Abstract
Coronavirus disease 2019 (COVID-19) has spread globally, and medical resources become insufficient in many regions. Fast diagnosis of COVID-19 and finding high-risk patients with worse prognosis for early prevention and medical resource optimisation is important. Here, we proposed a fully automatic deep learning system for COVID-19 diagnostic and prognostic analysis by routinely used computed tomography.
We retrospectively collected 5372 patients with computed tomography images from seven cities or provinces. Firstly, 4106 patients with computed tomography images were used to pre-train the deep learning system, making it learn lung features. Following this, 1266 patients (924 with COVID-19 (471 had follow-up for >5 days) and 342 with other pneumonia) from six cities or provinces were enrolled to train and externally validate the performance of the deep learning system.
In the four external validation sets, the deep learning system achieved good performance in identifying COVID-19 from other pneumonia (AUC 0.87 and 0.88, respectively) and viral pneumonia (AUC 0.86). Moreover, the deep learning system succeeded to stratify patients into high- and low-risk groups whose hospital-stay time had significant difference (p=0.013 and p=0.014, respectively). Without human assistance, the deep learning system automatically focused on abnormal areas that showed consistent characteristics with reported radiological findings.
Deep learning provides a convenient tool for fast screening of COVID-19 and identifying potential high-risk patients, which may be helpful for medical resource optimisation and early prevention before patients show severe symptoms.
Short abstract
A fully automatic deep learning system provides a convenient method for COVID-19 diagnostic and prognostic analysis, which can help COVID-19 screening and finding potential high-risk patients with worse prognosis https://bit.ly/3bRaxGw
Introduction
In December 2019, the novel coronavirus disease 2019 (COVID-19) occurred in Wuhan, China and became a global health emergency very fast with >170 000 people infected [1–3]. Due to its high infection rate, fast diagnosis and optimised medical resource assignment in epidemic areas are urgent. Accurate and fast diagnosis of COVID-19 can help isolating infected patients slow the spread of this disease. However, in epidemic areas insufficient medical resources have become a big challenge [4]. Therefore, finding high-risk patients with worse prognosis for prior medical resources and special care is crucial in the treatment of COVID-19.
Currently, reverse transcription (RT)-PCR is used as the gold truth for diagnosing COVID-19. However, the limited sensitivity of RT-PCR and the shortage of testing kits in epidemic areas increase the screening burden, and many infected people are thereby not isolated immediately [5, 6]. This accelerates the spread of COVID-19. Conversely, due to the lack of medical resources, many infected patients cannot receive immediate treatment. In this situation, finding high-risk patients with worse prognosis for prior treatment and early prevention is important. Consequently, fast diagnosis and finding high-risk patients with worse prognosis are very helpful for the control and management of COVID-19.
In recent studies, radiological findings demonstrated that computed tomography (CT) has great diagnostic and prognostic value for COVID-19. For example, CT showed much higher sensitivity than RT-PCR in diagnosing COVID-19 [5, 6]. For patients with COVID-19, bilateral lung lesions consisting of ground-glass opacities were frequently observed in CT images [6–8]. Even in asymptomatic patients, abnormalities and changes were observed in serial CT [9, 10]. As a common diagnostic tool, CT is easy and fast to acquire without adding much cost. Building a sensitive diagnostic tool using CT imaging can accelerate the diagnostic process and is complementary to RT-PCR. However, predicting personalised prognosis using CT imaging can identify the potential high-risk patients who are more likely to become severe and need urgent medical resources.
Deep learning (DL) as an artificial intelligence method has shown promising results in assisting lung disease analysis using CT images [11–15]. Benefiting from the strong feature learning ability, DL can mine features that are related to clinical outcomes from CT images automatically. Features learned by DL models can reflect high-dimensional abstract mappings which are difficult for humans to sense but are strongly associated with clinical outcomes. In contrast to the published DL models [16, 17], we aim to provide a fully automatic DL system for COVID-19 diagnostic and prognostic analysis. Without requiring any human-assisted annotation, this novel DL system is fast and robust in clinical use. Moreover, we collected a large multi-regional dataset for training and validating the proposed DL system, including 1266 patients (471 had follow-up) from six cities or provinces. Notably, different from many studies using transfer learning from natural images. We collected a large auxiliary dataset including 4106 patients with chest CT images and gene information to pre-train the DL system, aiming at making the DL system learn lung features that can reflect the association between micro-level lung functional abnormalities and chest CT images.
