Abstract
A broad range of fungi has been detected in molecular surveys of the oral mycobiome. However, knowledge is still lacking on interindividual variability of these communities and the ecologic and clinical significance of oral fungal commensals. In this cross-sectional study, we use internal transcribed spacer 1 amplicon sequencing to evaluate the salivary mycobiome in 59 subjects, 36 of whom were scheduled to receive cancer chemotherapy. Analysis of the broad population structure of fungal communities in the whole cohort identified 2 well-demarcated genus-level community types (mycotypes), with Candida and Malassezia as the main taxa driving cluster partitioning. The Candida mycotype had lower diversity than the Malassezia mycotype and was positively correlated with cancer and steroid use in these subjects, smoking, caries, utilizing a removable prosthesis, and plaque index. Mycotypes were also associated with metabolically distinct bacteria indicative of divergent oral environments, with aciduric species enriched in the Candida mycotype and inflammophilic bacteria increased in the Malassezia mycotype. Similar to their fungal counterparts, coexisting bacterial communities associated with the Candida mycotype showed lower diversity than those associated with the Malassezia mycotype, suggesting that common environmental pressures affected bacteria and fungi. Mycotypes were also seen in an independent cohort of 24 subjects, in which cultivation revealed Malassezia as viable oral mycobiome members, although the low-abundance Malassezia sympodialis was the only Malassezia species recovered. There was a high degree of concordance between the molecular detection and cultivability of Candida, while cultivation showed low sensitivity for detection of the Malassezia mycotype. Overall, our work provides insights into the oral mycobiome landscape, revealing 2 community classes with apparently distinct ecologic constraints and specific associations with coexisting bacteria and clinical parameters. The utility of mycotypes as biomarkers for oral diseases warrants further study.
Keywords: oral mycobiome, microbiome community classes, fungal-bacterial interactions, microbial ecology, saliva, salivary diagnostics
Introduction
Fungi have been documented as oral inhabitants for many decades, with cultivation reports showing high prevalence of Candida, Rhodotorula, Cryptococcus, Penicillium, Aspergillus, and Cladosporium (Young et al. 1951; Monteiro-da-Silva et al. 2014). Recent molecular surveys expanded the range of oral fungi, with individual samples sometimes showing hundreds of taxa (Ghannoum et al. 2010; Abusleme et al. 2018). Fungi are ubiquitous in food and the environment; therefore, it is unclear which of these taxa constitute functional oral mycobiome components. Malassezia, a cultivable fungus, is detected in high proportions in sequenced oral samples (Dupuy et al. 2014; Abusleme et al. 2018), but its role in the oral ecosystem is unknown. Most research has focused on the role that species from a single genus, Candida, play in disease states such as oral thrush (candidiasis) or caries (Falsetta et al. 2014; Abusleme et al. 2018; Xiao et al. 2018; Bertolini et al. 2019), but a broader view of oral mycobiome communities in states of symbiosis and dysbiosis is lacking. The ecologic factors that shape the mycobiome and the role that commensal fungi, other than Candida, play in oral ecosystem perturbations are still unknown.
Enhanced understanding of the oral mycobiome could have clinical implications. A common approach to evaluate microbiome populations involves unsupervised classification methods, such as clustering, to find groups of individuals who share communities of similar composition. Different bacteriome community classes associated with certain host characteristics have been reported in fecal, vaginal, and skin samples (Arumugam et al. 2011; Wu et al. 2011; Ding and Schloss 2014; Zhou et al. 2014). Oral commensal bacterial populations appear highly homogeneous (Zhou et al. 2014), but population distributions of oral fungi have not been evaluated.
Accordingly, the current study evaluated the population structure of salivary mycobiome communities in 59 subjects, 36 of whom were undergoing cancer chemotherapy. Using 3 unsupervised classification methods, we analyzed the community-wide distribution of salivary fungi to explore stratification patterns of ecologic or clinical significance. We uncovered 2 fungal community types and evaluated their relationships with coexisting bacteria and host medical and oral health characteristics. Community types were confirmed in an independent cohort of 24 subjects in which we pursued the cultivability of fungi driving mycobiome partitioning.
Methods
Subject Recruitment and Collection of Demographic and Clinical Information
Cohort 1 was part of a larger study (Hong et al. 2019) and consisted of subjects diagnosed with nonoral solid tumors who were scheduled to receive chemotherapy and noncancer controls. The current study included 59 of these subjects (36 cancer and 23 noncancer), who were selected per the availability of complete demographic, clinical, internal transcribed spacer 1 (ITS-1), and 16S rRNA gene amplicon data from a baseline visit (Hong et al. 2019).
Subjects were enrolled according to a protocol approved by the Institutional Review Board at UConn Health (IE-11-037 J-2). All participants provided written informed consent, and the study complied with STROBE guidelines. Inclusion and exclusion criteria were described by Hong et al. (2019) and appear in the Appendix Methods. Medical information was collected from questionnaires and medical charts. Subjects received an oral evaluation, including assessment of periodontal status via the Community Periodontal Index of Treatment Needs (Ainamo et al. 1982), presence and type of prosthetic restorations, presence of visible cavitated caries lesions according to the World Health Organization (WHO) criteria, and oral hygiene status per the plaque index (Silness and Loe 1964). An additional cohort (cohort 2; Institutional Review Board protocol 14-162-2), including 12 subjects receiving chemotherapy and 12 noncancer controls, was enrolled to evaluate the cultivability of Candida and Malassezia.
