Abstract
Commensal microbiomes exert critical functions at barrier sites. In particular, establishment of the commensal microbiome after birth dictates immune functionality and tissue homeostasis at mucosal surfaces. To investigate the establishment and stability of the oral mucosal microbiome in mice, we evaluated oral microbiome communities shortly after birth, through adulthood, and up to 1 y of life in a controlled manner, using sequential oral samples from the same mice over time. We further evaluated transmissibility of oral microbiomes from parents and during cohousing experiments and evaluated susceptibility to oral inflammatory disease in mice harboring distinct microbiomes. Our work reveals basic principles in the establishment and stability of a health-associated oral microbiome after birth and provides insights that may be important for host-microbiome experimentation in animal models.
Keywords: microbiome establishment, microbiome transfer, microbiome stability, oral microbial acquisition, oral mucosal microbiome, gingival microbiome
Introduction
Complex communities of microorganisms, known as the “commensal microbiome,” inhabit the body surfaces of virtually all vertebrates and are crucial for host physiology. In the gastrointestinal (GI) tract, microbes have an essential role for digestion, breaking down complex carbohydrates to provide vital nutrients to mammals. Beyond digestion, the tissue-associated commensal microbiota contributes profoundly to the development and function of tissues, regulating barrier integrity, homeostasis, and immunity (Hooper et al. 2012; Harrison et al. 2019). Indeed, studies in germ-free mice have revealed that in the absence of commensal microbiota, mice have defective development of peripheral immune organs and also impaired immune cell profile and functionality at barrier sites (Lee and Mazmanian 2010). In fact, commensal microbiota play key roles in the induction of innate anti-microbial defenses and adaptive immunity development (Lee and Mazmanian 2010). For example, particular microbiota have been shown to trigger specialized immune responses in the gastrointestinal mucosa, skin, and ocular surfaces (Ivanov et al. 2009; Naik et al. 2012; St Leger et al. 2017). Therefore, the characterization of commensal microbiota at barrier sites becomes essential toward the understanding of physiologic roles of microorganisms in the sites they inhabit.
Initial establishment and maturation of commensal microbial communities during infancy can be crucial for health later in life. Recent studies have shown microbial regulation of specific arms of immunity after birth and during infancy. Studies in mouse models have specifically demonstrated that microbial alterations can cause long-lasting immune effects that increase susceptibility to pathologies (Gensollen and Blumberg 2017; Constantinides et al. 2019). Notably, human epidemiological studies strongly support that factors altering bacterial communities in infants and/or during childhood will lead to an increased risk for several diseases, highlighting the importance of understanding early-life microbiome composition (Tamburini et al. 2016).
Establishment of the early microbiome is thought to depend on several factors, including vertical transmission of microbiota from parents, mode of delivery, feeding behaviors, and the environment (Stewart et al. 2018). In mice, establishment and stability of the commensal microbiomes can be additionally impacted by their coprophagic behavior, housing conditions, and variations in pathogen screening in animal facilities, among others (Levy et al. 2017). Importantly, stability and resilience of a health-associated microbiome is important for the health of the host. In recent years, many of the modern multifactorial diseases that show an increasing incidence are linked with disease-associated microbiome shifts, termed dysbiosis (Levy et al. 2017). Dysbiotic shifts in microbiome communities can be caused by various etiologies ranging from infection, inflammation, host genetics, and multiple environmental factors. Microbial dysbiosis has been linked to numerous human pathologies, including but not limited to inflammatory bowel disease, colorectal carcinogenesis, type I diabetes, and metabolic syndrome, and the oral disease periodontitis (Hajishengallis 2015; Levy et al. 2017).
In our current study, we aim to characterize factors which influence the establishment and stability of the health-associated oral microbiome from birth through adulthood. We therefore characterize oral microbiome communities in mice using sequential samples from the same mice shortly after birth, through adulthood, and up to 1 y of life. We further evaluate factors that can influence establishment and stability of oral microbiomes such as tooth eruption, weaning from mothers, vertical and horizontal transmission. Our findings provide insights into the dynamics of establishment of oral microbiome communities in murine models.
Materials and Methods
Mice
C57BL/6 mice included in this study were bred in our animal facility and for select experiments were purchased from Taconic Biosciences (Rensselaer, NY) and The Jackson Laboratory (Bar Harbor, ME). All animals were housed in a single room and rack, and were maintained under specific pathogen free (SPF) conditions at the National Institute of Dental and Craniofacial Research (NIDCR) veterinary resources core. Experimental procedures were approved by the NIDCR Animal Care and Use Committee (ASP # 15-787). Details of these experiments are further described in the Appendix.
Sample Collection and DNA Isolation
Oral mucosal samples were obtained by swabbing mucosal areas for 30 s using sterile ultra-fine cotton tips (Puritan Medical Products; Guilford, ME) and by dissecting palatal gingival tissues around molars. Samples were then individually placed in 150 µl of TE buffer and stored at −80°C until processing, as previously described (Abusleme et al. 2017). Briefly, DNA isolation was performed using a modified version of the DNeasy Blood and Tissue kit (Qiagen; Germantown, MD), that included an initial incubation with a chemical/enzymatic lysis buffer (Abusleme et al. 2014).
