Abstract
Aims
High-density lipoproteins (HDLs) are circulating micelles that transport proteins, lipids, and miRNAs. HDL-transported miRNAs (HDL-miRNAs) have lately received attention but their effects on vascular cells are not fully understood. Additionally, whether cardiovascular risk factors affect HDL-miRNAs levels and miRNA transfer to recipient cells remains equally poorly known. Here, we have investigated the changes induced by hypercholesterolaemia on HDL-miRNA levels and its effect on recipient endothelial cells (ECs).
Methods and results
Pigs were kept on a high-fat diet (HC; n = 10) or a normocholesterolaemic chow (NC; n = 10) for 10 days reaching cholesterol levels of 321.0 (229.7–378.5) mg/dL and 74.0 (62.5–80.2) mg/dL, respectively. HDL particles were isolated, purified, and quantified. HDL-miRNA profiling (n = 149 miRNAs) of HC- and NC-HDLs was performed by multipanel qPCR. Cell cultures of porcine aortic ECs were used to determine whether HDL-miRNAs were delivered to ECs. Potential target genes modulated by miRNAs were identified by bioinformatics and candidate miRNAs were validated by molecular analysis. In vivo effects in the coronary arteries of normocholesterolaemic swine administered HC- or NC-HDLs were analysed. Among the HDL-miRNAs, four were found in different amounts in HC- and NC-HDL (P < 0.05). miR-126-5p and -3p and miR-30b-5p (2.7×, 1.7×, and 1.3×, respectively) were found in higher levels and miR-103a-3p and miR-let-7g-5p (−1.6×, −1.4×, respectively) in lower levels in HC-HDL. miR-126-5p and -3p were transferred from HC-HDL to EC (2.5×; P < 0.05), but not from NC-HDL, by a SRB1-mediated mechanism. Bioinformatics revealed that HIF1α was the miR-126 target gene with the highest predictive value, which was accordingly found to be markedly reduced in HC-HDL-treated ECs and in miR126 mimic transfected ECs. In vivo validation confirmed that HIF1α was diminished in the coronary endothelial layer of NC pigs administered HC-HDL vs. those administered NC-HDL (P < 0.05).
Conclusion
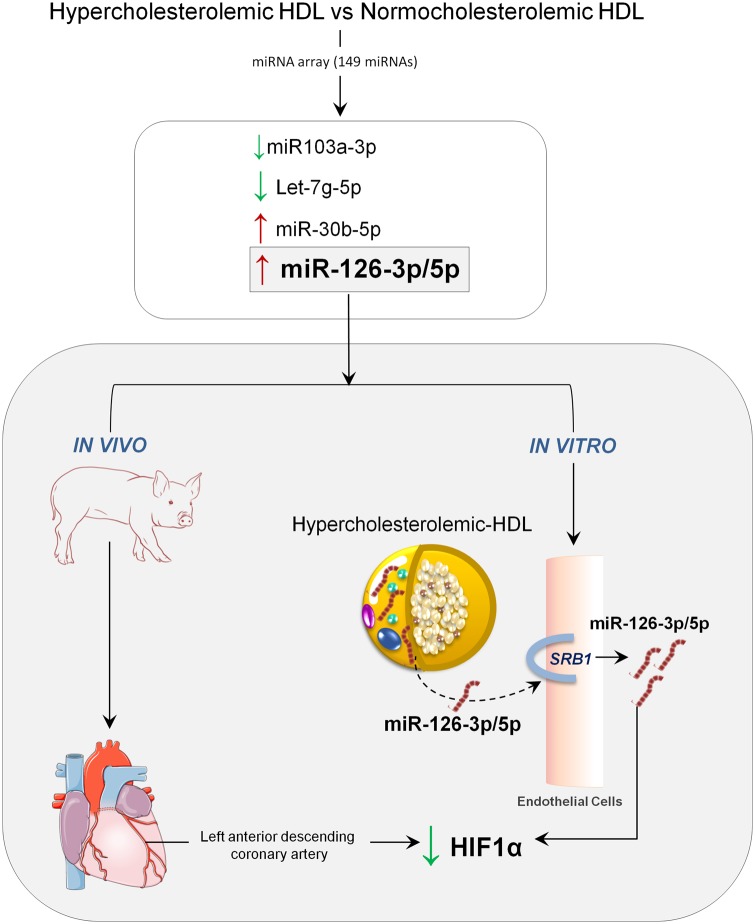
Hypercholesterolaemia induces changes in the miRNA content of HDL enhancing miR126 and its delivery to ECs with the consequent down-regulation of its target gene HIF1α.
Keywords: HDL, Dyslipidaemia, Translational research, miRNAs, Endothelial cells
Graphical Abstract
Graphical Abstract.
1. Introduction
MicroRNAs (miRNAs) are short, non-coding RNAs (∼22 nucleotide in length) that control gene expression through post-transcriptional regulation of target mRNAs. Extracellular miRNAs have been detected in multiple lipid-based carriers including exosomes, microparticles, and lipoproteins.1 Indeed, high-density lipoproteins (HDLs) are lipoproteins that in addition to cholesterol, transport other lipid species, proteins, and miRNAs.2 HDL particles exert multiple cardiovascular protective functions. Under physiological conditions HDL-cargo molecules that maintain cardiovascular homeostasis and function by promoting circulating cells, endothelial cells (ECs), vessel wall stabilization, and regulating reverse cholesterol transport and ischaemia/reperfusion injury.3–5 In this regard, HDL-mediated transport and delivery of miRNAs to recipient cells can regulate expression of genes involved in lipid metabolism, inflammation, angiogenesis, and apoptosis.6–9
Robust epidemiological data supports that increasing HDL-cholesterol (HDL-c) levels predict improved cardiovascular outcomes; however, HDL-c raising therapies in secondary prevention trials have failed to demonstrate cardiovascular benefits.10 As such, within the last decade, it has become increasingly evident that the measure of the cholesterol levels transported by HDL (HDL-c) is not a good read-out of HDL beneficial effects because the presence of excess cholesterol (hypercholesterolaemia) increases the transport of cholesterol in detriment of other beneficial cargos. Moreover, the presence of comorbidities modifies HDL particle composition and function hampering HDL-beneficial effects and even becoming deleterious.11–16 We have recently demonstrated that HDL in vivo remodelling is affected by low-density lipoprotein (LDL) cholesterol levels. Indeed, the presence of hypercholesterolaemia influences HDL lipidomic and proteomic profile and renders HDL particles dysfunctional.17–19 Particularly, HDL particles formed under hypercholesterolaemic conditions, as compared to HDL particles formed in a normocholesterolaemic milieu, have lower levels of phosphatidylcholine-lipid species and higher levels of cholesteryl esters and a lower cargo of several proteins with known cardioprotective functions including lipocalin retinol binding protein-4 and apolipoprotein-M. Interestingly, we have further proved that such adverse structural remodelling is associated with a marked reduction in HDL anti-oxidant and cholesterol efflux capacity and complete loss of HDL-induced cardioprotection.20 However, to date, there is no information on the impact of hypercholesterolaemia on HDL-transported miRNA (HDL-miRNA) content or on the possible effects of HDL-miRNAs on EC gene regulation. Accordingly, in the present study, we aimed to investigate the HDL-miRNA cargo of HDL formed in vivo in hypercholesterolaemic conditions and whether the tentative modifications were able to differentially regulate gene expression in target cells.
2. Methods
Experimental procedures were reviewed and approved by the Institutional Animal Care and Use Committees (CEEA-IR) and authorized by the Animal Experimental Committee of the local government (#5601) in accordance to the Spanish law (RD 53/2013) and European Directive 2010/63/EU. In addition, the investigation conforms to the Guide for the Care and Use of Laboratory Animals published by the US National Institutes of Health (NIH Publication No. 85-23, revised 1985), follows the ARRIVE guidelines and is committed to the 3Rs of laboratory animal research and consequently used the minimal number of animals to reach statistical significance.21 All animals were allowed to acclimate 7 days prior to any intervention and housed in individual cages under light-controlled conditions and room temperature.