Salivary Microbiome Evaluation
Saliva was collected and DNA extracted and amplified as previously described (Hong et al. 2019). Samples from cohort 1 were evaluated by sequencing of the bacterial 16S rRNA gene V1–V2 hypervariable region and the fungal ITS-1 region. Only ITS-1 sequencing was performed for cohort 2. Sequences are available in the National Center for Biotechnology Information’s Sequence Read Archive (PRJNA399163 and PRJNA593057).
Cultivation of Candida and Malassezia
Supplemented CHROMagar Malassezia was used for cultivation of Candida and Malassezia. Isolates were identified after Sanger ITS-1 sequencing.
Statistical Analyses
Three approaches were employed to evaluate community types: Dirichlet multinomial mixture (DMM) models, partitioning around medoids (PAM), and unsupervised hierarchical clustering. Linear discriminant analysis effect size (LEfSe; Segata et al. 2011) was used to evaluate differentially abundant taxa. Correlations were evaluated via Spearman rank tests and stepwise logistic regression. Benjamini-Hochberg false discovery rate was applied for multiple comparison adjustments. Additional details appear in the Appendix Methods.
Results
Identification of 2 Discrete Fungal Community Types (Mycotypes) in Saliva
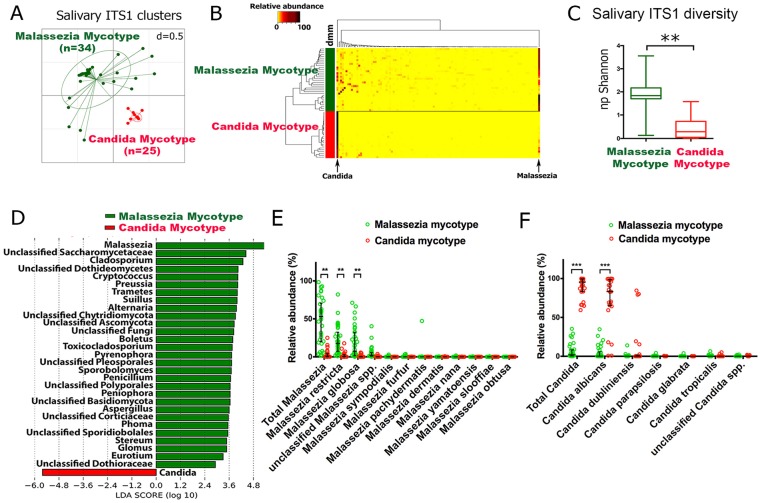
Cohort 1 characteristics are summarized in the Table, and fungal taxa detected are shown in Appendix Table 1. The existence of different fungal community classes in the whole cohort was explored through unsupervised methods. An analysis with DMM models suggested that the data set contained 2 genus-level clusters (Appendix Fig. 1). Visualization of these clusters via nonmetric multidimensional scaling ordination showed 1 highly cohesive cluster and 1 with greater sample spread (Fig. 1A). Unsupervised hierarchical clustering revealed 2 clusters, one more homogeneous than the other, and complete agreement with DMM in assignment of samples to clusters (Fig. 1B). Evaluation of clusters with PAM indicated 2 main clusters, although a small number of samples showed potential for forming separate groups (Appendix Fig. 2). The silhouette width, a measure of cluster cohesiveness and separation, was 0.57 for 1 PAM cluster and 0.64 for the second cluster, with an average silhouette width of 0.60, which is considered a moderately strong value (Wu et al. 2011; Koren et al. 2013). Overall, these results indicated the existence in saliva of 2 distinct fungal community types, which we refer to as mycotypes.
Figure 1.
Characterization of salivary mycotypes in 59 subjects (cohort 1). (A) Nonmetric multidimensional scaling plot based on thetaYC distances. Samples were colored according to Dirichlet multinomial mixture models analysis, which indicated that the data contained 2 clusters. (B) Unsupervised hierarchical clustering analysis also revealed 2 main clusters, as indicated in the dendrogram on the left side. The cluster to which each sample was assigned after Dirichlet multinomial mixture models analysis is indicated by the column bar. Genus-level relative abundances are shown in the heat map. (C) Differences in alpha diversity of the Candida and Malassezia mycotypes. Values are presented as median, interquartile range, and range. **P < 0.001 as determined by a Mann-Whitney rank test. (D) Linear discriminant analysis effect size evaluation of differences in relative abundances of fungal genera between mycotypes. Relative abundances of species of (E) Malassezia and (F) Candida according to mycotypes. Individual data points, median, and interquartile ranges are shown. **P < 0.001 and ***P < 0.0001 as determined by Mann-Whitney rank tests.
Table.