Methods
Study design and participants
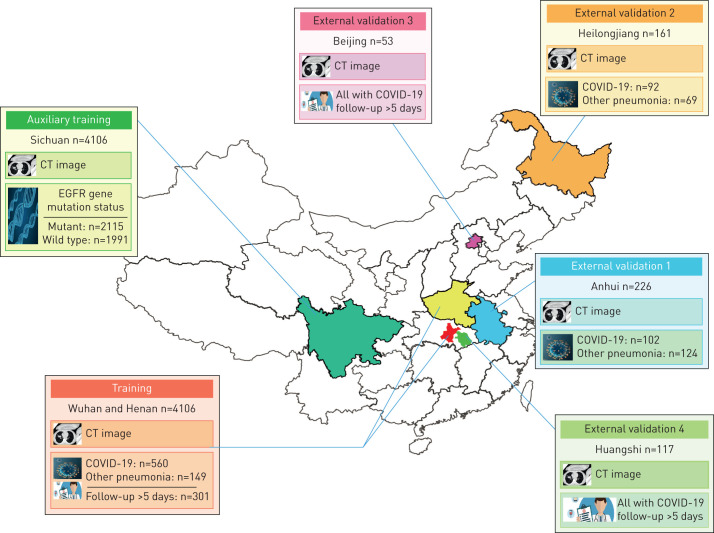
The institutional review board of the seven hospitals (supplementary methods S1) approved this multi-regional retrospective study and waived the need to obtain informed consent from the patients. In this study, we collected two datasets: COVID-19 dataset (n=1266) and CT-epidermal growth factor receptor (EGFR) dataset (n=4106). In the COVID-19 dataset, 1266 patients were finally included who met the following inclusion criteria: 1) RT-PCR confirmed COVID-19; 2) laboratory confirmed other types of pneumonia before December 2019; 3) have non-contrast enhanced chest CT at diagnosis time. Since RT-PCR has a relatively high false-negative rate, we collected other types of pneumonia before December 2019 when COVID-19 did not show up to guarantee the diagnoses of typical pneumonia are correct. In the COVID-19 dataset, patients from Wuhan city and Henan province formed the training set; patients from Anhui province formed the external validation set 1; patients from Heilongjiang province formed the validation set 2; patients from Beijing formed the validation set 3; and patients from Huangshi city formed the validation set 4 (figure 1).
FIGURE 1.
Datasets used in this study. A total of 5372 patients with computed tomography (CT) images from seven cities or provinces were enrolled in this study. The auxiliary training set included 4106 patients with lung cancer and epidermal growth factor receptor (EGFR) gene mutation status information, and is used to pre-train the COVID-19Net to learn lung features from CT images. The training set includes 709 patients from Wuhan city and Henan province. The external validation set 1 (226 patients) from Anhui province, and the external validation set 2 (161 patients) from Heilongjiang province are used to assess the diagnostic performance of the deep learning (DL) system. The external validation set 3 (53 patients with COVID-19) from Beijing, and the external validation set 4 (117 patients with COVID-19) from Huangshi city are used to evaluate the prognostic performance of the DL system.
In the CT-EGFR dataset, 4106 patients with lung cancer were finally included who met the following criteria: 1) EGFR gene sequencing was obtained; and 2) non-contrast enhanced chest CT data obtained within 4 weeks before EGFR gene sequencing. The CT-EGFR dataset was used for auxiliary training of the DL system, making the DL system learn lung features automatically. CT scanning parameters about the COVID-19 and CT-EGFR datasets are available in supplementary methods S1.
For prognostic analysis, 471 patients with COVID-19 and regular follow-up for at least 5 days were used. We defined the prognostic end event as the hospital stay time which was determined from the diagnosis of COVID-19 to the time when the patient was discharged from hospital (supplementary methods S2). A short hospital stay time corresponds to good prognosis, and a long hospital stay time means worse prognosis. Patients with long hospital stay time might take longer time to recover and are defined as high-risk patients in this study. These patients need prior medical resources and special care since they are more likely to become severe.
The training set was used to train the proposed DL system; validation sets 1 and 2 were used to evaluate the diagnostic performance of the DL system; and validation sets 3 and 4 were used for evaluating the prognostic performance of the DL system.
The fully automatic DL system for COVID-19 diagnostic and prognostic analysis
The proposed DL system includes three parts: automatic lung segmentation, non-lung area suppression, and COVID-19 diagnostic and prognostic analysis. In this DL system, two DL networks were involved: DenseNet121-FPN for lung segmentation in chest CT image, and the proposed novel COVID-19Net for COVID-19 diagnostic and prognostic analysis. DL is a family of hierarchical neural networks that aim at learning the abstract mapping between raw data to the desired clinical outcome. The computational units in the DL model are defined as layers and are integrated to simulate the inference process of the human brain. The main computational formulas are convolution, pooling, activation and batch normalisation as defined in supplementary methods S3.
Automatic lung segmentation
Routinely used chest CT images includes some non-lung areas (muscle, heart, etc.) and blank space outside body. To focus on analysing lung area we used a fully automatic DL model (DenseNet121-FPN) [18, 19] to segment lung areas in chest CT images. This model was pre-trained using ImageNet dataset, and fine-tuned on the VESSEL12 dataset (supplementary methods S4) [20].
Through this automatic lung segmentation procedure, we acquired the lung mask on CT images. However, some inflammatory tissues attaching to the lung wall may be falsely excluded by the DenseNet121-FPN model. To increase the robustness of the DL system, we used the cubic bounding box of the segmented lung mask to crop lung areas in CT images, and defined this cubic lung area as lung-region of interest (ROI) (figure 2). In this lung-ROI, all inflammatory tissues and the whole lung were correctly reserved, and most areas outside the lung were eliminated.
FIGURE 2.