2.1 Experimental model of hypercholesterolaemia and HDL isolation
2.1.1 Experimental animal model
Four-month-old pigs (N = 20; weight: ≈40kg) randomly received either regular normocholesterolaemic chow (NC; n = 10) or a high-fat/high-cholesterol diet (HC; n = 10) for 10 days (composition of the diet is detailed in Supplementary material online, Table S1). HC animals developed high LDL-cholesterol levels similar to those found in hypercholesterolaemic humans, 321.0 (229.7–378.5) mg/dL, while NC-animals had 74.0 (62.5–80.2) mg/dL (P < 0.0001; Supplementary material online, Table S2).22
2.1.2 HDL isolation and purification
On Day 10, animals were tranquilized by intramuscular injection of Azoperone (40 mg/mL; 7 mg/kg), anaesthesia was induced by propofol (2%; 3 mg/kg), and maintained by isofluorane inhalation (3%). Animals were then sacrificed by propofol overdose and potassium chloride (2 M). HDLs were isolated by standard sequential preparative ultracentrifugation techniques from plasma-EDTA (ethylenediaminetetra-acetic acid) before sacrifice as previously reported.19 Protein levels in HDL were quantified by standard Bradford protein assay (Pierce™ BCA Protein Assay Kit, Thermo Fisher Scientific, Waltham, Massachusetts, USA). Lipoprotein cholesterol content was assessed by the Cholesterol Quantification Kit (MAK043-Sigma-Aldrich; MO, USA). The purity of the HDL fraction was confirmed by excluding the presence of ApoB (by-agarose gel electrophoresis), microvesicles (FACS for Anexin V+ microvesicles), and exosomes (western blots for transpanins CD63 and CD81; exosomes isolated by ExoMir™ Kit).23
2.1.3 HDL miRNA isolation
HDL samples were thawed on ice and centrifuged at 3000 g for 5 min at 4°C. An aliquot of 200 μL per sample (protein concentration HC: 5.7 µg/µL and NC: 5.6 µg/µL) was transferred to a FluidX tube and 60 μL of lysissolution buffer containing 1 μg carrier-RNA per 60 μL lysis solution buffer and RNA spike-in template mixture were added to the sample and mixed for 1 min and incubated for 7 min at room temperature, followed by addition of 20 μL protein precipitation solution buffer. Total RNA was extracted from the samples using miRCURY RNA isolation Kit through a high throughput bead-based protocol v.1 (Exiqon, Vedbaek, Denmark) in an automated 96-well format. The purified total RNA was eluted in a final volume of 50 μL and its optimal quality checked.
2.1.4 HDL miRNA analysis by qPCR
Seven microlitres of RNA were reverse transcribed in 35 μL reactions using the miRCURY LNA™ Universal RT microRNA PCR, Polyadenylation and cDNA synthesis kit (Exiqon). cDNA was diluted 50× and assayed in 10 µL PCR reactions according to the protocol for miRCURY LNA™ Universal RT microRNA PCR. A panel of 149 miRNAs homologous for pig and human were analysed in HDL (Supplementary material online, Table S3). Each miRNA was assayed by qPCR (microRNA Ready-to-Use PCR, Custom Pick and Mix panel using ExiLENT SYBR® Green master mix) and negative controls were run in parallel. The amplification was performed in a LightCycler® 480 Real-Time PCR System (Roche) in 384-well plates. The amplification curves were analysed using the Roche LC software, both for Cq determination (following the 2nd derivative method) and for melting curve analysis.
2.1.5 HDL miRNA data analyses
The amplification efficiency was calculated (LinReg software). All assays were inspected for distinct melting curves and the Melt temperature (Tm) was checked to be within known specifications for the assay. Assays had to be detected beyond 5 Cqs of the negative control and with Cq < 37 in order to be included in the data analyses. The NormFinder helped to find out the best possible normalizer which resulted to be the average of the best assays detected in all samples (n = 20). The formula used to calculate the normalized Cq values was the following: Normalized Cq = average Cq (n = 20) − assay Cq (sample). We then performed a comparative analysis between NC and HC isolated HDL-miRNAs. Data were expressed as fold-change.
2.2 Cell culture studies
2.2.1 Porcine aortic endothelial cells isolation, expansion and treatment
Aortic ECs were obtained from regular-fed pigs by collagenase digestion as previously described.24 Cells were cultured in growth M199 (Gibco) medium supplemented with 10% FBS and antibiotics (0.1 mg/mL streptomycin, 100 U/mL penicillin) until confluence. Cells were detached by trypsin/EDTA treatment and seeded into larger flasks for expansion. Forty-eight hours after plating 300 000 cells in six well plates, 0.1% FBS was added to the medium for 24 h and then EC were incubated with NC-HDL (50 µg/mL protein), HC-HDL (50 µg/mL protein), or PBS/control for 18 h (in duplicates; n = 5) as previously reported.24,25 EC viability was determined by trypan blue exclusion. ECs were always used between the third and seventh passage.
2.2.2 ECs and plasma miRNA isolation and data analysis
ECs were collected and total RNA was obtained with mirVANA miRNA kit. The RNA was transcribed using TaqMan® Advanced miRNA cDNA Synthesis Kit (Applied Biosystems; CA, EU) protocol. All assays were performed in duplicates for the differentially expressed miRNAs including miR-126-5p and -3p, let7g-5p, miR-103a-3p and miR-30b-5p (Life Technologies; CA, EU) (n = 5). For normalization miR-423-3p and let-7a-5p means were used as endogenous miRNA controls.26 The amplification was performed in ABI PRISM 7900HT Sequence Detection System (Applied Biosystems; CA, EU) in 384-well plates. The amplification curves were analysed using the Applied Biosystems Sequence Detection System 2.4.1 software. Ct values were obtained and processed by the standard curve method and normalized with the endogen. To obtain fold difference vs. control the ratio sample/control was calculated. Networks, functional analyses, detection of miRNAs targets and interactions were generated by Ingenuity Pathway Analysis software (IPA) and RNA hybrid tool.
2.2.3 Assessment of HDL-miRNA transfer to ECs
To investigate whether the scavenger receptor class B member 1 (SRB1) was involved in HDL-miRNA transfer to ECs a specific SRB1 blocker, BLT1 (EMD Millipore Corp, USA; 8 µM), was added to ECs prior to HDL incubation.25 In addition, and taking into consideration that miR-126 (both 5p and 3p) is encoded in the 7th intron of the EGFL7 gene, we assessed the expression of both EFGL7 and the pre-miRNA sequence (pre-mir-126; n = 5) in ECs in order to ascertain whether the potential intracellular changes detected in miR-126-5p and -3p expression were related to an up-regulation of its cellular synthesis or because of transfer from HDL. Pri-miR126 could not be investigated because there is no commercial assay available for Sus scrofa so far.
2.3 miR126 target gene analysis
2.3.1 In silico analysis
Potential target genes were obtained by using databases TargetScan, miRwalk (5UTR, CDS, 3UTR) and the Ingenuity Pathway Analysis (IPA; http://www.ingenuity.com/), according to their search algorithms. The functionality of target genes in cellular processes was predicted by IPA Core analysis. Furthermore, RNAhybrid (Bielefeld University Bioinformatics server) was used for calculating minimum free energy (MFE), which indicates the accessibility of miRNAs at the target gene binding sites. Target genes were selected by comparison of MFE values.27
2.3.2 Gene expression validation
RNA was transcribed using the High Capacity cDNA Archive kit (Applied Biosystems, Foster City, CA, USA) protocol for genomic RNA. qPCRs were performed in duplicates for SCARB1, mir-126-5p and -3p, HIF1α, vascular cell adhesion protein 1 (VCAM1), vascular endothelial growth factor A (VEGFA) (Life Technologies; CA, EU), and EGFL7 (Bio-Rad Laboratories, CA, EU). 18SrRNA was used as endogenous control for tissue samples and GAPDH (Life Technologies; CA, EU) for EC samples. The amplification was performed in ABI PRISM 7900HT Sequence Detection System (Applied Biosystems; CA, EU) in 384-well plates. The amplification curves were analysed using the Applied Biosystems Sequence Detection System 2.4.1 software. Ct values were obtained and processed by the standard curve method and normalized with the endogenous gene.
