Abstract
Aims
Circular RNAs (circRNAs) are involved in gene regulation in a variety of physiological and pathological processes. The present study aimed to investigate the effect of circRNA_000203 on cardiac hypertrophy and the potential mechanisms involved.
Methods and results
CircRNA_000203 was found to be up-regulated in the myocardium of Ang-II-infused mice and in the cytoplasma of Ang-II-treated neonatal mouse ventricular cardiomyocytes (NMVCs). Enforced expression of circRNA_000203 enhances cell size and expression of atrial natriuretic peptide and β-myosin heavy chain in NMVCs. In vivo, heart function was impaired and cardiac hypertrophy was aggravated in Ang-II-infused myocardium-specific circRNA_000203 transgenic mice (Tg-circ203). Mechanistically, we found that circRNA_000203 could specifically sponge miR-26b-5p, -140-3p in NMVCs. Further, dual-luciferase reporter assay showed that miR-26b-5p, -140-3p could interact with 3′-UTRs of Gata4 gene, and circRNA_000203 could block the above interactions. In addition, Gata4 expression is transcriptionally inhibited by miR-26b-5p, -140-3p mimic in NMVCs but enhanced by over-expression of circRNA_000203 in vitro and in vivo. Functionally, miR-26b-5p, -140-3p, and Gata4 siRNA, could reverse the hypertrophic growth in Ang-II-induced NMVCs, as well as eliminate the pro-hypertrophic effect of circRNA_000203 in NMVCs. Furthermore, we demonstrated that NF-κB signalling mediates the up-regulation of circRNA_000203 in NMVCs exposed to Ang-II treatment.
Conclusions
Our data demonstrated that circRNA_000203 exacerbates cardiac hypertrophy via suppressing miR-26b-5p and miR-140-3p leading to enhanced Gata4 levels.
Keywords: Circular RNA, Cardiac hypertrophy, CircRNA_000203, MicroRNA-26b-5p, MicroRNA-140-3p
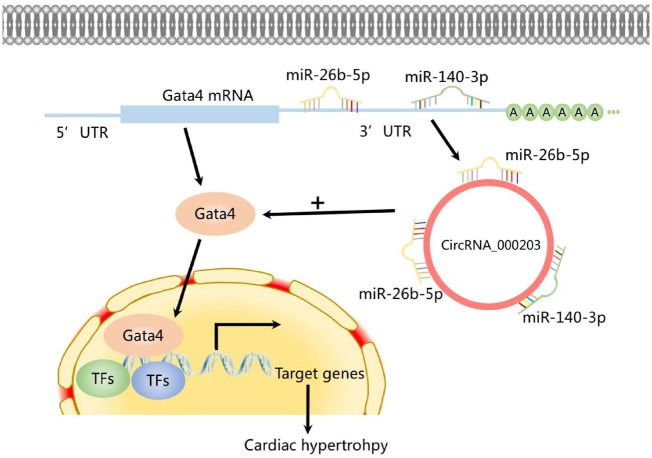
Graphical Abstract
Graphical Abstract.
1. Introduction
Cardiac hypertrophy is classified as physiological and pathological when associated with normal cardiac function and impaired cardiac function, respectively. It is well known that the pathological cardiac hypertrophy is an independent risk factor for myocardial infarction, arrhythmia, heart failure (HF), and even sudden death.1–3 Pathological hypertrophy is induced by factors such as myocardial infarction, mechanical stimulus, hormones, cytokines, growth factors, and pressure overload. Due to such complexity, no efficient therapeutic approaches are currently available for the management of cardiac hypertrophy. Understanding the molecular mechanisms associated with cardiac hypertrophy and the transition to HF is crucial to identify new targets to reverse maladaptive cardiac remodelling and even HF.
Transcription factors (TFs) are nuclear proteins that bind to the promoter regulatory elements to activate/repress the downstream gene expression. Some pro-hypertrophic TFs, such as GATA4, myocyte enhancer factor 2 (MEF2), NK family of TF 2.5 (Nkx2.5), nuclear factor of activated T-cells (NFAT), and Ca2+/cAMP response element-binding protein (CREB),4–6 have been known implicated as signal-responsive mediators of the cardiac transcriptional programme in cardiac hypertrophy.
Accumulating evidence indicates that non-coding RNAs, including microRNAs (miRNAs) and circular RNAs (circRNAs), are critical contributors to cardiovascular pathophysiology.7,8 MiRNAs are endogenous, 20–23 nucleotide RNAs that negatively regulate target genes involved in various physiological processes and diseases. For example, microRNA-1, -208a, -195, 214-3p, -92 b-3p were verified involved in cardiac hypertrophy.9–13
CircRNAs form covalently closed continuous loops with high tissue-specific expression.13 Compared with linear RNAs, circRNAs have the remarkable characteristic of non-canonical splicing without a free 3′ or 5′ end. They mainly arise from exons or introns and are differentially generated by back splicing or lariat introns. CircRNAs could specifically function as microRNA (miRNA) sponges, regulate alternative splicing, and modulate the expression of the parental gene, as well as possessing the potential in proteins translation.14,15 Increasing evidence reveals that circRNAs may participate in the pathogenesis of the cardiovascular system, such as vascular networks,16 atherosclerosis,17 cardiomyopathy,18–20 and HF.21–23
Our previous study revealed that circRNA_000203 and its parental gene of Myo9a were up-regulated in the diabetic mouse myocardium with pro-fibrotic effect by sponging microRNA-26b-5p (miR-26b-5p).18 However, the expression and role of circRNA_000203 have not been elucidated in cardiac hypertrophy, such as hypertension-induced cardiac hypertrophy. In this study, based on a mouse model of Angiotensin-II (Ang-II)-induced cardiac hypertrophy, we investigated the effect of circRNA_000203 on cardiac hypertrophy in vitro and in vivo and demonstrated the mechanism by which circRNA_000203 modulates cardiac hypertrophic growth.
2. Methods
2.1 Ethics statement
Male and female C57BL/6 mice weighing 20 ± 3 g [License number SCXK (YUE) 2004–0011, Department of Experimental Animal Research Center, Sun Yat-sen University, Guangzhou, China], and male myocardium-specific circRNA_000203 transgenic mice (Tg-circ203; License number T003447, Nanjing Biomedical Research Institute of Nanjing University, Nanjing, China) and wide-type (WT) control mice (License number XM100635, Nanjing Biomedical Research Institute of Nanjing University) weighing 20 ± 3 g were used in the current studies. The adult mice were housed under a 12 h light/dark cycle under pathogen-free conditions and with free access to standard mouse chow and tap water. This study conformed to the Guide for the Care and Use of Laboratory Animals published by the US National Institutes of Health (8th Edition, National Research Council, 2011). The present programme was also approved by the research ethics committee of Guangdong General Hospital (the approval number: No. GDREC2014066A).
2.2 Animal studies
According to our previous report,12 we established a mouse model of Ang-II (1.46 mg/kg/d, 28 days) infusion-induced cardiac hypertrophy. Mice were anaesthetized through the intraperitoneal application of sodium pentobarbital (50 mg/kg), followed by implantation of the Ang-II mini-osmotic pump (alzet model 2002, Cupertino, CA, USA). The adequacy of anaesthesia was confirmed by the absence of reflex response to foot squeeze. Body temperature was maintained at 37 ± 0.5°C during surgery. At the end of the experiments, mice were killed with the intraperitoneal injection of an overdose of sodium pentobarbital (200 mg/kg).