Illustration of the proposed deep learning (DL) system. Using the chest computed tomography (CT) scanning of a patient, the DL system predicts the probability the patient has COVID-19 and the prognosis of this patient directly without any human annotation. The DL system includes three parts: automatic lung segmentation (DenseNet121-FPN), non-lung area suppression, and COVID-19 diagnostic and prognostic analysis (COVID-19Net). To let the COVID-19Net learn lung features from the large dataset we used the auxiliary training process for pre-training, which trained the DL network to predict epidermal growth factor receptor (EGFR) gene mutation status using CT images of 4106 patients. The dense connection in this figure means each convolutional layer is connected to all of its previous convolutional layers inside the same dense block.
Non-lung area suppression
After the above processing, some non-lung tissues or organs (e.g. spine and heart) inside the lung-ROI may also exist. Consequently, we proposed a non-lung area suppression operation to suppress the intensities of non-lung areas inside the lung-ROI (supplementary methods S4). Finally, the lung-ROI was standardised by z-score normalisation, and resized to the size of 48×240×360 voxel for further process.
DL model for COVID-19 diagnosis and prognosis
After the non-lung area suppression operation, the standardised lung-ROI was sent to the COVID-19Net for diagnostic and prognostic analysis. Figure 2 illustrates the topological structure of the proposed novel COVID-19Net (table S1). This DL model used a DenseNet-like structure [18], consisting of four dense blocks, where each dense block was multiple stacks of convolution, batch normalisation and ReLU activation layers. Inside each dense block, we used dense connection to consider multi-level image information. At the end of the last convolutional layer, we used global average pooling to generate the 64-dimensional DL features. Finally, the output neuron was fully connected to the DL features to predict the probability the input patient had COVID-19.
To enable the COVID-19Net to learn discriminative features associated with COVID-19, a large training set was needed. Consequently, we proposed a two-step transfer learning process. Firstly, we proposed an auxiliary training process using the large CT-EGFR dataset (4106 patients) as illustrated in figure 2. In this auxiliary training process, we trained the COVID-19Net to predict EGFR mutation status (EGFR-mutant or EGFR wild-type) using the lung-ROI [11]. Benefitting from the large CT-EGFR dataset, the COVID-19Net learned CT features that can reflect the associations between micro-level lung functional abnormality and macro-level CT images.
In the second training process, we transferred the pre-trained COVID-19Net to the COVID-19 dataset to specifically mine lung characteristics associated with COVID-19. After an iterative training process in the COVID-19 dataset (supplementary methods S5), the COVID-19Net can predict the probability of the input patient being infected with COVID-19; this probability was defined as DL score in this study.
To explore the prognostic value of the DL features, we extracted the 64-dimensional DL feature from the COVID-19Net for prognostic analysis. Firstly, we combined the 64-dimensional DL feature and clinical features (age, sex and comorbidity) to construct a combined feature vector. Afterwards, we used a stepwise method to select prognostic features. These selected features were then used to build a multivariate Cox proportional hazard model [21] to predict the risk of the patient needing a long hospital stay time to recover.
Visualisation of lung features learnt by the DL system
Through the two-step transfer learning technique, the DL system learnt lung features from CT images of 4815 patients. To further understand the inference process of the DL system, we used a DL visualisation algorithm to analyse features learnt by the COVID-19Net from two perspectives: 1) visualising the DL-discovered suspicious lung area that contributes most to identifying COVID-19 for the DL system; 2) visualising the feature patterns extracted by hierarchical convolutional layers in the COVID-19Net (supplementary methods S6 and S7).
Statistical analysis
Area under the receiver operating characteristic (ROC) curve, accuracy, sensitivity, specificity, F1-score, calibration curves and Hosmer-Lemeshow test were used to assess the performance of the DL system in diagnosing COVID-19. Kaplan–Meier analysis and log-rank test were used to evaluate the performance of the DL system for prognostic analysis. The implementation of the DL system used the Keras 2.3.1 toolkit and Python 3.7 (https://github.com/wangshuocas/COVID-19).
Results
Clinical characteristics of patients in the COVID-19 dataset are presented in table 1. This dataset was collected from six cities or provinces including Wuhan city in China.
TABLE 1.
Clinical characteristics of patients
| Training set | Validation 1 | Validation 2 | Validation 3 | Validation 4 | |
| Subjects | 709 | 226 | 161 | 53 | 117 |
| City or province | Wuhan city and Henan | Anhui | Heilongjiang | Beijing | Huangshi city |
| Type | |||||
| COVID-19 | 560 | 102 | 92 | 53 | 117 |
| Bacterial pneumonia | 127 | 119 | 25 | 0 | 0 |
| Mycoplasma pneumonia | 11 | 5 | 15 | 0 | 0 |
| Viral pneumonia | 0 | 0 | 29 | 0 | 0 |
| Fungal pneumonia | 11 | 0 | 0 | 0 | 0 |
| Sex | |||||
| Male | 337 | 131 | 108 | 25 | 60 |
| Female | 372 | 95 | 53 | 28 | 57 |
| Age years | 50.52±18.91 | 49.15±18.44 | 58.44±16.19 | 50.26±19.29 | 47.67±14.20 |
| Comorbidity | |||||
| Any | 204 | NA | NA | 16 | 27 |
| Diabetes | 45 | 2 | 12 | ||
| Hypertension | 120 | 10 | 12 | ||
| Cerebrovascular disease | 18 | 1 | 0 | ||
| Cardiovascular disease | 21 | 5 | 9 | ||
| Malignancy | 19 | 0 | 1 | ||
| COPD | 10 | 1 | 2 | ||
| Pulmonary tuberculosis | 6 | 1 | 0 | ||
| Chronic kidney disease | 10 | 0 | 2 | ||
| Chronic liver disease | 16 | 3 | 2 | ||
| Follow-up >5 days | 301 | NA | NA | 53 | 117 |
Data are presented as n or mean±sd. COVID: coronavirus 2019: NA: not available.