2.3.3 Protein expression validation
Cell lysis was performed according to the previous established protocol.28 The cell protein lysates were stored at −80°C until use. Protein concentrations were assessed using the BSA assay and then SDS–polyacrylamide gel electrophoresis (SDS-PAGE) was run. Blots were blocked with BSA at room temperature for 1 h and incubated with the appropriate primary antibody [HIF1α (sc-10790, Santa Cruz Biotechnology, Dallas, TX, USA)] at 4°C overnight. Membranes were then incubated with a horseradish peroxidase-conjugated anti-mouse antibody for 2 h. Densitometry analyses were performed using commercially available quantitative software (ImageLab Software, Bio-Rad Laboratories, CA, USA) with the control representing 1.0-fold.
2.3.4 Mir126 target validation by miR126-mimic and miR126 inhibitor
ECs were transfected with miR126-mimic or miR126 inhibitor (both for -5p and -3p) (Life Technologies; CA, EU) with the Lipofectamine™ RNAiMAX Transfection Reagent (Life Technologies; CA, EU) following the supplier recommendations. Then, ECs were treated with HDL-HC or HDL-NC as reported above. A miRNA Mimic Negative Control #1 (Life Technologies; CA, EU) was run in parallel.
2.4 In vivo validation of miR-126 target genes by confocal microscopy
We assessed HIF1α protein expression in the coronary arteries of normocholesterolaemic pigs (n = 12) administered HC-HDL (n = 6) or NC-HDL (n = 6). In this regard, we processed the coronary arteries from pigs of our previous published study19 which had been kept in our tissue biobank at −80°C until use. Accordingly, we fixed the proximal portion of the coronary artery of all pigs in 4% paraformaldehyde and the paraffin-embedded for confocal microscopy analysis. Serial sections (5 µm thick) were incubated with blocking buffer and then with antibody against HIF1α (H1alpha67, Novus Biologicals, Littleton, CO, USA). Preparations were counterstained with fluorescently labelled secondary antibody (Invitrogen) and mounted with fluorescent medium. Fluorescent images were acquired in a scan format of 1024 × 1024 pixels in a spatial data set (x y z). Controls with no primary antibody showed no fluorescence labelling.
2.5 Statistics
The Shapiro–Wilk test was employed to analyse data normal or non-normal distribution; data are reported as the mean ± standard deviation and median (interquartile range), respectively. For the biochemical descriptive data and SRB1 in vivo analysis, we performed non-parametric test Kruskal–Wallis with Dunn’s multiple comparisons corrected test. Tukey’s honest significance corrected test was used in conjunction with an ANOVA (post hoc analysis) for multiple comparisons to find significantly difference between group means due to its condition of independent samples. Accordingly, t-test was used for two group independent samples mean comparison. A two-sided P-value less than 0.05 was considered statistically significant.
Sample size: For the EC approach accepting α risk of 0.05 and β risk of 0.2 in a two-sided test, an n = 3 was necessary to recognize as statistically significant difference ≥0.5 units. Yet, we performed n = 5 in all conditions to make our conclusions more robust. The common standard deviation is assumed to be 0.2 and it we anticipated a dropout rate of 0% (GRANMO software, Spain). All statistical analyses and graphics were performed using GraphPad Prism 7.0 version software.
3. Results
3.1 HDL-characterization
Cholesterol content was two-fold higher in HC- than in NC-HDLs (Supplementary material online, Table S4). This observation is in line with our previous findings regarding the effects of hypercholesterolaemia on modifying HDL-lipid composition.20 The isolated HDL samples were free from ApoB (Supplementary material online, Figure S1A), Anexin V+ microvesicles (Supplementary material online, Figure S1B), and exosomes (Supplementary material online, Figure S1C), supporting the purity of the HDL preparations. The protein concentration was similar in HC-HDL (5.7 µg/µL) and in NC-HDL (5.6 µg/µL).
3.2 HDL-related miRNA profile
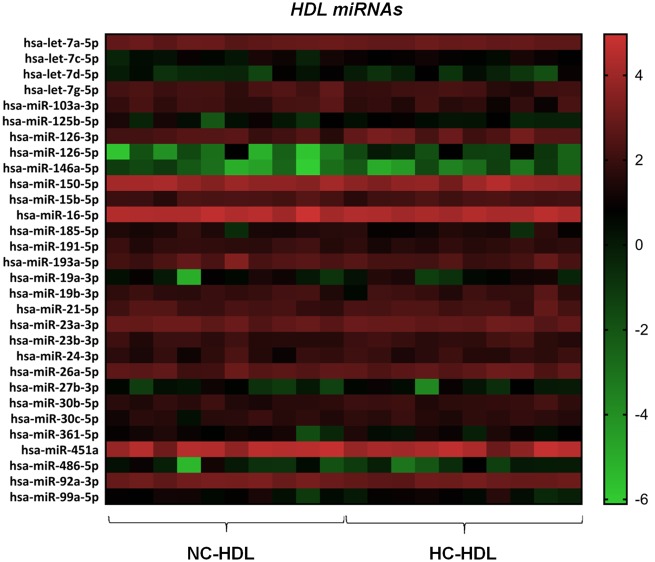
Among the 149 miRNAs analysed, 30 miRNAs were detected in all HDL samples (Figure 1 and Supplementary material online, Table S5). Among these 30 miRNA, the amounts of five miRNAs were significantly different in HC- and in NC-HDL particles (P < 0.05). Particularly, there was a higher content of miR126-5p, miR126-3p, and miR-30b-5p in HC-HDL particles as compared to NC-HDL (2.7×, 1.7×, and 1.3×, respectively), whereas, conversely, the amounts of miR103a-3p and let7g-5p were significantly lower (−1.6×, −1.4×, respectively).
Figure 1.
HeatMap including the miRNAs detected in all HDL samples isolated from hypercholesterolaemic (HC; n = 10) and normocholesterolaemic (NC; n = 10) pigs.
3.3 HC-HDL particles transfer miR-126 to ECs through a SRB1-dependent mechanism
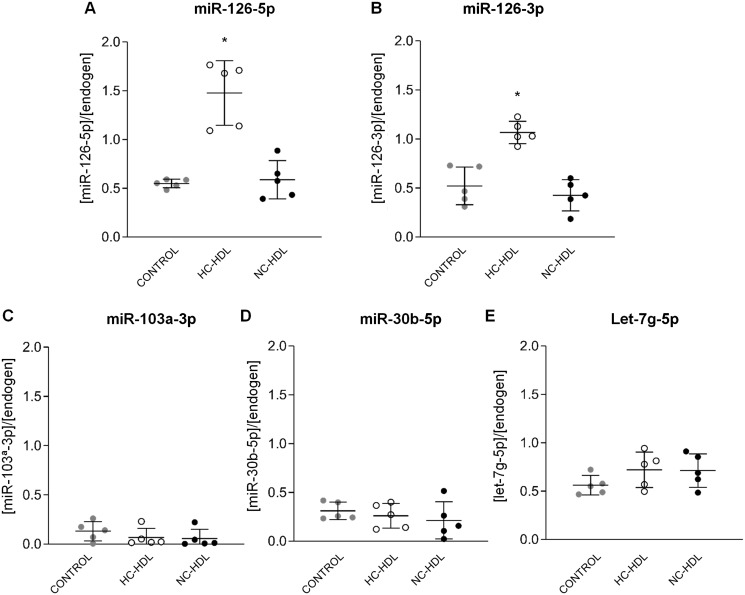
ECs incubated with HC-HDL showed significantly higher intracellular levels of miR-126-5p (P < 0.0001) and miR126-3p (P < 0.0001), while levels of both miR-126s (5p and 3p) were similar in control ECs and ECs incubated with NC-HDL (Figure 2A and B). No differences were observed for miR103a-3p (Figure 2C), miR30b-5p (Figure 2D), and let7g-5p (Figure 2E).
Figure 2.
Endothelial cells incubated with hypercholesterolaemic-HDL show higher miR-126-5p and 3p levels as compared to normocholesterolaemic-HDL [miR126-5p (A) and miR126-3p (B)], whereas no differences were observed for 103a-3p (C), miR-30b-5p (D), or let-7g-5p (E) levels. n = 5. Data are reported as mean ± SD. *ANOVA test P < 0.05 vs. Control and NC-HDL groups. HC: hypercholesterolaemic; NC: normocholesterolaemic.