2.3 Echocardiographic study
Transthoracic echocardiography was performed to assess left ventricular (LV) function variables. After the induction of light general anaesthesia, the rats underwent transthoracic two dimensional (2D) guided M-mode echocardiography with an 8.5 MHz transducer (Acuson, Mountain View, CA, USA). From the cardiac short axis (papillary level), the LV anterior wall end-diastolic thickness (LVAWd), the systolic LV anterior wall thickness (LVAWs), the LV internal dimension at end-diastole (LVIDd), the LV internal dimension at end-systole (LVIDs), the LV posterior wall end-diastolic thickness (LVPWd), the LV posterior wall end-systolic thickness (LVPWs), the ejection fraction (EF), and fractional shortening (FS) were measured. Echocardiographic measurements were averaged from at least three separate cardiac cycles.
2.4 Histological analysis
Formalin-fixed mouse myocardium specimens were embedded in paraffin and cut into 4 μm thick sections. Tissue sections were mounted on the regular glass slides and stained with 1.0 mg/mL Alexa Fluor® 488 conjugate of wheat germ agglutinin (WGA) solution (MolecularProbes, Eugene, OR, USA) to demonstrate the size of cardiomyocytes in mouse myocardium.
2.5 Neonatal mouse ventricular cardiomyocytes isolation, culture, and treatment
Neonatal mouse ventricular cardiomyocytes (NMVCs) were isolated from the hearts of 1- to 3-day-old newborn C57BL6 mice as described previously.12 NMVCs were incubated with 10−8 M Ang-II for 48 h to induce the hypertrophic phenotype. The NF-κB inhibitor JSH23 (5 μM) or QNZ (5 nM) was used to treat NMVCs. Cells were transfected with 50 nM scramble or miR-26b-5p, -140-3p, or 50 nM siRNA for Gata4 or p65 NFkB (Ribobio, Guangzhou, China) by oligofectamine reagent (Invitrogen, Carlsbad, CA, USA), respectively. Plasmid DNA was delivered into NMVCs by using Namipo reagent (Transheep Bio, Shanghai, China). As indicated, NMVCs were infected with the following recombinant adenovirus, respectively: rAd-GFP, rAd-Gata4 (Vigene Biosciences, MD, USA) and rAd-circRNA_000203 adenovirus (MOI 10).18
2.6 Fluorescence in situ hybridization
BaseScope™ Probe for circRNA_000203 was designed and synthesized by ACD (Advanced Cell Diagnostics, CA, USA), and the probe sequence covers the specific junction region of circRNA_000203. NMVCs were fixed by 10% neutral buffered formalin on slides for detection of the sub-cellular location of circRNA_000203. The signals of the circRNA_000203 probe were detected by BaseScope™ Detection Reagent Kit (ACD, USA) according to the manufacturer’s instructions. The images were acquired on Leica SP5 Spectral scanning laser confocal microscope (Leica Microsystems, Wetzlar, Germany).
2.7 RNA pull-down assay
One hundred micrograms total RNA extracted from HEK293 cells with plasmid vector-mediated overexpression of circRNA_000203 was used to incubate with 500 μg streptavidin magnetic beads (S1421S, NEB, Ipswich, MA, USA) which were previously incubated with 200 pmol biotin-miR-26b-5p or biotin-miR-140-3p, respectively. Eventually, the binding RNA was eluted and used for circRNA_000203 detection by quantitative reverse-transcription PCR (RT–qPCR) assay.
Adenovirus-mediated overexpression of circRNA_000203 was achieved in NMVCs. Three biotin-labelled circRNA_000203 probes (Ribobio, China) were incubated with cell lysates of NMVCs with enforced overexpression of circRNA_000203 at 37°C for 3–4 h, followed with addition of streptavidin beads (Bersinbio, Guangzhou, China) at 25°C for 40 min. After washing with the wash buffer, the RNA complexes bound to the beads were eluted and extracted with RNeasy Mini Kit (QIAGEN) and used for detection of circRNA_000203, miR-26b-5p, -140-3p by RT–qPCR assay, respectively.
2.8 FITC-phalloidin staining
To assess the cell size of NMVCs, the cultured NMVCs were fixed in 3.7% formaldehyde and permeabilized in 0.1% Triton X-100 for 10 min, respectively, followed by incubation with blocking solution for 40 min and subsequently with FITC-phalloidin (10 μg/ml, Sigma-Aldrich) at 37°C. Confocal micrographs were obtained using a Leica SP5.
2.9 Dual-luciferase assays
Dual-luciferase assay for interactions between miR-26b-5p, -140-3p, and circRNA_000203. As our previous report,24 the recombinant luciferase reporter plasmids containing the potential miR-26b-5p and miR-140-3p binding site sequences in circRNA_000203 were constructed. Human embryonic kidney (HEK) 293 cells (3 × 105 cells per well in 12 well plate) were co-transfected with 200 ng of recombinant luciferase reporter plasmid, 20 ng of pRL-TK as an internal control (Promega, Madison, WI, USA), 50 nM miR-26b-5p, -140-3p mimic, respectively. Activities of firefly luciferase (FL) and Renilla luciferase (RL) were measured 24 h after transfection, and the relative ratio of the FL/RL was used to reveal the interactions between miR-26b-5p, -140-3p, and circRNA_000203.
Dual-luciferase assay for Gata4 target identification. The recombinant luciferase reporter plasmids containing the potential miR-26b-5p and miR-140-3p binding site sequences in the 3ʹ-UTR of Gata4 gene were constructed. HEK293 cells were co-transfected with 200 ng of recombinant luciferase reporter plasmid, 20 ng of pRL-TK as an internal control, 200 ng of pAd-circ203, 50 nM miR-26b-5p, -140-3p mimic, respectively. Activities of FL and RL were measured, and the FL/RL ratio was used to indicate the miR-26b-5p, -140-3p-mediated knockdown of Gata4 gene.
Dual-luciferase assay for TF NF-κB p65 participating in Myo9a transcription. Three potential NF-κB p65 recognition elements, TTCCCAGAGC, AAAGTCCTCT, and TGAAAAGTCT, were screened in the promoter region of Myo9a gene by AliBaba2.1 programme (http://gene-regulation.com/pub/programs/alibaba2/index.html). The recombinant-luciferase reporter plasmids containing the above potential NF-κB p65 binding site sequences were constructed, respectively. NMVCs were co-transfected with 200 ng of recombinant luciferase reporter plasmid, 20 ng of pRL-TK as an internal control by using Namipo reagent, followed with Ang-II treatment. Activities of FL and RL were measured, and the relative ratio of the FL/RL was used to indicate the transcription activity of Myo9a gene.
2.10 Quantitative mRNA and miRNA measurements
To detect the expression of circRNA_000203 and coding genes, the first-strand cDNA was generated from 1.5 μg total RNA using a mixture of oligo (dT)15 and random primers with superscript reverse transcriptase (Invitrogen, Carlsbad, CA, USA). RT–qPCR for miR-26b-5p and -140-3p was performed on cDNA generated from 0.5 μg total RNA according to the manufacturer’s protocol (Ribobio, China). U6 was used for miRNAs template normalization and GAPDH was used for circRNA_000203 and coding genes template normalization, respectively. PCR was performed with the ViiA7 Quantitative PCR System (Applied Biosystems, Carlsbad, CA, USA). The 2−ΔCt and 2−ΔΔCt methods were used to calculate relative expression levels of the concerned coding genes and miRNAs, respectively. PCR primers, as well as the size of fragments amplified, used in this study are shown in Supplementary material online, Table S1.
2.11 Western-blot assay
Protein extracts (40 μg) were separated using 12% (wt./vol.) SDS-PAGE, transferred onto a polyvinylidene fluoride (PVDF) membrane, and probed with antibodies for atrial natriuretic peptide (ANP) and β-myosin heavy chain (β-MHC; Abcam), GATA4 (Proteintech, Chicago, IL, USA), p-NF-кB p65, NF-кB p65 (Cell Signaling Technology, Beverly, MA, USA) overnight at 4°C. Membranes were then washed extensively with TBS/T and incubated with a horseradish peroxidase-conjugated secondary antibody (Santa Cruz) for 1 h at room temperature. Protein was visualized using the ECL Plus detection system (GE Healthcare, WI, USA).
