +/− | − |
| High nasal root | + | − | − | + | + | +/− |
| High narrow palate | + | + | + | − | − | +/− |
| Open mouth | + | + | + | + | − | − |
| Frontal hair upsweep | − | + | − | − | − | − |
| Hand | ||||||
| Minor hand anomalies | + | + | + | + | − | +/− |
| Neurological | ||||||
| Congenital hypotonia | + | + | + | +/− | +/− | +/− |
| Intellectual disability | + | + | ++ | ++ | + | + |
| Little or no language | − | − | + | + | +/− | − |
| Hypernasal voice | + | − | − | − | − | − |
| Behavior disturbances | + | + | + | +/− | +/− | + |
| Autism spectrum disorder | +/− | − | +/− | +/− | +/− | +/− |
| Agenesis/hypoplasia of corpus callosum | + | + | − | +/− | − | + |
| Gastro-intestinal | ||||||
| Anal anomalies | − | + | − | − | − | − |
| Chronic constipation | − | + | + | − | +/− | +/− |
Herein, we demonstrate that MED12L fibroblasts for individual 1 and 2 show a defect in transcriptional activity as measured by recovery of RNA synthesis after UV irradiation. This defect is similar to the defect observed in a MED12 cell line and milder than the defect seen in a MED13L cell line derived from an individual who showed a more severe neurological phenotype. It can be hypothesized that the pathogenic variants in Mediator kinase module subunits could impair the transcriptional co-activation functions of the complex under specific conditions, during development and during postnatal life, leading to the phenotype observed in the individuals. It has already been similarly postulated that neurodevelopmental defects in individuals affected with CS 37–38 and trichothiodystrophy 39 could be due to transcriptional defects mediated by variants in the other transcriptional coactivation complexes TFIIH or TFIIE 40.
In conclusion, we report here the involvement of MED12L in human disease as has been seen for other subunits of the kinase module of the mediator complex, through transcriptional defect. Even if many intellectual disability syndromes are challenging to differentiate based on clinical phenotype alone, a common clinical pattern seems to emerge for the disorders related to transcriptional defect linked to Mediator complex.
Supplementary Material
AKNOWLEDGEMENTS
We thank the patients and their families for participating in this study.
Footnotes
CONFLICT OF INTEREST STATEMENT
Department of Molecular and Human Genetics at Baylor College of Medicine receives revenue from clinical genetic testing conducted at Baylor Genetics. Megan Truitt Cho and Kirsty McWalter are employees of GeneDx, Inc., a wholly owned subsidiary of OPKO Health, Inc.
The other authors declare no conflict of interest.
ACCESSION NUMBERS
The accession numbers for the SNPs and CNVs reported in this paper are DECIPHER: 284908, 280845 and 277489; and ClinVar: SCV000611598, SCV000853261, SCV000853262 and SCV000853263.
SUPPLEMENTAL DATA
Supplemental data includes one text file with detailed clinical data.
REFERENCES
- 1.Kornberg RD (2005). Mediator and the mechanism of transcriptional activation. Trends Biochem. Sci. 30, 235–239. [DOI] [PubMed] [Google Scholar]
- 2.Conaway RC, and Conaway JW (2011). Origins and activity of the Mediator complex. Semin. Cell Dev. Biol. 22, 729–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allen BL, and Taatjes DJ (2015). The Mediator complex: a central integrator of transcription. Nat. Rev. Mol. Cell Biol. 16, 155–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berk AJ (2012). Yin and yang of mediator function revealed by human mutants. Proc. Natl. Acad. Sci. U. S. A. 109, 19519–19520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jeronimo C, and Robert F (2017). The Mediator Complex: At the Nexus of RNA Polymerase II Transcription. Trends Cell Biol. 27, 765–783. [DOI] [PubMed] [Google Scholar]
- 6.Risheg H, Graham JM, Clark RD, Rogers RC, Opitz JM, Moeschler JB, Peiffer AP, May M, Joseph SM, Jones JR, et al. (2007). A recurrent mutation in MED12 leading to R961W causes Opitz-Kaveggia syndrome. Nat. Genet. 39, 451–453. [DOI] [PubMed] [Google Scholar]
- 7.Rump P, Niessen RC, Verbruggen KT, Brouwer OF, de Raad M, and Hordijk R (2011). A novel mutation in MED12 causes FG syndrome (Opitz-Kaveggia syndrome). Clin. Genet. 79, 183–188. [DOI] [PubMed] [Google Scholar]