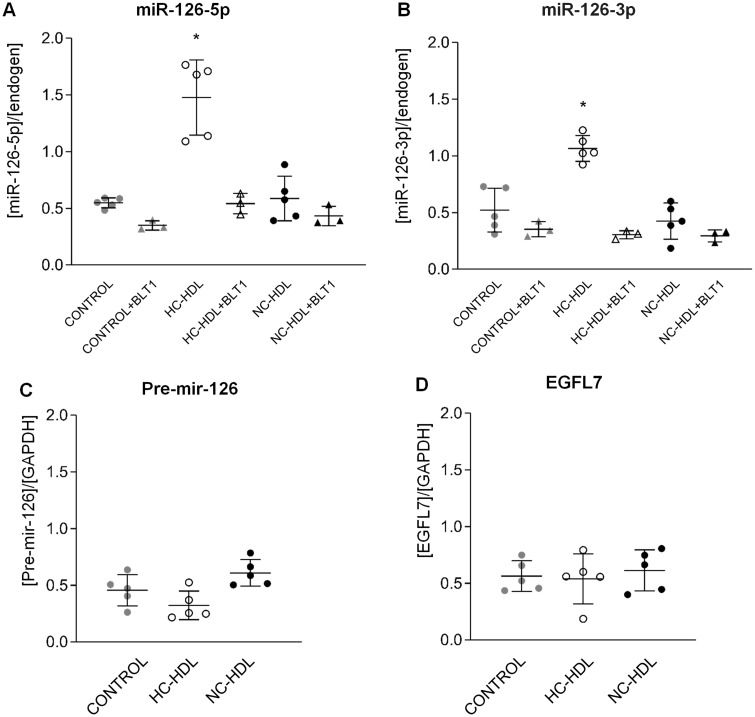
Interestingly, the internalization of miRNA-126 in ECs was mediated by SRB1.We found that SRB1 blockade by BLT-1 abolished HC-HDL-induced increase in miR-126 (both 5p and 3p) (Figure 3A and B) content. SRB1 expression in ECs did not differ by incubation with either NC- or HC-HDLs (Supplementary material online, Figure S2A). Moreover, comparable SRB1 transcript levels were detected in pig coronary arteries and liver (Supplementary material online, Figure S2B). Therefore, SRB1 was found abundantly expressed in pig ECs and its expression was not influenced by incubation with HDL. Finally, blockade of SRB1 in HDL- and PBS-incubated cells led to comparable up-regulation in SRB1 levels (Supplementary material online, Figure S3).
Figure 3.
Hypercholesterolaemic HDL particles transfer miR-126-5p and -3p (A and B, respectively) to endothelial cells through a SRB1-dependent mechanism. No differences are detected for Pre-mir-126 (C) or for miR-126 encoding gene EGFL7 (D) levels among the different groups. n = 5. Data are reported as mean ± SD. *ANOVA test P < 0.05 vs. any other group. HC: hypercholesterolaemic; NC: normocholesterolaemic.
To further confirm the transfer of miR-126-5p and -3p from HC-HDL to the ECs, we analysed pre-mir-126 and EGFL7 levels. As shown in Figure 3, both pre-mir-126 (Figure 3C) and EGFL7 (Figure 3D) were similarly expressed among all the three groups excluding an enhanced intracellular transcription of miRNA-126 in HC-HDL-treated cells and confirming transfer from HC-HDL.
3.4 miR-126 target genes on ECs
We used different computational databases (Supplementary material online, Figure S4 and Table S6) to identify potential target genes of miR-126-5p and -3p in ECs, and only those identified in at least two databases were further investigated. We identified a total of 7597 candidate target genes potentially regulated by miR-126 (3p-strands: 454; 5p-strands: 7143). Among them, common target genes were identified from at least two databases (51 for miR-126-3p and 942 for miR-126-5p). HIF1α was the potential best target gene for miR-126-5p regulation given the low MFE values (−8.3, −11.2, −12.6, −12.1, −8.5, −13.1 Kcal/mol) and its six tentative binding sites for miR126-5p (Table 1). The best potential target for miR126-3p was DR5 but it could not be validated because its sequence is not yet available in S. Scrofa.
Table 1.
miR-126 target genes by in silico analysis
| miR-126-5p target genes | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Top-10 miR-126-5p target genes | Cell cycle | Cell morphology | Lipid metabolism | Cellular movement | Cellular assembly and organization | Cellular development | Cellular growth and proliferation | Cell-to-cell signalling and interaction | Total score | MFE values | ||
| HGF | 2 | 9 | 1 | 9 | 3 | 8 | 1 | 7 | 40 | −9.4 | ||
| IL6 | 6 | 4 | 0 | 2 | 3 | 10 | 2 | 2 | 29 | −12.5 | −13.3 | |
| CD44 | 2 | 4 | 0 | 7 | 0 | 5 | 1 | 4 | 23 | −12.6 | −9.6 | −12.5 |
| ADIPOQ | 1 | 1 | 0 | 2 | 4 | 5 | 3 | 1 | 17 | −8.9 | ||
| BDNF | 0 | 4 | 0 | 4 | 3 | 4 | 0 | 1 | 16 | −9.7 | ||
| BCL2 | 3 | 4 | 0 | 0 | 0 | 3 | 3 | 1 | 14 | −13.1 | −14 | |
| NRG1 | 3 | 0 | 0 | 2 | 3 | 5 | 0 | 1 | 14 | −11 | ||
| HIF1A | 4 | 0 | 0 | 3 | 0 | 5 | 0 | 0 | 12 | −8.3 | −11.2 | −12.6 |
| ITGA4 | 0 | 2 | 0 | 6 | 3 | 0 | 0 | 1 | 12 | −12.1 | −8.5 | −13.1 |
| MAD2L1 | 9 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 12 | −17.9 | ||
| P-value | 0.000046 | 0.000048 | 0.000295 | 0.000597 | 0.000756 | 0.00179 | 0.00179 | 0.00197 | −10.9 | |||
The potential target genes for miR-126-5p related with the top 10 cellular functions by core analysis, a tool from IPA (Ingenuity Pathway Analysis). The cellular functions are showed by P-values according to score. Minimum Free Energy (MFE) values were obtained by RNAhybrid. Bold HIF1α MFE highlighted in bold.
