Abstract
During the last years, an extraordinary effort has been made to identify biomarkers as potential tools for improving prevention, diagnosis, drug response and drug development in psychiatric disorders. Contrary to other diseases, mental illnesses are classified by diagnostic categories with a broad variety list of symptoms. Consequently, patients diagnosed from the same psychiatric illness present a great heterogeneity in their clinical presentation. This fact together with the incomplete knowledge of the neurochemical alterations underlying mental disorders, contribute to the limited efficacy of current pharmacological options. In this respect, the identification of biomarkers in psychiatry is becoming essential to facilitate diagnosis through the developing of markers that allow to stratify groups within the syndrome, which in turn may lead to more focused treatment options. In order to shed light on this issue, this review summarizes the concept and types of biomarkers including an operational definition for therapeutic development. Besides, the advances in this field were summarized and sorted into five categories, which include genetics, transcriptomics, proteomics, metabolomics, and epigenetics. While promising results were achieved, there is a lack of biomarker investigations especially related to treatment response to psychiatric conditions. This review includes a final conclusion remarking the future challenges required to reach the goal of developing valid, reliable and broadly-usable biomarkers for psychiatric disorders and their treatment. The identification of factors predicting treatment response will reduce trial-and-error switches of medications facilitating the discovery of new effective treatments, being a crucial step towards the establishment of greater personalized medicine.
Keywords: biomarkers, neuropsychiatry, personalized medicine, lymphocytes, peripheral biomarkers, central biomarkers
Introduction
According to World Health Organization mental illness presented devastating rates of prevalence, mortality, morbidity and disability. Suffering a serious mental illness reduces average life expectancy in 13 to 32 years (1, 2). Aside from mortality, in most Western countries, mental disorders are the leading cause of disability, responsible for 30-40% of chronic sick leave and costing approximately a 4% of gross domestic product (3). Besides, for all types of mental illness, pharmacological treatment options are scarce and present limited efficacy. Several studies highlighted that, in terms of recovery and remission, current pharmacological interventions showed significant limitations. A series of effectiveness trials sponsored by the National Institute of Mental Health (NIMH) in USA provided relevant data in this regard. In CATIE (Clinical Antipsychotic Trials of Intervention Effectiveness) study, 74% of patients suffering from chronic schizophrenia (SCZ) experienced problems of treatment adherence within 18 months (4). In addition, in the Sequenced Treatment Alternatives to Relieve Depression (STAR*D) study only 31% of patients with major depressive disorder (MDD) were in remission after being treated with a selective serotonin reuptake inhibitor for a total of 14 weeks (5). An additional study carried out in patients diagnosed with bipolar disorder (BD) (STEP-BD, Systematic Treatment Enhancement Program for Bipolar Disorder) revealed that only 24% of patients experienced a remission of depression during 8 consecutive weeks, outcome similar to those observed in the vehicle group (6).
Several factors contribute to this clinical reality. On one hand, the heterogeneity/complexity of mental disorders. Patients suffering from a mental illness displayed several symptoms related with behavior, thinking, feelings and/or social interaction. To facilitate the diagnoses, mental disorders are classified by diagnostic categories with a broad variety list of symptoms according to the Diagnostic and Statistical Manual of Mental Disorders, Fifth Edition (DSM-IV), or International Statistical Classification of Diseases and Related Health Problems, Tenth Edition (ICD-11). Consequently, patients diagnosed with the same psychiatric illness present a great heterogeneity in their clinical presentation. In addition, several mental illnesses present symptoms in common, that can often make the diagnosis difficult.
On the other hand, psychiatric diseases present high comorbidity. Approximately 85%–90% of patients with depression also experience symptoms of anxiety, and vice versa (7, 8). Among schizophrenic patients, psychiatric comorbidities are common. Around 50% of patients suffer from depression and more than 47% have a lifetime diagnosis of comorbid substance use disorders (9–11). The simultaneous presence of two or more psychiatric diseases are associated with greater severity, worse response to the pharmacological treatment and have a greater risk of suicide than either condition alone.
Despite these sobering facts, progress in human brain research and the advent of new technologies, such as ‘omics’ technologies, offers the opportunity for change mental health treatment and outcomes in a near future. In this respect, the identification of biomarkers has become a new promising tool for guiding diagnosis, predicting clinical outcome and, therefore, improving the understanding of the pathophysiology of mental disorders. This review provides an overview about the current state of biomarkers in neuropsychiatry, with the ultimate aim of remarking some goals achieved up to date and the future challenges needed to develop valid, reliable and broadly-usable biomarkers for psychiatric disorders and their treatment. For this purpose, the review includes a definition of biomarker’s concept throughout history, describes the different types of biomarkers and their potential role in clinical practice, and emphasizes the samples and techniques commonly used. The role of ‘omics’ is described in greater detail due to its huge progress in the recent years. A final conclusion remarks the difficulties and limitations of current biomarkers strategies in neuropsychiatry and the future challenges needed to progress in this field.
What Are Biomarkers? Evolution of Biomarkers Through History
During the last 50 years, the definition of biomarker has been modified according to scientific and clinical progress. The term “biomarker” was used for the first time in 1973 to indicate the presence or absence of biological material. However, the concept is older, referenced as a “biochemical marker” in 1949 (12) and “biological marker” in 1957 (13).
In 2000, the Biomarker Definition Working Group, supported by the U.S. National Institute of Health (NIH), defined a biomarker as “a characteristic that is objectively measured and evaluated as an indication of normal biological processes, pathogenic processes, or pharmacologic responses to a therapeutic intervention” (14). This definition has two major limitations. The first one lies in the fact that sometimes a biomarker is measured by subjective parameters. The second one is the fact that additional processes or responses beyond those covered by the definition are excluded.
In 2016, Fitzgerald and colleagues redefined the concept of biomarker as “a functional variant or quantitative index of a biological process that predicts or reflects the evolution of or predisposition to a disease or a response to a therapy” (15). Nevertheless, this description lacks the consideration of structural variants and qualitative index as potential biomarkers.
In order to harmonize the term of biomarker, the Food and Drug Administration (FDA) in collaboration with the NIH Joint Leadership Council convened the FDA-NIH Biomarker Working Group in 2016. This group simplified the biomarker definition being considered as “a defined characteristic that is measured as an indicator of normal biological processes, pathogenic processes or responses to an exposure or intervention” (16). This definition, clearer and more concise, defines a biomarker specifying its principal applications without any unnecessary complexity or contradictory information. Besides, to ensure its clinical use, a good biomarker should be measured with high reproducibility, present a sizeable signal-to-noise ratio and, more importantly, meet the condition of being modified in a dynamic and reliable way as the clinical condition progress. In addition, a biomarker should be accessible for its detection and measurement, as would be the case of a plasmatic parameter or a genetic marker, or being detected by histological or image/neuroimaging techniques (17).
Types and Role of Biomarkers in the Clinical Practice
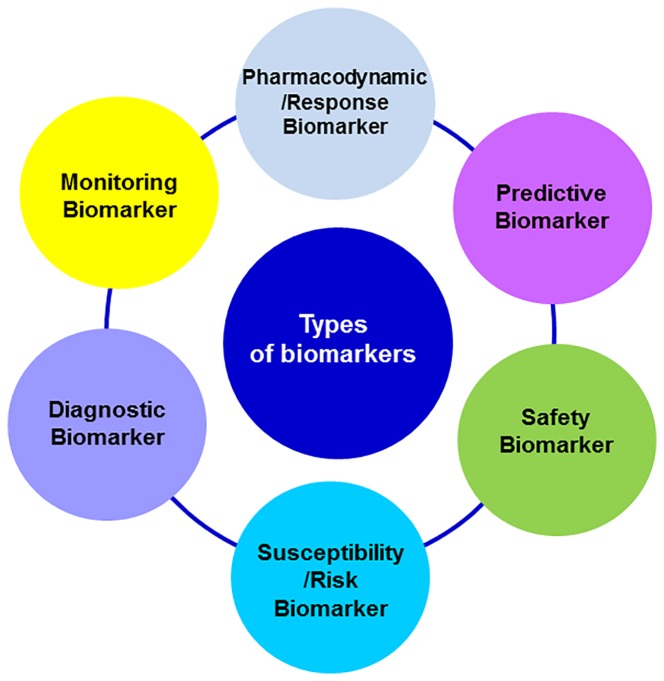
According to their applications, biomarkers can provide complementary information about the disease or the intervention under consideration. Biomarkers may be identified at any event occurring since the pathogenesis, the onset of first clinical manifestations, diagnosis, treatment outcome or recovery. The FDA-NIH Biomarker Working Group distinguished several types of biomarkers based on their main clinical application: diagnostic, monitoring, pharmacodynamic/response, predictive r, prognostic, safety, and susceptibility/risk biomarkers (Figure 1). A biomarker may meet multiple criteria for different uses or present specific features that enable its particular use (18).
Figure 1.
Classification of biomarkers based on its main clinical application.
Diagnostic Biomarker
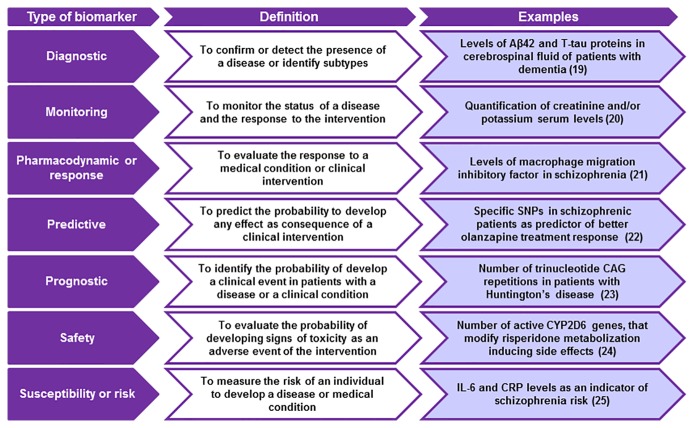
Encompasses a variety of biomarkers used to detect or confirm the presence of a disease or medical condition. This type of biomarker can be used to identify disease subtypes. The advent of the era of the precision medicine emphasizes the fact that diagnostic biomarkers are useful not only to identify patients with a disease, but also to redefine its classification. This is an important feature, because many diseases have subtypes with different prognosis or treatment responses. Thus, diagnostic biomarkers would contribute to improve personalized medicine increasing the effectiveness of the therapeutic response. These biomarkers may play also a critical role as prognostic biomarkers or predictive treatment outcome biomarkers (16, 17). An example could be the concentrations of Aβ42 and total tau (T-tau) in cerebrospinal fluid of patients with dementia as diagnostic biomarker for Alzheimer's disease (19) (Figure 2).