16S rRNA Sequencing and Bioinformatic Processing
DNA from oral samples was subjected to 16S rRNA gene sequencing. Details of library preparation and sequencing are included in the Appendix. Sequencing reads were processed using the software mothur version.1.41.1 (Schloss et al. 2009). Briefly, reads were quality filtered, assembled into contigs, and filtered by size, keeping those of 200 to 400 bp in length. Then, bioinformatic processing was performed as described in the MiSeq SOP pipeline (Kozich et al. 2013). We defined operational taxonomic units (OTUs) at a 97% similarity and classified up to genus-level when possible. A comprehensive description of these analyses together with a summary of read counts is available in the Appendix. All sequence data have been uploaded to NCBI SRA database under SRA accession number PRJNA589593.
Microbiome Data Analysis and Statistics
Principal coordinates analyses (PCoAs) of community structure were generated using mothur based on Yue–Clayton theta distances (θYC) (Yue and Clayton 2005). Analysis of molecular variance (AMOVA) was utilized to test for differences in community structure, as implemented in mothur. PCoAs showing discriminant OTUs were based on the correlations between the relative abundance of OTUs and the 2 principal components explaining microbial variability in PCoAs were determined using bivariate Spearman’s rank-order correlation tests. Depicted OTUs remained significant (P < 0.001) upon correcting for multiple comparisons using the false discovery rate method. Dissimilarities of microbial communities were compared using Wilcoxon Rank test or Friedman test and Dunn’s multiple comparisons test. Differences in bone loss measurements were evaluated via Mann-Whitney test. All software utilized for statistics and data visualization is described in the Appendix.
Alveolar Bone Loss Measurements
Mice skulls were defleshed and stained with 1% methylene blue for bone height evaluation. Bone measurements were evaluated after imaging using AxioVision 4.9.1 software (White Plains, NY). The distance between the cemento-enamel junction and alveolar bone crest (CEJ-ABC distance) was measured at 6 predetermined sites and merged, as previously described (Eskan et al. 2012).
Results
Evaluation of Oral Microbiome after Birth Highlights Tooth Eruption as a Major Event-driver in the Establishment of Oral Microbial Communities
To evaluate the acquisition and early establishment of oral microbial communities, we examined microbial communities in mice shortly after birth, during tooth eruption, after weaning (4 wk), and up to adulthood (8 wk of age). For our studies, oral microbiome samples were collected longitudinally from the same mice over time, and bacterial communities were characterized by 16S rRNA sequencing.
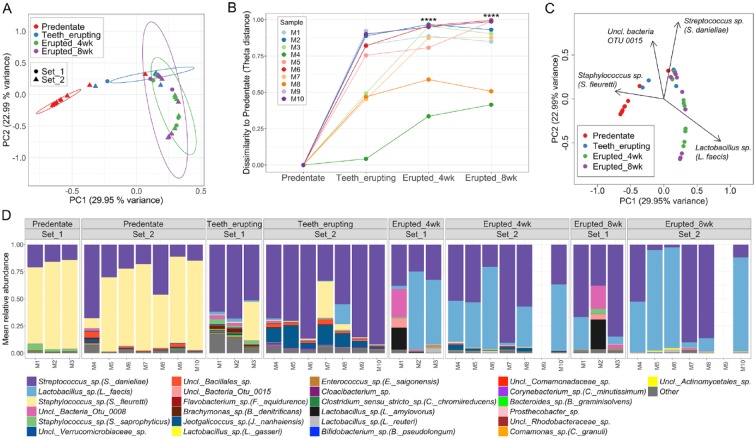
Our first, most striking observation was that postnatal microbial communities (postnatal day 7 to 9), before tooth eruption, significantly differ from oral microbial communities after teeth have erupted (postnatal day 16 to 17). In fact, predentate oral microbial communities significantly separate from communities during and after tooth eruption, based on community structure measurements (Fig. 1A). Predentate communities are also significantly dissimilar from posttooth eruption microbial communities based on the theta-dissimilarity index (Fig. 1B). Interestingly, the main microbial taxon driving the separation of predentate communities was a Staphylococcus sp. (S. fleuretii) (Fig. 1C), which was also a highly abundant constituent of predentate communities (Fig. 1D). As staphylococci are primarily members of the skin microbiome, these data suggest that the initial source of oral bacteria in early postnatal days is the skin.
Figure 1.