2.12 Statistical analysis
In each experiment, all determinations were performed at least in triplicate. Comparisons between two groups of data were made by one-way ANOVA followed by the post hoc test (Bonferroni) was chosen for the comparisons of interest without adjustment for multiple comparisons. A P-value of <0.05 was considered to be significant. Data are presented as the mean ± the standard error of the mean (SEM). Analyses were performed in IBM SPSS Statistics 25 (IBM Corp, Armonk, NY, USA).
3. Results
3.1 Up-regulation of circRNA_000203 in the hypertrophic myocardium of Ang-II-infused mice
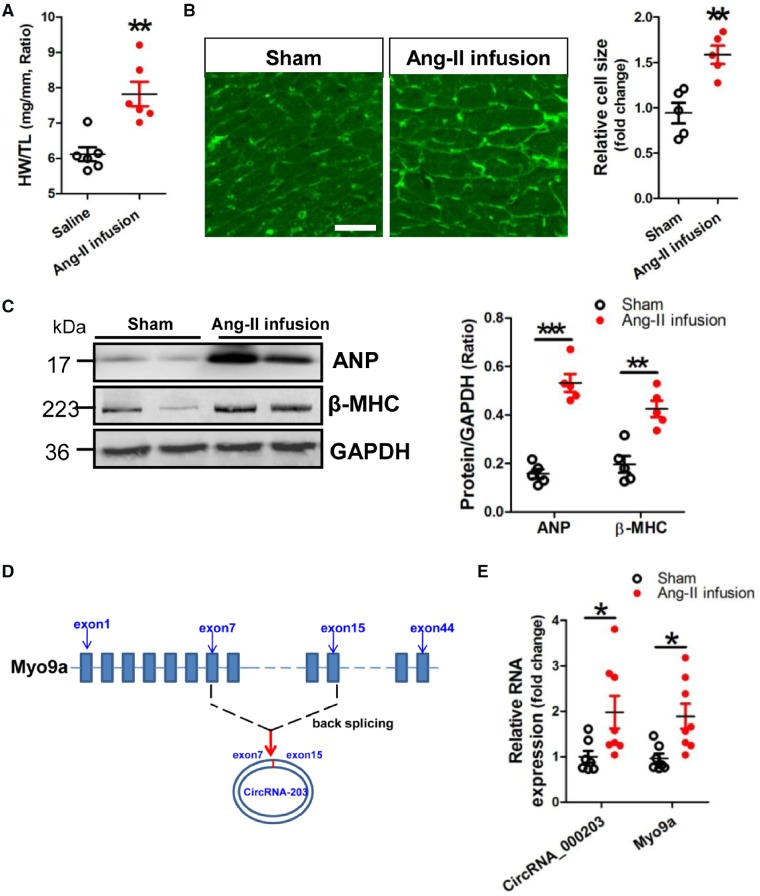
An animal model of cardiac hypertrophy was established in mice received Ang-II infusion for 4 weeks, with a significant increase in the ratio of weight/tibia length (HW/TL) (Figure 1A). Results of WGA staining showed that the cell size of cardiomyocytes was significantly increased in the myocardium of the Ang-II infusion mouse model (Figure 1B). Meanwhile, significant increase of ANP and β-MHC protein expression was observed in the hypertrophic mouse myocardium (Figure 1C). Results of RT–qPCR verified that circRNA_000203 and its parental gene of Myo9a mRNA (Figure 1D) were consistently up-regulated in the myocardium of Ang-II-infused mice (Figure 1E). Consistently, we further set up a female mouse model of Ang-II-infusion-induced cardiac hypertrophy, with a significant increase in the ratio of HW/TL. We also observed significant and consistent increases of circRNA_000203 and Myo9a mRNA in the hypertrophic myocardium of female mice (Supplementary material online, Figure S1). Moreover, we also demonstrated that human hsa-circ_0036167 (homologue of circRNA_000203) and its host gene of Myo9a were obviously up-regulated in the myocardium of HF patients (Supplementary material online, Figure S2).
Figure 1.
CircRNA_000203 is up-regulated in the myocardium of Ang-II-infused mice. (A) The ratio of HW/TL. (B) WGA staining assay of cardiomyocytes in the hypertrophic myocardium of a mouse model of Ang-II infusion-induced hypertrophy. The scale bar is 100 μm. (C) Protein expression of ANP and β-MHC in the myocardium of Ang-II-infused mice by western blot assay. (D) CircRNA_000203 sequence is derived from exon7 to exon 15 of Myo9a gene. (E) Detection of circRNA_000203 and Myo9a in the myocardium of Ang-II-infused mice by RT–qPCR assay. Data are shown as mean ± SEM, *P < 0.05, **P < 0.01, ***P < 0.001 vs. Sham group (ANOVA with post hoc tests, as indicated). N = 6, 5, 5, and 7–8 in (A), (B), (C), and (E), respectively.
3.2 CircRNA_000203 is up-regulated in Ang-II-induced NMVCs
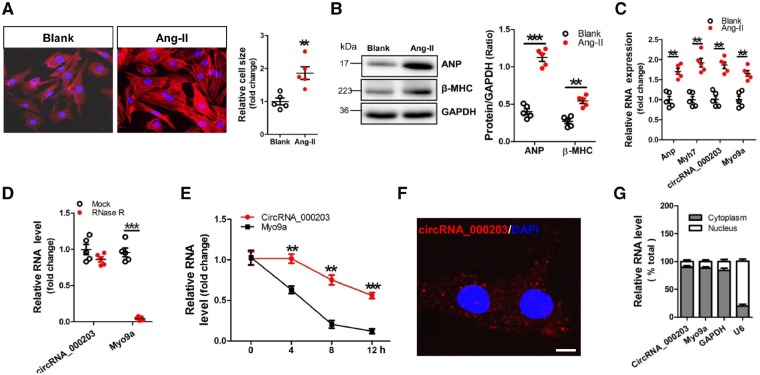
A cell model of Ang-II-induced NMVC hypertrophy was used in this study. Obvious increases of cell size of NMVCs and protein expression of ANP and β-MHC were observed in Ang-II-treated NMVCs (Figure 2A, B). Consistently, significant up-regulations of circRNA_000203, Myo9a, Anp and Myh7 mRNA were also found in Ang-II-treated NMVCs (Figure 2C). We detected the stability of circRNA_000203 in NMVCs, RT–qPCR results showed that circRNA_000203, but not Myo9a mRNA, was resistant to RNase-R treatment (Figure 2D). We further performed RNase-R resistance assay based on the total RNA from the myocardium of C57BL/6 mice, and circRNA_000203 was also more stable compared with Myo9a mRNA (Supplementary material online, Figure S3). Moreover, we used actinomycin D to inhibit RNA transcription and measured the levels of circRNA_000203 and Myo9a mRNA at different time points in NMVCs post-actinomycin D treatment. The relative level of circRNA_000203 was markedly higher than that of Myo9a mRNA at 4, 8, and 12 h after actinomycin D treatment (Figure 2E). In addition, fluorescence in situ hybridization (FISH) and RT–qPCR assay showed the predominant cytoplasmic distribution of circRNA_000203 in NMVCs (Figure 2F, G).
Figure 2.