Demographic and Clinical Characteristics of Study Participants Included in Cohort 1.
| Variable | Cancer (n = 36) | Noncancer (n = 23) | P Value |
|---|---|---|---|
| Age | 57.64 ± 12.05 | 48.65 ± 14.42 | 0.0171 a |
| Male | 50.0 | 26.1 | 0.103b |
| White | 91.7 | 100.0 | 0.274b |
| Current smokerc | 16.7 | 10.5 | 0.700b |
| Proton pump inhibitor use | 25.0 | 8.7 | 0.174b |
| Inhaler steroid use | 8.3 | 0.0 | 0.274b |
| Steroid premedication | 16.7 | 0.0 | 0.072b |
| Inhaler steroid or premedication | 25 | 0.0 | 0.009 b |
| No. of teeth | 26 (0 to 32) [21 to 28] | 28 (0 to 32) [27 to 28] | 0.007 d |
| Prosthetic teeth or oral appliance | 80.6 | 69.6 | 0.363b |
| No. of teeth replaced by prostheses | 2 (0 to 32) [1 to 5] | 2 (0 to 28) [0 to 7] | 0.671d |
| Removable prosthesis | 11.1 | 8.7 | >0.999b |
| Visible cavitated caries lesions | 33.3 | 21.7 | 0.391b |
| No. of teeth with visible caries lesions | 0 (0 to 11) [0 to 1] | 0 (0 to 2) [0 to 0] | 0.202d |
| Plaque index | 1.05 (0.0 to 2.5)[0.6 to 1.5] | 0.60 (0.1 to 1.3)[0.5 to 1.0] | 0.007 d |
| At least 1 periodontal pocket >5.5 mm | 5.6 | 4.5 | >0.999b |
| Salivary flow rate, mL/min | 0.38 (0.05 to 1.46)[0.29 to 0.54] | 0.45 (0.07 to 0.95)[0.23 to 0.59 ] | 0.810d |
| Peripheral absolute neutrophil count, ×1,000/mm3 (blood) | 6.29 (2.23 to 18.41)[3.56 to 11.99] | 3.25 (1.87 to 5.35)[2.88 to 4.02] | <0.001 d |
| Cancer diagnosis | NA | NA | |
| Squamous cell carcinoma | 44.4 | ||
| Breast cancer | 30.6 | ||
| Adenocarcinoma | 19.4 | ||
| Other | 5.6 |
Values are presented as follows: for normally distributed continuous variables, mean ± SD; for nonnormally distributed continuous variables, median (range) [interquartile range]. Statistical tests for continuous data were applied according to data distribution. For nominal variables, data are presented as percentage of subjects positive. Bold indicates P < 0.05.
NA, not applicable.
Independent sample t test.
Fisher’s exact test.
Smoking descriptive statistics for the noncancer group are based on n = 19, as there were 3 cases with missing smoking data.
Mann-Whitney U test.
The composition of mycotypes was then examined. As shown in Figure 1B, 1 mycotype was characterized by high Malassezia proportions (1.5% to 98%), while Candida was the dominant taxon in the second mycotype (59% to 99%), suggesting that these genera are the main drivers of mycotype partitioning. The Malassezia-enriched mycotype was more diverse than Candida-enriched communities (Fig. 1C). Consistent with these findings, LEfSe evaluation of taxa that differed between mycotypes showed that Malassezia and several other taxa were enriched in 1 cluster, while only Candida was enriched in the second, less diverse mycotype (Fig. 1D).
Distributions of Malassezia and Candida species were then evaluated. Figure 1E shows that the most abundant Malassezia species and the most discriminative between mycotypes were Malassezia restricta and Malassezia globosa, while Candida albicans was the most abundant and differentially enriched species in the Candida mycotype (Fig. 1F). Mycotype clustering was due to differences in relative abundances of Candida and Malassezia rather than in presence/absence. As seen in Figure 1E and F, some Candida mycotype subjects had low levels of Malassezia and vice versa.
Associations between Mycotypes and Host Characteristics
Correlations of mycotypes with demographic, medical, and oral health characteristics of subjects were then evaluated. Although both mycotypes occurred in subjects with cancer and controls, the Candida mycotype was more frequent in the cancer group (P = 0.015; Fig. 2A). Variables that differed between groups (Table) and could explain this difference were then examined. Age and recent history of chemotherapy were not correlated with mycotypes. However, as seen in Figure 2B, receiving steroid premedication (administered to certain subjects with cancer 1 to 5 d prior to sampling) and having higher peripheral neutrophil counts as a consequence of steroid intake (Mishler and Emerson 1977) were positively correlated with the Candida mycotype. Other variables that possibly explained the higher Candida mycotype frequency in subjects with cancer were inhaler steroid use and plaque index, which showed higher values in subjects with cancer (Table), and were positively correlated with the Candida mycotype (Fig. 2B). In a partial correlation analysis, the association of cancer and mycotypes became nonsignificant (P = 0.098) after controlling for steroid intake (via any route) but remained significant when adjusting for the plaque index (P = 0.049), suggesting that steroid use was the main factor explaining the higher prevalence of the Candida mycotype in cancer subjects. Variables that did not differ between cancer and noncancer groups but were associated with mycotypes included smoking, utilizing a removable prosthesis, and number of teeth with visible caries lesions, which showed a positive correlation with the Candida mycotype, while number of teeth was negatively correlated (Fig. 2B). A stepwise logistic regression model was then constructed incorporating cancer, steroids (via any route), number of teeth, plaque index, removable prosthesis, caries, and smoking as predictors and mycotype as outcome. Only steroid and plaque index were retained in the model, with steroid showing an odds ratio of 10.23 (95% CI, 0.98 to 106.28) and borderline significance (P = 0.05) and with plaque index showing an odds ratio of 6.23 (95% CI, 1.15 to 33.65) and P = 0.033 (Appendix Fig. 3). The prediction accuracy of this model for the Candida mycotype was 78.9%.