Tooth eruption is a critical event during establishment of oral microbiome communities. (A) Principal coordinates analysis (PCoA) plot of community structure, based on θYC distances, depicting that predentate oral microbial communities cluster apart from communities at later timepoints when teeth are present. Additionally, oral communities belonging to the teeth erupting timepoint are significantly different to those of 4 wk and 8 wk of age. Each sphere represents 1 sample from an individual mouse. ***P < 0.001 as determined by analysis of molecular variance (AMOVA) comparing all groups (P < 0.001 predentate vs. all other timepoints, and P < 0.001 teeth erupting vs. erupted 4 wk and vs. 8 wk). 95% confidence ellipses are also shown. (B) Changes of microbiome similarity for each mouse pup. Points indicate theta dissimilarity at each time point compared to predentate for each mouse. Lines connect points from same mouse. ****P < 0.0001, determined using Friedman Test and Dunn’s multiple comparisons test. (C) PCoA plot of microbial community structure (based on θYC distances), in which black arrows denote operational taxonomical units (OTUs) significantly correlated with principal coordinate axes, and hence, are major segregation drivers for these microbial communities. Depicted OTUs remained significant (P value < 0.001) upon correcting for multiple comparisons using the false discovery rate (FDR) correction. (D) Relative abundance plot showing main OTUs across communities at predentate, teeth erupting, erupted 4 wk, and erupted 8 wk timepoints. Set 1 and 2 refer to two independent mouse litters analyzed for these experiments. Species level taxonomy is reported in parentheses when >97% similarity was achieved. Each bar represents an individual mouse. Empty bars represent missing samples.
During tooth eruption, microbial communities become significantly different from the predentate stage (Fig. 1A). Streptococcus sp. (S. danieliae), becomes the most abundant OTU at this stage (Fig. 1C and D), consistent with a higher representation of taxa typically associated with the oral microbiome (Costello et al. 2009; Abusleme et al. 2017). During tooth eruption, microbial communities also appear most diverse compared to both pre- and posteruption communities (Appendix Fig. 1A).
Finally, after weaning and tooth eruption (weeks 4 and 8), oral communities become most dissimilar to that of the predentate stage (Fig. 1B). At this stage, microbiome communities are dominated by both Streptococcus sp. (S. danieliae) and Lactobacillus sp. (L. faecis) (Fig. 1C, D, and Appendix Fig. 1B). Interestingly, these data were replicated in 2 independent sets of parents/pups, suggesting that these phenomena are not particular to an experiment or cage.
Vertical Transmission from Parents Influences Oral Microbiome Acquisition
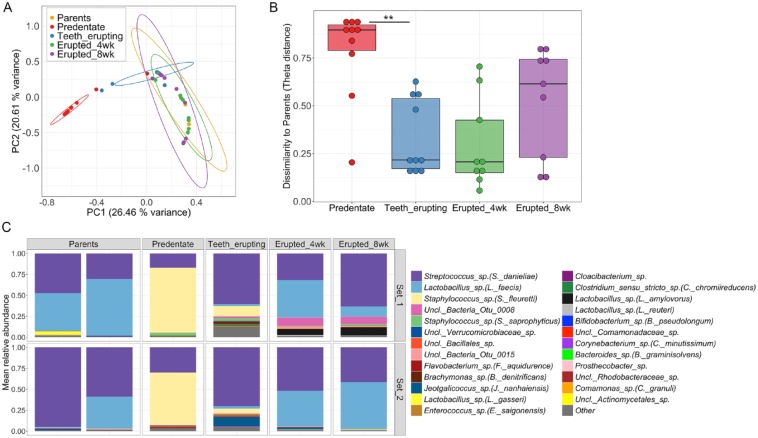
Next, we aimed to evaluate the influence of the parental oral microbiomes in the acquisition and establishment of offspring oral microbial communities. For these studies, longitudinal microbial samples from pups across timepoints were compared to that of their parents. Our data revealed that samples from pups at all dentate timepoints were clustered together with the microbial communities of their parents, indicating shared community structure (Fig. 2A). Indeed, only samples from pups at the predentate, and during tooth eruption stages separated significantly from the parent oral microbiome, based on community structure measures (Fig. 2A). To complement this analysis, we calculated theta-dissimilarity distances between parent samples and each timepoint for all mice. These data showed that predentate oral communities of pups are significantly more dissimilar than communities during tooth eruption are to that of parents (Fig. 2B). Taxonomical examination of parental communities revealed that Streptococcus sp. (S. danieliae) and Lactobacillus sp. (L. faecis) were the main OTUs dominating oral microbiome samples in both sets of parents (Fig. 2C). These were the same bacterial taxa that emerged as the most abundant in the pups’ oral microbiota at 4 and 8 wk, evidencing the influence of vertical transmission in the assembly of these communities, particularly after weaning of pups (after week 3).
Figure 2.
Vertical transmission of the oral microbiome. (A) Principal coordinates analysis (PCoA) plot of community structure, based on θYC distances, shows that oral communities of mouse pups at dentate timepoints cluster together with microbial samples from their parents (Analysis of Molecular Variance (AMOVA), P = 0.27 (ns), all pup timepoints vs. parents’ samples). Within pup timepoints, parents’ oral communities are significantly different to that of predentate and teeth-erupting samples (AMOVA, ***P < 0.001). 95% confidence ellipses are also depicted. (B) Theta dissimilarities of mouse pups’ microbial communities at each time point compared to that of parents. **P = 0.0097, determined using Friedman Test and Dunn’s multiple comparisons test. (C) Relative abundance plot depicting main OTUs across communities of parents’ samples and of pups’ across timepoints. Set 1 and 2 refer to 2 independent mouse litters analyzed for these experiments. Species level taxonomy is reported in parenthesis when >97% similarity was achieved. For parents’ oral communities, each bar represents an individual mouse (mother and father) and for pups’ oral communities each bar represents the average of all samples from offspring.
