Abstract
Exposure to fine particulate matter (PM2.5), of which secondary organic aerosol (SOA) is a major constituent, is linked to adverse health outcomes, including cardiovascular disease, lung cancer, and preterm birth. Atmospheric oxidation of isoprene, the most abundant nonmethane hydrocarbon emitted into Earth’s atmosphere primarily from vegetation, contributes to SOA formation. Isoprene-derived SOA has previously been found to alter inflammatory/oxidative stress genes. MicroRNAs (miRNAs) are epigenetic regulators that serve as post-transcriptional modifiers and key mediators of gene expression. To assess whether isoprene-derived SOA alters miRNA expression, BEAS-2B lung cells were exposed to laboratory-generated isoprene-derived SOA constituents derived from the acid-driven multiphase chemistry of authentic methacrylic acid epoxide (MAE) or isomeric isoprene epoxydiols (IEPOX) with acidic sulfate aerosol particles. These IEPOX- and MAE-derived SOA constituents have been shown to be measured in large quantities within PM25 collected from isoprene-rich areas affected by acidic sulfate aerosol particles derived from human activities. A total of 29 miRNAs were identified as differentially expressed when exposed to IEPOX-derived SOA and 2 when exposed to MAE-derived SOA, a number of which are inflammatory/oxidative stress associated. These results suggest that miRNAs may modulate the inflammatory/oxidative stress response to SOA exposure, thereby advancing the understanding of airway cell epigenetic response to SOA.
Graphical Abstract
1. INTRODUCTION
Exposure to atmospheric fine particulate matter (PM2.5, aerosol with aerodynamic diameter <2.5 μm) is associated with significant adverse health outcomes such as cardiovascular disease,1–3 respiratory system damage and lung cancer4–11 as well as preterm birth.12–15 PM25 is a heterogeneous mixture of organic and inorganic solid and/or liquid particles suspended in air.15,16 A dominant component of PM2.5 is secondary organic aerosol (SOA), the majority of which is formed through gas-phase photooxidation (i.e., hydroxyl radical (OH)-initiated oxidation) of isoprene (2-methyl-1,3-butadiene).16–19 Isoprene, released primarily from broad-leaf trees, is the most abundant nonmethane hydrocarbon emitted into Earth’s atmosphere (~600 Tg year−1).20 The interaction of gas-phase isoprene-derived oxidation products with anthropogenic pollutants, in particular acidic sulfate aerosol derived from combustion sources, produces isoprene-derived SOA.17 Thus, the composition of isoprene-derived SOA is strongly influenced by controllable anthropogenic emissions.21 Intermediates in isoprene-derived SOA formation, including isomeric isoprene epoxydiols (IEPOX), isoprene hydroxyhy-droperoxide (ISOPOOH), and methacrylic acid epoxide (MAE), are critical gas-phase SOA precursors.21
PM2.5-induced oxidative stress plays a key role in respiratory system damage caused by air pollution.22,23 Oxidative stress, an oxidant/antioxidant imbalance in favor of oxidants, is a key driver of injury and inflammatory response in respiratory diseases, including asthma and chronic obstructive pulmonary disease.24,25 Given its size, PM2.5 can completely penetrate the lungs, resulting in direct exposure of lung cells.22 Our previous work has shown that isoprene-derived SOA induced the expression of oxidative stress and inflammation genes in human lung cells.16,21,26,27 Of the SOA components tested in these previous studies, MAE-derived SOA was identified as being a more potent inducer of inflammatory and oxidative stress gene expression response than IEPOX-derived SOA or ISOPOOH-derived SOA.21 While the gene activation studies are important in providing insight into the cellular response to isoprene-derived SOA, other aspects of the regulatory pathway remain to be elucidated’ namely post-transcriptional regulation of mRNA.
MicroRNAs (miRNAs) are noncoding single-stranded RNA molecules approximately 22 nucleotides in length.28 miRNAs serve as post-transcriptional modifiers and key mediators of gene expression. Thus, they act as epigenetic regulators of genomic response to environmental exposures. In general, miRNAs interact with complementary regions of target mRNAs to induce mRNA degradation and repress translation.29 In some cases, however, miRNAs have been shown to activate translation.29
We hypothesized that exposure to isoprene-derived SOA would alter the expression of miRNAs that modulate genes previously found to be responsive to isoprene-derived SOA exposure. To test this hypothesis, we exposed BEAS-2B cells to both MAE- and IEPOX-derived SOA and evaluated sub-sequent miRNA expression relative to a control exposure. These results provide molecular level evidence linking isoprene-derived SOA exposure and observed system level respiratory outcomes.