Figure 2.
Association of mycotypes with host characteristics. (A) Prevalence of mycotypes according to cancer status. Bars show percentage of subjects in which the specific mycotype was detected. Differences evaluated via chi-square. (B) Correlogram depicts correlations (Spearman) between mycotypes (0, Malassezia; 1, Candida) and host parameters. Only those host variables that showed a significant correlation with mycotypes after multiple-test adjustment are shown. Colors indicate correlation coefficients. *P < 0.01 and **P < 0.001. Inh. Steroid, use of corticosteroids as an inhaler or nasal spray; PNC, peripheral neutrophil counts; Premed. Steroid, patients who received intravenous corticosteroids days prior to sampling. (C) Correlations (Spearman) between relative abundances of Candida and Malassezia (genus-level totals and individual species abundances) and host characteristics. Only taxa that showed significant correlations are depicted. Colors indicate correlation coefficients, and diagonal crossed lines indicate nonsignificant correlations (after multiple-comparison adjustment). As a reference, correlations of the salivary abundances of the caries-associated bacterium Streptococcus mutans and the periodontitis-associated bacterium Porphyromonas gingivalis are included. PD, periodontal probing depth.
Correlations of Salivary Candida and Malassezia Proportions with Host Characteristics
We next evaluated correlations of Malassezia and Candida individual proportions and clinical characteristics. Candida was higher in subjects with cancer (Appendix Fig. 4) and positively correlated with steroids, smoking, removable prosthesis, presence of caries, number of teeth with caries lesions, and plaque index (Fig. 2C). At a species level, C. albicans positively correlated with steroids, smoking, removable prosthesis, and proton pump inhibitor use. Confirming their exclusive relationship, Malassezia, including M. restricta and M. globosa individual proportions, negatively correlated with the same variables. Malassezia proportions were also positively correlated with the presence of at least 1 periodontal pocket >5.5 mm, although this finding should be interpreted with caution as only 3 individuals had this clinical characteristic.
Bacterial Communities Coexisting with Mycotypes
DMM analysis indicated that salivary bacterial communities contained 2 weakly separated clusters (Appendix Fig. 1, Fig. 3A). PAM analysis showed a low silhouette width (0.13 for k = 2), confirming the homogeneity of salivary bacterial communities (Appendix Fig. 5). However, if bacterial communities associated with each mycotype were contrasted, communities associated with the Malassezia mycotype were more diverse than those associated with the Candida mycotype (Fig. 3B). These results paralleled the fungal diversity differences seen between mycotypes (Fig. 1C), suggesting that a common environmental pressure affected bacteria and fungi. LEfSe showed that, indeed, metabolically distinct bacteria were differentially enriched in mycotypes, with aciduric bacterial species, such as Lactobacillus salivarius, Lactobacillus ultunensis, and Propionibacterium acidifaciens, enriched in subjects harboring the Candida mycotype, while for the Malassezia mycotype, subjects had higher proportions of anaerobic species of Fusobacterium, Porphyromonas, Prevotella, Treponema, and Leptotrichia, among others. These results were confirmed in a correlation analysis (Fig. 3D), which showed that Candida proportions positively correlated with bacteria known to have high acid tolerance, such as lactobacilli and Veillonella spp., while Malassezia proportions positively correlated with bacterial anaerobes typically associated with gingival inflammation, that rely on amino acid catabolism and prefer more basic environmental pH (e.g., Porphyromonas spp., Fusobacterium spp., Treponema lecithinolyticum, Selenomonas sputigena, and Leptotrichia shahii, among others). Altogether, our data show that although salivary bacterial communities do not contain discrete clusters, specific bacterial species with distinct metabolic requirements are enriched in each mycotype.
Figure 3.
Bacterial communities associated with mycotypes. (A) Clusters contained in the species-level bacterial community data. Graph is a nonmetric multidimensional scaling ordination plot based on thetaYC distances with samples colored according to Dirichlet multinomial mixture models analysis. (B) Differences in diversity of bacterial communities associated with mycotypes. Values are presented as median, interquartile range, and range. *P < 0.01 as determined by Mann-Whitney rank test. (C) Linear discriminant analysis effect size evaluation of differences in relative abundances of bacterial species coexisting with each mycotype. (D) Correlations (Spearman) between relative abundances of Candida and Malassezia (genus-level totals and individual species abundances) and bacterial relative abundances. Only taxa that showed significant correlations with Candida and Malassezia are depicted. Colors indicate correlation coefficients, and diagonal crossed lines indicate nonsignificant correlations (after multiple-comparison adjustment).