Stability of the Oral Microbiome with Age
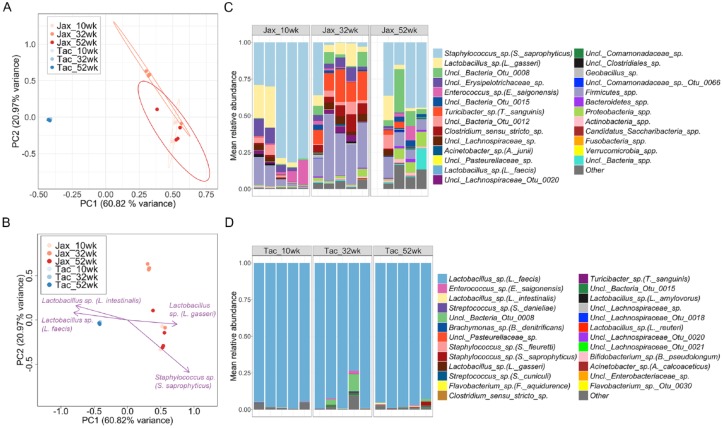
Next, we wanted to interrogate the stability of oral microbiome communities. For these experiments we purchased C57BL/6 (WT) mice from 2 different vendors known to harbor distinct commensal microbiota in their vivaria, Taconic Biosciences (Tac) and The Jackson Laboratory (Jax). In fact, mice from these vendors have been previously shown to exhibit distinct oral and gut microbial communities (Ivanov et al. 2009; Dutzan et al. 2017). We imported these mice and first confirmed that they did indeed harbor significantly different oral microbiome communities at 10 wk (Fig. 3A). We next investigated whether these mice with distinct microbiomes will retain their communities over time by longitudinal sampling of the oral cavity of Jax and Tac mice from week 10 and for up to a year (52 wk). We found that microbial communities from the 2 vendors remain significantly different between each other at all timepoints examined (Fig. 3A). Next, we investigated the main constituents of Jax and Tac oral communities driving their separation. Lactobacillus spp. (L. intestinalis and L. faecis) were found to guide the segregation of Tac samples, while another Lactobacillus sp. (L. gasseri) and Staphylococcus sp. (S. saprophyticus) were drivers for separation for Jax communities (Fig. 3B). Examination of the microbial composition of Jax communities revealed these were highly diverse, with Staphylococcus sp. (S. saprophyticus) being more prominent at 10 and 52 wk, followed by Lactobacillus sp. (L. gasseri) and other Firmicutes spp. (Fig. 3C). In contrast, oral communities from Tac mice were almost exclusively dominated by a Lactobacillus sp. (L. faecis) at all timepoints (Fig. 3D), displaying significantly decreased microbial diversity compared to Jax mice (Fig. 3C and D, Appendix Fig. 2). Microbial communities from Tac mice were also remarkably stable, exhibiting no differences in microbial structure over time (Appendix Fig. 3A and B). Microbial communities from Jax mice displayed a shift in community structure at 32 wk that was restored by 52 wk, becoming more similar to that of 10 wk (Appendix Fig. 3C and D). Our data indicate that Jax and Tac mice exhibit different oral microbiomes, whose imprint is largely preserved over time as they remain distinct upon aging.
Figure 3.
Stability of oral microbial communities with age. (A) Principal coordinates analysis (PCoA) plot of community structure, based on θYC distances, showing oral microbial communities of longitudinally sampled mice obtained from Taconic Biosciences (Tac) and The Jackson Laboratory (Jax). ***P < 0.001 as determined by Analysis of Molecular Variance (AMOVA), comparing Tac versus Jax samples at all timepoints. 95% confidence ellipses are also depicted. (B) PCoA plot of microbial community structure (based on θYC distances), in which purple arrows denote operational taxonomical units (OTUs) significantly correlated with principal coordinate axes, and hence, are major segregation drivers for these microbial communities. Depicted OTUs remained significant (P value < 0.001) upon correcting for multiple comparisons using the false discovery rate (FDR) correction. (C) Relative abundance plot showing main OTUs across Jax oral microbial communities at 10, 32, and 52 wk of age. Species level taxonomy is reported in parenthesis when >97% similarity was achieved. Each bar represents an individual mouse. Empty bars represent missing samples. (D) Relative abundance plot showing main OTUs across Tac oral microbial communities at 10, 32, and 52 wk of age. Species level taxonomy is reported in parenthesis when >97% similarity was achieved. Each bar represents an individual mouse.
Distinct Oral Microbiome Communities (from Tac and Jax Mice) Are Consistent with Oral Health
We next wanted to investigate whether distinct oral microbiome communities present in Tac and Jax mice are both health-associated communities, or one may render mice more susceptible to microbiome-triggered oral disease. Mice from both vendors had no overt signs of oral disease or issues with feeding and grooming throughout the observation period (1 y). We thus investigated whether mice from the 2 vendors may have distinct susceptibility to periodontitis. Mice are known to develop naturally occurring periodontal disease with age (Hajishengallis et al. 2011), which is exaggerated in the presence of genetic susceptibility (Eskan et al. 2012; Moutsopoulos et al. 2014).