2. EXPERIMENTAL METHODS
Synthesis of SOA Precursors.
Experimental procedures for the synthesis and characterization of SOA precursors trans-β-IEPOX and MAE and extraction of SOA constituents have been published previously.16,30,31 Identity and purity (>99%) of the precursors was confirmed by 1H and 13C nuclear magnetic resonance (NMR) as well as by gas chromatography/electron ionization mass spectrometry (GC/EI-MS) analysis with prior trimethylsilylation (TMS) or ultraperformance liquid chromatography coupled to electrospray ionization high-resolution quadrupole time-of flight mass spectrometry (UPLC/ESI-HR-QTOFMS).30,31
Generation and Chemical Characterization of IEPOX- and MAE-Derived SOA.
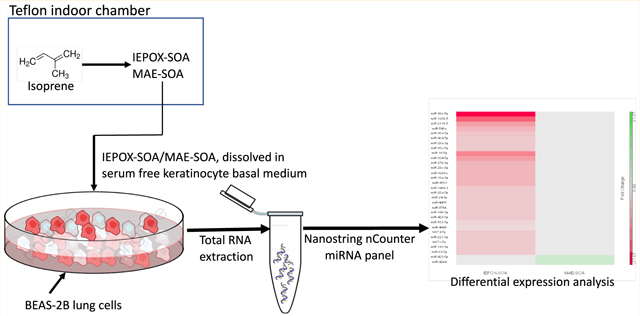
To generate SOA, authentic trans-β-IEPOX or MAE was injected into a 10-m3 flexible Teflon indoor chamber in the presence of acidic sulfate aerosol. This was conducted under dark conditions; therefore, radicals and peroxides were not expected to be present. Detailed operating procedures for this chamber facility for the purpose of in vitro exposures have been described previously.16,30,32 Filter extraction procedures have been described elsewhere.32 In brief, efficiency of removal of IEPOX- and MAE-derived SOA from filters was estimated to be over 90%. Filter samples were chemically characterized by GC/EI-MS and UPLC/ESI-HR-QTOFMS as described previously.16 Detailed sample preparation, column conditions, and operating parameters for GC/EI-MS and UPLC/ESI-HR-QTOFMS have been published elsewhere.33,34
Cell Culture.
BEAS-2B cells were cultured in keratinocyte growth medium (KGM) (KGM BulletKit, Lonza), which contains serum-free KBM, bovine pituitary extract, human epidermal growth factor, insulin, hydrocortisone, and GA-1000 (gentamincin, amphotericin B) and grown at 37 °C and 5% CO2 in a humidified incubator. A serum-free medium was chosen to maximize comparability to other studies evaluating in vitro exposure to isoprene-derived SOA, to follow ACCT recommendations, and because studies have noted that serum-exposed BEAS-2B cells are not an ideal model of the epithelial phenotype.16,21,26,35,36
Extraction of SOA Constituents for Cell Exposure.
Sonication in high-purity methanol was used to extract the Teflon filter membranes. Several filter extracts were combined to achieve the desired dose level, then dried under a gentle stream of nitrogen and redissolved in growth factor-deprived serum-free keratinocyte basal medium (KBM). The acidified sulfate aerosol-only control filters were processed in the same way. SOA constituents are known to be highly methanol and water-soluble, and thus, the SOA constituents can be assumed to have been fully dissolved in the cell medium.37
Cell Exposure.
For the exposure, cells were seeded in 24-well plates at a density of 2.5 × 104 cells/well in 250 μL of KGM 2 days before exposure. At 60–70% confluence, cells were washed twice with the phosphate-buffered saline (PBS) and then exposed for 24 h to KBM medium containing 0.01, 0.1, and 1 mg/mL SOA. This procedure was repeated for extracts of the acidic sulfate aerosol control filters. Experiments were conducted in triplicate per treatment group.
Assessment of Cytotoxicity.
To ensure that toxicity of exposure levels would not affect gene expression, toxicity at each exposure level was assayed using a lactate dehydrogenase (LDH) cytotoxicity detection kit (Takara Bio, Mountain View, California) according to the manufacturer’s protocol. After being exposed for 24 h, the supernatants were collected to assess LDH levels. Cells exposed to filter extracts from acidified sulfate aerosol-only experiments and cells maintained in KBM alone were treated as control groups.
miRNA Extraction.