Figure 2.
Aims and examples of the main types of biomarkers. SNPs, single nucleotide polymorphisms; CRP, c-reactive protein; IL-6, interleukin 6.
Monitoring Biomarker
This category includes biomarkers that are analyzed at different time points to monitor the status of a disease or medical condition, and as a marker of the response to an intervention, including exposure to a medical product or an environmental agent (Figure 2) (16). Changes in biomarkers values are considered as indicators of the progression of the clinical condition and as measurements of the pharmacological response and other types of clinical interventions (17). An example of a monitoring biomarker is the elevation of serum creatinine and/or potassium concentrations after a pharmacological or medical intervention, parameters that are commonly used as an indicator of the probability to develop side effects (20). Monitoring biomarkers can be applied in different situations including clinical care or clinical trials, at the beginning of a treatment, for medical product development purposes, as a measure of the risk of developing a disease, or to evaluate the pharmacodynamics of a clinical intervention (Table 1).
Table 1.
Potential role of monitoring biomarkers in neuropsychiatry.
| Type of intervention | Utility | References |
|---|---|---|
| Clinical care or clinical trial | To evaluate patient's clinical situation during treatment or at the end of the intervention | (21) |
| Before treatment initiation | To detect signs and/or symptoms of a disease or medical condition as an indicative parameter of the prognosis To determine the need for prompt treatment |
(22) |
| Medical product development | To provide information about the safety and effectiveness of a drug | (23) |
| Public health | To provide information about the risk of developing any disease or medical condition among the population | (24) |
| Pharmacodynamics studies | To provide evidences about therapeutic response | (25) |
Pharmacodynamic or Response Biomarker
Proposed to be a potential useful tool in clinical practice providing useful information for patient management. A pharmacodynamic biomarker is modified in response to a medical condition or clinical intervention, including drug treatments (16). Because of the serial nature of their assessment, this type of biomarker is frequently considered as a monitoring biomarker (20). The main utility of this biomarker is to guide the clinical management, providing crucial information for deciding whether or not to continue the treatment. Thus, pharmacodynamic biomarkers determine the progression of the treatment (26) (Figure 2).
Another area in which these biomarkers are of special interest is in early therapeutic drug development, being useful to establish the proof that a drug induces pharmacodynamic changes in humans related to its clinical benefit and to guide dose-response studies (18).
Predictive Biomarker
A marker is considered as a predictive biomarker when its presence or modification allows predicting which patient or group of patients are more likely to experience an effect as consequence of being exposed to a medical product or environmental agent (16). This effect could be a symptomatic benefit, an increase in survival rates or an adverse event. These biomarkers are frequently used in randomized controlled clinical trials of new therapies. In this context, the biomarker is used to select patients for participation or to stratify them into intervention groups. If the biomarker predicts a favorable outcome, its presence may indicate a greater effect of the new therapy compared to the control therapy (20). Thus, the use of predictive biomarkers facilitates the selection of specific patients more likely to respond or not to therapy (Figure 2). An example of a predictive biomarker is the presence of 12 single nucleotide polymorphisms (SNPs) in Had Chinese schizophrenic population, that were correlated with greater olanzapine effectiveness (27).
Prognostic Biomarker
Commonly used to identify the probability of developing a clinical event in patients diagnosed with a disease or medical condition (16). These events include death, disease progression or recurrence, or the development of a new medical condition. In clinical trials, prognostic biomarkers are used to identify patients more likely to develop a clinical event or disease progression, allowing to identify populations at higher risk. In this context, prognostic biomarkers are used as inclusion or exclusion criteria (17). An example of a prognostic biomarker is the number of trinucleotide CAG repetitions in patients with Huntington's disease. A high number of CAG nucleotides repetitions are correlated with a greater threshold of disease's severity (Figure 2) (28).
An additional utility of prognostic biomarkers is in treatment selection. They can provide information about treatment safety, guiding patient hospitalization or their entrance in intensive care units.
Several factors influence the clinical outcome, including the clinical condition severity, the effects induced by all treatments and the intrinsic characteristics of patients. Some of these characteristics may be used as a prognostic biomarker, allowing to identify patients more likely to experience a clinical event, disease recurrence or progression, and any effect (favorable or unfavorable) induced by a medical product or environmental agent (16, 20).
Safety Biomarker
Is any measure that can be assessed before and after the exposure to a medical intervention, or an environmental agent, allowing to identify the probability of developing signs of toxicity as an adverse event, to detect the presence of toxicity, and for monitoring its extension (Figure 2) (16).
For many therapies, monitoring hepatic, renal and cardiovascular functions are critical to detect toxicity ensuring the safety of the therapy under study. All safety biomarkers have in common its ability to detect or predict toxicity prior to theNBSP;onset of clinical signs and before irreversible damage. The toxicity can be determined by the detection or changes in the biomarker level.
Another usefulness of safety biomarkers is the identification of patients in which particular therapies should not be initiated because of significant safety risks. For example, genetic variations in CYP2D6 enzymes modify the response to certain drugs commonly used in psychiatry such as almost 50% of antipsychotics drugs. Alterations in the metabolism of drugs can modify its effectiveness, decreasing the response to the treatment or enhancing toxicity risk in patients (15). In case of the antipsychotic risperidone, there is a correlation between the number of active CYP2D6 genes and its cardiac toxicity. QTc interval is longer in subjects with one active CYP2D6 gene compared to patients with two. The study revealed that the number of CYP2D6 active genes was related with the corrected plasma concentration of risperidone (29). Safety biomarkers are used with this purpose in public health or in epidemiological interventions aimed to control or mitigate risk exposure.
Susceptibility or Risk Biomarker
Is used as a risk measure to develop a disease or medical condition (8). An example is a genetic biomarker that can be detected many years or decades before the onset of clinical signs and/or symptoms of the disease (Figure 2) (10). Susceptibility/risk biomarkers are essential for the development of epidemiological studies aimed to evaluate the risk of developing a disease, contributing to establish preventive strategies in clinical practice. In this line, some studies suggested a potential correlation between interleukin-6 (IL-6) and C-reactive protein (CRP) levels and the risk of developing SCZ. Lower CRP levels together with the blockade of IL-6 signaling significantly increase SCZ risk, being proposed as a potential susceptibility/risk biomarkers for this neuropsychiatric disorder (30).
Samples and Techniques Used for the Searching of Biomarkers
Biomarkers should be easily measurable, in easily accessible samples and using affordable techniques to ensure its inclusion in the routine clinical practice. Historically, plasma together with tissues obtained from biopsies were one of the most common samples used in the searching for biomarkers. Besides, based on the disease of interest, additional body fluids readily available in large amounts as urine, saliva, tear fluid, sweat, amniotic, cerebrospinal and pleural fluids, cervicovaginal secretion and wound efflux can be used for this purpose (31).
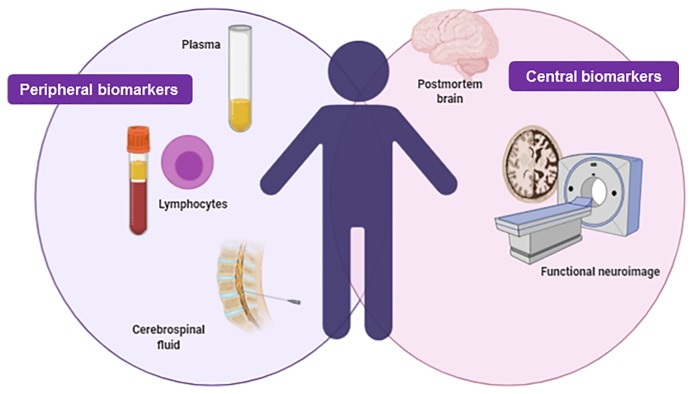
In the case of diseases of the central nervous system (CNS), such as psychiatric and neurological disorders, access to brain samples is of particular interest. In this respect, brain human post-mortem samples, usually provided by brain banks, play a crucial role. However, systematic biochemical investigations using these samples are scarce, limited and unrealistic mainly to the fact that the course of the disease cannot be monitored. In this respect, the progress of functional neuroimaging has allowed to study some neuronal functions including alterations of local cerebral flow, energy metabolism and neurotransmitter receptor density and occupation over the course of disease. Nevertheless, functional neuroimaging fails to provide information at cellular biochemistry level and the access to this technique is limited due to its high economic costs.
In this context, blood lymphocytes have gained special attention in the searching of peripheral biomarkers (32). Lymphocytes can be isolated easily from blood samples and studied on a daily basis allowing to monitor the course of the disease. This is possible due to the fact that receptor properties and transduction processes of lymphocytes are similar to those observed in the CNS. Several studies pointed out a close bidirectional interaction between the CNS and the immune system, in particular with lymphocytes (33). For instance, peripheral cytokines released by lymphocytes modify CNS functions including its autonomic control as well as neuroendocrine and behavioral responses. Besides, several evidences suggested that alterations in neurotransmitters and hypothalamic-pituitary-adrenal (HPA) axis in the CNS are concomitant with alterations in the function and metabolism of lymphocytes.