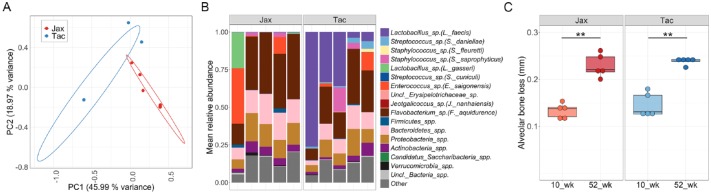
We first examined the gingival communities of Tac and Jax mice as these tooth-adherent microbiome communities trigger mucosal inflammation in periodontitis (Hajishengallis 2014; Moutsopoulos and Konkel 2018). We found that Jax and Tac gingival communities, were significantly separated from each other based on structure measurements and relative abundance of species detected (Fig. 4A–C). However, it is worth mentioning that microbial structure of these gingival communities was significantly different to that of the overall oral mucosal communities, highlighting the importance of selecting the appropriate type of sample when studying distinct oral niches (Appendix Fig. 4A and B). Jax and Tac mice were colonized by different gingival microbial communities, both groups exhibited very similar levels of periodontal bone loss at both early and late timepoints evaluated (10 and 52 wk of age) (Fig. 4C). These data indicate that despite significantly distinct microbiome communities, neither Jax nor Tac mice where particularly susceptible to periodontal bone loss, but rather developed naturally occurring bone loss consistent with their age. These findings suggest that very different oral microbiomes are compatible with oral health.
Figure 4.
Jax and Tac mice have significantly different microbiomes, but comparable susceptibility to periodontal bone loss. (A) Principal coordinates analysis (PCoA) plot of community structure, based on θYC distances, showing gingival microbial communities of Jax and Tac mice at 52 wk of age. ***P < 0.001 as determined by Analysis of Molecular Variance (AMOVA), comparing Tac versus Jax samples. Each sphere represents a sample from an individual mouse, some data points are not visible due to tight clustering. 95% confidence ellipses are also depicted. (B) Relative abundance plot showing main operational taxonomic units (OTUs) in Jax and Tac gingival communities at 52 wk of age. Species level taxonomy is reported in parenthesis when >97% similarity was achieved. Each bar represents an individual mouse. (C) Alveolar (periodontal) bone loss levels measured as the distances from the cemento-enamel junction to alveolar bone crest distances in the maxilla of 52 wk-old Jax and Tac mice. **P = 0.0079, as determined by Mann-Whitney test.
Horizontal Transfer of Health-Associated Oral Microbial Communities
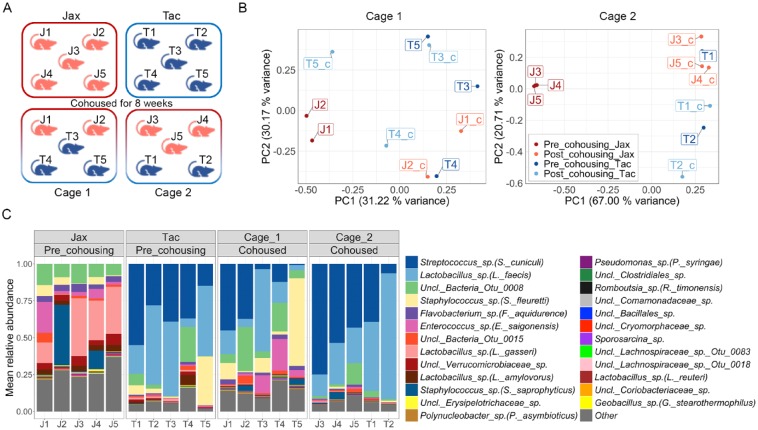
To investigate the potential for horizontal transfer of oral microbial communities through cohousing we obtained mice from Jax and Tac vendors and cohoused them for 8 wk, and characterized their microbiomes before and after cohousing (Experimental scheme, Fig. 5A). As shown previously, microbiomes from Jax and Tac were significantly separated (Fig. 5B). However, following 8 wk of cohousing almost all cohoused communities clustered together with the precohousing samples of Tac mice (Fig. 5B and Appendix Fig. 5A). Indeed, Tac samples were significantly more similar to their baseline communities after cohousing compared to Jax samples, in which communities had a virtually completely different microbial structure to that of their own baseline (Appendix Fig. 5B). Notably, microbial shifts upon cohousing seen in Tac and Jax mice are distinct, and not attributable to time-dependent variation, since microbial communities from sentinel cages (noncohoused) of both Tac and Jax mice did not change with time (Appendix Fig. 5C). Lastly, taxonomical analyses of these communities revealed that after cohousing, the majority samples from both cohoused cages were dominated by Lactobacillus sp. (L. faecis) and Streptococcus sp. (S. cuniculi), which were the most abundant taxa in the precohousing Tac communities (Fig. 5C). Hence, our data demonstrate horizontal transfer of the oral microbiome with cohousing. However, we find that communities from Tac mice displayed a greater capability of being horizontally transferred, as cohoused mice exhibited a microbial structure and taxonomical profile that resembles more that of Tac than Jax mice.