Diagnostic performance of the DL system
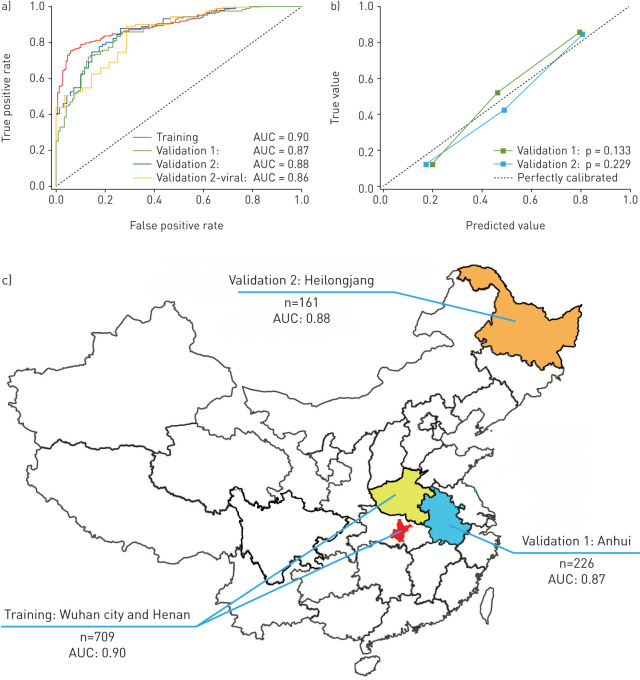
Table 2 and figure 3 illustrated the diagnostic performance of the DL system. In the training set, the DL system showed good diagnostic performance (AUC: 0.90, sensitivity: 78.93%, specificity: 89.93%). This performance was further confirmed in the two external validation sets (AUC: 0.87 and 0.88; sensitivity: 80.39% and 79.35%; specificity: 76.61% and 81.16%, respectively). The DL score revealed a significant difference between COVID-19 and other pneumonia groups in the three datasets (p<0.0001). The good performance in the validation sets indicated that the DL system generalised well on diagnosing COVID-19 of unseen new patients. Meanwhile, we illustrated the ROC curves of the DL system in the three datasets in figure 3a, and the calibration curves of the DL system in the two validation sets in figure 3b. The good calibration in figure 3b indicated that the DL system did not systematically under predict or over predict COVID-19 probability because the Hosmer–Lemeshow test yielded a nonsignificant statistic to the perfect model (p=0.133 and p=0.229, respectively, in the two validation sets). Benefiting from the auxiliary training process in the large CT-EGFR dataset, the generalisation ability of the DL system improved largely compared with the DL system without auxiliary training (table S2).
TABLE 2.
Diagnostic performance of the deep learning system
| Training | Validation 1 | Validation 2 | Validation 2-viral# | |
| Subjects n | 709 | 226 | 161 | 121 |
| AUC 95% CI | 0.90 (0.89–0.91) | 0.87 (0.86–0.89) | 0.88 (0.86–0.90) | 0.86 (0.83–0.89) |
| Accuracy % | 81.24 | 78.32 | 80.12 | 85.00 |
| Sensitivity % | 78.93 | 80.39 | 79.35 | 79.35 |
| Specificity % | 89.93 | 76.61 | 81.16 | 71.43 |
| F1-score | 86.92 | 77.00 | 82.02 | 90.11 |
AUC: area under the receiver operating characteristic curve. #: a stratified analysis using the patients with coronavirus 2019 and viral pneumonia in validation set 2.
FIGURE 3.
Diagnostic performance of the deep learning (DL) system. a) Receiver operating characteristic curves of the DL system in the training set and the two independent external validation sets. Validation 2-viral is a stratified analysis using the patients with coronavirus 2019 and viral pneumonia in the validation set 2. b) Calibration curves of the DL system in the two external validation sets. c) Area under the curve and distribution of the training set and the two external validation sets.
In other types of pneumonia, viral pneumonia has similar radiological characteristics to COVID-19, and therefore is more difficult to identify. Consequently, we performed a stratified analysis in the validation set 2. Table 1 indicated that the DL system also achieved good results in distinguish COVID-19 to other viral pneumonia (AUC=0.86).