CircRNA_000203 is up-regulated in Ang-II-induced NMVCs. (A) Morphologies of Ang-II-induced NMVCs by FITC-phalloidin staining assay. The scale bar is 50 μm. (B) Protein expression of ANP and β-MHC in Ang-II-induced NMVCs by western blot assay. (C) Expression of circRNA_000203, Myo9a, Anp, and Myh7 mRNA in Ang-II-induced NMVCs by RT–qPCR assay. (D) Levels of circRNA_000203 and Myo9a mRNA in RNase-R-treated total RNA from NMVCs (20 U/µg RNA) were detected by RT–qPCR and normalized to the value detected in the mock group. Data are shown as mean ± SEM, **P < 0.01, ***P < 0.001 (ANOVA with post hoc tests, as indicated). N = 5. (E) Levels of circRNA_000203 and Myo9a mRNA in NMVCs after treatment with 2 mg/ml Actinomycin D at the indicated time points. Data are shown as mean ± SEM, **P < 0.01, ***P < 0.001 vs. Myo9a mRNA group (ANOVA with post hoc tests, as indicated). N = 5. (F) RNA FISH for circRNA_000203 in NMVCs. CircRNA_000203 was shown in red and nuclei were stained with DAPI. The scale bar is 5 μm. (G) circRNA_000203 and Myo9a mRNA are abundant in the cytoplasm of NMVCs. Gapdh mRNA and U6 were applied as positive controls in the cytoplasm and nucleus, respectively.
3.3 CircRNA_000203 enhances hypertrophy of NMVCs
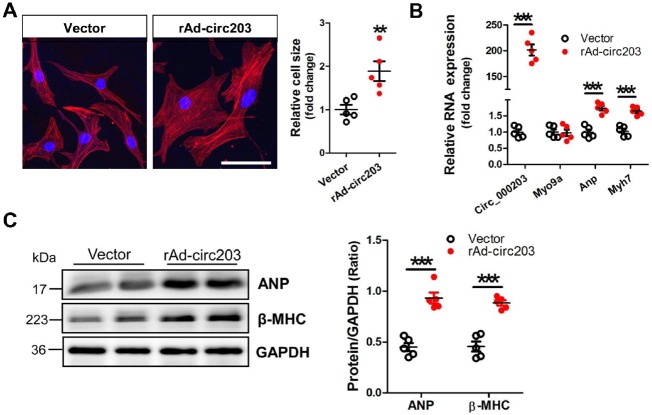
In this study, the recombinant circRNA_000203 adenovirus, rAd-circ203, was used to mediate the overexpression of circRNA_000203 in NMVCs. The FITC-Phalloidin staining showed that the cell size was significantly increased in NMVCs infected with rAd-circ203 (Figure 3A). The level of circRNA_000203, but not Myo9a mRNA, was highly increased in NMVCs, along with significant up-regulation of Anp and Myh7 mRNA (Figure 3B). Consistently, protein expression of ANP and β-MHC was also markedly enhanced in NMVCs with enforced expression of circRNA_000203 (Figure 3C).
Figure 3.
CircRNA_000203 enhances hypertrophy in NMVCs. (A) Morphologies of NMVCs with overexpression of circRNA_000203 by FITC-phalloidin staining assay. The scale bar is 50 μm. (B) Expression of circRNA_000203, Myo9a, Anp, and Myh7 mRNA in NMVCs with overexpression of circRNA_000203 by RT–qPCR assay. (C) Protein expression of ANP and β-MHC in NMVCs with overexpression of circRNA_000203 by western blot assay. Data are shown as mean ± SEM, **P < 0.01, ***P < 0.001 (ANOVA with post hoc tests, as indicated). N = 5.
3.4 Overexpression of circRNA_000203 exacerbates Ang-II-induced cardiac hypertrophy in vivo
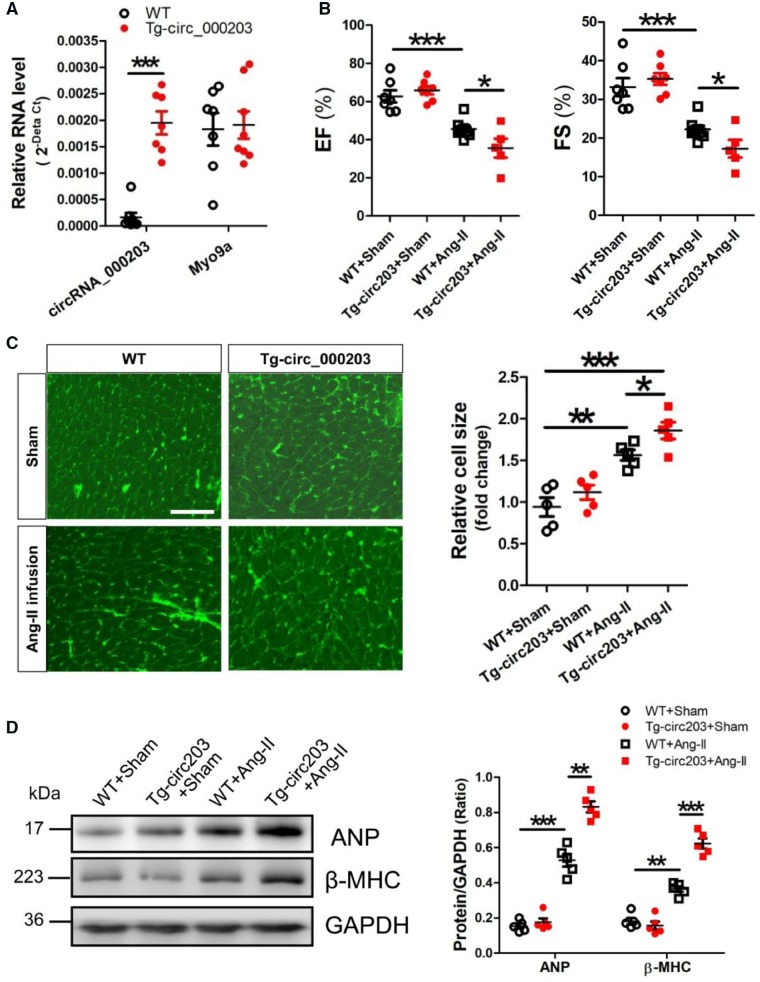
We successfully established the Tg-circ203 mice with cardiac-specific overexpression of circRNA_000203 (Supplementary material online, Figures S4 and S5). RT–qPCR results showed that circRNA_000203, but not Myo9a mRNA, was highly expressed in the myocardium of Tg-circ203 mice (Figure 4A). In the present study, an animal model of hypertrophy was established in mice with Ang-II infusion for 4 weeks. Echocardiography was performed to reveal the changes of cardiac structure and function in Ang-II-infused Tg-circ203 mice. Significant decreases of EF and FS were observed in Ang-II-infused Tg-circ203 mice (Figure 4B). Meanwhile, the LV internal diameters (LVIDd, LVIDs) were also markedly decreased, but without significant changes of HW/TL and the LV walls (LVPWd, LVPWs) in Ang-II infused Tg-circ203 mice (Supplementary material online, Table S2). In addition, the WGA staining results showed that the cell size of cardiomyocytes in the myocardium was significantly increased in Ang-II-infused Tg-circ203 mice (Figure 4C). Western blot results demonstrated that expression of ANP and β-MHC was also significantly enhanced in the myocardium of Ang-II-infused Tg-circ203 mice (Figure 4D).
Figure 4.
Overexpression of circRNA_000203 exacerbates Ang-II-induced cardiac hypertrophy in vivo. (A) Expression of circRNA_000203 and Myo9a mRNA in the myocardium of Tg-circ203 mice by RT–qPCR assay. (B) The representative variables of EF and FS by echocardiography. (C) WGA staining assay of cardiomyocytes in mouse myocardium. (D) Protein expression of ANP and β-MHC in mouse myocardium by western blot assay. Data are shown as mean ±SEM, *P < 0.05, **P < 0.01, ***P < 0.001 (ANOVA with post hoc tests, as indicated), N = 7–8, 5–8, 5, 5 in (A), (B), (C), and (D), respectively.