To date, some genes such as c-fos, interleukins (IL-2, IL-4, IL-6, IL-10), nerve growth factor (NGF), brain-derived neurotrophic factor (BDNF), cannabinoid receptors, acetylcholine, GABAA receptors, B2-adrenergic receptors, glucocorticoid receptors, mineralocorticoid receptors, D3 dopaminergic receptor, and serotonin receptors have been analyzed in lymphocytes from psychiatric patients, such as schizophrenic and depressive patients, with promising results as peripheral biomarkers (34–41). Thus, gene expression studies in lymphocytes of psychiatric patients at different stages of the disease, that may reflect alterations in the CNS, would allow to further characterize the mechanisms underlying the pathogenesis of the disease and may contribute to predict the pharmacological treatment response (biomarkers of treatment outcome) (Figure 3).
Figure 3.
Integrative figure regarding the main samples used in the searching of biomarkers in neuropsychiatry: peripheral and central biomarkers. Created with BioRender.com.
Another crucial factor for the searching of biomarkers is the techniques used. These techniques should have a high-throughput for the application of analytical data through robust dimensional data obtained in high performance tests (42). In this respect, ‘omics' technologies, including genomics, proteomics, transcriptomics, metabolomics, and epigenetics, have contributed to the rapid discovery of many potential biomarkers.
‘Omics’ Biomarkers and Neuropsychiatry
This section summarizes the main advantages of each ‘omics’ technology in the search of biomarkers for assessing risk, diagnosis, monitoring progression and prediction of treatment response in neuropsychiatry disorders.
Genomic Biomarkers
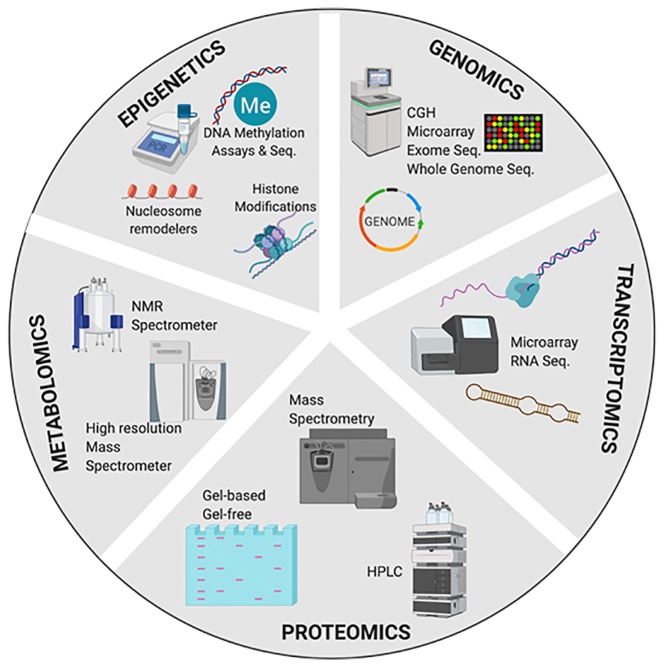
Genomic biomarkers are expanding knowledge for the understanding of disease pathogenesis providing new targets for disease characterization, early diagnosis, and better-targeted treatment (drug discovery, drug development and adverse drug responses) to direct patients towards a more likely benefit based on their unique profile (43). According to the European Medicines Agency (EMEA), a genomic biomarker is defined as “a measurable DNA and/or RNA characteristic that is an indicator of normal biologic processes, pathogenic processes, and/or response to therapeutic or other interventions” (44). These measurable features include the expression, function and regulation of a particular gene. In the DNA, these features can be characterized by single nucleotide polymorphisms (SNPs), variability of short sequence repeats, haplotypes, deletions or insertions of (a) single nucleotide (s), copy number variations and cytogenetic rearrangements (translocations, duplications, deletions or inversions) (45). The use of genetic techniques allowed the analysis of candidate genes, genome-wide studies and polygenetic risk score analysis to understand multiple psychiatric disorders such as SCZ (46, 47). These techniques include Comparative Genomic Hybridization (CGH), microarray, exome sequencing, and whole genome sequence. Specifically, pharmacogenomics is crucial to identify genetic polymorphisms in drug-metabolizing enzymes, transporters, receptors, and other drug targets, being essential to drug discovery and drug therapy optimization [for review (48, 49)] (Figure 4).
Figure 4.
Multi-omics approach for the discovery and validation of biomarkers to probe multidimensional phases of the disease. CGH, comparative genomic hybridization; Seq, sequencing; qRT-PCR, quantitative real time PCR; qPCR, semiquantitative real time PCR; HPLC, high performance liquid chromatography; NMR, nuclear magnetic resonance spectroscopy. Created with BioRender.com.
Genome-wide association studies have allowed the identification of potential genomic biomarkers in different neuropsychiatric (50–52) and drug-use disorders (53). For example, the Collaborative Study of the Genetics of Alcoholism (COGA) have correlated the SNPs rs4780836 [A > C; chromosome 16:19974071 (GRCh38.p12)], rs2605140 [A > G; chromosome 17: 18253061 (GRCh38.p12)], rs11690265 [chromosome 2: C > T; chr2:27418655 (GRCh38.p12)], rs692854 [non-functional Se (FUT2) gene; alleles C > A; chromosome 19: 48706207 (GRCh38.p12)], and rs13380649 (alleles A > G; chromosome 16: 19999778 (GRCh38.p12)] with greater vulnerability and predisposition to develop alcohol use disorders (AUD) in European American and African American. Besides, the study has demonstrated that there is a correlation between these SNPs and alterations in electroencephalograms, such as lower posterior gamma, higher slow wave connectivity (delta, theta, alpha), higher frontal gamma ratio and higher beta correlation in the parietal area of patients with AUD (54).
In bipolar disorder (BD), the SNP rs17026688 in the gene encoding glutamate decarboxylase-like protein 1 (GADL1) has been associated with the response to lithium in Chinese patients (55). This SNP has been related to immune dysfunctions in BD patients, such as higher percentages of total T cells, CD4+T cells, activated B cells and monocytes. Besides, treatment of BD patients-derived peripheral blood mononuclear cells (PBMCs) with lithium in vitro increases the immune response (CD4+ cells). These findings suggest that the immune imbalance might not only be a biomarker for diagnosis but also a biomarker of the disease progression and therapeutic response in BD
In addition, a large study carried out through Europe, North America and Australia identified 30 genome-wide significant loci for BD in patients of European descent. These loci contain genes that encode for neurotransmitters transporters, synaptic components, and ion channels, including calcium voltage-gated channel subunit alpha1 C (CACNA1C) and other voltage-gated calcium channel genes. Among the 30 loci identified in BD patients, eight have also been described in SCZ patients (56–58); however, conditional analyses performed in this study suggested that BD and SCZ associations are independent for three of the eight shared loci, providing information that may be useful for understanding the genetics mechanisms underlying these psychiatric disorders that in some cases present symptoms in common that make its diagnosis difficult. Furthermore, the BD subtype polygenic risk score analyses performed in the study supported the nosological distinction between bipolar I (BD1) and bipolar II disorder (BD2) and the importance of psychosis beyond DSM subtypes. One limitation of the study is the genetic heterogeneity of the samples that may contribute to inconsistent replication in some of the results (59).
Besides, DNA genomic biomarkers as a useful indicator of the state of the disease (severity) also presented relevant consequences for the clinical management of neuropsychiatric diseases. In this respect, a recent study revealed a close relationship between the SNPs rs1360780 in the FK506-binding protein 5 (FKBP5) gene and rs17689918 in the corticotrophin-releasing hormone receptor 1 (CRHR1) gene and greater severity of the disease in posttraumatic stress disorder (PTSD) patients (60).
Transcriptomic Biomarkers in Neuropsychiatry
The transcriptome is defined as the complete set of all RNA molecules in one cell or a population of cells at a specific developmental stage or physiological condition (61). Thus, transcriptome is dynamic and reflects the cellular state. Measuring the expression of an organism’s genes as a snapshot in different tissues, conditions, or time points provides information on how genes are regulated and would contribute to a better understanding of human diseases and their pharmacological treatment, allowing to identify potential therapeutic biomarkers when variations in treatment outcomes occur (62, 63). Although first studies for transcriptome began in the early 1990s, technological advances have spread throughout this time. There are two key contemporary techniques in the field: microarrays, which quantify a set of predetermined sequences, and RNA sequencing (RNA-Seq), which uses high-throughput sequencing to capture all RNA sequences (63).
For instance, transcriptomics studies identified that the efficacy of antidepressants is related to gene expression changes at transcriptome-wide scale. In a microarray study, alterations of MMO28 and KXD1 genes encoding for matrix metallopeptidase 28 and KxDL motif-containing protein 1, respectively, were associated with better response to nortriptyline in depressive patients (64). This data could contribute to improve the characterization of the molecular pathways underlying the efficacy of antidepressants.
In addition, a clinical study associated new miRNAs (miR-146a-5p, miR-146b-5p, miR-24-3p, and miR-425-3p) with the effectiveness of antidepressant drugs such as duloxetine, escitalopram, and nortriptyline, in patients with MDD. These miRNAs are ubiquitously expressed and highly correlated in blood and in brain tissue, playing an important role in the regulation of the mitogen-activated protein kinase (MAPK) and Wnt signaling pathways, closely related with stress response and MDD (65). Interestingly, an additional study revealed that MDD patients responders to antidepressant treatment present a significant reduction of miR-1202 baseline levels compared to non-responders. Moreover, miR-1202 increases as the efficacy of the antidepressant treatment is observed (66).
Besides, a total of 25 miRNAs have been modified in the amygdala of rats exposed to the learned helpless animal model of depression being the miR-128-3p the most affected. Also, a reduction of Wnt signaling genes has been also detected. Accordingly, an increase of miR-128-3p expression along with a significant downregulation of key target genes from Wnt pathway signaling (WNT5B, DVL, and LEF1) has been identified in the AMY of MDD patients (67).