Figure 5.
Oral microbiome changes during cohousing of Jax and Tac mice. (A) Diagram representing the experimental scheme utilized to evaluate horizontal transfer of oral microbiome through cohousing of Tac and Jax mice for 8 wk (Jax mice indicated by label and red color, Tac mice by label and blue color). (B) Principal coordinates analysis (PCoA) plots of community structure (based on θYC distances), depicts microbial communities of Jax and Tac mice before and after 8 wk of cohousing per cage. Each sphere represents a sample from an individual mouse, labeled with their unique identifier (J1 to J5 for Jax mice and T1 to T5 for Tac mice). ***P < 0.001 as determined by Analysis of Molecular Variance (AMOVA), comparing pre- and postcohoused samples for Jax and Tac. Only significant comparison was postcohousing Jax versus postcohousing Tac, and postcohousing Tac versus precohousing Jax. (C) Relative abundance plot showing main operational taxonomic units (OTUs) in Jax and Tac oral communities before and after 8 wk of cohousing. Species level taxonomy is reported in parenthesis when >97% similarity was achieved. Each bar represents an individual mouse, labeled with their unique identifier (J1 to J5 for Jax mice and T1 to T5 for Tac mice).
Discussion
Our study reveals important principles relevant to the establishment of the oral microbiome in mice. We document that the initial communities observed in oral mucosa resemble skin-associated microbiome communities, suggesting initial transmission from maternal skin during feeding. Indeed, the most abundant taxa observed by day 7 to 9 in the oral mucosa are Staphylococcus spp., which are known skin commensals (Grice et al. 2009; Parlet et al. 2019). These findings are in agreement with earlier culture-based reports, indicating the early dominance of staphylococci followed by lactobacilli in BALB/c mice (Rodrigue et al. 1993). Coincidently, studies on the establishment of the GI tract microbiome also demonstrate initial and transient colonization of the gut with skin commensals, while eventual colonization with niche-specific microbes is more stable and will dictate eventual establishment of the community (Ferretti et al. 2018).
We find that vertical transmission from oral microbiome of parents becomes evident days after mice are weaned. In fact, oral microbiome communities from pups become similar to that of parents after tooth eruption and particularly after 4 wk of age. At this stage, coprophagic behavior of mice increases postweaning; that may be the route by which oral and gastrointestinal microbiota are shared between parents and pups. Regarding the ecological processes that govern the assembly of murine oral microbial communities, we hypothesize that homogeneous selection and homogenizing dispersal are likely to play an important role in shaping these communities due to the highly controlled conditions inherently associated with laboratory animals.
We document that tooth eruption is a key event which sets the stage for assembly of oral-associated communities, since after this event the microbiota dramatically changes. Communities become more diverse and oral-associated species (such as streptococci) become detected. We theorize that the presence of the tooth structure allows for the colonization of oral-related species and the initiation of the tooth-adherent biofilm, a complex multi-species formation adherent to the tooth surface (Valm et al. 2011; Mark Welch et al. 2016). Indeed, studies in humans investigating the establishment of the oral microbiome in children, clearly show that eruption of primary teeth is a “turning point” in the establishment of oral microbial communities which following this stage become considerably more rich and diverse (Dzidic, Abrahamsson, et al. 2018; Dzidic, Collado, et al. 2018; Mason et al. 2018).
We observe that following establishment of the oral microbiome, communities remain largely stable and retain key initial features over time. In fact, we imported mice with significantly different oral microbial communities from 2 different vendors (Taconic and Jackson) and found that they largely retain their distinct microbiota for up to 1 y.
Interestingly, despite dramatically different oral microbiota (both mucosa and tooth-associated) Tac and Jax mice were not increasingly susceptible to oral inflammatory disease (periodontitis), suggesting that very distinct microbiota can be associated with health in mice. These remarkably distinct communities we hypothesize are not due to genetic differences, as all were C57BL/6 mice, but theorize that are rather due to conditions during early establishment of communities in their respective facilities.
Finally, we document horizontal transmission of health-associated oral microbiome communities. Horizontal transmission of dysbiotic microbial communities has been recently reported (Xiao et al. 2017; Payne et al. 2019). Our findings now confirm that this can occur between health-associated microbiomes. However, transfer of communities was very biased, with mice from Jax altering their oral communities towards a “Tac” microbiome and Tac mice not significantly shifting their communities. Despite the fact that in our cohousing experiments mice harbored their own resident microbiota, Tac communities are distinctively transferred, similarly to what has been observed in cohousing of SPF and germ-free mice (Wu et al. 2018). Taconic communities are of very limited diversity and were more stable both through time and during cohousing. In contrast, we hypothesize that Jax oral microbial communities are more susceptible to environmental perturbations as they appear less stable than Tac communities. We speculate that the shift seen in Jax mice at 32 wk could be due to a transient colonization of gut-associated commensals (increased abundance of Turicibacter sp. and Clostridium senso stricto sp.), that is no longer evident by 52 wk. This reduced stability of Jax communities was also evident in the shift seen in Jax mice’s shift towards a Tac microbiome upon cohousing.