3.5 CircRNA_000203 sponges miR-26b-5p and miR-140-3p in NMVCs
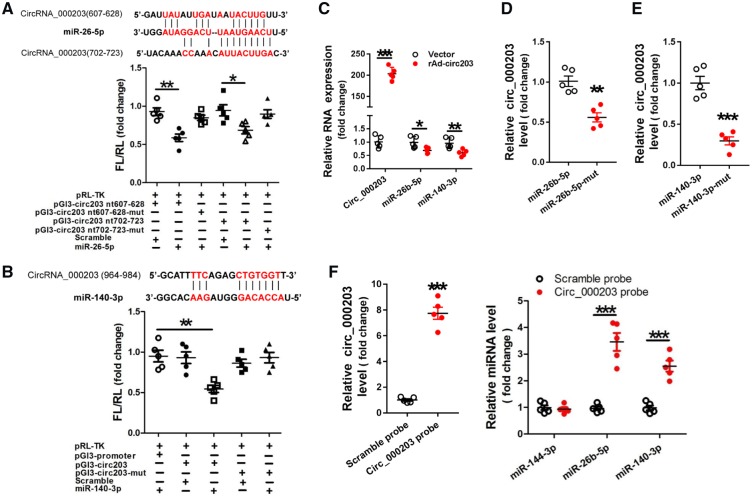
Sequence analysis indicated that there are two potential binding sites (nt 607–628 and nt 702–723) for miR-26b-5p (Figure 5A), and one potential binding site (nt 964–984) for miR-140-3p (Figure 5B) in circRNA_000203. The dual-luciferase assay indicated that miR-26b-5p can interact with two sites (nt 607–628 and nt 702–723), and miR-140-3p can interact with one site of nt 964–984 in circRNA_000203. And the interactions of either miR-26b-5p or miR-140-3p with circRNA_000203 could be abolished after the corresponding binding sites were mutated (Figure 5A, B). RT–qPCR results revealed that levels of miR-26b-5p and miR-140-3p were markedly decreased in NMVCs with overexpression of circRNA_000203 (Figure 5C), as well as in the myocardium of Ang-II-infusion mice and in Ang-II-treated NMVCs (Supplementary material online, Figure S6). Results of RNA pull-down and RT–qPCR assay showed that the amount of circRNA_000203 pulled down by miR-26b-5p-mut was significantly less than that by miR-26b-5p (Figure 5D), and the amount of circRNA_000203 pulled down by miR-140-3p-mut was also significantly less than that by miR-140-3p (Figure 5E). CircRNA_000203 was efficiently isolated from NMVCs lysate by biotin-labelled circRNA_000203 probe (Figure 5F). Meanwhile, miR-26b-5p and miR-140-3p, but not the irrelevant miR-144-3p control, could be specifically pulled down with circRNA_000203 by using circRNA_000203 probe (Figure 5F).
Figure 5.
Identification of miR-26b-5p and miR-140-3p as sponged targets of circRNA_000203. Verification of miR-26b-5p (A) and miR-140-3p (B) as sponged targets of circRNA_000203 by the dual-luciferase reporter assay. The predicted binding sites of miR-26b-5p and miR-140-3p in circRNA_000203 were shown in red. Data are shown as mean ±SEM, *P < 0.05, **P < 0.01 (ANOVA with post hoc tests, as indicated), N = 5. (C) Expression of circRNA_000203, miR-140-3p, -26 b-5p in NMVCs with overexpression of circRNA_000203 by RT–qPCR assay. Data are shown as mean ±SEM, *P < 0.05, **P < 0.01, ***P < 0.001 vs. vector control (ANOVA with post hoc tests, as indicated), N = 5. The pulled down circRNA_000203 by miR-26b-5p (D), and by miR-140-3p (E) was detected by RT–qPCR assay, respectively. Data are shown as mean ±SEM, **P < 0.01 vs. miR-26b-5p, ***P < 0.001 vs. miR-140-3p (ANOVA with post hoc tests, as indicated), N = 5. (F) MiR-26b-5p, miR-140-3p was pulled down from NMVCs lysate by biotin-labelled circRNA_000203 probe, and RT–qPCR was performed to detect the relative level of miR-144-3p, -26 b-5p, -140-3p. Data are shown as mean ±SEM, ***P < 0.001 vs. Scramble probe (ANOVA with post hoc tests, as indicated), N = 5.
3.6 CircRNA_000203 abolishes the interactions of miR-26b-5p, -140-3p with the common target gene of Gata4
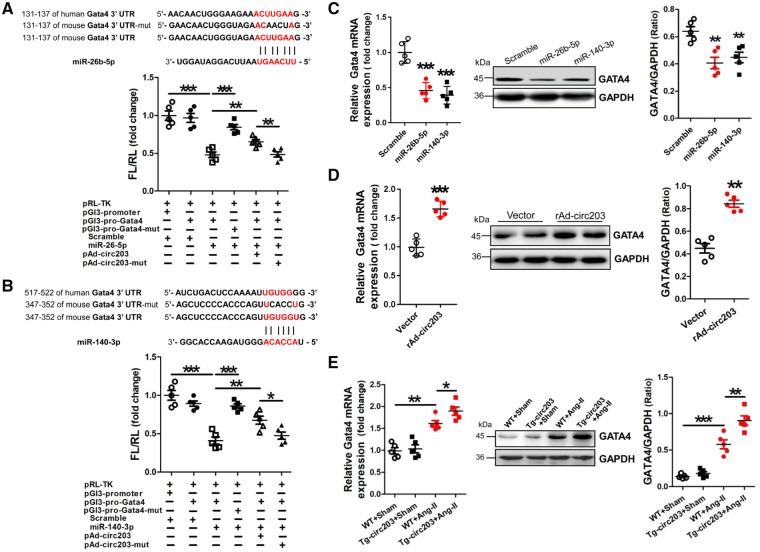
Analyses of the databases Mirdb (www.mirdb.org) and TargetScan-Vert (www.targetscan.org) showed that Gata4 was the common target gene of miR-26b-5p, -140-3p. The matching positions for miR-26b-5p, -140-3p within 3′-UTRs of Gata4 mRNA are shown in Figure 6A, B. Results of the dual-luciferase assay indicated that miR-26b-5p, -140-3p mimic can specifically interact with the 3′-UTRs of Gata4. In addition, the interactions of miR-26b-5p, -140-3p with the 3′-UTRs of Gata4 could be blocked by circRNA_000203 (Figure 6A, B). Compared with the scramble negative control, the mRNA and protein expression of Gata4 could be significantly reduced in NMVCs after transfection with miR-26b-5p, -140-3p mimic (P < 0.05, P < 0.01, respectively; Figure 6C). As expected, the mRNA and protein expression of Gata4 was found significantly increased in NMVCs with overexpression of circRNA_000203 (P < 0.01, respectively; Figure 6D). Moreover, Gata4 mRNA and protein expression were also observed markedly increased in the myocardium of Ang-II-infused Tg-circ203 mice (P < 0.05, respectively; Figure 6E).
Figure 6.