Cultivability of Mycotypes
The observed mycotypes and their cultivability were evaluated in a second cohort (cohort 2; Appendix Table 2). All but 1 subject in cohort 2 yielded ITS-1 amplicons. Mycobiome analysis showed 2 clusters with distinct levels of Candida and Malassezia (Fig. 4A). Similar to cohort 1, C. albicans, M. restricta, and M. globosa were the most abundant species (Appendix Table 3). Molecular mycotypes did not differ in frequency between control and cancer groups (chi-square, P = 0.795; Fig. 4A). However, as observed in cohort 1, the Candida mycotype positively correlated with number of teeth with visible caries lesions (r = 0.556, P = 0.006).
Figure 4.
Cultivability of Candida and Malassezia in relation to mycotypes. A second cohort of 24 subjects was enrolled, and their salivary mycobiomes were characterized by ITS-1 (internal transcribed spacer 1) sequencing and cultivation. (A) Unsupervised hierarchical clusters and heat map show genus-level relative abundances of salivary fungi. Notice 2 clusters with Malassezia and Candida as the main distinctive taxa. (B) Total fungal, Candida, and Malassezia salivary cultivable load according to molecular mycotype classification. (C) Ability of cultivation to detect molecular mycotypes. (D) Heat map shows loads of fungi detected via cultivation in relation to cancer and mycotype groupings. (E) Correlation (Spearman) between number of teeth with visible cavitated caries lesions and salivary cultivable load of Candida.
Although Malassezia have been observed in oral samples by molecular methods (Dupuy et al. 2014; Abusleme et al. 2018), to our knowledge, oral Malassezia have not been cultivated. Therefore, the cultivability of Malassezia, as compared with that of Candida, was evaluated with a microbiological medium that allows growth of both genera. Figure 4B shows Malassezia and Candida load in subjects harboring each mycotype. Candida was principally cultivated from the Candida mycotype group, while 3 individuals—2 classified in the Malassezia mycotype group and 1 undetermined due to a negative polymerase chain reaction result—yielded Malassezia colonies. Figure 4C demonstrates concordance between the molecular Candida mycotype classification and cultivability of Candida (sensitivity, 1; specificity, 0.82). However, cultivation did not allow detection of subjects harboring the Malassezia mycotype (sensitivity, 0.14; specificity, 1).
All fungi recovered by cultivation are shown in Figure 4D and Appendix Table 4. Only 1 species of Malassezia, M. sympodialis, was recovered on agar. Colonies of Pichia, Exophiala, Rhodotorula, Cryptococcus, and Clavispora were also observed. Colony morphologies of these fungi on CHROMagar Malassezia are shown in Appendix Figure 6.
Confirming the association of Candida and caries, Candida cultivable load showed a strong positive correlation with number of teeth with caries (r = 0.616, P = 0.001; Fig. 4E) and caries presence (r = 0.458, P = 0.024). At the species level,C. albicans and Candida parapsilosis cultivable load positively correlated with number of teeth with caries (r = 0.446,P = 0.029; r = 0.431, P = 0.035).
Discussion
Despite considerable intersubject variability in the human microbiome composition, different community types have been described (Arumugam et al. 2011; Gajer et al. 2012; Zhou et al. 2014). Compositional analyses of fecal communities have revealed 3 community types, known as enterotypes (Arumugam et al. 2011), although others have questioned the status of enterotypes as discrete clusters (Koren et al. 2013; Knights et al. 2014). The oral mycotypes found in this study represent clusters with a degree of separation much higher than that of enterotypes. By comparison, the silhouette width for enterotypes has been reported at ~0.2 (Zhou et al. 2014; Costea et al. 2018), while that for salivary mycotypes was 0.6. Moreover, different partitioning methods agreed in the optimal number of clusters and sample assignment, and an independently enrolled cohort sequenced with a slightly different approach (different primers and sequencing platform) showed similar fungal distribution patterns. Our data strongly suggest the existence of 2 discrete salivary fungal communities. Due to the high dominance of Candida, the Candida mycotype was more cohesive and less diverse than the Malassezia mycotype. When the LEfSe alpha value (Fig. 1D) was relaxed to 0.1, only 1 other taxon, Saccharomyces, appeared enriched in the Candida mycotype. In contrast, the Malassezia mycotype was more diverse, showing Malassezia as the dominant community member but accompanied by other co-occurring fungi (Fig. 1B, D). Analysis of a larger and more diverse population of subjects could reveal whether this binary partitioning and the specific community structures are maintained. The temporal stability of mycotypes within subjects also warrants further investigation.