Cells were lysed with 350 μL of Trizol Reagent (Life Technologies, Carlsbad, California) after 24 hours for total RNA isolation.16 Spin column-based Direct-zol RNA MiniPrep kit (Zymo Research, Irvine, California) was used to purify the RNA samples. RNA quantity was determined with Nanodrop (Thermo Scientific, Wilmington, Delaware) and quality with Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, California). Total RNA was hybridized to the nCounter Human v3 miRNA panel comprising 798 human miRNA targets and 5 housekeeping genes (NanoString Technologies, Inc., Seattle, Washington). The Nano-String hybridization was performed by the Translational Genomics Laboratory at the UNC at Chapel Hill Lineberger Comprehensive Cancer Center.
Statistical Analysis.
Raw expression data were extracted using the nSolver Analysis Software 4.0 (NanoString Technologies, Inc., Seattle, Washington) and normalized both to positive controls and housekeeping genes as recommended by the manufacturer.38 miRNA expression changes in cells exposed to SOA constituents were compared to changes in cells exposed to the extracts from acidic sulfate aerosol controls to assess the effects induced solely by the extracted SOA constituents using log fold changes calculated via an ANOVA model. Log fold changes are defined as the value of the geometric mean of the normalized miRNA expression in the cells exposed to 0.1 mg/mL IEPOX- or MAE-derived SOA, divided by the geometric mean of the normalized miRNA expression in the cells exposed to 0.1 mg/mL acidified sulfate aerosol. This was conducted in Partek Genomics Suite software (Partek Inc., St. Louis, MO, United States) and significant differential expression was defined as a log fold change with an associated false discovery rate adjusted q < 0.05. Ingenuity Pathway Analysis (IPA) software miRNA Target Filter tool (Ingenuity Systems, Inc., Redwood City, CA, United States) was used to identify two comprehensive lists of computationally predicted gene targets of the miRNAs differentially expressed in response to IEPOX- and MAE-derived SOA, respectively. Expression pairs were defined as miRNA-mRNA pairs in which the miRNA’s computationally predicted gene targets were also found to be differentially expressed in gene expression studies of isoprene-SOA exposure.16,21 To validate the predicted gene target, we queried the European Bioinformatics Institute’s Expression Atlas’ “RNA-seq analysis of 2 cell line models of lung disease (A549 and BEAS-2B) and primary bronchial epithelial cells in Project 3 of Open Targets 020”, which contained mRNA baseline data.39 Additionally, network analysis to identify enriched biological pathways associated with the miRNAs differentially expressed was conducted using IPA software (Ingenuity Systems, Inc., Redwood City, CA, United States). Cellular networks representing perturbed pathways were identified through enrichment analysis performed using the Fisher’s Exact test as detailed previously40. Over-represented diseases and/or biological functions were defined as those associated with more gene product targets than expected by chance using a p-value <0.01.
3. RESULTS
Exposure Characterization.
Detailed exposure conditions have been described elsewhere, including description of aerosol chemical composition analysis.16 SOA mass yields from the reactive uptake of trans-β-IEPOX onto acidified sulfate aerosol are substantially larger than those from reactive uptake of MAE under the same experimental conditions (<10% RH) and time scale (2 hour reaction time), which is consistent with ambient measures from the southeastern United States.41 Additionally, the SOA constituents found in the filter extracts of IEPOX- and MAE-derived SOA were also observed in ambient PM2.5 samples collected from isoprene-rich regions like the southeastern United States, which further supports the validity of the chamber experiments as representative of ambient SOA composition.16,27,41 The amount of SOA material the cells were exposed to was also relevant to a human ambient acute exposure. Ambient isoprene-derived SOA mass concentrations can range from 500 ng/m3 to 10–16 μg/m3.42,43 If a worst-case exposure scenario of 16 μg/m3 for 8 hours is assumed, with an inhalation rate of 6 L/min (i.e., 12 breaths per minute and the tidal volume ~0.5 L), then ambient exposure for 8 hours is 16 μg/m3 X 6 L/min X60 min/h X8 h X 0.001 m3/L = 46.08 μg = 0.046 mg.
Cytotoxicity Measurements.