Especially, RNA studies provide promising results in the searching for biomarkers of suicide. Alterations of RNA editing on the cyclic nucleotide phosphodiesterase (PDE, particularly PDE8A involved in the hydroxylation of cAMP and cGMP) were found in the dorsolateral prefrontal cortex (DLPFC) and the anterior cingulate cortex (ACC) of suicide completers. These alterations have been proposed as a biomarker of risk for attempting suicide in patients with depressive symptoms (68).
Other example of the potential role of these biomarkers in neuropsychiatry is a genome-wide expression study in patients who met the DSM-IV criteria for methamphetamine dependence. The results revealed that treatment with topiramate significantly modified the gene expression of specified genes GRINA, PRKACA, PRKCI, SNAP23 and TRAK2 involved in severe pathways underlying drug addiction and other relevant physiological functions, including neuronal function/synaptic plasticity, signal transduction, cardiovascular function, and inflammation/immune response (69).
Likewise, the microRNA-124 (miR-124) and microRNA-181 (miR-181) were pointed out as potential biomarkers for cocaine use disorder (CUD) (70). The study revealed that these two microRNAs were upregulated in the blood samples of females CUD compared with healthy female controls.
Proteomic Biomarkers in Neuropsychiatry
Proteomics approaches using blood, plasma or serum constitutes a highly desired method for biomarker profiling of psychiatric disorders, due to the fact that these biological samples are used for routine diagnostic analyses in clinical practice, making easier to obtain samples. Besides, in neuropsychiatry, the cerebrospinal fluid (CSF) is a sample of particular interest for the identification of potential proteomic biomarkers due to its proximity to the brain. Although its collecting is very complex, due to the invasive procedure involved, it contains much less proteins than plasma. Thus, the “buffering” of protein composition is much weaker and tend to lead in a reduction of chances to identify potential proteomic biomarkers.
In proteomics, the separation of proteins using gel-based or gel free techniques, commonly followed by mass spectrometry are the mainly techniques used. The strategies for obtaining biological samples are diverse, but it is recommended to reduce the complexity of the sample, and sometimes to employ enrichment techniques improving the levels of certain subcellular fractions of interest or for specific types of proteins (glycoproteins, membrane, secreted, nuclear matrix and phosphorylated proteins) (71).
Diagnostic complications and timely treatment in neuropsychiatric disorders are frequent. Such is the case of SCZ, diagnosed by certain signs and symptoms but not by measurable and identifiable biological characteristics. In this respect, proteome studies carried out in blood plasma, serum and postmortem brain tissue from SCZ patients identified alterations in proteins that play a significant role in neuronal transmission and synaptic function, calcium homeostasis and signalling, energy metabolism, oxidative stress, cytoskeleton and in immune system and inflammation. These proteins have been proposed as biomarker candidates for prognosis, diagnosis, and medication monitoring in SCZ (72, 73). One of these proteins is zinc finger protein 729 that was found significantly down-regulated in patients with SCZ compared to healthy individuals and patients diagnosed with depression or BD (74). Another example is the study that showed reduced plasma levels of glia maturation factor beta (GMF-β), the brain-derived neurotrophic factor (BDNF), and the 115-kDa isoform of the Rab3 GTPase-activating protein catalytic subunit (RAB3GAP1) in SCZ patients. These biological markers have been proposed as potential biomarkers in this pathology (75).
Besides, the acetyl-l-carnitine (LAC) has been proposed as a proteomic biomarker in MDD. LAC plays an important role in several behavioral features. The reduction of LAC concentrations was associated with abnormal hippocampal glutamatergic function and plasticity. Such alterations suggested that the degree of LAC deficiency was directly proportional with the severity, the age of MDD onset, and the clinical history of treatment-resistant depression (TRD). These findings suggest that LAC may be useful as a diagnostic and prognosis biomarker for MDD (76).
Recently, neurofilaments light chains (NF-L) have been proposed as potential biomarkers for neuronal damage in certain psychiatric diseases. In the plasma of female patients affected by anorexia nervosa, levels of NF-L were significantly elevated, being associated with the neuronal damage observed in AN patients, that partially normalizes with weight recovery (77). An additional study pointed out the potential role of NF-L as a discriminative biomarker between primary psychiatric disorders and neurodegenerative clinical conditions with wide-ranging of behavioral, psychiatric, and cognitive symptoms (78). Interestingly, a reduction of NF-L has been identified in the hippocampus of rodents exposed to an animal model of depression (inescapable stress). In this study, treatment with valproic acid reduces depressive-like behavior and reverses NF-L reduction (79). Besides, elevated concentrations of NF-L have been observed in the CSF of BD patients. Authors demonstrated that there is a positive correlation between CSF NF-L levels and the response to antipsychotics and lithium (80). However, another prospective study failed to observe any association between the high baseline NF-L levels in CSF and clinical outcomes in BD (81). Further studies are needed to identify the role of neurofilaments as biomarkers for psychiatric disorders.
In conclusion, to consider a potential proteomic biomarker, it is necessary to evaluate its sensitivity, specificity and positive or negative predictive values upon the disease of interest (31). In this respect, modern proteomics workflows that enable high throughput studies with large cohorts of well-defined samples represent the opportunity to solve the limited reproducibility of past proteomic workflows (73).
Metabolomics Biomarkers in Neuropsychiatry
Metabolomics biomarkers for drug development are growing. This technology focuses on the presence of small molecules metabolites in various complex matrices like CSF, blood, urine, saliva, and other human fluids. The metabolome is inherently more dynamic and time sensitive than proteome and genome, providing a direct functional measure of cellular activity and physiological status (82, 83). Changes in metabolome are the consequence of the interaction between lifestyle, environmental, genetic, developmental, and pathological factors. Consequently, metabolomics are of particular interest because, in contrast to genomics, captures the dynamic nature of the disease, and in contrast to proteomics, metabolomics measure the final products produced by complex interactions between proteins, signalling cascades and cellular environments.
Metabolomics biomarkers are not characterized by one single metabolite. Rather, they are a set of correlated metabolites defining a specific state of disease or the response to a clinical or pharmacological intervention (84). Currently, gas chromatography-mass spectrometry (GC-MS), liquid chromatography-mass spectrometry (LC-MS), and nuclear magnetic resonance (NMR) are the main types of analytical platforms used in the searching for metabolomics biomarkers.
Several studies have focused on the identification of potential metabolomics biomarkers in different psychiatric diseases [for review see (85–88)]. For example, urinary metabolomics have been proposed to be potential useful tools for the identification of pathways that may be involved in the mechanism of action of specific treatments. Such is the case of the study in which 77 urinary metabolites were identified in children with autism spectrum disorder treated with sulforaphane, a supplement that significantly improved the social responsiveness. Some of these metabolites play a role in the regulation of neurotransmitters, hormones, oxidative stress, amino acid/gut microbiome, and sphingomyelin metabolism (89).
Recently, a study conducted in patients with symptoms of depression, tried to found a predictor or a biological correlation of depression recovery after the administration of certain antidepressants including escitalopram, bupropion-escitalopram or venlafaxine-mirtazapine combinations. An increase of phosphatidylcholine C38:1 baseline plasma concentrations was associated with poorer outcome in patients. In contrast, an increased ratio in hydroxylated sphingomyelins after 8 weeks of treatment was linked to symptoms recovery (90).
However, few metabolomics biomarkers, especially in neuropsychiatry, have passed the regulatory standards for their use in clinical practice, mainly due to the lack of robust assays for the routine quantification of potential biomarkers and the heterogeneity of studies. The reduced sample size, particularly of some clinical subgroups, and the limited quantitative power of current mass spectroscopy technology, hampered the identification of robust metabolomics biomarkers, making necessary their validation trough additional assays. Besides, the complexity of samples, such as urine or blood, also contributes to that reality. In this respect, the use of chromatographic techniques for separation is needed to reduce the potential interferences associated with the complexity of human samples (91).
Epigenetic Biomarkers
Dynamic variations in the structure of chromatin, which do not change the sequence of DNA itself but modified the expression of genes, have been paid attention due to its potential implication in the development of human diseases, including psychiatric disorders (92, 93). Accordingly, epigenetics may provide a functional interface between genotype, environmental exposure and phenotype (94).
To date, different forms of epigenetic regulation have been identified, such as direct methylation of DNA, histone modifications (as methylation and acetylation ubiquitination), exchanges of histone molecules with related isoforms, modification on chromatin by nucleosome remodelers that modify the access to DNA, and additional mechanisms like non-coding RNAs, non-genic DNAs, and differential exosome expression (95). In this way, identifying the aberrant changes in the epigenetic scenery associated to neuropsychiatry diseases and the factors that promote such alterations may allow the identification of potential new biomarkers (96).
An epigenetic biomarker is defined as “any epigenetic mark or altered epigenetic mechanism that can be measured in the body fluids or tissues defining a disease (detection); predicts the outcome of disease (prognostic), responds to therapy (predictive); monitors responses to therapy or medication (therapy monitoring) and predicts risk of future disease development (risk)” (97). So far, several techniques have been designed to analyze not only epigenetic processes at the level of specific genes but also epigenetic changes that occur in defined regions of the genome by epigenome-wide association studies. DNA methylation assays and DNA methylation sequencing are the most employed techniques, but not exclusively. Novel epigenetic techniques, such as those provided by CRISPR/Cas9 system, represent new opportunities in the searching for epigenetic biomarkers (98).
Many of the findings achieved thus far are encouraging, revealing significant associations with epigenetic modulations of genes regulating neurotransmission, neurodevelopment, and immune function in psychiatric diseases (99). One example is the hypermethylation of BDNF gene identified in brain and peripheral blood samples of MDD, SCZ and BD patients (100, 101). Another similar example is the hypermethylation of FKBP5 gene, an important modulator of stress response, detected in peripheral blood samples of PTSD patients (102). In panic disorder, hypomethylation of monoamine oxidase A (MAOA) and glutamate decarboxylases 1 (GAD1) genes have been evident in recent studies (103).