Mycotype associations with host characteristics were explored to better understand ecologic and host constraints determining mycotypes. Our cohorts included subjects undergoing cancer chemotherapy, since we have an ongoing interest in predisposing factors leading to oral complications of cancer treatment (Diaz et al. 2019; Hong et al. 2019). In cohort 1, the Candida mycotype was more prevalent in subjects with cancer and correlated with their intake of corticosteroids, which have been associated with oral Candida colonization (Pereira Tdos et al. 2014). Several lines of evidence also pointed to the oral environment as a selective constrain for mycotypes. The relationship of plaque index, the only variable retained after logistic regression, and mycotypes may result from coadhesive interactions between Candida and dental plaque bacteria, which may facilitate the retention of the fungus, while Candida promotes greater bacterial biomass accumulation (Gregoire et al. 2011; Xu et al. 2017). It should be noted, however, that the number of covariates included in the logistic regression model was large given the sample size and some of the variables included showed collinearity. Therefore, a larger study is needed to validate the independent relationships between oral characteristics and mycotypes. Apart from dental plaque levels, caries also showed a relationship with mycotypes. Candida prefers glucose or lactate as carbon sources and has been shown to thrive in carbohydrate-rich polymicrobial communities (Chiew 1989; Ene et al. 2012; Koopman et al. 2015). Moreover, in agreement with our findings, levels of Candida and saccharolytic acidogenic bacteria have been shown to correlate (Kraneveld et al. 2012). Therefore, it appears that the Candida mycotype is associated with aciduric oral conditions. In contrast, Malassezia spp. are incapable of carbohydrate fermentation and depend on lipids (Senczek et al. 1999; Wu et al. 2015). Saliva, gingival crevicular fluid, and the host diet could represent lipid sources for Malassezia, which possess a diverse array of secretory lipases (Larsson et al. 1996; Wu et al. 2015). Moreover, the Malassezia mycotype was associated with bacteria that rely on amino acid fermentation and prefer slightly basic conditions for growth, again suggesting oral pH as a mycotype determinant.
The described mycotypes may have diagnostic utility. We recently reported a multivariate model able to predict oral candidiasis during chemotherapy (Diaz et al. 2019). Since Candida and Malassezia salivary proportions were significant predictors in the reported model, mycotypes could represent another discriminatory parameter. Mycotypes may be useful as screening tools for tooth-associated diseases, but further research is needed. Due to the site specificity of caries, the use of saliva as a diagnostic tool has been questioned (Mira 2018). However, our data show a correlation between salivary proportions of S. mutans and caries. The salivary Candida mycotype, Candida proportions, and cultivable Candida load positively correlated with caries. Additional research is required to assess the utility of mycotypes as caries biomarkers, in particular with more sensitive tools to document initial disease stages. The relationship of Malassezia and periodontitis needs further study in a larger cohort. It would also be important to discern whether Candida and Malassezia serve only as diagnostic indicators or if they participate in dysbiotic events at diseased sites.
We investigated whether Malassezia are viable oral mycobiome members. Using a lipid-containing medium, we cultivated M. sympodialis, a species present at <1% abundance according to ITS-1 sequencing in subjects that yielded colonies. The abundant M. restricta and M. globosa were not recovered, although skin derived–type strains of these species grew under the cultivation conditions used. This suggests that Malassezia are viable oral mycobiome components, but the cultivation requirements of oral M. restricta and M. globosa need additional study. Since the most abundant Malassezia species present were not recovered, it was not possible to estimate the total fungal load. Total fungi present in mycotypes may differ, but more work is needed to develop appropriate methods to measure load. We decided against employing universal ITS primers and quantitative polymerase chain reaction to estimate load, since the high inter- and intraspecies variability in the copy number of the rRNA gene cluster region among fungi is likely to bias load estimates (Rustchenko et al. 1993; Diaz et al. 2017; Lofgren et al. 2019). Such copy number variability could also bias relative abundance estimates, but unfortunately, a database does not yet exist that has information on the number of rRNA gene cluster copies in the genomes of oral mycobiome components and that could be used to normalize abundance estimates. A critical appraisal of which ITS-1 reads represent metabolically active fungi is also needed. For instance, Boletus are edible mushrooms. Saccharomyces, certain Aspergillus, Fusarium, and Phoma are associated with food. Alternaria and Cladosporium may represent inhaled spores from indoor environments. It is likely that only yeasts, such as Candida, Pichia, Clavispora, Malassezia, and Rhodotorula, are functional, metabolically active mycobiome components.
In summary, our work revealed 2 salivary fungal community types associated with specific host characteristics and metabolically distinct bacteria. Since mycotypes appear to be associated with distinct ecologic conditions, which in turn are related to specific oral diseases (Marsh 1994), the mycotype classification could represent a biomarker and warrants further evaluation in the context of salivary diagnostics.
Author Contributions
B.Y. Hong, A. Hoare, A. Cardenas, contributed to data acquisition and analysis, critically revised the manuscript; A.K. Dupuy, L. Choquette, A.L. Salner, P.K. Schauer, U. Hegde, contributed to data acquisition, critically revised the manuscript; D.E. Peterson, contributed to conception, design, and data acquisition, critically revised the manuscript; A. Dongari-Bagtzoglou, contributed to conception, design, and data interpretation, critically revised the manuscript; L.D. Strausbaugh, contributed to conception, design, data acquisition, and interpretation, critically revised the manuscript; P.I. Diaz, contributed to conception, design, data acquisition, analysis, and interpretation, drafted and critically revised the manuscript. All authors gave final approval and agree to be accountable for all aspects of the work.
Supplemental Material
Supplemental material, DS_10.1177_0022034520915879 for The Salivary Mycobiome Contains 2 Ecologically Distinct Mycotypes by B.Y. Hong, A. Hoare, A. Cardenas, A.K. Dupuy, L. Choquette, A.L. Salner, P.K. Schauer, U. Hegde, D.E. Peterson, A. Dongari-Bagtzoglou, L.D. Strausbaugh and P.I. Diaz in Journal of Dental Research
Footnotes
A supplemental appendix to this article is available online.