- 8.Graham JM, and Schwartz CE (2013). MED12 related disorders. Am. J. Med. Genet. A. 161A, 2734–2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schwartz CE, Tarpey PS, Lubs HA, Verloes A, May MM, Risheg H, Friez MJ, Futreal PA, Edkins S, Teague J, et al. (2007). The original Lujan syndrome family has a novel missense mutation (p.N1007S) in the MED12 gene. J. Med. Genet. 44, 472–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vulto-van Silfhout AT, de Vries BBA, van Bon BWM, Hoischen A, Ruiterkamp-Versteeg M, Gilissen C, Gao F, van Zwam M, Harteveld CL, van Essen AJ, et al. (2013). Mutations in MED12 cause X-linked Ohdo syndrome. Am. J. Hum. Genet. 92, 401–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Isidor B, Lefebvre T, Le Vaillant C, Caillaud G, Faivre L, Jossic F, Joubert M, Winer N, Le Caignec C, Borck G, et al. (2014). Blepharophimosis, short humeri, developmental delay and hirschsprung disease: expanding the phenotypic spectrum of MED12 mutations. Am. J. Med. Genet. A. 164A, 1821–1825. [DOI] [PubMed] [Google Scholar]
- 12.Donnio L-M, Bidon B, Hashimoto S, May M, Epanchintsev A, Ryan C, Allen W, Hackett A, Gecz J, Skinner C, et al. (2017). MED12-related XLID disorders are dose-dependent of immediate early genes (IEGs) expression. Hum. Mol. Genet. 26, 2062–2075. [DOI] [PubMed] [Google Scholar]
- 13.Mukhopadhyay A, Kramer JM, Merkx G, Lugtenberg D, Smeets DF, Oortveld MAW, Blokland EAW, Agrawal J, Schenck A, van Bokhoven H, et al. (2010). CDK19 is disrupted in a female patient with bilateral congenital retinal folds, microcephaly and mild mental retardation. Hum. Genet. 128, 281–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smol T, Petit F, Piton A, Keren B, Sanlaville D, Afenjar A, Baker S, Bedoukian EC, Bhoj EJ, Bonneau D, et al. (2018). MED13L-related intellectual disability: involvement of missense variants and delineation of the phenotype. Neurogenetics 19, 93–103. [DOI] [PubMed] [Google Scholar]
- 15.Snijders Blok L, Hiatt SM, Bowling KM, Prokop JW, Engel KL, Cochran JN, Bebin EM, Bijlsma EK, Ruivenkamp CAL, Terhal P, et al. (2018). De novo mutations in MED13, a component of the Mediator complex, are associated with a novel neurodevelopmental disorder. Hum. Genet 13, 675–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Firth HV, Richards SM, Bevan AP, Clayton S, Corpas M, Rajan D, Van Vooren S, Moreau Y, Pettett RM, and Carter NP (2009). DECIPHER: Database of Chromosomal Imbalance and Phenotype in Humans Using Ensembl Resources. Am. J. Hum. Genet. 84, 524–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sobreira N, Schiettecatte F, Valle D, and Hamosh A (2015). GeneMatcher: a matching tool for connecting investigators with an interest in the same gene. Hum. Mutat. 36, 928–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang Y, Muzny DM, Xia F, Niu Z, Person R, Ding Y, Ward P, Braxton A, Wang M, Buhay C, et al. (2014). Molecular findings among patients referred for clinical whole-exome sequencing. JAMA 312, 1870–1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bowling KM, Thompson ML, Amaral MD, Finnila CR, Hiatt SM, Engel KL, Cochran JN, Brothers KB, East KM, Gray DE, et al. (2017). Genomic diagnosis for children with intellectual disability and/or developmental delay. Genome Med. 9, 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Calmels N, Greff G, Obringer C, Kempf N, Gasnier C, Tarabeux J, Miguet M, Baujat G, Bessis D, Bretones P, et al. (2016). Uncommon nucleotide excision repair phenotypes revealed by targeted high-throughput sequencing. Orphanet J. Rare Dis. 11, 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Limsirichaikul S, Niimi A, Fawcett H, Lehmann A, Yamashita S, and Ogi T (2009). A rapid non-radioactive technique for measurement of repair synthesis in primary human fibroblasts by incorporation of ethynyl deoxyuridine (EdU). Nucleic Acids Res. 37, e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Epanchintsev A, Costanzo F, Rauschendorf M-A, Caputo M, Ye T, Donnio L-M, Proietti-de-Santis L, Coin F, Laugel V, and Egly J-M (2017). Cockayne’s Syndrome A and B Proteins Regulate Transcription Arrest after Genotoxic Stress by Promoting ATF3 Degradation. Mol. Cell 68, 1054–1066.e6. [DOI] [PubMed] [Google Scholar]
- 23.Olsen L, Sparsø T, Weinsheimer SM, Dos Santos MBQ, Mazin W, Rosengren A, Sanchez XC, Hoeffding LK, Schmock H, Baekvad-Hansen M, et al. (2018). Prevalence of rearrangements in the 22q11.2 region and population-based risk of neuropsychiatric and developmental disorders in a Danish population: a case-cohort study. Lancet Psychiatry 5, 573–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, O’Donnell-Luria AH, Ware JS, Hill AJ, Cummings BB, et al. (2016). Analysis of protein-coding genetic variation in 60,706 humans. Nature 536, 285–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cai D-C, Fonteijn H, Guadalupe T, Zwiers M, Wittfeld K, Teumer A, Hoogman M, Arias-Vásquez A, Yang Y, Buitelaar J, et al. (2014). A genome-wide search for quantitative trait loci affecting the cortical surface area and thickness of Heschl’s gyrus. Genes Brain Behav. 13, 675–685. [DOI] [PubMed] [Google Scholar]