In suicide, advances in epigenetic techniques have allow to characterize epigenetic alterations in key elements of the hypothalamus-pituitary-adrenal axis (HPA-axis), neurotrophic factors, serotoninergic and GABAergic systems, that have been proposed as epigenetic biomarkers for suicide, suicide ideation and suicide attempt (104).
Interestingly, epigenetic biomarkers have been pointed out as potential biomarkers for guiding treatment. Thus, antipsychotic drugs, such as olanzapine, induced DNA methylations alterations through the brain in SCZ patients, changes related with its efficacy (105, 106). For instance, reduced response to antidepressants has been associated with the absence of methylation at a specific CpG site in exon 4 of BDNF in MDD patients (107). Consequently, BDNF exon 4 methylation, and circulating BDNF protein levels may be used together as a predictive tool to personalize treatment of MDD (108).
More interestingly, histone deacetylases (HDAC), that have been demonstrated to control epigenetic programming associated with the modulation of behaviour and cognition, appears to be crucial for reversing dysfunctional epigenetic regulation induced by early life events exposure in preclinical models (109, 110). Additional studies have supported the potential role of HDAC as promising new therapeutic targets for the treatment of MDD (111). In this context, HDAC inhibitors, alone or in combination with current antidepressant drugs, are currently being explored (112–114).
Altogether, epigenetic studies highlight the importance of epigenetic mechanisms on controlling genes or gene complexes. In neuropsychiatry, despite huge advances were achieved, there are still far for providing a clear molecular mechanism underlying these disorders and effective treatment options. The heterogeneity of the techniques and methods used, with a range in sensitivity for detecting effects (115–117); the lack of adjusting the genome-wide results to account for cell specificity (118, 119); the confounding factors such as patient's treatment, population origin and phenotypes included (105, 120); and the lack of further studies to demonstrate the concordance between brain-blood data have hampered the clinical use of epigenetic biomarkers (121).
Summary and Conclusions
As set out in this review, there are several proteins, metabolites and genes that have been linked with certain neuropsychiatric diseases mainly due to the advance in ‘omics' technologies. However, none of them have demonstrated to be a real and useful biomarker in clinical practice.
Despite each ‘omic' presents its limitations and challenges (122–124), three essential key targets are in common to advance in the searching of biomarkers in neuropsychiatry: 1) accurate selection of the clinical population, 2) shortened sampling time and 3) standardization of procedures for sample processing. These items can be applied for any diseases, but are of special interest for psychiatric disorders. The broad spectrum of phenotypes in patients diagnosed from the same psychiatric disorder and the overlapping of some traits or clusters in different neuropsychiatric disorders, which can often make diagnosis difficult, increases the heterogeneity of the clinical population analyzed. To overcome this issue, emerged ‘omics' studies have focused on the identification of potential biomarkers for specific traits. However, the reduced number of samples analyzed per trait/phenotype has made difficult to achieve robust conclusions about the potential clinical use of the proposed biomarkers. In this respect, modern ‘omics’ workflows that enable high throughput studies with large cohorts of well-defined samples can solve this problem.
Besides, the heterogeneity of procedures for sample processing along with the differences in power and sensitivity of each ‘omics' technologies have contribute to that reality. In this respect, new ‘omics’ with better quantity power and sensitivity would contribute to find robust and realistic biomarkers.
One of the major challenges still lying ahead is the way to integrate the plethora of data obtained from each ‘omics’ to reach the holistic realization of a ‘systems biology’ understanding the biological question (125). In this context, bioinformatics tools have been designed to understand the potential of ‘omics’ technology (126).
Another concern is that current biomarker validation is a lengthy and complex process. In essence, this process includes the validation of the method, determined by the characteristics of the assay employed, and the clinical validation, to provide evidences that the biomarker is linked specifically with the disease or clinical end point under consideration. Is in this aspect in which future longitudinal integrative ‘omics’ studies can be crucial to provide a rigorously biomarkers validation ensuring its sensitivity, specificity, predictive value, and likelihood ratio, by its assessment in a large cohort (normal clinical population). It is expected that in the following years considerable breakthroughs will occur in these regards.
Author Contributions
MG-G and JM conceived the presented idea. MG-G took the lead in writing the manuscript. FN, FS, AG, and AA-O wrote the manuscript in consultation with MG-G. All authors provided critical feedback and helped shape the research, analysis, and manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank all participants in this study. We greatly appreciated to Biorender for helping to create Figures 3 and 4.
References
- 1. Ilyas A, Chesney E, Patel R. Improving life expectancy in people with serious mental illness: should we place more emphasis on primary prevention? Br J Psychiatry (2017) 211(4):194–7. 10.1192/bjp.bp.117.203240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chesney E, Goodwin GM, Fazel S. Risks of all-cause and suicide mortality in mental disorders: a meta-review. World Psychiatry (2014) 13(2):153–60. 10.1002/wps.20128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. OECD. Safety OfEC-oadECfHaF Health at a glance: Europe 2018. State of health in the EU cycle. Paris: OECD Publishing; (2018). [Google Scholar]
- 4. Swartz MS, Stroup TS, McEvoy JP, Davis SM, Rosenheck RA, Keefe RS, et al. What CATIE found: results from the schizophrenia trial. Psychiatr Serv (2008) 59(5):500–6. 10.1176/ps.2008.59.5.500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sinyor M, Schaffer A, Levitt A. The sequenced treatment alternatives to relieve depression (STAR*D) trial: a review. Can J Psychiatry (2010) 55(3):126–35. 10.1177/070674371005500303 [DOI] [PubMed] [Google Scholar]
- 6. Parikh SV, LeBlanc SR, Ovanessian MM. Advancing bipolar disorder: key lessons from the Systematic Treatment Enhancement Program for Bipolar Disorder (STEP-BD). Can J Psychiatry (2010) 55(3):136–43. 10.1177/070674371005500304 [DOI] [PubMed] [Google Scholar]
- 7. Gorman JM. Comorbid depression and anxiety spectrum disorders. Depression Anxiety (1996) 4(4):160–8. [DOI] [PubMed] [Google Scholar]
- 8. Angst J, Merikangas K. The depressive spectrum: diagnostic classification and course. J Affect Disord (1997) 45(1-2):31–9; discussion 9-40. 10.1016/s0165-0327(97)00057-8 [DOI] [PubMed] [Google Scholar]
- 9. Pincus HA, Tew JD, First MB. Psychiatric comorbidity: is more less? World Psychiatry (2004) 3(1):18–23. [PMC free article] [PubMed] [Google Scholar]
- 10. Green AI, Canuso CM, Brenner MJ, Wojcik JD. Detection and management of comorbidity in patients with schizophrenia. Psychiatr Clinics North America (2003) 26(1):115–39. 10.1016/s0193-953x(02)00014-x [DOI] [PubMed] [Google Scholar]
- 11. Buckley PF, Miller BJ, Lehrer DS, Castle DJ. Psychiatric comorbidities and schizophrenia. Schizophr Bull (2009) 35(2):383–402. 10.1093/schbul/sbn135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mundkur BD. Evidence excluding mutations, polysomy, and polyploidy as possible causes of non-mendelian segregations in Saccharomyces. Ann Messouri Botanical Garden (1949) 36(3):23. 10.2307/2394394 [DOI] [Google Scholar]
- 13. Porter KA. Effect of homologous bone marrow injections in x-irradiated rabbits. Br J Exp Pathol (1957) 38(4):401–12. [PMC free article] [PubMed] [Google Scholar]
- 14. Biomarkers Definitions Working G Biomarkers and surrogate endpoints: preferred definitions and conceptual framework. Clin Pharmacol Ther (2001) 69(3):89–95. 10.1067/mcp.2001.113989 [DOI] [PubMed] [Google Scholar]
- 15. FitzGerald GA. Measure for Measure: Biomarker standards and transparency. Sci Trans Med (2016) 8(343):343fs10. 10.1126/scitranslmed.aaf8590 [DOI] [PubMed] [Google Scholar]
- 16. Biomarker Working Group F-N. BEST (Biomarkers, Endpoints, and other Tools) Resource. In: Spring S, editor. BEST (Biomarkers, Endpoints, and other Tools) Resource. Silver Spring (MD): FDA-NIH; (2016). [Google Scholar]
- 17. Aronson JK, Ferner RE. Biomarkers-A General Review. Curr Protoc Pharmacol (2017) 76:9 23:1–9 17. 10.1002/cpph.19 [DOI] [PubMed] [Google Scholar]