This work was supported by grant R01DE021578 from the National Institute of Dental and Craniofacial Research, National Institutes of Health. We also acknowledge the support of pilot grant M01RR006192 from the UConn Health General Clinical Research Center.
The authors declare no potential conflicts of interest with respect to the authorship and/or publication of this article.
ORCID iDs: A.K. Dupuy  https://orcid.org/0000-0001-5626-6391
https://orcid.org/0000-0001-5626-6391
P.I. Diaz  https://orcid.org/0000-0002-9501-1145
https://orcid.org/0000-0002-9501-1145
References
- Abusleme L, Diaz PI, Freeman AF, Greenwell-Wild T, Brenchley L, Desai JV, Ng WI, Holland SM, Lionakis MS, Segre JA, et al. 2018. Human defects in STAT3 promote oral mucosal fungal and bacterial dysbiosis. JCI Insight. 3(17):122061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ainamo J, Barmes D, Beagrie G, Cutress T, Martin J, Sardo-Infirri J. 1982. Development of the World Health Organization (WHO) Community Periodontal Index of Treatment Needs (CPITN). Int Dent J. 32(3):281–291. [PubMed] [Google Scholar]
- Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, Fernandes GR, Tap J, Bruls T, Batto JM, et al. 2011. Enterotypes of the human gut microbiome. Nature. 473(7346):174–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertolini M, Ranjan A, Thompson A, Diaz PI, Sobue T, Maas K, Dongari-Bagtzoglou A. 2019. Candida albicans induces mucosal bacterial dysbiosis that promotes invasive infection. PLoS Pathog. 15(4):e1007717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiew YY. 1989. The dynamics of carbohydrate metabolism in Candida albicans. Exp Mycol. 13(1):49–60. [Google Scholar]
- Costea PI, Hildebrand F, Arumugam M, Backhed F, Blaser MJ, Bushman FD, de Vos WM, Ehrlich SD, Fraser CM, Hattori M, et al. 2018. Enterotypes in the landscape of gut microbial community composition. Nat Microbiol. 3(1):8–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz PI, Hong BY, Dupuy AK, Choquette L, Thompson A, Salner AL, Schauer PK, Hegde U, Burleson JA, Strausbaugh LD, et al. 2019. Integrated analysis of clinical and microbiome risk factors associated with the development of oral candidiasis during cancer chemotherapy. J Fungi (Basel). 5(2):E49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz PI, Hong BY, Dupuy AK, Strausbaugh LD. 2017. Mining the oral mycobiome: methods, components, and meaning. Virulence. 8(3):313–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding T, Schloss PD. 2014. Dynamics and associations of microbial community types across the human body. Nature. 509(7500):357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupuy AK, David MS, Li L, Heider TN, Peterson JD, Montano EA, Dongari-Bagtzoglou A, Diaz PI, Strausbaugh LD. 2014. Redefining the human oral mycobiome with improved practices in amplicon-based taxonomy: discovery of Malassezia as a prominent commensal. PLoS One. 9(3):e90899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ene IV, Adya AK, Wehmeier S, Brand AC, MacCallum DM, Gow NA, Brown AJ. 2012. Host carbon sources modulate cell wall architecture, drug resistance and virulence in a fungal pathogen. Cell Microbiol. 14(9):1319–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falsetta ML, Klein MI, Colonne PM, Scott-Anne K, Gregoire S, Pai CH, Gonzalez M, Watson G, Krysan DJ, Bowen WH, et al. 2014. Symbiotic relationship between Streptococcus mutans and Candida albicans synergizes the virulence of plaque-biofilms in vivo. Infect Immun. 82(5):1968–1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gajer P, Brotman RM, Bai G, Sakamoto J, Schutte UM, Zhong X, Koenig SS, Fu L, Ma ZS, Zhou X, et al. 2012. Temporal dynamics of the human vaginal microbiota. Sci Trans Med. 4(132):132ra152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghannoum MA, Jurevic RJ, Mukherjee PK, Cui F, Sikaroodi M, Naqvi A, Gillevet PM. 2010. Characterization of the oral fungal microbiome (mycobiome) in healthy individuals. PLoS Pathog. 6(1):e1000713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregoire S, Xiao J, Silva BB, Gonzalez I, Agidi PS, Klein MI, Ambatipudi KS, Rosalen PL, Bauserman R, Waugh RE, et al. 2011. Role of glucosyltransferase b in interactions of Candida albicans with Streptococcus mutans and with an experimental pellicle on hydroxyapatite surfaces. Appl Environ Microbiol. 77(18):6357–6367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong BY, Sobue T, Choquette L, Dupuy AK, Thompson A, Burleson JA, Salner AL, Schauer PK, Joshi P, Fox E, et al. 2019. Chemotherapy-induced oral mucositis is associated with detrimental bacterial dysbiosis. Microbiome. 7(1):66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knights D, Ward TL, McKinlay CE, Miller H, Gonzalez A, McDonald D, Knight R. 2014. Rethinking “enterotypes.” Cell Host Microb. 16(4):433–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koopman JE, Roling WF, Buijs MJ, Sissons CH, ten Cate JM, Keijser BJ, Crielaard W, Zaura E. 2015. Stability and resilience of oral microcosms toward acidification and Candida outgrowth by arginine supplementation. Microb Ecol. 69(2):422–433. [DOI] [PubMed] [Google Scholar]