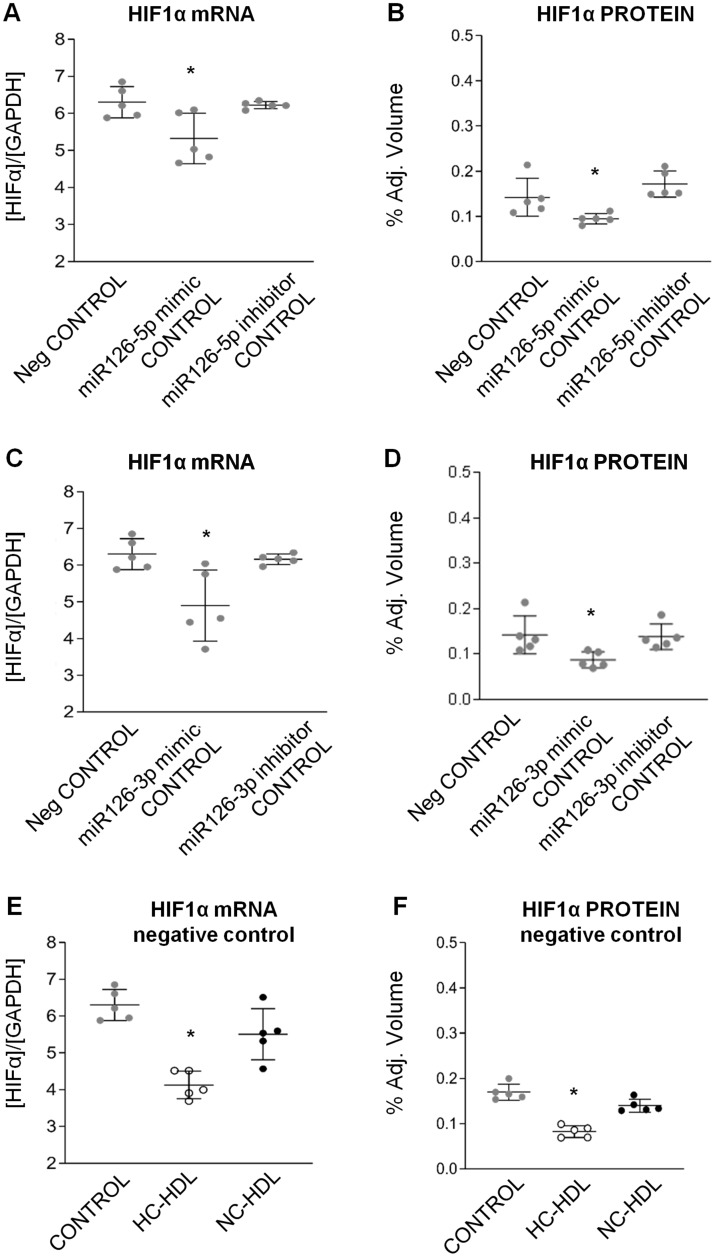
To further validate HIF1α as a target gene for miR126, we examined the effects of miR-126-mimics and miR-126-inhibitors (both -5p and -3p) in ECs (Figure 4). Indeed, control ECs transfected with both miR126-mimics showed lower HIF1α transcript levels and protein expression than ECs transfected with the negative control and both miR126-inhibitors (Figure 4A–D and Supplementary material online, Figure S5). HIF1α gene expression was lower in negative-control transfected ECs incubated with HC-HDLs as compared to NC-HDL and control (P < 0.05; Figure 4E and F and Supplementary material online, Figure S5). In the EC cultures treated with HDL the effects of HL-HDL on the down-regulation of HIF1a expression were abrogated by miR126-5p/3p inhibitors (Supplementary material online, Figure S6).
Figure 4.
Control ECs transfected with miR126-mimic show lower HIF1α gene levels as compared to ECs transfected with the negative control both at mRNA expression and protein level (A–D). HIF1α-mRNA expression was lower in negative-control transfected ECs incubated with HC- as compared to NC-HDLs and control (E and F). *ANOVA test P < 0.05. HC: hypercholesterolaemic; NC: normocholesterolaemic.
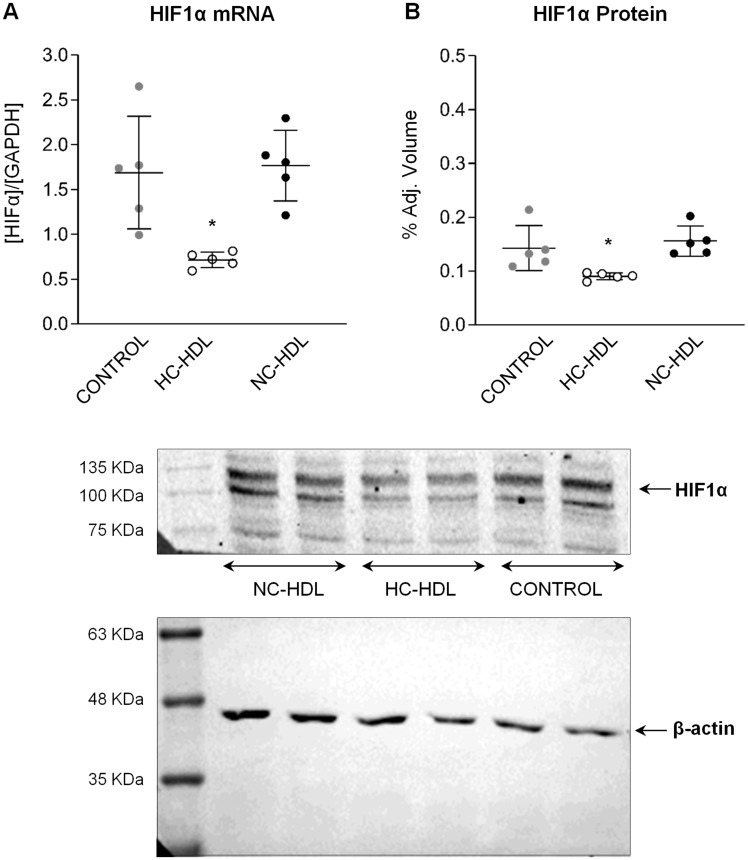
Based on these findings, we proceeded to validate HIF1α by qPCR (n = 5) and found that HIF1α transcript levels were significantly down-regulated in ECs treated with HC-HDL (P < 0.01 vs. PBS/control; P < 0.006 vs. NC-HDL; Figure 5A). HIF1α protein levels were also significantly reduced in HC-HDL treated ECs (Figure5B).
Figure 5.
mRNA expression and protein levels of miR126 target HIF1α in endothelial cells. n = 5. Data are reported as mean ± SD. *ANOVA test P < 0.05 vs. Control and NC-HDL groups. HC: hypercholesterolaemic; NC: normocholesterolaemic.
Mimic-MiR126 (-5p and -3p) induced down-regulation of VCAM1 mRNA expression (Supplementary material online, Figure S7A and B) and up-regulation of VEGFA mRNA expression (Supplementary material online, Figure S7C and D).
3.5 In vivo validation of HC-HDL-related miR-126 potential to modulate endothelial HIF1α
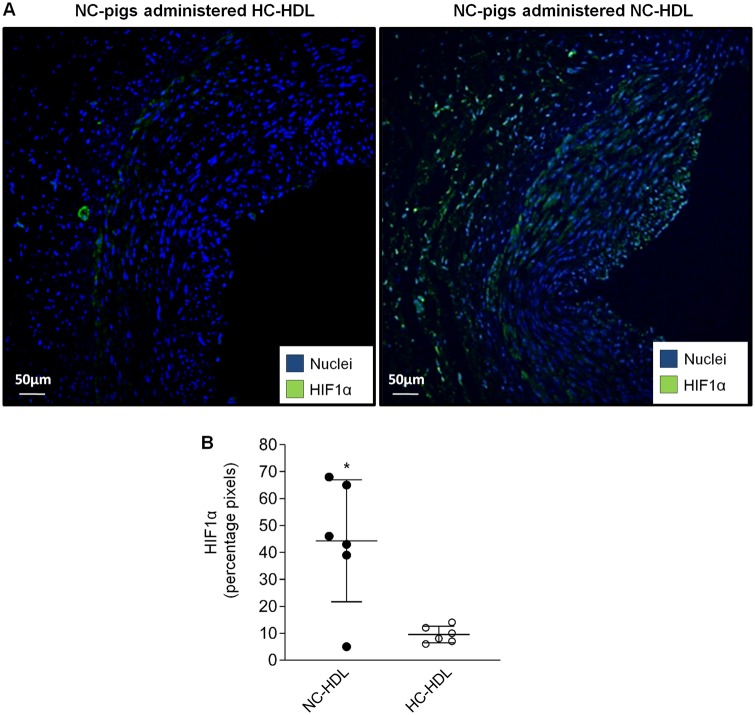
Finally, to determine whether our in vitro findings translated into the in vivo setting, we assessed HIF1α expression by confocal microscopy in the coronary arteries of NC pigs administered HC- or NC- HDLs. Interestingly, as shown in Figure 6, expression of HIF1α was markedly reduced in the intimal layer of the coronary arteries of those NC animals administered HC-HDLs as compared to those administered NC-HDLs.
Figure 6.
Confocal microscopy analysis of HIF-1α detection in the endothelial layer of the coronary artery of normocholesterolaemic (NC) pigs administered NC- or hypercholesterolaemic (HC)-HDLs. Representative images of HIF1α (green) detection (A) and bar-graph quantification (B). Data are reported as mean ± SD. n = 6 animals/group. *T-test P < 0.05 vs. NC pigs administered HC-HDL.
4. Discussion
In the present study, we report that: (i) HDL formed in hypercholesterolaemic conditions transport different amounts of miRNAs than those HDL formed in normocholesterolaemic conditions; (ii) miR-126-5p and -3p content is higher in HC-HDL particles; (iii) HC-HDL transfer miR-126 to ECs through a SRB1-dependent mechanism; (iv) internalized miRNA-126 regulates HIF1a in recipient cells. HIF1α down-regulation is observed in vivo as seen in the coronary arteries of animals treated with HL-HDL but not in animals treated with NL-HDL.