Prognostic value of the DL features
In the COVID-19 dataset, 471 patients had follow-up for >5 days. Through the stepwise prognostic feature selection, three features were selected (table S3). These selected prognostic features were fed into the multivariate Cox proportional hazard model to predict a hazard value for each patient. We used median value of the hazards in the training set as a cut-off value to stratify patients into high- and low-risk groups. This cut-off value was also applied to validation sets 3 and 4. Kaplan–Meier analysis in figure S1 demonstrated that patients in high- and low-risk groups had a significant difference in hospital stay time in the three datasets (p<0.0001, p=0.013 and p=0.014, respectively, log-rank test). These results suggested that the DL features have potential prognostic value for COVID-19.
Suspicious lung area discovered by the DL system
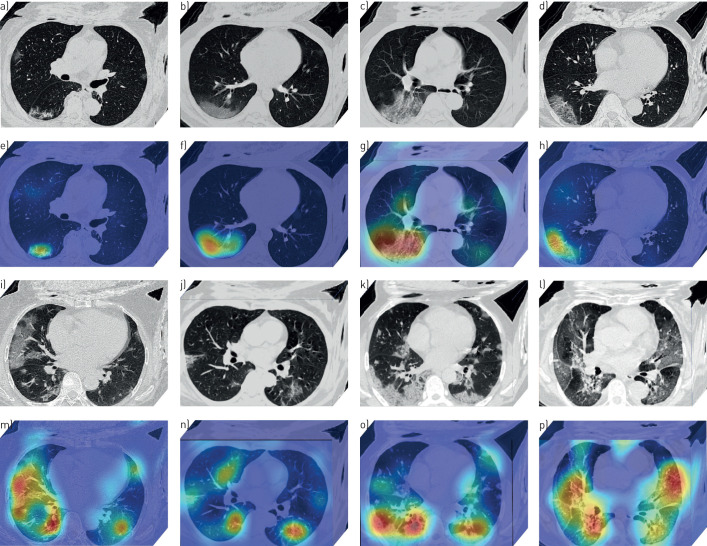
Through the DL visualisation algorithm [22, 23], we are able to visualise the lung area that draws most attention to the DL system. These DL discovered suspicious lung areas usually demonstrated abnormal characteristics consistent with radiologists’ findings. Figure 4 illustrated DL discovered suspicious lung areas of eight patients with COVID-19. From this figure, we can see that although the input lung-ROI to the DL system includes some non-lung tissues such as muscle and bones, the DL system can always focus on areas inside the lung for prediction instead of being disturbed by other tissues.
FIGURE 4.
Deep learning (DL) discovered suspicious lung area. a–p) Computed tomography (CT) images of eight patients with coronavirus 2019. a–d and i–l) CT images of the patients (these CT images are processed by the DL system). e–h and m–p) Heat maps of the DL discovered suspicious lung area. In the heat map, areas with bright red colour are more important than dark blue areas.
Moreover, the DL discovered suspicious lung areas showed high overlap with the actual inflammatory areas. Figure 4a–h shows that, although we did not involve any human annotation in the DL system, the DL system focused on the ground-glass opacity area automatically for inference. This is consistent with radiologists’ experiences that many COVID-19 patients illustrated ground-glass opacity features [6, 9]. In figure 4i–p, the DL discovered suspicious lung areas distributed on bilateral lung, and mainly focused on lesions with consolidation, ground-glass opacities, diffuse or mixture patterns. When comparing these DL discovered suspicious lung areas with actual abnormal lung areas, we found a high overlap and consistency.
Although we did not use human annotation (e.g. human annotated ROI) to tell the DL system where to watch, the DL system is capable of discovering the abnormal and important lung areas automatically. This phenomenon could come from the advantage of using the large CT-EGFR dataset and the large COVID-19 dataset for training.
DL feature visualisation
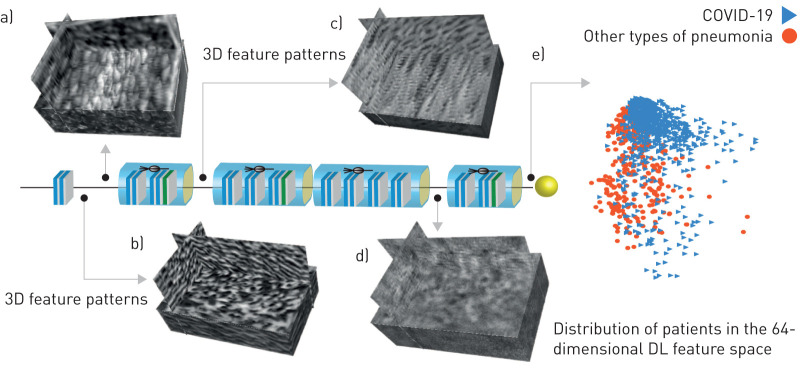
Since DL is an end-to-end prediction model that learns abstract mappings between lung CT images and COVID-19 directly, it is helpful to explain the inference process of the DL system. The most important component of the DL model is the convolutional filter. Therefore, we visualised the 3-dimensional feature patterns extracted by hierarchical convolutional layers in figure 5. The shallow convolutional layer learnt low-level simple features, such as spindle edges (figure 5a) and wave-like edges (figure 5b). A deeper convolutional layer learnt more complex and detailed features (figure 5c). When going deeper, the feature pattern became more abstract and lacked visual characteristics (figure 5d). However, these high-level feature patterns are more related to COVID-19 information.