- 18. Cagney DN, Sul J, Huang RY, Ligon KL, Wen PY, Alexander BM. The FDA NIH Biomarkers, EndpointS, and other Tools (BEST) resource in neuro-oncology. Neuro Oncol (2018) 20(9):1162–72. 10.1093/neuonc/nox242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Olsson B, Lautner R, Andreasson U, Ohrfelt A, Portelius E, Bjerke M, et al. CSF and blood biomarkers for the diagnosis of Alzheimer's disease: a systematic review and meta-analysis. Lancet Neurol (2016) 15(7):673–84. 10.1016/S1474-4422(16)00070-3 [DOI] [PubMed] [Google Scholar]
- 20. Califf RM. Biomarker definitions and their applications. Exp Biol Med (Maywood) (2018) 243(3):213–21. 10.1177/1535370217750088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kraus VB. Biomarkers as drug development tools: discovery, validation, qualification and use. Nat Rev Rheumatol (2018) 14(6):354–362. 10.1038/s41584-018-0005-9 [DOI] [PubMed] [Google Scholar]
- 22. Prata J, Santos SG, Almeida MI, Coelho R, Barbosa MA. Bridging Autism Spectrum Disorders and Schizophrenia through inflammation and biomarkers - pre-clinical and clinical investigations. J Neuroinflammation (2017) 14(1):179. 10.1186/s12974-017-0938-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pu S, Setoyama S, Noda T. Association between cognitive deficits and suicidal ideation in patients with major depressive disorder. Sci Rep 2017. 7(1):11637. 10.1038/s41598-017-12142-8. PMID: 28912439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rosen C, Zetterberg H. Cerebrospinal fluid biomarkers for pathological processes in Alzheimer's disease. Curr Opin Psychiatry 2013. 26(3):276–282. 10.1097/YCO.0b013e32835f6747 [DOI] [PubMed] [Google Scholar]
- 25. Wiecki TV, Antoniades CA, Stevenson A, Kennard C, Borowsky B, Owen G, et al. A Computational Cognitive Biomarker for Early-Stage Huntington's Disease. PLoS One 2016. 11(2):e0148409. 10.1371/journal.pone.0148409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Okazaki S, Hishimoto A, Otsuka I, Watanabe Y, Numata S, Boku S, et al. Increased serum levels and promoter polymorphisms of macrophage migration inhibitory factor in schizophrenia. Prog Neuropsychopharmacol Biol Psychiatry (2018) 83:33–41. 10.1016/j.pnpbp.2018.01.001 [DOI] [PubMed] [Google Scholar]
- 27. Zhou W, Xu Y, Lv Q, Sheng YH, Chen L, Li M, et al. Genetic Association of Olanzapine Treatment Response in Han Chinese Schizophrenia Patients. Front Pharmacol (2019) 10:177. 10.3389/fphar.2019.00177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ghosh R, Tabrizi SJ. Huntington disease. Handb Clin Neurol (2018) 147:255–78. 10.1016/B978-0-444-63233-3.00017-8 [DOI] [PubMed] [Google Scholar]
- 29. Llerena A, Berecz R, Dorado P, de la Rubia A. QTc interval, CYP2D6 and CYP2C9 genotypes and risperidone plasma concentrations. J Psychopharmacol (2004) 18(2):189–93. 10.1177/0269881104042618 [DOI] [PubMed] [Google Scholar]
- 30. Hartwig FP, Borges MC, Horta BL, Bowden J, Davey Smith G. Inflammatory Biomarkers and Risk of Schizophrenia: A 2-Sample Mendelian Randomization Study. JAMA Psychiatry (2017) 74(12):1226–33. 10.1001/jamapsychiatry.2017.3191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lescuyer P, Hochstrasser D, Rabilloud T. How shall we use the proteomics toolbox for biomarker discovery? J Proteome Res (2007) 6(9):3371–6. 10.1021/pr0702060 [DOI] [PubMed] [Google Scholar]
- 32. Gladkevich A, Kauffman HF, Korf J. Lymphocytes as a neural probe: potential for studying psychiatric disorders. Prog Neuropsychopharmacol Biol Psychiatry (2004) 28(3):559–76. 10.1016/j.pnpbp.2004.01.009 [DOI] [PubMed] [Google Scholar]
- 33. Quan N, Herkenham M. Connecting cytokines and brain: a review of current issues. Histol Histopathol (2002) 17(1):273–88. 10.14670/HH-17.273 [DOI] [PubMed] [Google Scholar]
- 34. Elenkov IJ, Chrousos GP, Wilder RL. Neuroendocrine regulation of IL-12 and TNF-alpha/IL-10 balance. Clinical implications. Ann New York Acad Sci (2000) 917:94–105. 10.1111/j.1749-6632.2000.tb05374.x [DOI] [PubMed] [Google Scholar]
- 35. Peng S, Li W, Lv L, Zhang Z, Zhan X. BDNF as a biomarker in diagnosis and evaluation of treatment for schizophrenia and depression. Discovery Med (2018) 26(143):127–36. [PubMed] [Google Scholar]
- 36. Chu CS, Chu CL, Wu CC, Lu T. Serum nerve growth factor beta, brain- and glial-derived neurotrophic factor levels and psychopathology in unmedicated patients with schizophrenia. J Chin Med Assoc : JCMA (2018) 81(6):577–81. 10.1016/j.jcma.2017.11.010 [DOI] [PubMed] [Google Scholar]
- 37. Annunziata P, Cioni C, Mugnaini C, Corelli F. Potent immunomodulatory activity of a highly selective cannabinoid CB2 agonist on immune cells from healthy subjects and patients with multiple sclerosis. J Neuroimmunol (2017) 303:66–74. 10.1016/j.jneuroim.2016.12.009 [DOI] [PubMed] [Google Scholar]
- 38. de Campos-Carli SM, Araujo MS, de Oliveira Silveira AC, de Rezende VB, Rocha NP, Ferretjans R, et al. Cannabinoid receptors on peripheral leukocytes from patients with schizophrenia: Evidence for defective immunomodulatory mechanisms. J Psychiatr Res (2017) 87:44–52. 10.1016/j.jpsychires.2016.12.001 [DOI] [PubMed] [Google Scholar]
- 39. Ilani T, Ben-Shachar D, Strous RD, Mazor M, Sheinkman A, Kotler M, et al. A peripheral marker for schizophrenia: Increased levels of D3 dopamine receptor mRNA in blood lymphocytes. Proc Natl Acad Sci U S A (2001) 98(2):625–8. 10.1073/pnas.021535398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lima L, Urbina M. Serotonin transporter modulation in blood lymphocytes from patients with major depression. Cell Mol Neurobiol (2002) 22(5-6):797–804. 10.1023/a:1021869310702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hernandez E, Lastra S, Urbina M, Carreira I, Lima L. Serotonin, 5-hydroxyindoleacetic acid and serotonin transporter in blood peripheral lymphocytes of patients with generalized anxiety disorder. Int Immunopharmacol (2002) 2(7):893–900. 10.1016/s1567-5769(02)00025-5 [DOI] [PubMed] [Google Scholar]
- 42. Matsui S. Genomic biomarkers for personalized medicine: development and validation in clinical studies. Comput Math Methods Med (2013) 2013:865980. 10.1155/2013/865980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Novelli G, Ciccacci C, Borgiani P, Papaluca Amati M, Abadie E. Genetic tests and genomic biomarkers: regulation, qualification and validation. Clin cases Miner Bone Metab (2008) 5(2):149–54. [PMC free article] [PubMed] [Google Scholar]
- 44. European Medicines Agency (EMA) ICH Topic E15 Definitions for genomic biomarkers, pharmacogenomics, pharmacogenetics, genomic data and sample coding categories. (2007). [Google Scholar]
- 45. European Medicines Agency (EMA) Definitions for genomic biomarkers, pharmacogenomics, pharmacogenetics, genomic data and sample coding categories. (2007). [Google Scholar]
- 46. Jiang W, King TZ, Turner JA. Imaging Genetics Towards a Refined Diagnosis of Schizophrenia. Front Psychiatry (2019) 10:494. 10.3389/fpsyt.2019.00494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Martin AR, Daly MJ, Robinson EB, Hyman SE, Neale BM. Predicting Polygenic Risk of Psychiatric Disorders. Biol Psychiatry (2019) 86(2):97–109. 10.1016/j.biopsych.2018.12.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. van Westrhenen R, Aitchison KJ, Ingelman-Sundberg M, Jukic MM. Pharmacogenomics of Antidepressant and Antipsychotic Treatment: How Far Have We Got and Where Are We Going? Front Psychiatry (2020) 11:94. 10.3389/fpsyt.2020.00094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Corponi F, Fabbri C, Serretti A. Pharmacogenetics in Psychiatry. Adv Pharmacol (2018) 83:297–331. 10.1016/bs.apha.2018.03.003 [DOI] [PubMed] [Google Scholar]
- 50. Baresic A, Nash AJ, Dahoun T, Howes O, Lenhard B. Understanding the genetics of neuropsychiatric disorders: the potential role of genomic regulatory blocks. Mol Psychiatry (2020) 25(1):6–18. 10.1038/s41380-019-0518-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Maul S, Giegling I, Fabbri C, Corponi F, Serretti A, Rujescu D. Genetics of resilience: Implications from genome-wide association studies and candidate genes of the stress response system in posttraumatic stress disorder and depression. Am J Med Genet Part B Neuropsychiatr Genet : Off Publ Int Soc Psychiatr Genet (2020) 183(2):77–94. 10.1002/ajmg.b.32763 [DOI] [PubMed] [Google Scholar]
- 52. Ikeda M, Saito T, Kondo K, Iwata N. Genome-wide association studies of bipolar disorder: A systematic review of recent findings and their clinical implications. Psychiatry Clin Neurosci (2018) 72(2):52–63. 10.1111/pcn.12611 [DOI] [PubMed] [Google Scholar]
- 53. Sanchez-Roige S, Palmer AA, Clarke TK. Recent Efforts to Dissect the Genetic Basis of Alcohol Use and Abuse. Biol Psychiatry (2020) 87(7):609–18. 10.1016/j.biopsych.2019.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kinreich S, Meyers JL, Maron-Katz A, Kamarajan C, Pandey AK, Chorlian DB, et al. Predicting risk for Alcohol Use Disorder using longitudinal data with multimodal biomarkers and family history: a machine learning study. Mol Psychiatry (2019). 10.1038/s41380-019-0534-x [DOI] [PMC free article] [PubMed]