Collectively, our work reveals new information regarding the establishment and stability of oral microbiome communities in mice and provides insights which can inform experimentation in murine animal models.
Author Contributions
L. Abusleme, contributed to conception, design, data acquisition, analysis, and interpretation, drafted and critically revised the manuscript; H. O’Gorman, N. Dutzan, T. Greenwell-Wild, contributed to data acquisition, critically revised the manuscript; N.M. Moutsopoulos, contributed to conception, design, data analysis, and interpretation, drafted and critically revised the manuscript. All authors gave final approval and agree to be accountable for all aspects of the work.
Supplemental Material
Supplemental material, DS_10.1177_0022034520915485 for Establishment and Stability of the Murine Oral Microbiome by L. Abusleme, H. O’Gorman, N. Dutzan, T. Greenwell-Wild and N.M. Moutsopoulos in Journal of Dental Research
Acknowledgments
We also thank the NIDCR Veterinary Research Core, NCI microbiome sequencing core, W. Yuan, and V. Thovarai for assistance with 16S rRNA sequencing and data deposition.
Footnotes
A supplemental appendix to this article is available online.
This work was funded in part by the Intramural Program of the National Institute of Dental and Craniofacial Research (NIDCR)/National Institutes of Health (NIH) (to N.M.M.) and by the Chilean National Fund for Scientific and Technologic Development (FONDECYT), grants # 11180505 (to L.A.) and # 11180389 (to N.D.).
The authors declare no potential conflicts of interest with respect to the authorship and/or publication of this article.
ORCID iDs: L. Abusleme  https://orcid.org/0000-0002-7210-1236
https://orcid.org/0000-0002-7210-1236
N. Dutzan  https://orcid.org/0000-0001-8343-0214
https://orcid.org/0000-0001-8343-0214
References
- Abusleme L, Hong BY, Dupuy AK, Strausbaugh LD, Diaz PI. 2014. Influence of DNA extraction on oral microbial profiles obtained via 16s rRNA gene sequencing. J Oral Microbiol. 6. doi: 10.3402/jom.v6.23990. eCollection 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abusleme L, Hong BY, Hoare A, Konkel JE, Diaz PI, Moutsopoulos NM. 2017. Oral microbiome characterization in murine models. Bio Protoc. 7(24). pii: e2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constantinides MG, Link VM, Tamoutounour S, Wong AC, Perez-Chaparro PJ, Han SJ, Chen YE, Li K, Farhat S, Weckel A, et al. 2019. Mait cells are imprinted by the microbiota in early life and promote tissue repair. Science. 366(6464). pii: eaax6624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI, Knight R. 2009. Bacterial community variation in human body habitats across space and time. Science. 326(5960):1694–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutzan N, Abusleme L, Bridgeman H, Greenwell-Wild T, Zangerle-Murray T, Fife ME, Bouladoux N, Linley H, Brenchley L, Wemyss K, et al. 2017. On-going mechanical damage from mastication drives homeostatic th17 cell responses at the oral barrier. Immunity. 46(1):133–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzidic M, Abrahamsson TR, Artacho A, Collado MC, Mira A, Jenmalm MC. 2018. Oral microbiota maturation during the first 7 years of life in relation to allergy development. Allergy. 73(10):2000–2011. [DOI] [PubMed] [Google Scholar]
- Dzidic M, Collado MC, Abrahamsson T, Artacho A, Stensson M, Jenmalm MC, Mira A. 2018. Oral microbiome development during childhood: an ecological succession influenced by postnatal factors and associated with tooth decay. ISME J. 12(9):2292–2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eskan MA, Jotwani R, Abe T, Chmelar J, Lim JH, Liang S, Ciero PA, Krauss JL, Li F, Rauner M, et al. 2012. The leukocyte integrin antagonist Del-1 inhibits IL-17-mediated inflammatory bone loss. Nat Immunol. 13(5):465–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferretti P, Pasolli E, Tett A, Asnicar F, Gorfer V, Fedi S, Armanini F, Truong DT, Manara S, Zolfo M, et al. 2018. Mother-to-infant microbial transmission from different body sites shapes the developing infant gut microbiome. Cell Host Microbe. 24(1):133–145.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gensollen T, Blumberg RS. 2017. Correlation between early-life regulation of the immune system by microbiota and allergy development. J Allergy Clin Immunol. 139(4):1084–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grice EA, Kong HH, Conlan S, Deming CB, Davis J, Young AC, Program NCS, Bouffard GG, Blakesley RW, Murray PR, et al. 2009. Topographical and temporal diversity of the human skin microbiome. Science. 324(5931):1190–1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajishengallis G. 2014. Immunomicrobial pathogenesis of periodontitis: keystones, pathobionts, and host response. Trends Immunol. 35(1):3–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajishengallis G. 2015. Periodontitis: from microbial immune subversion to systemic inflammation. Nat Rev Immunol. 15(1):30–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajishengallis G, Liang S, Payne MA, Hashim A, Jotwani R, Eskan MA, McIntosh ML, Alsam A, Kirkwood KL, Lambris JD, et al. 2011. Low-abundance biofilm species orchestrates inflammatory periodontal disease through the commensal microbiota and complement. Cell Host Microbe. 