FIGURE 5.
Deep learning (DL) feature visualisation. a–d) Four 3-dimensional (3D) convolutional filters from different convolutional layers. e) Distribution of patients in the 64-dimensional DL feature space. For display convenience, the 64-dimensional DL feature space is reduced to 2-dimensional by a principle component analysis algorithm.
At the end of the DL model, the outputs of convolutional filters were compressed into a 64-dimensional vector, which was defined as a DL feature. In figure 5e, we reduced the 64-dimensional DL feature into two-dimensional space to see the DL feature distribution in the two classes (COVID-19 versus other types of pneumonia). This figure demonstrated that the two classes distributed separately in the DL feature space, which means the DL features are discriminative to identify COVID-19 from other types of pneumonia.
Discussion
In this study, we proposed a novel fully automatic DL system using raw chest CT image to help COVID-19 diagnostic and prognostic analysis. To let the DL system mine lung features automatically without involving any time-consuming human annotation, we used a two-step transfer learning strategy. Firstly, we collected 4106 lung cancer patients with both CT image and EGFR gene sequencing. Through training in this large CT-EGFR dataset, the DL system learned hierarchical lung features that can reflect the associations between chest CT image and micro-level lung functional abnormality. Afterwards, we collected a large multi-regional COVID-19 dataset (n=1266) from six cities or provinces to train and validate the diagnostic and prognostic performance of the DL system.
The good diagnostic and prognostic performance of the DL system illustrates that DL could be helpful in the epidemic control of COVID-19 without adding much cost. Given a suspected patient, CT scanning can be acquired within minutes. Afterwards, this DL system can be applied to predict the probability the patient has COVID-19. If the patient is diagnosed as COVID-19, the DL system also predicts their prognostic situation simultaneously, which can be used to find potential high-risk patients who need urgent medical resources and special care. More importantly, this DL system is fast and does not require human-assisted image annotation, which increases its clinical value and becomes more robust. For a typical chest CT scan of a patient, the DL system takes less than 10 s for prognostic and diagnostic prediction.
During building and training the DL system, we did not involve any human annotation to tell the system where the inflammatory area was. However, the DL system managed to automatically discover the important features that are strongly associated with COVID-19. In figure 4, we visualised the DL discovered suspicious lung areas that were used by the DL system for inference. These DL discovered suspicious lung areas had high overlap with the actual inflammatory areas that are used by radiologists for diagnosis. In previous studies, some radiological features such as ground-glass opacities, crazy-paving pattern and bilateral involvement have been reported to be important for diagnosing CVOID-19 [7]. In the DL discovered suspicious lung areas, we also observed these radiological features. This demonstrates that the high-dimensional features mined by the DL system can probably reflect these reported radiological finding.
Recently, deep learning methods with different processes and models were reported to diagnose COVID-19 using CT images. These methods can be classified into three types. 1) Using manually or automatically segmented lesions for diagnosis. Wang et al. [16] used manually annotated lesions as ROI, and a modified ResNet34 model combined with decision tree and AdaBoost classifiers was used to diagnose COVID-19. To avoid time-consuming lesion annotation by radiologists, automatic lesion segmentation models [17, 24] were used in further studies. Afterwards, 3-dimensional CNN models such as 3DResNet were used to diagnose COVID-19 using the lesion images. 2) Using 2-dimensional lung image slices to train the DL model. Since lesions can be distributed in many locations in lungs, and automatic lesion segmentation may not guarantee very high precision. More studies used the whole lung image slices for analysis. In the study by Song et al. [25], a feature pyramid network using ResNet50 as the backbone was used to analyse 2-dimensional image slices of the whole lung area. Similarly, Jin et al. [26] used DeepLabv1 and Li et al. [27] used U-Net to segment lung from CT images, and then used the 2-dimensional ResNet model to analyse image slices of lung area. 3) Using a 3-dimensional DL model to analyse whole lung in CT images. To consider 3-dimensional information of the whole lung, Zheng et al. [28] used the 3DResNet model to analyse the 3-dimensional lung area in CT images.
Compared with these studies, our study has three main differences. 1) We used the 3-dimensional bounding box of the whole lung as ROI instead of only using lesions or segmented lung fields. Since lesion segmentation may not guarantee a very high accuracy, inaccurate lesion segmentation may cause information loss. Compared with segmenting lung lesion, lung segmentation is easier, and analysing the whole lung can mine more information. However, different with the methods using only the segmented lung areas [25, 27], we used the 3-dimensional bounding box of lung as ROI. In figure S2, we illustrated the lung segmentation results. In most situations, the lung segmentation method generated good results. However, for some patients with severe symptoms and consolidation lesions, the performance of the lung segmentation method may be affected. Consequently, we used the 3-dimensional bounding box of the segmented lung mask as ROI, which ensures the lung-ROI covering the complete lung area. Combined with the non-lung area suppression strategy, the lung-ROI can reserve complete lung area, and suppress images outside lung area. 2) We used a large auxiliary dataset including chest CT images of 4106 patients to pre-train the proposed COVID-19Net, making it learn lung features. Many existed studies used DL models pre-trained in ImageNet dataset, this may increase the generalisation ability of the DL model. However, natural images in the ImageNet dataset have large difference to chest CT images. Consequently, using a chest CT dataset for auxiliary training (pre-training) enables the DL model learn features that are more specific to chest CT images. 3) Most studies used a small dataset and randomly selected data for validation. To assess the generalisation ability of the deep learning model, we used a large dataset and two independent validation sets from different regions.