- 55. Wu TN, Lee CS, Wu BJ, Sun HJ, Chang CH, Chen CY, et al. Immunophenotypes associated with bipolar disorder and lithium treatment. Sci Rep (2019) 9(1):17453. 10.1038/s41598-019-53745-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Schizophrenia Working Group of the Psychiatric Genomics Consortium Biological insights from 108 schizophrenia-associated genetic loci. Nature (2014) 511(7510):421–7. 10.1038/nature13595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ripke S, O'Dushlaine C, Chambert K, Moran JL, Kahler AK, Akterin S, et al. Genome-wide association analysis identifies 13 new risk loci for schizophrenia. Nat Genet (2013) 45(10):1150–9. 10.1038/ng.2742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Goes FS, McGrath J, Avramopoulos D, Wolyniec P, Pirooznia M, Ruczinski I, et al. Genome-wide association study of schizophrenia in Ashkenazi Jews. Am J Med Genet Part B Neuropsychiatr Genet : Off Publ Int Soc Psychiatr Genet (2015) 168(8):649–59. 10.1002/ajmg.b.32349 [DOI] [PubMed] [Google Scholar]
- 59. Stahl EA, Breen G, Forstner AJ, McQuillin A, Ripke S, Trubetskoy V, et al. Genome-wide association study identifies 30 loci associated with bipolar disorder. Nat Genet (2019) 51(5):793–803. 10.1038/s41588-019-0397-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Jaksic N, Sabic Dzananovic E, Aukst Margetic B, Rudan D, Cima Franc A, Bozina N, et al. A Candidate Gene Association Study of FKBP5 and CRHR1 Polymorphisms in Relation to War-Related Posttraumatic Stress Disorder. Psychiatr Danubina (2019) 31(2):269–75. 10.24869/psyd.2019.269 [DOI] [PubMed] [Google Scholar]
- 61. Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet (2009) 10(1):57–63. 10.1038/nrg2484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Karuturi SAJGaRKM Transcriptome Analysis. In: Ranganathan S, Nakai K, Schonbach C. editors. Encyclopedia of Bioinformatics and Computational Biology. Elsevier; (2019) p. 792–805. [Google Scholar]
- 63. Lowe R, Shirley N, Bleackley M, Dolan S, Shafee T. Transcriptomics technologies. PloS Comput Biol (2017) 13(5):e1005457. 10.1371/journal.pcbi.1005457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Hodgson K, Tansey KE, Powell TR, Coppola G, Uher R, Zvezdana Dernovsek M, et al. Transcriptomics and the mechanisms of antidepressant efficacy. Eur Neuropsychopharmacol (2016) 26(1):105–12. 10.1016/j.euroneuro.2015.10.009 [DOI] [PubMed] [Google Scholar]
- 65. Lopez JP, Fiori LM, Cruceanu C, Lin R, Labonte B, Cates HM, et al. MicroRNAs 146a/b-5 and 425-3p and 24-3p are markers of antidepressant response and regulate MAPK/Wnt-system genes. Nat Commun (2017) 8:15497. 10.1038/ncomms15497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Fiori LM, Lopez JP, Richard-Devantoy S, Berlim M, Chachamovich E, Jollant F, et al. Investigation of miR-1202, miR-135a, and miR-16 in Major Depressive Disorder and Antidepressant Response. Int J Neuropsychopharmacol / Off Sci J Collegium Internationale Neuropsychopharmacol (CINP) (2017) 20(8):619–23. 10.1093/ijnp/pyx034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Roy B, Dunbar M, Agrawal J, Allen L, Dwivedi Y. Amygdala-Based Altered miRNome and Epigenetic Contribution of miR-128-3p in Conferring Susceptibility to Depression-Like Behavior via Wnt Signaling. Int J Neuropsychopharmacol / Off Sci J Collegium Internationale Neuropsychopharmacol (CINP) (2020) 23(3):165–77. 10.1093/ijnp/pyz071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Chimienti F, Cavarec L, Vincent L, Salvetat N, Arango V, Underwood MD, et al. Brain region-specific alterations of RNA editing in PDE8A mRNA in suicide decedents. Trans Psychiatry (2019) 9(1):91. 10.1038/s41398-018-0331-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Li MD, Wang J, Niu T, Ma JZ, Seneviratne C, Ait-Daoud N, et al. Transcriptome profiling and pathway analysis of genes expressed differentially in participants with or without a positive response to topiramate treatment for methamphetamine addiction. BMC Med Genomics (2014) 7:65. 10.1186/s12920-014-0065-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Viola TW, Heberle BA, Zaparte A, Sanvicente-Vieira B, Wainer LM, Fries GR, et al. Peripheral blood microRNA levels in females with cocaine use disorder. J Psychiatr Res (2019) 114:48–54. 10.1016/j.jpsychires.2019.03.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Frantzi M, Bhat A, Latosinska A. Clinical proteomic biomarkers: relevant issues on study design & technical considerations in biomarker development. Clin Transl Med (2014) 3(1):7. 10.1186/2001-1326-3-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Nascimento JM, Martins-de-Souza D. The proteome of schizophrenia. NPJ Schizophr (2015) 1:14003. 10.1038/npjschz.2014.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Comes AL, Papiol S, Mueller T, Geyer PE, Mann M, Schulze TG. Proteomics for blood biomarker exploration of severe mental illness: pitfalls of the past and potential for the future. Trans Psychiatry (2018) 8(1):160. 10.1038/s41398-018-0219-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Xu R, Liang J, Luo Y, Wan X, Li K, Qi L, et al. Mass spectrometry identification of potential biomarker proteins in the 150-kD electrophoretic band in patients with schizophrenia. Med (Baltimore) (2018) 97(51):e13553. 10.1097/MD.0000000000013553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Rodrigues-Amorim D, Rivera-Baltanas T, Vallejo-Curto MDC, Rodriguez-Jamardo C, de Las Heras E, Barreiro-Villar C, et al. Proteomics in Schizophrenia: A Gateway to Discover Potential Biomarkers of Psychoneuroimmune Pathways. Front Psychiatry (2019) 10:885. 10.3389/fpsyt.2019.00885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Nasca C, Bigio B, Lee FS, Young SP, Kautz MM, Albright A, et al. Acetyl-l-carnitine deficiency in patients with major depressive disorder. Proc Natl Acad Sci U S A (2018) 115(34):8627–32. 10.1073/pnas.1801609115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Nilsson IAK, Millischer V, Karrenbauer VD, Jureus A, Salehi AM, Norring C, et al. Plasma neurofilament light chain concentration is increased in anorexia nervosa. Trans Psychiatry (2019) 9(1):180. 10.1038/s41398-019-0518-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Katisko K, Cajanus A, Jaaskelainen O, Kontkanen A, Hartikainen P, Korhonen VE, et al. Serum neurofilament light chain is a discriminative biomarker between frontotemporal lobar degeneration and primary psychiatric disorders. J Neurol (2020) 267(1):162–7. 10.1007/s00415-019-09567-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Ferrero AJ, Cereseto M, Sifonios LL, Reines A, Peixoto E, Rubio MC, et al. Cytoskeleton of hippocampal neurons as a target for valproic acid in an experimental model of depression. Prog Neuropsychopharmacol Biol Psychiatry (2007) 31(7):1419–28. 10.1016/j.pnpbp.2007.06.014 [DOI] [PubMed] [Google Scholar]
- 80. Jakobsson J, Bjerke M, Ekman CJ, Sellgren C, Johansson AG, Zetterberg H, et al. Elevated concentrations of neurofilament light chain in the cerebrospinal fluid of bipolar disorder patients. Neuropsychopharmacology (2014) 39(10):2349–56. 10.1038/npp.2014.81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Isgren A, Sellgren C, Ekman CJ, Holmen-Larsson J, Blennow K, Zetterberg H, et al. Markers of neuroinflammation and neuronal injury in bipolar disorder: Relation to prospective clinical outcomes. Brain Behav Immun (2017) 65:195–201. 10.1016/j.bbi.2017.05.002 [DOI] [PubMed] [Google Scholar]
- 82. Quinones MP, Kaddurah-Daouk R. Metabolomics tools for identifying biomarkers for neuropsychiatric diseases. Neurobiol Dis (2009) 35(2):165–76. 10.1016/j.nbd.2009.02.019 [DOI] [PubMed] [Google Scholar]
- 83. Martins-de-Souza D. Proteomics, metabolomics, and protein interactomics in the characterization of the molecular features of major depressive disorder. Dialogues Clin Neurosci (2014) 16(1):63–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Marchand CR, Farshidfar F, Rattner J, Bathe OF. A Framework for Development of Useful Metabolomic Biomarkers and Their Effective Knowledge Translation. Metabolites (2018) 8(4):59. 10.3390/metabo8040059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Shih PB. Metabolomics Biomarkers for Precision Psychiatry. Adv Exp Med Biol (2019) 1161:101–13. 10.1007/978-3-030-21735-8_10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Glinton KE, Elsea SH. Untargeted Metabolomics for Autism Spectrum Disorders: Current Status and Future Directions. Front Psychiatry (2019) 10:647. 10.3389/fpsyt.2019.00647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Konjevod M, Tudor L, Svob Strac D, Nedic Erjavec G, Barbas C, Zarkovic N, et al. Metabolomic and glycomic findings in posttraumatic stress disorder. Prog Neuropsychopharmacol Biol Psychiatry (2019) 88:181–93. 10.1016/j.pnpbp.2018.07.014 [DOI] [PubMed] [Google Scholar]
- 88. Davison J, O'Gorman A, Brennan L, Cotter DR. A systematic review of metabolite biomarkers of schizophrenia. Schizophr Res (2018) 195:32–50. 10.1016/j.schres.2017.09.021 [DOI] [PubMed] [Google Scholar]