Cells exposed to a level of 0.1 mg/mL were selected for miRNA expression analysis as this dose was determined to be noncytotoxic. Significant cell death was observed in acidified sulfate aerosol-only controls at a concentration of 1 mg/mL (cell death, ~27%; p = 0.02), while acidified sulfate aerosol-only controls at concentrations of ≤0.1 mg/mL were not cytotoxic (cell death, ≤10%; p > 0.05).
Altered Expression of miRNAs Induced by Exposure to Isoprene-Derived SOA.
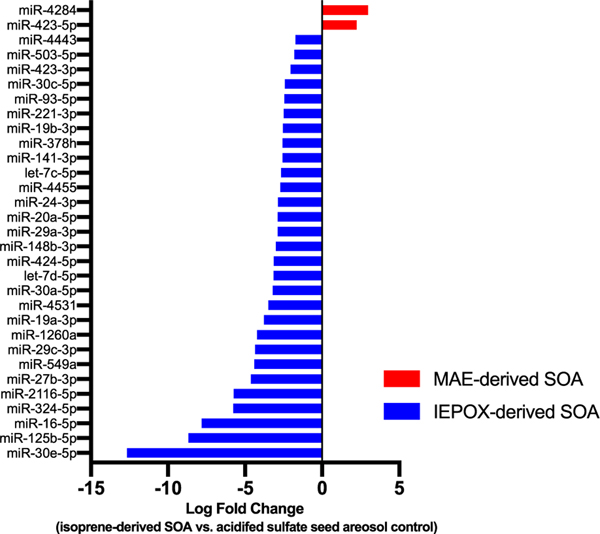
A total of 29 miRNAs were identified that displayed differential expression when exposed to IEPOX-derived SOA (Table 1). Two miRNAs were identified that were differentially expressed when exposed to MAE-derived SOA (Table 1). IEPOX-derived SOA exhibited a more robust response in absolute magnitude of expression log fold changes and number of miRNAs differentially expressed (Figure 1). All miRNAs differentially expressed in response to IEPOX-derived SOA were down-regulated (negative log fold change), while the two miRNAs differentially expressed in response to MAE-derived SOA were up-regulated (positive log fold change). miRNAs differentially expressed in response to IEPOX-derived SOA exhibited greater absolute log fold changes than those in response to MAE-derived SOA. The largest log fold change in response to MAE-derived SOA was +2.99, versus −12.67 in response to IEPOX-derived SOA.
Table 1.
Thirty-One miRNAs Identified with Significant Differential Expression Associated with Exposure to Either IEPOX-Derived SOA or MAE-Derived SOAa
| SOA type | miRNA | fold change | FDR-adjusted q-value | p-value | chromosome |
|---|---|---|---|---|---|
| IEPOX-derived | miR-30e-5p | −12.67 | 0.04 | 0.004 | 1 |
| miR-125b-5p | −8.69 | 0.04 | 0.003 | 11 | |
| miR-2116-5p | −5.73 | 0.04 | 0.005 | 15 | |
| miR-549a | −4.41 | 0.04 | 0.007 | 15 | |
| miR-30a-5p | −3.2 | 0.04 | 0.007 | 6 | |
| miR-424-5p | −3.14 | 0.04 | 0.002 | X | |
| miR-29a-3p | −2.9 | 0.04 | 0.006 | 7 | |
| miR-30c-5p | −2.42 | 0.04 | 0.005 | 6 | |
| miR-16-5p | −7.82 | 0.041 | 0.021 | 13 | |
| miR-324-5p | −5.78 | 0.041 | 0.021 | 17 | |
| miR-27b-3p | −4.64 | 0.041 | 0.011 | 9 | |
| miR-29c-3p | −4.35 | 0.041 | 0.013 | 1 | |
| miR-1260a | −4.24 | 0.041 | 0.013 | 14 | |
| miR-19a-3p | −3.78 | 0.041 | 0.019 | 13 | |
| miR-4531 | −3.49 | 0.041 | 0.016 | 19 | |
| miR-148b-3p | −3.02 | 0.041 | 0.021 | 12 | |
| miR-20a-5p | −2.9 | 0.041 | 0.02 | 13 | |
| miR-24-3p | −2.86 | 0.041 | 0.022 | 9 | |
| miR-4455 | −2.73 | 0.041 | 0.022 | 4 | |
| miR-378h | −2.58 | 0.041 | 0.011 | 5 | |
| miR-19b-3p | −2.55 | 0.041 | 0.012 | 13 | |
| miR-423-3p | −2.05 | 0.041 | 0.018 | 17 | |
| miR-503-5p | −1.81 | 0.041 | 0.014 | X | |
| miR-4443 | −1.73 | 0.041 | 0.022 | 3 | |
| let-7d-5p | −3.16 | 0.043 | 0.026 | 9 | |
| miR-221-3p | −2.5 | 0.043 | 0.026 | X | |
| let-7c-5p | −2.67 | 0.049 | 0.032 | 21 | |
| miR-141-3p | −2.58 | 0.049 | 0.033 | 12 | |
| miR-93-5p | −2.47 | 0.049 | 0.033 | 7 | |
| MAE-derived | miR-423-5p | 2.24 | 0.03 | 0.0004 | 17 |
| miR-4284 | 2.99 | 0.03 | 0.0009 | 7 |
Significance defined as q < 0.05.