- 26.Wan C, Borgeson B, Phanse S, Tu F, Drew K, Clark G, Xiong X, Kagan O, Kwan J, Bezginov A, et al. (2015). Panorama of ancient metazoan macromolecular complexes. Nature 525, 339–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martin C-A, Murray JE, Carroll P, Leitch A, Mackenzie KJ, Halachev M, Fetit AE, Keith C, Bicknell LS, Fluteau A, et al. (2016). Mutations in genes encoding condensin complex proteins cause microcephaly through decatenation failure at mitosis. Genes Dev. 30, 2158–2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clark AD, Oldenbroek M, and Boyer TG (2015). Mediator kinase module and human tumorigenesis. Crit. Rev. Biochem. Mol. Biol. 50, 393–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jeronimo C, Langelier M-F, Bataille AR, Pascal JM, Pugh BF, and Robert F (2016). Tail and Kinase Modules Differently Regulate Core Mediator Recruitment and Function In Vivo. Mol. Cell 64, 455–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang Y, Li W, Yao X, Lin Q-J, Yin J-W, Liang Y, Heiner M, Tian B, Hui J, and Wang G (2012). Mediator complex regulates alternative mRNA processing via the MED23 subunit. Mol. Cell 45, 459–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lai F, Orom UA, Cesaroni M, Beringer M, Taatjes DJ, Blobel GA, and Shiekhattar R (2013). Activating RNAs associate with Mediator to enhance chromatin architecture and transcription. Nature 494, 497–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Graham JM, Visootsak J, Dykens E, Huddleston L, Clark RD, Jones KL, Moeschler JB, Opitz JM, Morford J, Simensen R, et al. (2008). Behavior of 10 patients with FG syndrome (Opitz-Kaveggia syndrome) and the p.R961W mutation in the MED12 gene. Am. J. Med. Genet. A. 146A, 3011–3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Muncke N, Jung C, Rüdiger H, Ulmer H, Roeth R, Hubert A, Goldmuntz E, Driscoll D, Goodship J, Schön K, et al. (2003). Missense mutations and gene interruption in PROSIT240, a novel TRAP240-like gene, in patients with congenital heart defect (transposition of the great arteries). Circulation 108, 2843–2850. [DOI] [PubMed] [Google Scholar]
- 34.Asadollahi R, Oneda B, Sheth F, Azzarello-Burri S, Baldinger R, Joset P, Latal B, Knirsch W, Desai S, Baumer A, et al. (2013). Dosage changes of MED13L further delineate its role in congenital heart defects and intellectual disability. Eur. J. Hum. Genet. 21, 1100–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Adegbola A, Musante L, Callewaert B, Maciel P, Hu H, Isidor B, Picker-Minh S, Le Caignec C, Delle Chiaie B, Vanakker O, et al. (2015). Redefining the MED13L syndrome. Eur. J. Hum. Genet. 23, 1308–1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huttlin EL, Ting L, Bruckner RJ, Gebreab F, Gygi MP, Szpyt J, Tam S, Zarraga G, Colby G, Baltier K, et al. (2015). The BioPlex Network: A Systematic Exploration of the Human Interactome. Cell 162, 425–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kristensen U, Epanchintsev A, Rauschendorf M-A, Laugel V, Stevnsner T, Bohr VA, Coin F, and Egly J-M (2013). Regulatory interplay of Cockayne syndrome B ATPase and stress-response gene ATF3 following genotoxic stress. Proc. Natl. Acad. Sci. U. S. A. 110, E2261–2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang Y, Chakravarty P, Ranes M, Kelly G, Brooks PJ, Neilan E, Stewart A, Schiavo G, and Svejstrup JQ (2014). Dysregulation of gene expression as a cause of Cockayne syndrome neurological disease. Proc. Natl. Acad. Sci. U. S. A. 111, 14454–14459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Compe E, Malerba M, Soler L, Marescaux J, Borrelli E, and Egly J-M (2007). Neurological defects in trichothiodystrophy reveal a coactivator function of TFIIH. Nat. Neurosci. 10, 1414–1422. [DOI] [PubMed] [Google Scholar]
- 40.Kuschal C, Botta E, Orioli D, Digiovanna JJ, Seneca S, Keymolen K, Tamura D, Heller E, Khan SG, Caligiuri G, et al. (2016). GTF2E2 Mutations Destabilize the General Transcription Factor Complex TFIIE in Individuals with DNA Repair-Proficient Trichothiodystrophy. Am. J. Hum. Genet. 98, 627–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.