- 89. Bent S, Lawton B, Warren T, Widjaja F, Dang K, Fahey JW, et al. Identification of urinary metabolites that correlate with clinical improvements in children with autism treated with sulforaphane from broccoli. Mol Autism (2018) 9:35. 10.1186/s13229-018-0218-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Czysz AH, South C, Gadad BS, Arning E, Soyombo A, Bottiglieri T, et al. Can targeted metabolomics predict depression recovery? Results from the CO-MED trial. Trans Psychiatry (2019) 9(1):11. 10.1038/s41398-018-0349-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Khamis MM, Adamko DJ, El-Aneed A. Strategies and Challenges in Method Development and Validation for the Absolute Quantification of Endogenous Biomarker Metabolites Using Liquid Chromatography-Tandem Mass Spectrometry. Mass Spectrometry Rev (2019). 10.1002/mas.21607 [DOI] [PubMed]
- 92. Margueron R, Reinberg D. Chromatin structure and the inheritance of epigenetic information. Nat Rev Genet (2010) 11(4):285–96. 10.1038/nrg2752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Kular L, Kular S. Epigenetics applied to psychiatry: Clinical opportunities and future challenges. Psychiatry Clin Neurosci (2018) 72(4):195–211. 10.1111/pcn.12634 [DOI] [PubMed] [Google Scholar]
- 94. van Os J, Rutten BP, Poulton R. Gene-environment interactions in schizophrenia: review of epidemiological findings and future directions. Schizophr Bull (2008) 34(6):1066–82. 10.1093/schbul/sbn117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Focking M, Doyle B, Munawar N, Dillon ET, Cotter D, Cagney G. Epigenetic Factors in Schizophrenia: Mechanisms and Experimental Approaches. Mol Neuropsychiat (2019) 5(1):6–12. 10.1159/000495063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Dupont C, Armant DR, Brenner CA. Epigenetics: definition, mechanisms and clinical perspective. Semin Reprod Med (2009) 27(5):351–7. 10.1055/s-0029-1237423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Garcia-Gimenez JL, Seco-Cervera M, Tollefsbol TO, Roma-Mateo C, Peiro-Chova L, Lapunzina P, et al. Epigenetic biomarkers: Current strategies and future challenges for their use in the clinical laboratory. Crit Rev Clin Lab Sci (2017) 54(7-8):529–50. 10.1080/10408363.2017.1410520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Xie N, Zhou Y, Sun Q, Tang B. Novel Epigenetic Techniques Provided by the CRISPR/Cas9 System. Stem Cells Int (2018) 2018:7834175. 10.1155/2018/7834175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Smigielski L, Jagannath V, Rossler W, Walitza S, Grunblatt E. Epigenetic mechanisms in schizophrenia and other psychotic disorders: a systematic review of empirical human findings. Mol Psychiatry (2020). 10.1038/s41380-019-0601-3 [DOI] [PubMed]
- 100. Ikegame T, Bundo M, Murata Y, Kasai K, Kato T, Iwamoto K. DNA methylation of the BDNF gene and its relevance to psychiatric disorders. J Hum Genet (2013) 58(7):434–8. 10.1038/jhg.2013.65 [DOI] [PubMed] [Google Scholar]
- 101. Angelucci F, Brene S, Mathe AA. BDNF in schizophrenia, depression and corresponding animal models. Mol Psychiatry (2005) 10(4):345–52. 10.1038/sj.mp.4001637 [DOI] [PubMed] [Google Scholar]
- 102. Kang JI, Kim TY, Choi JH, So HS, Kim SJ. Allele-specific DNA methylation level of FKBP5 is associated with post-traumatic stress disorder. Psychoneuroendocrinology (2019) 103:1–7. 10.1016/j.psyneuen.2018.12.226 [DOI] [PubMed] [Google Scholar]
- 103. Kim EJ, Kim YK. Panic disorders: The role of genetics and epigenetics. AIMS Genet (2018) 5(3):177–90. 10.3934/genet.2018.3.177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Cheung S, Woo J, Maes MS, Zai CC. Suicide epigenetics, a review of recent progress. J Affect Disord (2020) 265:423–38. 10.1016/j.jad.2020.01.040 [DOI] [PubMed] [Google Scholar]
- 105. Ovenden ES, McGregor NW, Emsley RA, Warnich L. DNA methylation and antipsychotic treatment mechanisms in schizophrenia: Progress and future directions. Prog Neuropsychopharmacol Biol Psychiatry (2018) 81:38–49. 10.1016/j.pnpbp.2017.10.004 [DOI] [PubMed] [Google Scholar]
- 106. Melka MG, Castellani CA, Rajakumar N, O'Reilly R, Singh SM. Olanzapine-induced methylation alters cadherin gene families and associated pathways implicated in psychosis. BMC Neurosci (2014) 15:112. 10.1186/1471-2202-15-112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Tadic A, Muller-Engling L, Schlicht KF, Kotsiari A, Dreimuller N, Kleimann A, et al. Methylation of the promoter of brain-derived neurotrophic factor exon IV and antidepressant response in major depression. Mol Psychiatry (2014) 19(3):281–3. 10.1038/mp.2013.58 [DOI] [PubMed] [Google Scholar]
- 108. Lieb K, Dreimuller N, Wagner S, Schlicht K, Falter T, Neyazi A, et al. BDNF Plasma Levels and BDNF Exon IV Promoter Methylation as Predictors for Antidepressant Treatment Response. Front Psychiatry (2018) 9:511. 10.3389/fpsyt.2018.00511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Machado-Vieira R, Ibrahim L, Zarate CA., Jr. Histone deacetylases and mood disorders: epigenetic programming in gene-environment interactions. CNS Neurosci Ther (2011) 17(6):699–704. 10.1111/j.1755-5949.2010.00203.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Zhao WN, Ghosh B, Tyler M, Lalonde J, Joseph NF, Kosaric N, et al. Class I Histone Deacetylase Inhibition by Tianeptinaline Modulates Neuroplasticity and Enhances Memory. ACS Chem Neurosci (2018) 9(9):2262–73. 10.1021/acschemneuro.8b00116 [DOI] [PubMed] [Google Scholar]
- 111. Schroeder M, Hillemacher T, Bleich S, Frieling H. The epigenetic code in depression: implications for treatment. Clin Pharmacol Ther (2012) 91(2):310–4. 10.1038/clpt.2011.282 [DOI] [PubMed] [Google Scholar]
- 112. Kv A, Madhana RM, Js IC, Lahkar M, Sinha S, Naidu VGM. Antidepressant activity of vorinostat is associated with amelioration of oxidative stress and inflammation in a corticosterone-induced chronic stress model in mice. Behav Brain Res (2018) 344:73–84. 10.1016/j.bbr.2018.02.009 [DOI] [PubMed] [Google Scholar]
- 113. Deussing JM, Jakovcevski M. Histone Modifications in Major Depressive Disorder and Related Rodent Models. Adv Exp Med Biol (2017) 978:169–83. 10.1007/978-3-319-53889-1_9 [DOI] [PubMed] [Google Scholar]
- 114. Fuchikami M, Yamamoto S, Morinobu S, Okada S, Yamawaki Y, Yamawaki S. The potential use of histone deacetylase inhibitors in the treatment of depression. Prog Neuropsychopharmacol Biol Psychiatry (2016) 64:320–4. 10.1016/j.pnpbp.2015.03.010 [DOI] [PubMed] [Google Scholar]
- 115. Olova N, Krueger F, Andrews S, Oxley D, Berrens RV, Branco MR, et al. Comparison of whole-genome bisulfite sequencing library preparation strategies identifies sources of biases affecting DNA methylation data. Genome Biol (2018) 19(1):33. 10.1186/s13059-018-1408-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Walker DL, Bhagwate AV, Baheti S, Smalley RL, Hilker CA, Sun Z, et al. DNA methylation profiling: comparison of genome-wide sequencing methods and the Infinium Human Methylation 450 Bead Chip. Epigenomics (2015) 7(8):1287–302. 10.2217/EPI.15.64 [DOI] [PubMed] [Google Scholar]
- 117. Timmons JA, Szkop KJ, Gallagher IJ. Multiple sources of bias confound functional enrichment analysis of global -omics data. Genome Biol (2015) 16:186. 10.1186/s13059-015-0761-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Kinoshita M, Numata S, Tajima A, Ohi K, Hashimoto R, Shimodera S, et al. Aberrant DNA methylation of blood in schizophrenia by adjusting for estimated cellular proportions. Neuromol Med (2014) 16(4):697–703. 10.1007/s12017-014-8319-5 [DOI] [PubMed] [Google Scholar]
- 119. Montano C, Taub MA, Jaffe A, Briem E, Feinberg JI, Trygvadottir R, et al. Association of DNA Methylation Differences With Schizophrenia in an Epigenome-Wide Association Study. JAMA Psychiatry (2016) 73(5):506–14. 10.1001/jamapsychiatry.2016.0144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Rahmani E, Shenhav L, Schweiger R, Yousefi P, Huen K, Eskenazi B, et al. Genome-wide methylation data mirror ancestry information. Epigenet Chromatin (2017) 10:1. 10.1186/s13072-016-0108-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Edgar RD, Jones MJ, Meaney MJ, Turecki G, Kobor MS. BECon: a tool for interpreting DNA methylation findings from blood in the context of brain. Trans Psychiatry (2017) 7(8):e1187. 10.1038/tp.2017.171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Kim SH, Weiss C, Hoffmann U, Borggrefe M, Akin I, Behnes M. Advantages and Limitations of Current Biomarker Research: From Experimental Research to Clinical Application. Curr Pharmaceut Biotechnol (2017) 18(6):445–55. 10.2174/1389201018666170601091205 [DOI] [PubMed] [Google Scholar]
- 123. Lippolis JD, Powell EJ, Reinhardt TA, Thacker TC, Casas E. Symposium review: Omics in dairy and animal science-Promise, potential, and pitfalls. J dairy Sci (2019) 102(5):4741–54. 10.3168/jds.2018-15267 [DOI] [PubMed] [Google Scholar]
- 124. Jiang D, Armour CR, Hu C, Mei M, Tian C, Sharpton TJ, et al. Microbiome Multi-Omics Network Analysis: Statistical Considerations, Limitations, and Opportunities. Front Genet (2019) 10:995. 10.3389/fgene.2019.00995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Misra BB, Langefeld CD, Olivier M, Cox LA. Integrated Omics: Tools, Advances, and Future Approaches. J Mol Endocrinol (2018). 10.1530/JME-18-0055 [DOI] [PubMed]
- 126. Liang Y, Kelemen A. Dynamic modeling and network approaches for omics time course data: overview of computational approaches and applications. Briefings Bioinf (2018) 19(5):1051–68. 10.1093/bib/bbx036 [DOI] [PubMed] [Google Scholar]