MiR-26b-5p and miR-140-3p negatively modulate Gata4 expression. (A) Verification of Gata4 as a target gene of miR-26b-5p by the dual–luciferase reporter assay. Predicted miR-26b-5p seed sequence matches to the sequence in the 3′-UTRs of Gata4 mRNA. The seed sequence of miR-26b-5p is UCAAGU, and the complementary nucleotide sequences are shown in red words. Data are shown as mean ±SEM, **P < 0.01, ***P < 0.001 (ANOVA with post hoc tests, as indicated), N = 5. (B) Verification of Gata4 as a target gene of miR-140-3p by the dual-luciferase reporter assay. Predicted miR-140-3p seed sequence matches to the sequence in the 3′-UTRs of Gata4 mRNA. The seed sequence of miR-140-3p is ACCACA, and the complementary nucleotide sequences are shown in red words. Data are shown as mean ±SEM, *P < 0.05, **P < 0.01, ***P < 0.001 (ANOVA with post hoc tests, as indicated), N = 5. Gata4 mRNA and protein expression in NMVCs transfected with miR-26b-5p and miR-140-3p mimic, respectively (C), and in NMVCs with overexpression of rAd-circ203 (D) were assessed by RT–qPCR and western blot assay, respectively. Data are shown as mean ± SEM, **P < 0.01, ***P < 0.001 (ANOVA with post hoc tests, as indicated), N = 5. (E) Gata4 mRNA and protein expression in the myocardium of Ang-II-infused Tg-circ203 mice. Data are shown as mean ± SEM, *P < 0.05, **P < 0.01, ***P < 0.001 (ANOVA with post hoc tests, as indicated), N = 5.
3.7 MiR-26b-5p and miR-140-3p eliminate the pro-hypertrophy effect of circRNA_000203 in NMVCs
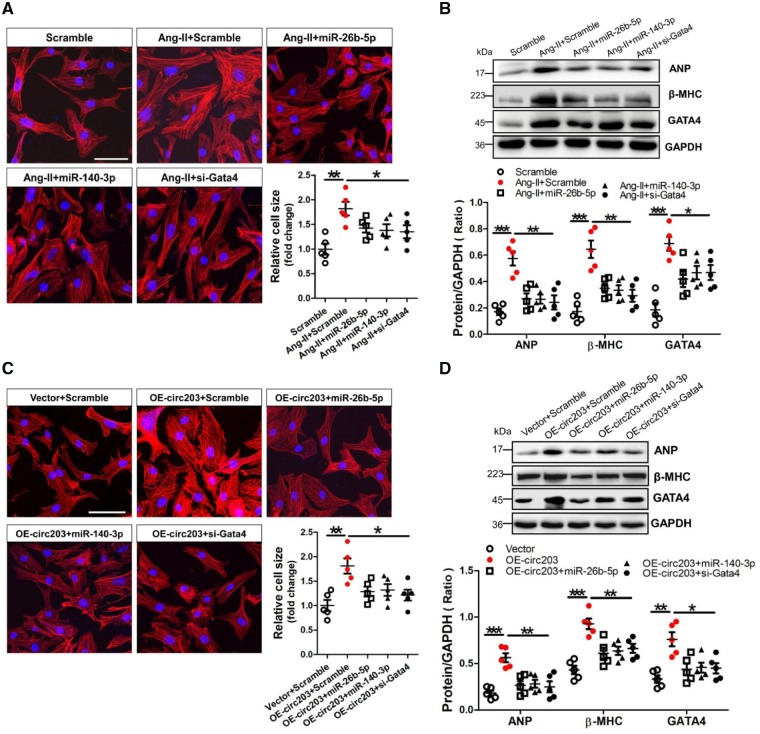
We investigated the consistent effects of miR-26b-5p, -140-3p, and Gata4 siRNA on hypertrophy in Ang-II-treated NMVCs. The FITC-Phalloidin staining showed that the cell size was significantly increased in Ang-II-treated NMVCs but could be reversed after transfection with miR-26b-5p, -140-3p, and Gata4 siRNA, respectively (Figure 7A). Western blot results demonstrated that protein expression of ANP, β-MHC, and GATA4 was significantly enhanced in Ang-II-treated NMVCs but could also be alleviated after transfection with miR-26b-5p, -140-3p, and Gata4 siRNA, respectively (Figure 7B). In addition, western blot results showed that adenovirus-mediated overexpression of GATA4 could significantly enhance ANP and β-MHC expression in NMVCs but could be efficiently reversed after transfection with miR-26b-5p, -140-3p, respectively (Supplementary material online, Figure S7). We also investigated the effects of miR-26b-5p, -140-3p, and Gata4 siRNA on eliminating the pro-hypertrophic growth effect of circRNA_000203 in NMVCs. Results of FITC-Phalloidin staining showed that the cell size was significantly increased in NMVCs with overexpression of circRNA_000203 but could be abolished by miR-26b-5p, -140-3p, and Gata4 siRNA, respectively (Figure 7C). Moreover, Western blot results demonstrated that protein expression of ANP, β-MHC, and GATA4 was markedly increased in circRNA_000203-modified NMVCs but could be ameliorated after transfection with miR-26b-5p, -140-3p, and Gata4 siRNA, respectively (Figure 7D).
Figure 7.
MiR-26b-5p and miR-140-3p alleviate the pro-hypertrophy effect of circRNA_000203 on NMVCs. (A) Morphologies of Ang-II-treated NMVCs after transfection with miR-26b-5p, miR-140-3p, and Gata4 siRNA as revealed by FITC-phalloidin staining assay, respectively. ANP, β-MHC and GATA4 protein expression in NMVCs was assessed by western blot assay (B, D). Data are shown as mean ± SEM, *P < 0.05, **P < 0.01, ***P < 0.001 (ANOVA with post hoc tests, as indicated). N = 5. (C) Morphologies of circRNA_000203-modified NMVCs after transfection with miR-26b-5p, miR-140-3p, and Gata4 siRNA as revealed by FITC-phalloidin staining assay, respectively. The scale bar is 50 μm. Data are shown as mean ± SEM, *P < 0.05, **P < 0.01 (ANOVA with post hoc tests, as indicated). N = 5.
3.8 Activation of NF-κB mediates Ang-II-up-regulated expression of circRNA_000203 in NMVCs
To investigate the effect of Ang-II on NF-κB activation, p-NF-κB p65 level was detected in NMVCs at 5, 10, 20, and 60 min after Ang-II treatment. Significant increase of p-NF-κB p65 level was observed in NMVCs at 10 and 20 min in response to Ang-II treatment (Supplementary material online, Figure S8A). Sequence analysis indicated that three potential NF-κB recognition elements were located in the promoter region of Myo9a gene (Supplementary material online, Figure S8B). Results of the dual-luciferase assay showed that one of the three potential NF-κB recognition elements participated in Ang-II-induced Myo9a transcription in NMVCs (Supplementary material online, Figure S8B). We pre-treated NMVCs with NF-κB p65 inhibitor JSH23 or QNZ for 0.5 h before Ang-II treatment, followed by RT–qPCR analysis. Our data demonstrated that treatment with either JSH23 or QNZ could prevent Ang-II-induced circRNA_000203 and Myo9a mRNA expression in NMVCs (Supplementary material online, Figure S8C). In addition, knockdown of NF-κB p65 could also efficiently inhibit circRNA_000203 and Myo9a mRNA expression in Ang-II-treated NMVCs (Supplementary material online, Figure S8D). Collectively, our results suggested that up-regulation of circRNA_000203 in Ang-II-induced cardiomyocytes resulted from activation of the NF-κB signal.