During the last years, it has become increasingly evident that HDL can transport multiple miRNAs including miR-12629, miR-10330, let-7g, miR-222, and miR-2236 among others. However, it remained to be determined whether the presence of cardiovascular risk factors altered the HDL transported miRNA signature. In the present study, we demonstrate that HDL formed under hypercholesterolaemic conditions, because of high dietary fat intake, show a higher abundance of the miRNAs miR-126-5p and -3p, and miR-30b-5p and, conversely, a lower amount of miRNAs miR-103a-3p and let-7g-5p as compared to those HDL formed in a normocholesterolaemic milieu. Interestingly, Vickers et al.6 had reported a differential HDL-miRNA signature (particularly miR223 levels) in familial hypercholesterolaemia patients that have high LDL-cholesterol levels during life, due to the inherited condition of the disease. So far, a positive association between miR-126 levels in plasma and in HDL has been described, as well as in coronary artery disease and myocardial infarction.29,31,32 Furthermore, higher amount of miR-126 was found in HDL particles of overweight and obese subjects,33,34 and we have recently shown that miR-126 regulates monocyte/macrophage differentiation under a rich LDL-cholesterol environment.28 Indeed, a positive association between miR-126 expression and LDL-cholesterol levels has been demonstrated.35 Here, we demonstrate that HDL particles formed in a diet-induced hypercholesterolaemic milieu with high LDL-cholesterol levels carry a high content of miR-126, which is delivered into ECs via a SRB1, leading to the down-regulation of its target gene HIF-1α. As such, SRB1 blockade inhibited miR-126 (both -5p and -3p) delivery into ECs pointing to the critical role of this receptor in HDL-miRNA internalization into target recipient cells. This finding is further supported by the fact that no changes are detected in pre-mir-126 (mir-126) or its encoding gene EGFL7.36 SRB1 is a membrane receptor involved in the selective uptake of cholesteryl ester from HDL. SRB1 is highly expressed in the liver and other cells including monocytes/macrophages, ECs, smooth muscle cells, adipocytes, and steroidogenic cells.37–39 The ability of HDL particles to transfer cargo-miRNAs into recipient cells via SRB1 had already been described in macrophages and hepatocytes.6 In fact, Vickers et al.6 reported that SRB1 over expression associated with the greatest cellular uptake of HDL-miRNAs.
Whereas in rodents SRB1 is expressed most abundantly in tissues that efficiently utilize cholesterol ester (biliary cholesterol secretion or steroid hormone production),40 in humans SRB1 is homogenously distributed in all cells.41 In the present study, we show that in pigs there are comparable SRB1 expression levels in the liver and arterial beds. It is possible that SRB1 serves as docking receptor for HDL to transfer their miRNA-cargo into target cells. Although the SRB1 is the endothelial receptor mainly involved in HDL-miRNA transfer, we cannot exclude that other HDL receptors (ABCA1, ABCG1) may also be involved in HDL-miR126 transfer to ECs.42
The role of miR-126 on the cardiovascular system is controversial, with opposed results in different studies. miR-126 has shown to regulate angiogenesis and blood vessel integrity by directly repressing the Sprouty Related EVH1 Domain Containing 1 (SPRED1)43 gene and the phosphoinositide-3-kinase regulatory subunit 244,45 with the consequent increase in VEGFA.46 Moreover, miR-126 has also shown to reduce leucocyte adherence and vascular inflammation by targeting VCAM-1.47 On the other hand, LDL-cholesterol and miRNA-126 levels are positively associated in patients with asymptomatic coronary artery disease and have been suggested to exert prognostic value for future cardiovascular events.48 In line with previous work, we further confirm the ability of miR-126 to down-regulate endothelial VCAM-1 expression and indirectly enhance VEGFA transcript levels; yet, we further demonstrate the ability of both miR-126s’ (5p and 3p) to reduce endothelial HIF1α gene levels. Moreover, our in vitro findings are further recapitulated in vivo where HIF1α protein levels are lower in the intima of the coronary arteries of hypercholesterolaemic pigs. Although the complexity of the in vivo model does not allow to exclude the existence of other potential mechanisms modulating HIF-1α, all animals were kept under identical conditions except for the type of infused HDL. It is, therefore, plausible to consider that the changes observed in endothelial HIF1α may derive from HC-HDL-miR126 transfer. HIF1α is a transcription factor that serves as a hypoxia sensor to induce the expression of angiogenic genes. Yet, under normoxic conditions, HIF1α is known to modulate compensatory and adaptive mechanisms. In this regard, Liu et al.49 have recently described in cancer cells that suppression of miR-126 increases HIF1α expression promoting metastasis and therapeutic resistance. We evidence, in ECs, that overexpression of miR-126 reduces HIF1α expression likely interfering with metabolic adaptation, innate immune response, survival and apoptosis.50 On the other hand, HIF1α has been described to be modulated, under hypoxic conditions, by miR-155,51 miR-18a, miR-199a, miR-429, miR-433,52 and miR-497∼195.53,54 The exact mechanism by which HDL are loaded with miR-126 under hypercholesterolaemic condition is not known. Biophysical studies have suggested that dipolar lipids facilitate small RNAs incorporation within HDL particles.55 In this regard, we could speculate that the reported changes in lipid composition observed in HC-HDL particles20 may have favoured miR-126 association to circulating HDLs. Yet, further studies are needed to confirm this hypothesis and determine the relevance of modulating miR-126 expression in the vascular wall. In this regard, the in vivo functional impact of miR-126/HIF1α deserves to be investigated.
In conclusion, we demonstrate that diet-induced hypercholesterolaemia leads to alterations in HDL-miRNA profile favouring an enrichment in miR-126 that can be delivered to ECs interfering with key vascular functions. Collectively, our data provides further insights as to the impact of hypercholesterolaemia on HDL biogenesis, HDL biology, and HDL composition as well as on the potential outcomes on HDL-miRNA intercellular communication. Whether miR-126 may serve as a potential therapeutic target or whereas reversing hypercholesterolaemia (statin treatment, diet approaches) may restore HDL-related miRNA normal profile deserve to be investigated.
Supplementary material
Supplementary material is available at Cardiovascular Research online.
Conflict of interest: none declared.
Supplementary Material
Acknowledgements
We gratefully acknowledge the valuable help and support of M.A. Canovas, P. Catalina, O.García, J. Moreno, L. Nasarre, F.J. Rodriguez, and M. Pescador with animal handling and for the proper conduct of all the experimental and molecular work.
Funding
This work was supported by the Plan Nacional de Salud (PNS) [PGC2018-094025-B-I00 to G.V and SAF2016-76819-R to L.B.] from the Spanish Ministry of Science and Innovation and funds FEDER ‘Una Manera de Hacer Europa’; a grant from the Spanish Society of Cardiology (Beca FEC Investigación Básica/2016 to G.V); the Instituto de Salud Carlos III (ISCIII) [P17/01321 to G.A]; and CIBERCV (to L.B). We thank the support of the Generalitat of Catalunya (Secretaria d’Universitats i Recerca del Departament d’Economia i Coneixement de la Generalitat, 2017 SGR 1480) and the Fundación Investigación Cardiovascular-Fundación Jesus Serra for their continuous support.
Translational perspectives
Clinical trials aimed at increasing high-density lipoprotein (HDL)-cholesterol in secondary prevention have resulted disappointing. Furthermore, studies have reported on the negative impact of cardiovascular disease on HDL composition and function. The structural/functional changes that suffer HDL particles under cardiovascular comorbid pathological conditions need to be investigated because they render them dysfunctional and even deleterious. We evidence that diet-induced hypercholesterolaemia (the most prevalent cardiovascular risk factor) alters HDL miRNA signature and these altered miRNAs directly affect vascular homeostasis, adversely regulating the vascular response. Identifying the remodelling changes affecting HDL particles due to pathological conditions will open new avenues for therapeutic intervention.