Figure 1.
Log fold changes of miRNA expression when exposed to IEPOX- or MAE-derived SOA compared to acidified sulfate aerosol control.
Network Analysis and miRNA-mRNA Expression Pairing.
There was substantial overlap in the computationally predicted gene targets of miRNAs that were differentially expressed in the current study and genes that we have previously found to be differentially expressed in response to isoprene-derived SOA exposure.16,21 For IEPOX-derived SOA, 52 gene target-miRNA expression pairings were identified (Table S1). Forty-five of these pairs showed an inverse expression pattern, suggestive of miRNA repression of mRNA levels. For MAE-derived SOA, 7 gene target-miRNA expression pairings were identified, 6 of which involved miR-423–5p (Table S2). For these expression pairs responsive to MAE-derived SOA exposure, both miRNA and mRNA demonstrated increased expression in response to exposure, counter to the expected pattern. Of the 59 total miRNA–mRNA expression pairs identified, 55 of the mRNA genes are known to be expressed in BEAS-2B cells according to baseline RNA-sequencing, thus providing confidence in the computationally predicted parings (Tables S1 and S2).
Network analysis revealed that of the 59 total miRNA–mRNA pairings identified as responsive to IEPOX-derived SOA, 33 of the pairings had NRF2-mediated oxidative stress response as one of their associated pathways. Given that there were only 2 miRNAs responsive to MAE-derived SOA, network analysis was not performed. However, the 29 miRNAs differentially expressed when exposed to IEPOX-derived SOA were analyzed for enrichment within biological pathways and networks. Two biological networks were highly enriched. Network 1 (Supplementary Figure S1) is associated with inflammatory disease, inflammatory response, organismal injuries and abnormalities (p-value = 10−34). Network 2 (Supplementary Figure S2) is associated with cancer, organismal injury and abnormalities, and reproductive system disease (p-value = 10−10).
4. DISCUSSION
Our previous work found that isoprene-derived SOA induced the expression of oxidative response and inflammatory genes.16,21,26,27 Here, we demonstrate that inflammatory and oxidative stress associated miRNAs are also differentially expressed in response to isoprene-derived SOA. There were three major findings from this study: first, miRNAs are likely involved in the modulation of the genomic response to isoprene-derived SOA; second, marked differences exist between the miRNA response to IEPOX-derived SOA and MAE-derived SOA; third, miRNAs may specifically modulate inflammatory response to IEPOX-derived SOA exposure.
The present study suggests that miRNAs are functioning as genomic regulators in relation to SOA in lung cells. There was substantial overlap in the gene targets predicted from the miRNAs differentially expressed in the present study and genes previously identified as differentially expressed in response to isoprene-derived SOA. Of the 52 miRNA–gene target parings identified in response to IEPOX-derived SOA, 45 of them showed the expected inverse relationship (i.e., miRNA reduced expression, mRNA increased expression) suggestive of miRNA suppression of mRNA expression. On the other hand, all pairings identified in response to MAE-derived SOA demonstrated an upregulation of both miRNA and mRNA, which is not indicative of miRNA suppression. Positive correlation between upregulated miRNA levels and gene expression may be explained by the action of the miRNA blocking a suppressor transcription factor or by an indirect pathway, as has been observed with other miRNAs.44,45 Therefore, the current study suggests that miRNAs are involved in the genomic response to SOA and perhaps more extensively involved in the epigenetic regulation of the response to IEPOX-derived SOA than the response to MAE-derived SOA.