4. Discussion
In this study, we found that circRNA_000203 and its host gene of Myo9a were markedly up-regulated in the myocardium of Ang-II-infused both male and female mice, suggesting that expression of circRNA_000203 may not affected by sex differences. Obvious increases of circRNA_000203 (homologue of human hsa-circ_0036167) and Myo9a mRNA were also observed in the myocardium of HF patients, as well as in Ang-II-treated mouse cardiomyocytes. The level of circRNA_000203 was correlated with markers of cardiac hypertrophy in vivo and in vitro. To investigate the role of circRNA_000203 in cardiac hypertrophy, we firstly detected the phenotypes of NMVCs with adenovirus-mediated overexpression of circRNA_000203. We found that overexpression of circRNA_000203 induced obvious cellular hypertrophy in NMVCs. Then we adopted a mouse model of Ang-II infusion-induced cardiac hypertrophy. We found that cardiac hypertrophy was markedly aggravated in Ang-II infusion Tg-circ203 mice with myocardium-specific overexpression of circRNA_000203. We demonstrated that circRNA_000203 sponged miR-26b-5p, -140-3p, abolished the suppression of Gata4 by miR-26b-5p, -140-3p, resulting in the increase of GATA4 in NMVCs. Since GATA4 is a TF, it plays an important role in cardiac hypertrophy.25 The present study revealed that up-regulation of GATA4 by circRNA_000203 contributes to the pro-hypertrophy effect of circRNA_000203 in vivo and in vitro.
CircRNAs can be produced through back splicing of a precursor mRNA (pre-mRNA), and the spliceosome has been demonstrated involved in the exon circularization process.26,27 CircRNA_000203 contains the exon sequence from exon 7 to 15 of Myo9a gene, and circRNA_000203 was shown mainly localized in cytoplasma of NMVCs in this study. Consistently, the in vitro and in vivo experimental data indicated that circRNA_000203, but not Myo9a, could specifically enhance cardiac hypertrophy. To reveal the mechanism underlying the pro-hypertrophy effect of circRNA_000203 on NMVCs, we proposed that circRNA_000203 may sponge specific miRNAs to aggravate cardiac hypertrophy. Bioinformatic analysis indicated that there are potential binding sites of miR-26b-5p, -140-3p in circRNA_000203, meanwhile, the negative correlation between the expression of circRNA_000203 and miR-26b-5p, -140-3p was observed in the myocardium of Ang-II-infused mice and in Ang-II-induced NMVCs, as well as in NMVCs with enforced expression of circRNA_000203. Moreover, the results of dual-luciferase reporter assay, RNA pull-down and RT–qPCR assay confirmed the specific interactions between circRNA_000203 and miR-26b-5p, -140-3p in NMVCs. Our data are partially consistent with previous report that circRNA_000203 sponged miR-26b-5p in mouse cardiac fibroblasts.18 Functionally, miR-26b-5p, -140-3p could inhibit the increases of cell size and the expression of ANP and β-MHC in Ang-II-treated NMVCs, and eliminate the pro-hypertrophy effect of circRNA_000203 on NMVCs.
Our current study has provided several lines of evidence to support the notion that miR-26b-5p, -140-3p inhibit cardiac hypertrophy through targeting Gata4. First, the in silico prediction indicated that Gata4 was a potential common target of miR-26b-5p, -140-3p, and the dual-luciferase assay revealed that miR-26b-5p, -140-3p specifically bound to the 3′-UTRs of Gata4 at different sites. In addition, miR-26b-5p, -140-3p mimic inhibited Gata4 expression at the transcriptional level in NMVCs. Importantly, our data validated that circRNA_000203 specifically eliminates the interactions of miR-26b-5p, -140-3p with the 3′-UTRs of Gata4, and transcriptionally enhances Gata4 expression in NMVCs and in the myocardium of Ang-II-infused mice with overexpression of circRNA_000203. MiR-26b-5p, -140-3p could alleviate the increases of ANP and β-MHC in NMVCs with overexpression of Gata4. In parallel to miR-26b-5p, -140-3p, Gata4 siRNA decreased the cell size of cardiomyocytes and inhibited the expressions of ANP and β-MHC in Ang-II-induced NMVCs and in NMVCs with enforced expression of circRNA_000203. The present conclusion has been partially supported by previous studies showing that Gata4 could be regulated by miR-26a, -26b during cardiac hypertrophy.28,29
The mechanism leading to the increased circRNA_000203 expression is currently unknown. In consistence with its parental gene of Myo9a, circRNA_000203 was observed up-regulated in cardiac hypertrophy in vivo and in vitro. Previous reports suggested a role for the NF-κB signalling in cardiac hypertrophy.30–32 Our present data have confirmed that NF-κB p65 was activated in Ang-II-treated NMVCs, and one potential NF-κB p65 recognition element participated in Ang-II-induced Myo9a mRNA transcription in NMVCs. We used NF-κB p65 inhibitor JSH23 and QNZ, p65 NF-kB siRNA to further verify the role of the NF-κB p65 signalling in Ang-II-promoted up-regulation of circRNA_000203 and Myo9a in NMVCs. Collectively, our results suggest that up-regulation of circRNA_000203 in cardiac hypertrophy results from activation of the NF-κB signalling pathway.
In summary, our study demonstrated that circRNA_000203 was up-regulated in cardiac hypertrophy, enhancing the cardiac hypertrophic growth in vivo and in vitro. CircRNA_000203 could sponge miR-26b-5p, -140-3p to increase GATA4 expression, contributing to enhancing the cardiac hypertrophy. We also conclude that activation of the NF-κB signalling pathway participates in the up-regulation of circRNA_000203 in cardiac hypertrophy (Supplementary material online, Figure S9).
Supplementary material
Supplementary material is available at Cardiovascular Research online.
Conflict of interest: none declared.
Funding
This work was supported by the following grants: National Science Foundation of China (Grant numbers: 91649109, 81770264, and 81470439). Guangzhou science and technology programme project (201804010045) and High-level Hospital Construction Project of Guangdong Provincial People’s Hospital (DFJH201902 and DFJH201807).