- Koren O, Knights D, Gonzalez A, Waldron L, Segata N, Knight R, Huttenhower C, Ley RE. 2013. A guide to enterotypes across the human body: meta-analysis of microbial community structures in human microbiome datasets. PLoS Comput Biol. 9(1):e1002863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraneveld EA, Buijs MJ, Bonder MJ, Visser M, Keijser BJ, Crielaard W, Zaura E. 2012. The relation between oral Candida load and bacterial microbiome profiles in Dutch older adults. PLoS One. 7(8):e42770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson B, Olivecrona G, Ericson T. 1996. Lipids in human saliva. Arch Oral Biol. 41(1):105–110. [DOI] [PubMed] [Google Scholar]
- Lofgren LA, Uehling JK, Branco S, Bruns TD, Martin F, Kennedy PG. 2019. Genome-based estimates of fungal rDNA copy number variation across phylogenetic scales and ecological lifestyles. Mol Ecol. 28(4):721–730. [DOI] [PubMed] [Google Scholar]
- Marsh PD. 1994. Microbial ecology of dental plaque and its significance in health and disease. Adv Dent Res. 8(2):263–271. [DOI] [PubMed] [Google Scholar]
- Mira A. 2018. Oral microbiome studies: potential diagnostic and therapeutic implications. Adv Dent Res. 29(1):71–77. [DOI] [PubMed] [Google Scholar]
- Mishler JM, Emerson PM. 1977. Development of neutrophilia by serially increasing doses of dexamethasone. Br J Haematol. 36(2):249–257. [DOI] [PubMed] [Google Scholar]
- Monteiro-da-Silva F, Araujo R, Sampaio-Maia B. 2014. Interindividual variability and intraindividual stability of oral fungal microbiota over time. Med Mycol. 52(5):498–505. [DOI] [PubMed] [Google Scholar]
- Pereira Tdos S, Silva Alves Jde F, Gomes CC, Rocha do, Nascimento A, Stoianoff MA, Gomez RS. 2014. Kinetics of oral colonization by Candida spp. during topical corticotherapy for oral lichen planus. J Oral Pathol Med. 43(8):570–575. [DOI] [PubMed] [Google Scholar]
- Rustchenko EP, Curran TM, Sherman F. 1993. Variations in the number of ribosomal DNA units in morphological mutants and normal strains of Candida albicans and in normal strains of Saccharomyces cerevisiae.J Bacteriol. 175(22):7189–7199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, Huttenhower C. 2011. Metagenomic biomarker discovery and explanation. Genome Biol. 12(6):R60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senczek D, Siesenop U, Bohm KH. 1999. Characterization of Malassezia species by means of phenotypic characteristics and detection of electrophoretic karyotypes by pulsed-field gel electrophoresis (PFGE). Mycoses. 42(5–6):409–414. [DOI] [PubMed] [Google Scholar]
- Silness J, Loe H. 1964. Periodontal disease in pregnancy: II. Correlation between oral hygiene and periodontal condtion. Acta Odontol Scand. 22:121–135. [DOI] [PubMed] [Google Scholar]
- Wu G, Zhao H, Li C, Rajapakse MP, Wong WC, Xu J, Saunders CW, Reeder NL, Reilman RA, Scheynius A, et al. 2015. Genus-wide comparative genomics of Malassezia delineates its phylogeny, physiology, and niche adaptation on human skin. PLoS Genet. 11(11):e1005614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu GD, Chen J, Hoffmann C, Bittinger K, Chen YY, Keilbaugh SA, Bewtra M, Knights D, Walters WA, Knight R, et al. 2011. Linking long-term dietary patterns with gut microbial enterotypes. Science. 334(6052): 105–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao J, Grier A, Faustoferri RC, Alzoubi S, Gill AL, Feng C, Liu Y, Quivey RG, Kopycka-Kedzierawski DT, Koo H, et al. 2018. Association between oral Candida and bacteriome in children with severe ECC. J Dent Res. 97(13):1468–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Sobue T, Bertolini M, Thompson A, Vickerman M, Nobile CJ, Dongari-Bagtzoglou A. 2017. S. oralis activates the Efg1 filamentation pathway in C. albicans to promote cross-kingdom interactions and mucosal biofilms. Virulence. 8(8):1602–1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young G, Resca HG, Sullivan MT. 1951. The yeasts of the normal mouth and their relation to salivary acidity. J Dent Res. 30(3):426–430. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Mihindukulasuriya KA, Gao H, La Rosa PS, Wylie KM, Martin JC, Kota K, Shannon WD, Mitreva M, Sodergren E, et al. 2014. Exploration of bacterial community classes in major human habitats. Genom Biol. 15(5):R66. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, DS_10.1177_0022034520915879 for The Salivary Mycobiome Contains 2 Ecologically Distinct Mycotypes by B.Y. Hong, A. Hoare, A. Cardenas, A.K. Dupuy, L. Choquette, A.L. Salner, P.K. Schauer, U. Hegde, D.E. Peterson, A. Dongari-Bagtzoglou, L.D. Strausbaugh and P.I. Diaz in Journal of Dental Research