The striking differences in miRNA response between the IEPOX- and MAE-derived SOA is consistent with previous studies that show variations in gene expression response based on the specific SOA; however, the exact interplay between miRNAs and gene expression in this context remains to be elucidated fully. Previous studies have shown that MAE-derived SOA induces a gene expression response more robust than that of IEPOX-derived SOA.16,21 The present study finds that the miRNA response is more robust in response to IEPOX-derived SOA. In fact, while previous studies have shown specifically inflammatory related genes to be upregulated in response to MAE-derived SOA more than IEPOX-derived SOA, here we find that inflammatory-related miRNAs are downregulated in response to IEPOX-derived SOA, which would lead to upregulation of their respective gene targets. Thus, it is likely that other epigenetic mechanisms are also at play in the control of genomic response to isoprene-derived SOA.
IEPOX-derived SOA responsive miRNAs were found to be associated with inflammatory function and disease. Two biological networks with inflammatory and cancer functions were identified in network analysis of IEPOX-derived SOA responsive miRNAs. The first of these contained insulin as a central node. Increased exposure to air pollution has been linked to insulin resistance in meta-analyses of epidemiologic evidence46 as well as in experimental studies.47 Additionally, insulin resistance is a known risk factor of asthma.48 The second network contained TP53, which represents a potential newly identified pathway not previously found in our gene activation studies of isoprene-derived SOA exposure.16,21,27 TP53 encodes the tumor suppressor p53, which is thought to mediate PM-induced epithelial cell mitochondria-regulated apoptosis, which is relevant to the pathogenesis of lung cancer.49 Moreover, NRF2-mediated oxidative stress response was an associated pathway of 33 of the target gene-miRNA pairings responsive to IEPOX-derived SOA NRF2, a transcription factor that activates various antioxidant and detoxification enzymes to reduce reactive oxygen species, has been linked in many studies to PM25 exposure.50–53 Notably, NRF2-mediated oxidative stress response was shown to be enriched in the gene sets previously found to be responsive to IEPOX- and MAE-derived SOA16 as well as total isoprene-derived SOA.27 Hence, the networks identified here provide further evidence that the identified miRNAs play a role in IEPOX-derived SOA inflammatory/oxidative stress related epigenetic changes and resultant health outcomes. This is of particular importance to public health because IEPOX-derived SOA has been shown to be the largest contributor to global isoprene-derived SOA, which is up to 30–40% of the total organic material in PM2.5.54–56
While this study provides novel information regarding cellular response to isoprene-derived SOA, it is not without limitations. The use of an immortalized cell line as well as the limitation of delivering PM to submerged cells should be noted. To address these limitations, future studies could explore PM deposition using in vitro air–liquid interface technologies or utilize in vivo inhalation designs to investigate transcriptional and epigenetic changes induced by isoprene-derived SOA exposure. Additionally, future studies are warranted to assess whether specific miRNAs or a set of miRNAs identified here can act as biomarkers of exposure to isoprene-derived SOA and to evaluate the role of other epigenetic machinery in the genomic response to isoprene-derived SOA.
In summary, in areas such as the southeastern United States, where isoprene emissions are high, understanding the specific cellular responses to isoprene-derived SOA is crucial to public health.42,43,54,57 This study provides evidence that miRNAs play a role in isoprene-derived SOA-induced morbidity and do so in a manner that is specific to chemical composition of the SOA.
Supplementary Material
Table S1: IEPOX-responsive miRNA–mRNA pairings from the target filter analysis
Table S2: MAE-responsive miRNA–mRNA pairings from the target filter analysis (XLSX)
Acknowledgments
Funding
Research described in this article was conducted (in part) under contract to the Health Effects Institute (HEI), an organization jointly funded by the United States Environmental Protection Agency (EPA) (Assistance Award R-82811201) and certain motor vehicle and engine manufacturers. The contents of this article do not necessarily reflect the views of HEI or its sponsors, nor do they necessarily reflect the views and policies of the EPA or the motor vehicle and engine manufacturers. The research was also supported in part by a grant from the National Institute of Environmental Health Sciences (T32ES007018). This research was additionally supported in part by an National Institutes of Health Center Core grant (P30 ES010126).