References
- 1. He L, Hannon GJ.. Erratum: microRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet 2004;5:522–531. [DOI] [PubMed] [Google Scholar]
- 2. Kontush A, Lindahl M, Lhomme M, Calabresi L, Chapman MJ, Davidson WS.. Structure of HDL: particle subclasses and molecular components In Barrett, JE. (ed.). Handbook of Experimental Pharmacology. Switzerland: Springer Nature; 2015. p. 3–51. [DOI] [PubMed] [Google Scholar]
- 3. Badimon L, Vilahur G.. LDL-cholesterol versus HDL-cholesterol in the atherosclerotic plaque: inflammatory resolution versus thrombotic chaos. Ann N Y Acad Sci 2012;1254:18–32. [DOI] [PubMed] [Google Scholar]
- 4. Theilmeier G, Schmidt C, Herrmann J, Keul P, Schäfers M, Herrgott I, Mersmann J, Larmann J, Hermann S, Stypmann J, Schober O, Hildebrand R, Schulz R, Heusch G, Haude M, von Wnuck Lipinski K, Herzog C, Schmitz M, Erbel R, Chun J, Levkau B.. High-density lipoproteins and their constituent, sphingosine-1-phosphate, directly protect the heart against ischemia/reperfusion injury in vivo via the S1P3 lysophospholipid receptor. Circulation 2006;114:1403–1409. [DOI] [PubMed] [Google Scholar]
- 5. Kontush A. HDL-mediated mechanisms of protection in cardiovascular disease. Cardiovasc Res 2014;103:341–349. [DOI] [PubMed] [Google Scholar]
- 6. Vickers KC, Palmisano BT, Shoucri BM, Shamburek RD, Remaley AT.. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat Cell Biol 2011;13:423–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Michell DL, Vickers KC.. Lipoprotein carriers of microRNAs. Biochim Biophys Acta 2016;1861:2069–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Condorelli G, Latronico MVG, Cavarretta E.. microRNAs in cardiovascular diseases: current knowledge and the road ahead. J Am Coll Cardiol 2014;63:2177–2187. [DOI] [PubMed] [Google Scholar]
- 9. Urbich C, Kuehbacher A, Dimmeler S.. Role of microRNAs in vascular diseases, inflammation, and angiogenesis. Cardiovasc Res 2008;79:581–588. [DOI] [PubMed] [Google Scholar]
- 10. Doggrell SA. No cardiovascular benefit with evacetrapib—is this the end of the road for the ‘cetrapibs’? Expert Opin Pharmacother 2017;18:1439–1442. [DOI] [PubMed] [Google Scholar]
- 11. Fisher EA, Feig JE, Hewing B, Hazen SL, Smith JD.. High-density lipoprotein function, dysfunction, and reverse cholesterol transport. Arterioscler Thromb Vasc Biol 2012;32:2813–2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nicholls SJ, Lundman P, Harmer JA, Cutri B, Griffiths KA, Rye K-A, Barter PJ, Celermajer DS.. Consumption of saturated fat impairs the anti-inflammatory properties of high-density lipoproteins and endothelial function. J Am Coll Cardiol 2006;48:715–720. [DOI] [PubMed] [Google Scholar]
- 13. Kontush A, Chapman MJ.. Functionally defective high-density lipoprotein: a new therapeutic target at the crossroads of dyslipidemia, inflammation, and atherosclerosis. Pharmacol Rev 2006;58:342–374. [DOI] [PubMed] [Google Scholar]
- 14. Navab M, Reddy ST, Lenten BJVan, Fogelman AM.. HDL and cardiovascular disease: atherogenic and atheroprotective mechanisms. Nat Rev Cardiol 2011;8:222–232. [DOI] [PubMed] [Google Scholar]
- 15. Holy EW, Besler C, Reiner MF, Camici GG, Manz J, Beer JH, Lüscher TF, Landmesser U, Tanner FC.. High-density lipoprotein from patients with coronary heart disease loses anti-thrombotic effects on endothelial cells: impact on arterial thrombus formation. Thromb Haemost 2014;112:1024–1035. [DOI] [PubMed] [Google Scholar]
- 16. Sorrentino SA, Besler C, Rohrer L, Meyer M, Heinrich K, Bahlmann FH, Mueller M, Horváth T, Doerries C, Heinemann M, Flemmer S, Markowski A, Manes C, Bahr MJ, Haller H, von Eckardstein A, Drexler H, Landmesser U.. Endothelial-vasoprotective effects of high-density lipoprotein are impaired in patients with type 2 diabetes mellitus but are improved after extended-release niacin therapy. Circulation 2010;121:110–122. [DOI] [PubMed] [Google Scholar]
- 17. Riwanto M, Rohrer L, Roschitzki B, Besler C, Mocharla P, Mueller M, Perisa D, Heinrich K, Altwegg L, von Eckardstein A., Luscher TF, Landmesser U.. Altered activation of endothelial anti- and proapoptotic pathways by high-density lipoprotein from patients with coronary artery disease: role of high-density lipoprotein-proteome remodeling. Circulation 2013;127:891–904. [DOI] [PubMed] [Google Scholar]
- 18. Zewinger S, Kleber ME, Rohrer L, Lehmann M, Triem S, Jennings RT, Petrakis I, Dressel A, Lepper PM, Scharnagl H, Ritsch A, Thorand B, Heier M, Meisinger C, de las Heras Gala T, Koenig W, Wagenpfeil S, Schwedhelm E, Böger RH, Laufs U, von Eckardstein A, Landmesser U, Lüscher TF, Fliser D, März W, Meinitzer A, Speer T.. Symmetric dimethylarginine, high-density lipoproteins and cardiovascular disease. Eur Heart J 2017;38:1597–1607. [DOI] [PubMed] [Google Scholar]
- 19. Vilahur G, Gutiérrez M, Casaní L, Cubedo J, Capdevila A, Pons-Llado G, Carreras F, Hidalgo A, Badimon L.. Hypercholesterolemia abolishes high-density lipoprotein-related cardioprotective effects in the setting of myocardial infarction. J Am Coll Cardiol 2015;66:2469–2470. [DOI] [PubMed] [Google Scholar]
- 20. Padró T, Cubedo J, Camino S, Béjar MT, Ben-Aicha S, Mendieta G, Escolà-Gil JC, Escate R, Gutiérrez M, Casani L, Badimon L, Vilahur G.. Detrimental effect of hypercholesterolemia on high-density lipoprotein particle remodeling in pigs. J Am Coll Cardiol 2017;70:165–178. [DOI] [PubMed] [Google Scholar]
- 21. Kilkenny C, Browne WJ, Cuthill IC, Emerson M, Altman DG.. Improving bioscience research reporting: the ARRIVE guidelines for reporting animal research. PLoS Biol 2010;8:e1000412.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Vilahur G, Casani L, Peña E, Juan-Babot O, Mendieta G, Crespo J, Badimon L.. HMG-CoA reductase inhibition prior reperfusion improves reparative fibrosis post-myocardial infarction in a preclinical experimental model. Int J Cardiol 2014;175:528–538. [DOI] [PubMed] [Google Scholar]
- 23. Cremer SE, Krogh AKH, Hedström MEK, Christiansen LB, Tarnow I, Kristensen AT.. Analytical validation of a flow cytometric protocol for quantification of platelet microparticles in dogs. Vet Clin Pathol 2018;47:186–196. [DOI] [PubMed] [Google Scholar]
- 24. Rodríguez C, Martínez-González J, Sánchez-Gómez S, Badimon L.. LDL downregulates CYP51 in porcine vascular endothelial cells and in the arterial wall through a sterol regulatory element binding protein-2-dependent mechanism. Circ Res 2001;88:268–274. [DOI] [PubMed] [Google Scholar]
- 25. Yu M, Romer KA, Nieland TJF, Xu S, Saenz-Vash V, Penman M, Yesilaltay A, Carr SA, Krieger M.. Exoplasmic cysteine Cys384 of the HDL receptor SR-BI is critical for its sensitivity to a small-molecule inhibitor and normal lipid transport activity. Proc Natl Acad Sci USA 2011;108:12243–12248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Das MK, Andreassen R, Haugen TB, Furu K.. Identification of endogenous controls for use in miRNA quantification in human cancer cell lines. Cancer Genomics Proteomics 2016;13:63–68. [PubMed] [Google Scholar]