Supplementary Material
References
- 1. Dorn GW, Robbins J, Sugden PH.. Phenotyping hypertrophy: eschew obfuscation. Circ Res 2003;92:1171–1175. [DOI] [PubMed] [Google Scholar]
- 2. Aaronson KD, Sackner-Bernstein J.. Risk of death associated with nesiritide in patients with acutely decompensated heart failure. J Am Med Assoc 2006;296:1465–1466. [DOI] [PubMed] [Google Scholar]
- 3. Rajabi M, Kassiotis C, Razeghi P, Taegtmeyer H.. Return to the fetal gene program protects the stressed heart: a strong hypothesis. Heart Fail Rev 2007;12:331–343. [DOI] [PubMed] [Google Scholar]
- 4. Putt ME, Hannenhalli S, Lu Y, Haines P, Chandrupatla HR, Morrisey EE, Margulies KB, Cappola TP.. Cappola TP. Evidence for coregulation of myocardial gene expression by MEF2 and NFAT in human heart failure. Circ Cardiovasc Genet 2009;2:212–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nishida W, Nakamura M, Mori S, Takahashi M, Ohkawa Y, Tadokoro S, Yoshida K, Hiwada K, Hayashi K, Sobue K.. A triad of serum response factor and the GATA and NK families governs the transcription of smooth and cardiac muscle genes. J Biol Chem 2002;277:7308–7317. [DOI] [PubMed] [Google Scholar]
- 6. Ozgen N, Obreztchikova M, Guo J, Elouardighi H, Dorn GW 2nd, Wilson BA, Steinberg SF.. Protein kinase D links Gq-coupled receptors to cAMP response element-binding protein (CREB)-Ser133 phosphorylation in the heart. Biol J Chem 2008;283:17009–17019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Viereck J, Thum T.. Circulating noncoding RNAs as biomarkers of cardiovascular disease and injury. Circ Res 2017;120:381–399. [DOI] [PubMed] [Google Scholar]
- 8. Zhang Y, Ren J.. Epigenetics and obesity cardiomyopathy: from pathophysiology to prevention and management. Pharmacol Ther 2016;161:52–66. [DOI] [PubMed] [Google Scholar]
- 9. Callis TE, Pandya K, Seok HY, Tang RH, Tatsuguchi M, Huang ZP, Chen JF, Deng Z, Gunn B, Shumate J, Willis MS, Selzman CH, Wang DZ.. MicroRNA-208a is a regulator of cardiac hypertrophy and conduction in mice. J Clin Invest 2009;119:2772–2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. van Rooij E, Sutherland LB, Liu N, Williams AH, McAnally J, Gerard RD, Richardson JA, Olson EN.. A signature pattern of stress-responsive microRNAs that can evoke cardiac hypertrophy and heart failure. Proc Natl Acad Sci USA 2006;103:18255–18260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sayed D, Hong C, Chen IY, Lypowy J, Abdellatif M.. MicroRNAs play an essential role in the development of cardiac hypertrophy. Circ Res 2007;100:416–424. [DOI] [PubMed] [Google Scholar]
- 12. Tang CM, Liu FZ, Zhu JN, Fu YH, Lin QX, Deng CY, Hu ZQ, Yang H, Zheng XL, Cheng JD, Wu SL, Shan ZX.. Myocyte-specific enhancer factor 2C: a novel target gene of miR-214-3p in suppressing angiotensin II-induced cardiomyocyte hypertrophy. Sci Rep 2016;6:36146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hu ZQ, Luo JF, Yu XJ, Zhu JN, Huang L, Yang J, Fu YH, Li T, Xue YM, Feng YQ, Shan ZX.. Targeting myocyte-specific enhancer factor 2D contributes to the suppression of cardiac hypertrophic growth by miR-92b-3p in mice. Oncotarget 2017;8:92079–92089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, Loewer A, Ziebold U, Landthaler M, Kocks C, Le Noble F, Rajewsky N.. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013;495:333–338. [DOI] [PubMed] [Google Scholar]
- 15. Zhang Z, Yang T, Xiao J.. Circular RNAs: promising biomarkers for human diseases. EBioMedicine 2018;34:267–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Boeckel JN, Jae N, Heumuller AW, Chen W, Boon RA, Stellos K, Zeiher AM, John D, Uchida S, Dimmeler S.. Identification and characterization of hypoxia-regulated endothelial circular RNA. Circ Res 2015;117:884–890. [DOI] [PubMed] [Google Scholar]
- 17. Holdt LM, Stahringer A, Sass K, Pichler G, Kulak NA, Wilfert W, Kohlmaier A, Herbst A, Northoff BH, Nicolaou A, Gäbel G, Beutner F, Scholz M, Thiery J, Musunuru K, Krohn K, Mann M, Teupser D.. Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nat Commun 2016;7:12429.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tang CM, Zhang M, Huang L, Hu ZQ, Zhu JN, Xiao Z, Zhang Z, Lin QX, Zheng XL, Yang M, Wu SL, Cheng JD, Shan ZX.. CircRNA_000203 enhances the expression of fibrosis-associated genes by derepressing targets of miR-26b-5p, Col1a2 and CTGF, in cardiac fibroblasts. Sci Rep 2017;7:40342.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Khan MAF, Reckman YJ, Aufiero S, van den Hoogenhof MMG, van der Made I, Beqqali A, Koolbergen DR, Rasmussen TB, van der Velden J, Creemers EE, Pinto YM.. RBM20 regulates circular RNA production from the titin gene. Circ Res 2016;119:996–1003. [DOI] [PubMed] [Google Scholar]
- 20. Shan K, Liu C, Liu BH, Chen X, Dong R, Liu X, Zhang YY, Liu B, Zhang SJ, Wang JJ, Zhang SH, Wu JH, Zhao C, Yan B.. Circular noncoding RNA HIPK3 mediates retinal vascular dysfunction in diabetes mellitus. Circulation 2017;136:1629–1642. [DOI] [PubMed] [Google Scholar]
- 21. Wang K, Long B, Liu F, Wang JX, Liu CY, Zhao B, Zhou LY, Sun T, Wang M, Yu T, Gong Y, Liu J, Dong YH, Li N, Li PF.. A circular RNA protects the heart from pathological hypertrophy and heart failure by targeting miR-223. Eur Heart J 2016;37:2602–2611. [DOI] [PubMed] [Google Scholar]
- 22. Wang K, Gan TY, Li N, Liu CY, Zhou LY, Gao JN, Chen C, Yan KW, Ponnusamy M, Zhang YH, Li PF.. Circular RNA mediates cardiomyocyte death via miRNA-dependent upregulation of MTP18 expression. Cell Death Differ 2017;24:1111–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zeng Y, Du WW, Wu Y, Yang Z, Awan FM, Li X, Yang W, Zhang C, Yang Q, Yee A, Chen Y, Yang F, Sun H, Huang R, Yee AJ, Li RK, Wu Z, Backx PH, Yang BB.. A circular RNA binds to and activates AKT phosphorylation and nuclear localization reducing apoptosis and enhancing cardiac repair. Theranostics 2017;7:3842–3855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yuan W, Tang C, Zhu W, Zhu J, Lin Q, Fu Y, Deng C, Xue Y, Yang M, Wu S, Shan Z.. CDK6 mediates the effect of attenuation of miR-1 on provoking cardiomyocyte hypertrophy. Mol Cell Biochem 2016;412:289–296. [DOI] [PubMed] [Google Scholar]
- 25. Pikkarainen S, Tokola H, Kerkelä R, Ruskoaho H.. GATA transcription factors in the developing and adult heart. Cardiovasc Res 2004;63:196–207. [DOI] [PubMed] [Google Scholar]
- 26. Chao CW, Chan DC, Kuo A, Leder P.. The mouse formin (Fmn) gene: abundant circular RNA transcripts and gene-targeted deletion analysis. Mol Med 1998;4:614–628. [PMC free article] [PubMed] [Google Scholar]
- 27. Hansen TB, Wiklund ED, Bramsen JB, Villadsen SB, Statham AL, Clark SJ, Kjems J.. miRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA. EMBO J 2011;30:4414–4422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Liu Y, Wang Z, Xiao W.. MicroRNA-26a protects against cardiac hypertrophy via inhibiting GATA4 in rat model and cultured cardiomyocytes. Mol Med Rep 2016;14:2860–2866. [DOI] [PubMed] [Google Scholar]
- 29. Han M, Yang Z, Sayed D, He M, Gao S, Lin L, Yoon S, Abdellatif M.. GATA4 expression is primarily regulated via a miR-26b-dependent post-transcriptional mechanism during cardiac hypertrophy. Cardiovasc Res 2012;93:645–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Qi HP, Wang Y, Zhang QH, Guo J, Li L, Cao YG, Li SZ, Li XL, Shi MM, Xu W, Li BY, Sun HL.. Activation of peroxisome proliferator-activated receptor γ (PPARγ) through NF-κ B/Brg1 and TGF-β 1 pathways attenuates cardiac remodeling in pressure-overloaded rat hearts. Cell Physiol Biochem 2015;35:899–912. [DOI] [PubMed] [Google Scholar]
- 31. Zhang S, Tang F, Yang Y, Lu M, Luan A, Zhang J, Yang J, Wang H.. Astragaloside IV protects against isoproterenol-induced cardiac hypertrophy by regulating NF-κB/PGC-1α signaling mediated energy biosynthesis. PLoS One 2015;10:e0118759.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cau SB, Guimaraes DA, Rizzi E, Ceron CS, Gerlach RF, Tanus-Santos JE.. The nuclear factor kappaB inhibitor pyrrolidine dithiocarbamate prevents cardiac remodelling and matrix metalloproteinase-2 up-regulation in renovascular hypertension. Basic Clin Pharmacol Toxicol 2015;117:234–241. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.