ABBREVIATIONS
- SOA
secondary organic aerosol
- MAE
methacrylic acid epoxide
- IEPOX
isomeric isoprene epoxydiols
- miRNA
micro-RNA
- PM2.5
fine particulate matter (aerosol with aerodynamic diameter <2.5 μm)
- KBM
keratinocyte basal medium
- KGM
keratinocyte growth medium
- LDH
lactate dehydrogenase
- PBS
phosphate-buffered saline
Footnotes
The authors declare no competing financial interest.
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.chemrestox.9b00322.
Supplemental figures depicting molecular networks (PDF)
REFERENCES
- (1).Du Y, Xu X, Chu M, Guo Y, and Wang J (2016) Air particulate matter and cardiovascular disease: the epidemiological, biomedical and clinical evidence. J. Thorac Dis. 8 (1), E8–E19 PubMed PMID: 26904258.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Kim H, Kim J, Kim S, Kang S-H, Kim H-J, Kim H, Heo J, Yi S-M, Kim K, Youn T-J, and Chae I-H. Cardiovascular Effects of Long-Term Exposure to Air Pollution: A Population-Based Study With 900 845 Person-Years of Follow-up. J. Am. Heart Assoc. 2017; 6 (11). DOI: 10.1161/JAHA.117.007170. PubMed PMID: 29118034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Dominici F, Peng RD, Bell ML, Pham L, McDermott A, Zeger SL, and Samet JM (2006) Fine Particulate Air Pollution and Hospital Admission for Cardiovascular and Respiratory Diseases. JAMA 295 (10), 1127–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Pope CA, Burnett RT, Thun MJ, Calle EE, Krewski D, Ito K, and Thurston GD (2002) Lung Cancer, Cardiopulmonary Mortality, and Long-term Exposure to Fine Particulate Air Pollution. JAMA 287 (9), 1132–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Turner MC, Krewski D, Pope CA, Chen Y, and Gapstur SM (2011) Long-term Ambient Fine Particulate Matter Air Pollution and Lung Cancer in a Large Cohort of Never-Smokers. Am. J. Respir. Crit. Care Med. 184 (12), 1374–81. [DOI] [PubMed] [Google Scholar]
- (6).Raaschou-Nielsen O, Andersen ZJ, Beelen R, Samoli E, Stafoggia M, Weinmayr G, Hoffmann B, Fischer P, Nieuwenhuijsen MJ, Brunekreef B, Xun WW, Katsouyanni K, Dimakopoulou K, Sommar J, Forsberg B, Modig L, Oudin A, Oftedal B, Schwarze PE, Nafstad P, De Faire U, Pedersen NL, Östenson C-G, Fratiglioni L, Penell J, Korek M, Pershagen G, Eriksen KT, Sørensen M, Tjønneland A, Ellermann T, Eeftens M, Peeters PH, Meliefste K, Wang M, Bueno-de-Mesquita B, Key TJ, de Hoogh K, Concin H, Nagel G, Vilier A, Grioni S, Krogh V, Tsai M-Y, Ricceri F, Sacerdote C, Galassi C, Migliore E, Ranzi A, Cesaroni G, Badaloni C, Forastiere F, Tamayo I, Amiano P, Dorronsoro M, Trichopoulou A, Bamia C, Vineis P, and Hoek G (2013) Air pollution and lung cancer incidence in 17 European cohorts: prospective analyses from the European Study of Cohorts for Air Pollution Effects (ESCAPE). Lancet Oncol. 14, 813–22. [DOI] [PubMed] [Google Scholar]
- (7).Katanoda K, Sobue T, Satoh H, Tajima K, Suzuki T, Nakatsuka H, Takezaki T, Nakayama T, Nitta H, Tanabe K, and Tominaga S (2011) An association between long-term exposure to ambient air pollution and mortality from lung cancer and respiratory diseases in Japan. Journal of epidemiology. 21 (2), 132–43 Epub 2011/02/18. PubMed PMID: 21325732; PMCID: PMC3899505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Nafstad P, Haheim L, Oftedal B, Gram F, Holme I, Hjermann I, and Leren P (2003) Lung cancer and air pollution: a 27 year follow up of 16 209 Norwegian men. Thorax 58, 1071–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Hystad P, Demers PA, Johnson KC, Carpiano RM, and Brauer M (2013) Long-term residential exposure to air pollution and lung cancer risk. Epidemiology 24 (5), 762–72 Epub 2013/05/17. PubMed PMID: 23676262. [DOI] [PubMed] [Google Scholar]