Despite the good performance of the DL system, this study has several limitations. Firstly, there are other prognostic end events such as death or admission to an intensive care unit, and they were not considered in this study. Secondly, the management of severe and mild COVID-19 are different, thereby, exploring the prognosis of COVID-19 in these two groups separately should be helpful. However, CT images of different slice thickness were included in this study. In the future, we will use a generative adversarial network to convert CT images of different slice thickness into CT images with a unified slice thickness, which may further improve the diagnostic performance of the DL system.
Supplementary material
Please note: supplementary material is not edited by the Editorial Office, and is uploaded as it has been supplied by the author.
Supplementary material ERJ-00775-2020 (1,000.1KB, pdf)
Shareable PDF
Supplementary Material
Acknowledgments
We thank all the collaborative hospitals for data collection, and especially thank Shuang Yan from Cancer Hospital Chinese Academy of Medical Sciences.
Footnotes
This article has supplementary material available from erj.ersjournals.com
Author contributions: Y. Zha and J. Tian conceived and designed the study. S. Wang implemented the DL system and wrote the paper. Q. Wu, Y. Zhu and L. Wang contributed to the data process and analysis. M. Niu, H. Yu, W. Gong, Y. Bai, X. Qiu, L. Li, X. Li, M. Wang, H. Li and W. Li contributed to data collection.
Conflict of interest: S. Wang has nothing to disclose.
Conflict of interest: Y. Zha has nothing to disclose.
Conflict of interest: W. Li has nothing to disclose.
Conflict of interest: Q. Wu has nothing to disclose.
Conflict of interest: X. Li has nothing to disclose.
Conflict of interest: M. Niu has nothing to disclose.
Conflict of interest: M. Wang has nothing to disclose.
Conflict of interest: X. Qiu has nothing to disclose.
Conflict of interest: H. Li has nothing to disclose.
Conflict of interest: H. Yu has nothing to disclose.
Conflict of interest: W. Gong has nothing to disclose.
Conflict of interest: Y. Bai has nothing to disclose.
Conflict of interest: L. Li has nothing to disclose.
Conflict of interest: Y. Zhu has nothing to disclose.
Conflict of interest: L. Wang has nothing to disclose.
Conflict of interest: J. Tian has nothing to disclose.
Support statement: This paper is supported by the National Natural Science Foundation of China under grant numbers 81930053, 81227901, 81871332, 61936013 and 81771806, the National Key R&D Program of China under grant number 2017YFA0205200, Novel Coronavirus Pneumonia Emergency Key Project of Science and Technology of Hubei Province under grant number 2020FCA015. Funding information for this article has been deposited with the Crossref Funder Registry. Funding was also received from the Fundamental Research Funds for the Central Universities under grant number 2042020kfxg10, Anhui Natural Science Foundation under grant number 202004a07020003, Hubei Health Committee General Program and Anti-schistosomiasis Fund during 2019–2020 under grant number WJ2019M043, Beijing Municipal Commission of Health under grant number 2020-TG-002, Youan Medical Development Fund under grant number BJYAYY-2020YC-03 and China Postdoctoral Science Special Foundation under grant number 2019TQ0019.