- 27. Kruger J, Rehmsmeier M.. RNAhybrid: microRNA target prediction easy, fast and flexible. Nucleic Acids Res 2006;34:W451–W454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Escate R, Padro T, Badimon L.. LDL accelerates monocyte to macrophage differentiation: effects on adhesion and anoikis. Atherosclerosis 2016;246:177–186. [DOI] [PubMed] [Google Scholar]
- 29. Choteau SA, Cuesta Torres LF, Barraclough JY, Elder AMM, Martínez GJ, Chen Fan WY, Shrestha S, Ong KL, Barter PJ, Celermajer DS, Rye K-A, Patel S, Tabet F.. Transcoronary gradients of HDL-associated MicroRNAs in unstable coronary artery disease. Int J Cardiol 2018;253:138–144. [DOI] [PubMed] [Google Scholar]
- 30. Michell DL, Vickers KC.. HDL and microRNA therapeutics in cardiovascular disease. Pharmacol Ther 2016;168:43–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wagner J, Riwanto M, Besler C, Knau A, Fichtlscherer S, Roxe T, Zeiher AM, Landmesser U, Dimmeler S.. Characterization of levels and cellular transfer of circulating lipoprotein-bound MicroRNAs. Arterioscler Thromb Vasc Biol 2013;33:1392–1400. [DOI] [PubMed] [Google Scholar]
- 32. Zampetaki A, Willeit P, Tilling L, Drozdov I, Prokopi M, Renard J-M, Mayr A, Weger S, Schett G, Shah A, Boulanger CM, Willeit J, Chowienczyk PJ, Kiechl S, Mayr M.. Prospective study on circulating MicroRNAs and risk of myocardial infarction. J Am Coll Cardiol 2012;60:290–299. [DOI] [PubMed] [Google Scholar]
- 33. Tabet F, Cuesta Torres LF, Ong KL, Shrestha S, Choteau SA, Barter PJ, Clifton P, Rye K-A.. High-density lipoprotein-associated miR-223 is altered after diet-induced weight loss in overweight and obese males. PLoS One 2016;11:e0151061.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Krause BJ, Carrasco-Wong I, Dominguez A, Arnaiz P, Farías M, Barja S, Mardones F, Casanello P.. Micro-RNAs Let7e and 126 in plasma as markers of metabolic dysfunction in 10 to 12 years old children. PLoS One 2015;10:e0128140.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sezer Zhmurov Ç, Timirci-Kahraman Ö, Amadou FZ, Fazlıoğulları O, Başaran C, Catal T, Zeybek Ü, Bermek H.. Expression of Egfl7 and miRNA-126-5p in symptomatic carotid artery disease. Genet Test Mol Biomarkers 2016;20:125–129. [DOI] [PubMed] [Google Scholar]
- 36. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004;116:281–297. [DOI] [PubMed] [Google Scholar]
- 37. Kratzer A, Giral H, Landmesser U.. High-density lipoproteins as modulators of endothelial cell functions: alterations in patients with coronary artery disease. Cardiovasc Res 2014;103:350–361. [DOI] [PubMed] [Google Scholar]
- 38. Nieland TJF, Chroni A, Fitzgerald ML, Maliga Z, Zannis VI, Kirchhausen T, Krieger M.. Cross-inhibition of SR-BI- and ABCA1-mediated cholesterol transport by the small molecules BLT-4 and glyburide. J Lipid Res 2004;45:1256–1265. [DOI] [PubMed] [Google Scholar]
- 39. Krieger M. Charting the fate of the “good cholesterol”: identification and characterization of the high-density lipoprotein receptor SR-BI. Annu Rev Biochem 1999;68:523–558. [DOI] [PubMed] [Google Scholar]
- 40. Acton S, Rigotti A, Landschulz KT, Xu S, Hobbs HH, Krieger M.. Identification of scavenger receptor SR-BI as a high density lipoprotein receptor. Science 1996;271:518.. [DOI] [PubMed] [Google Scholar]
- 41. Calvo D, Gómez-Coronado D, Lasunción MA, Vega MA.. CLA-1 is an 85-kD plasma membrane glycoprotein that acts as a high-affinity receptor for both native (HDL, LDL, and VLDL) and modified (OxLDL and AcLDL) lipoproteins. Arterioscler Thromb Vasc Biol 1997;17:2341.. [DOI] [PubMed] [Google Scholar]
- 42. Röhrl C, Stangl H.. HDL endocytosis and resecretion. Biochim Biophys Acta 2013;1831:1626–1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Mathiyalagan P, Liang Y, Kim D, Misener S, Thorne T, Kamide CE, Klyachko E, Losordo DW, Hajjar RJ, Sahoo S.. Angiogenic mechanisms of human CD34+ stem cell exosomes in the repair of ischemic hindlimb. Circ Res 2017;120:1466–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Chen L, Wang J, Wang B, Yang J, Gong Z, Zhao X, Zhang C, Du K.. MiR-126 inhibits vascular endothelial cell apoptosis through targeting PI3K/Akt signaling. Ann Hematol 2016;95:365–374. [DOI] [PubMed] [Google Scholar]
- 45. Fish JE, Santoro MM, Morton SU, Yu S, Yeh R-F, Wythe JD, Ivey KN, Bruneau BG, Stainier DYR, Srivastava D.. miR-126 regulates angiogenic signaling and vascular integrity. Dev Cell 2008;15:272–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Donnem T, Lonvik K, Eklo K, Berg T, Sorbye SW, Al-Shibli K, Al-Saad S, Andersen S, Stenvold H, Bremnes RM, Busund L-T.. Independent and tissue-specific prognostic impact of miR-126 in nonsmall cell lung cancer. Cancer 2011;117:3193–3200. [DOI] [PubMed] [Google Scholar]
- 47. Harris TA, Yamakuchi M, Ferlito M, Mendell JT, Lowenstein CJ.. MicroRNA-126 regulates endothelial expression of vascular cell adhesion molecule 1. Proc Natl Acad Sci USA 2008;105:1516–1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sun X, Zhang M, Sanagawa A, Mori C, Ito S, Iwaki S, Satoh H, Fujii S.. Circulating microRNA-126 in patients with coronary artery disease: correlation with LDL cholesterol. Thrombosis J 2012;10:16.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Liu W, Chen H, Wong N, Haynes W, Baker CM, Wang X.. Pseudohypoxia induced by miR-126 deactivation promotes migration and therapeutic resistance in renal cell carcinoma. Cancer Lett 2017;394:65–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kuschel A, Simon P, Tug S.. Functional regulation of HIF-1α under normoxia-is there more than post-translational regulation? J Cell Physiol 2012;227:514–524. [DOI] [PubMed] [Google Scholar]
- 51. Nallamshetty S, Chan SY, Loscalzo J.. Hypoxia: a master regulator of microRNA biogenesis and activity. Free Radic Biol Med 2013;64:20–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Serocki M, Bartoszewska S, Janaszak-Jasiecka A, Ochocka RJ, Collawn JF, Bartoszewski R.. miRNAs regulate the HIF switch during hypoxia: a novel therapeutic target. Angiogenesis 2018;21:183–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Yang M, Li C-J, Sun X, Guo Q, Xiao Y, Su T, Tu M-L, Peng H, Lu Q, Liu Q, He H-B, Jiang T-J, Lei M-X, Wan M, Cao X, Luo X-H.. MiR-497∼195 cluster regulates angiogenesis during coupling with osteogenesis by maintaining endothelial Notch and HIF-1α activity. Nat Commun 2017;8:16003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Bai R, Zhao A-Q, Zhao Z-Q, Liu W-L, Jian D-M.. MicroRNA-195 induced apoptosis in hypoxic chondrocytes by targeting hypoxia-inducible factor 1 alpha. Eur Rev Med Pharmacol Sci 2015;19:545–551. [PubMed] [Google Scholar]
- 55. Lu D, Rhodes DG.. Binding of phosphorothioate oligonucleotides to zwitterionic liposomes. Biochim Biophys Acta 2002;1563:45–52. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.