10(5):497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison OJ, Linehan JL, Shih HY, Bouladoux N, Han SJ, Smelkinson M, Sen SK, Byrd AL, Enamorado M, Yao C, et al. 2019. Commensal-specific T cell plasticity promotes rapid tissue adaptation to injury. Science. 363(6422). pii: eaat6280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper LV, Littman DR, Macpherson AJ. 2012. Interactions between the microbiota and the immune system. Science. 336(6086):1268–1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, Wei D, Goldfarb KC, Santee CA, Lynch SV, et al. 2009. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 139(3):485–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD. 2013. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the miseq illumina sequencing platform. Appl Environ Microbiol. 79(17):5112–5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YK, Mazmanian SK. 2010. Has the microbiota played a critical role in the evolution of the adaptive immune system? Science. 330(6012):1768–1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy M, Kolodziejczyk AA, Thaiss CA, Elinav E. 2017. Dysbiosis and the immune system. Nat Rev Immunol. 17(4):219–232. [DOI] [PubMed] [Google Scholar]
- Mark Welch JL, Rossetti BJ, Rieken CW, Dewhirst FE, Borisy GG. 2016. Biogeography of a human oral microbiome at the micron scale. Proc Natl Acad Sci USA. 113(6):E791–E800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason MR, Chambers S, Dabdoub SM, Thikkurissy S, Kumar PS. 2018. Characterizing oral microbial communities across dentition states and colonization niches. Microbiome. 6(1):67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moutsopoulos NM, Konkel J, Sarmadi M, Eskan MA, Wild T, Dutzan N, Abusleme L, Zenobia C, Hosur KB, Abe T, et al. 2014. Defective neutrophil recruitment in leukocyte adhesion deficiency type I disease causes local IL-17-driven inflammatory bone loss. Sci Transl Med. 6(229):229ra240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moutsopoulos NM, Konkel JE. 2018. Tissue-specific immunity at the oral mucosal barrier. Trends Immunol. 39(4):276–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naik S, Bouladoux N, Wilhelm C, Molloy MJ, Salcedo R, Kastenmuller W, Deming C, Quinones M, Koo L, Conlan S, et al. 2012. Compartmentalized control of skin immunity by resident commensals. Science. 337(6098):1115–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parlet CP, Brown MM, Horswill AR. 2019. Commensal staphylococci influence staphylococcus aureus skin colonization and disease. Trends Microbiol. 27(6):497–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne MA, Hashim A, Alsam A, Joseph S, Aduse-Opoku J, Wade W, Curtis MA. 2019. Horizontal and vertical transfer of oral microbial dysbiosis and periodontal disease. J Dent Res. 98(13):1503–1510. [DOI] [PubMed] [Google Scholar]
- Rodrigue L, Barras MJ, Marcotte H, Lavoie MC. 1993. Bacterial colonization of the oral cavity of the BALB/c mouse. Microb Ecol. 26(3):267–275. [DOI] [PubMed] [Google Scholar]
- Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, et al. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 75(23):7537–7541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Leger AJ, Desai JV, Drummond RA, Kugadas A, Almaghrabi F, Silver P, Raychaudhuri K, Gadjeva M, Iwakura Y, Lionakis MS, et al. 2017. An ocular commensal protects against corneal infection by driving an interleukin-17 response from mucosal γδ T cells. Immunity. 47(1):148–158.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart CJ, Ajami NJ, O’Brien JL, Hutchinson DS, Smith DP, Wong MC, Ross MC, Lloyd RE, Doddapaneni H, Metcalf GA, et al. 2018. Temporal development of the gut microbiome in early childhood from the teddy study. Nature. 562(7728):583–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamburini S, Shen N, Wu HC, Clemente JC. 2016. The microbiome in early life: implications for health outcomes. Nat Med. 22(7):713–722. [DOI] [PubMed] [Google Scholar]
- Valm AM, Mark Welch JL, Rieken CW, Hasegawa Y, Sogin ML, Oldenbourg R, Dewhirst FE, Borisy GG. 2011. Systems-level analysis of microbial community organization through combinatorial labeling and spectral imaging. Proc Natl Acad Sci USA. 108(10):4152–4157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu YY, Westwater C, Xiao E, Dias Correa J, Xiao WM, Graves DT. 2018. Establishment of oral bacterial communities in germ-free mice and the influence of recipient age. Mol Oral Microbiol. 33(1):38–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao E, Mattos M, Vieira GHA, Chen S, Correa JD, Wu Y, Albiero ML, Bittinger K, Graves DT. 2017. Diabetes enhances IL-17 expression and alters the oral microbiome to increase its pathogenicity. Cell Host Microbe. 22(1):120–128 e124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue J, Clayton MK. 2005. A similarity measure based on species proportions. Commun Stat Theory Methods. 34(11):2123–2131. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, DS_10.1177_0022034520915485 for Establishment and Stability of the Murine Oral Microbiome by L. Abusleme, H. O’Gorman, N. Dutzan, T. Greenwell-Wild and N.M. Moutsopoulos in Journal of Dental Research