References
- 1.Wang D, Hu B, Hu C, et al. . Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus–infected pneumonia in Wuhan, China. JAMA 2020; 323: 1061–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li Q, Guan X, Wu P, et al. . Early transmission dynamics in Wuhan, China, of novel coronavirus–infected pneumonia. N Engl J Med 2020; 382: 1199–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang X, Yu Y, Xu J, et al. . Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. Lancet Respir Med 2020; 8: 475–481. doi: 10.1016/S2213-2600(20)30079-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ji Y, Ma Z, Peppelenbosch MP, et al. . Potential association between COVID-19 mortality and health-care resource availability. Lancet Glob Health 2020; 8: e480. doi: 10.1016/S2214-109X(20)30068-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xie X, Zhong Z, Zhao W, et al. . Chest CT for typical 2019-nCoV pneumonia: relationship to negative RT-PCR testing. Radiology 2020; 296; E41–E45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ai T, Yang Z, Hou H, et al. . Correlation of chest CT and RT-PCR testing in coronavirus disease 2019 (COVID-19) in China: a report of 1014 cases. Radiology 2020; 296; E32–E40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang S, Li H, Huang S, et al. . High-resolution computed tomography features of 17 cases of coronavirus disease 2019 in Sichuan province, China. Eur Respir J 2020; 55; 2000334. doi: 10.1183/13993003.00334-2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang L, Gao Y-h, Zhang G-J. The clinical dynamics of 18 cases of COVID-19 outside of Wuhan, China. Eur Respir J 2020; 55; 2000398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shi H, Han X, Jiang N, et al. . Radiological findings from 81 patients with COVID-19 pneumonia in Wuhan, China: a descriptive study. Lancet Infect Dis 2020; 20; 425–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee EY, Ng M-Y, Khong P-L. COVID-19 pneumonia: what has CT taught us? Lancet Infect Dis 2020; 20: 384–385. doi: 10.1016/S1473-3099(20)30134-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang S, Shi J, Ye Z, et al. . Predicting EGFR mutation status in lung adenocarcinoma on computed tomography image using deep learning. Eur Respir J 2019; 53: 1800986. doi: 10.1183/13993003.00986-2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Walsh SL, Calandriello L, Silva M, et al. . Deep learning for classifying fibrotic lung disease on high-resolution computed tomography: a case-cohort study. Lancet Respir Med 2018; 6: 837–845. doi: 10.1016/S2213-2600(18)30286-8 [DOI] [PubMed] [Google Scholar]
- 13.Walsh SL, Humphries SM, Wells AU, et al. . Imaging research in fibrotic lung disease; applying deep learning to unsolved problems. Lancet Respir Med 2020; in press [ 10.1016/S2213-2600(20)30003-5]. [DOI] [PubMed] [Google Scholar]
- 14.Angelini E, Dahan S, Shah A. Unravelling machine learning: insights in respiratory medicine. Eur Respir J 2019; 54: 1901216. doi: 10.1183/13993003.01216-2019 [DOI] [PubMed] [Google Scholar]
- 15.Wang S, Zhou M, Liu Z, et al. . Central focused convolutional neural networks: developing a data-driven model for lung nodule segmentation. Med Image Anal 2017; 40: 172–183. doi: 10.1016/j.media.2017.06.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang S, Kang B, Ma J, et al. . A deep learning algorithm using CT images to screen for corona virus disease (COVID-19). medRxiv 2020; preprint [ 10.1101/2020.02.14.20023028]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu X, Jiang X, Ma C, et al. . Deep learning system to screen coronavirus disease 2019 pneumonia. arXiv 2020; preprint [https://arxiv.org/abs/2002.09334]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang G, Liu Z, Van Der Maaten L, et al. . Densely connected convolutional networks. In: Proceedings of the IEEE conference on computer vision and pattern recognition 2017; pp. 4700–4708. [Google Scholar]
- 19.Lin T-Y, Dollár P, Girshick R, et al. . Feature pyramid networks for object detection. In: Proceedings of the IEEE conference on computer vision and pattern recognition 2017; pp. 2117–2125. [Google Scholar]
- 20.Rudyanto RD, Kerkstra S, Van Rikxoort EM, et al. . Comparing algorithms for automated vessel segmentation in computed tomography scans of the lung: the VESSEL12 study. Med Image Anal 2014; 18: 1217–1232. doi: 10.1016/j.media.2014.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang S, Liu Z, Rong Y, et al. . Deep learning provides a new computed tomography-based prognostic biomarker for recurrence prediction in high-grade serous ovarian cancer. Radiother Oncol 2019; 132: 171–177. doi: 10.1016/j.radonc.2018.10.019 [DOI] [PubMed] [Google Scholar]
- 22.Selvaraju RR, Cogswell M, Das A, et al. . Grad-cam: visual explanations from deep networks via gradient-based localization. In: Proceedings of the IEEE international conference on computer vision. 2017; pp. 618–626. [Google Scholar]
- 23.Kotikalapudi R. 2017. Keras Visulization Toolkit. https://github.com/raghakot/keras-vis.
- 24.Jin S, Wang B, Xu H, et al. . AI-assisted CT imaging analysis for COVID-19 screening: building and deploying a medical AI system in four weeks. medRxiv 2020; preprint [ 10.1101/2020.03.19.20039354] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Song Y, Zheng S, Li L, et al. . Deep learning enables accurate diagnosis of novel coronavirus (COVID-19) with CT images. medRxiv 2020; preprint [ 10.1101/2020.02.23.20026930]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jin C, Chen W, Cao Y, et al. . Development and evaluation of an AI system for COVID-19 diagnosis. medRxiv 2020; preprint [ 10.1101/2020.03.20.20039834]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li L, Qin L, Xu Z, et al. . Artificial intelligence distinguishes COVID-19 from community acquired pneumonia on chest CT. Radiology 2020: in press [ 10.1148/radiol.2020200905]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zheng C, Deng X, Fu Q, et al. . Deep learning-based detection for COVID-19 from chest CT using weak label. medRxiv 2020; preprint [ 10.1101/2020.03.12.20027185]. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Please note: supplementary material is not edited by the Editorial Office, and is uploaded as it has been supplied by the author.
Supplementary material ERJ-00775-2020 (1,000.1KB, pdf)
This one-page PDF can be shared freely online.
Shareable PDF ERJ-00775-2020.Shareable (227.1KB, pdf)