- (10).Gotschi T, Heinrich J, Sunyer J, and Kunzli N (2008) Long-term effects of ambient air pollution on lung function: a review. Epidemiology. 19 (5), 690–701 Epub 2008/08/16. PubMed PMID: 18703932. [DOI] [PubMed] [Google Scholar]
- (11).Huang F, Pan B, Wu J, Chen E, and Chen L (2017) Relationship between exposure to PM2.5 and lung cancer incidence and mortality: A meta-analysis. Oncotarget. 8 (26), 43322–31 PubMed PMID: 28487493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Guo T, Wang Y, Zhang H, Zhang Y, Zhao J, Wang Q, Shen H, Xie X, Wang L, Xu Z, Yan D, He Y, Yang Y, Xu J, Peng Z, and Ma X (2018) The association between ambient PM2.5 exposure and the risk of preterm birth in China: A retrospective cohort study. Sci. Total Environ. 633, 1453–9 Epub 2018/05/16. PubMed PMID: 29758897. [DOI] [PubMed] [Google Scholar]
- (13).Trasande L, Malecha P, and Attina TM (2016) Particulate Matter Exposure and Preterm Birth: Estimates of U.S. Attributable Burden and Economic Costs. Environ. Health Perspect. 124 (12), 1913–8 ehp.1510810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Malley CS, Kuylenstierna JC, Vallack HW, Henze DK, Blencowe H, and Ashmore MR (2017) Preterm birth associated with maternal fine particulate matter exposure: A global, regional and national assessment. Environ. Int. 101, 173–82 Epub 2017/02/16. PubMed PMID: 28196630. [DOI] [PubMed] [Google Scholar]
- (15).Kim K-H, Kabir E, and Kabir S (2015) A review on the human health impact of airborne particulate matter. Environ. Int. 74, 136–43. [DOI] [PubMed] [Google Scholar]
- (16).Lin Y-H, Arashiro M, Martin E, Chen Y, Zhang Z, Sexton KG, Gold A, Jaspers I, Fry RC, and Surratt JD (2016) Isoprene-Derived Secondary Organic Aerosol Induces the Expression of Oxidative Stress Response Genes in Human Lung Cells. Environ. Sci. Technol. Lett. 3 (6), 250–4. [Google Scholar]
- (17).Carlton AG, Wiedinmyer C, and Kroll JH (2009) A review of Secondary Organic Aerosol (SOA) formation from isoprene. Atmos. Chem. Phys. 9 (14), 4987–5005. [Google Scholar]
- (18).Xu L, Guo H, Boyd CM, Klein M, Bougiatioti A, Cerully KM, Hite JR, Isaacman-VanWertz G, Kreisberg NM, Knote C, Olson K, Koss A, Goldstein AH, Hering SV, Gouw Jd, Baumann K, Lee S-H, Nenes A, Weber RJ, and Ng NL (2015) Effects of anthropogenic emissions on aerosol formation from isoprene and monoterpenes in the southeastern United States. Proc. Natl. Acad. Sci. U.S.A.112 (1), 37–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Li J, Wang G, Wu C, Cao C, Ren Y, Wang J, Li J, Cao J, Zeng L, and Zhu T (2018) Characterization of isoprene-derived secondary organic aerosols at a rural site in North China Plain with implications for anthropogenic pollution effects. Sci. Rep. 8 (1), 535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Guenther A, Karl T, Harley P, Wiedinmyer C, Palmer PI, and Geron C (2006) Estimates of global terrestrial isoprene emissions using MEGAN (Model of Emissions of Gases and Aerosols from Nature). Atmos. Chem. Phys. 6 (11), 3181–210. [Google Scholar]
- (21).Arashiro M, Lin Y-H, Zhang Z, Sexton KG, Gold A, Jaspers I, Fry RC, and Surratt JD (2018) Effect of secondary organic aerosol from isoprene-derived hydroxyhydroperoxides on the expression of oxidative stress response genes in human bronchial epithelial cells. Envrionmental Science: Processes & Impacts. 20 (2), 332–9. [DOI] [PubMed] [Google Scholar]
- (22).Xing Y-F, Xu Y-H, Shi M-H, and Lian Y-H (2016) The impact of PM2.5 on the human respiratory system. Journal of Thoracic Disease. 8 (1), E69–E74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Kelly F (2003) Oxidative stress: its role in air pollution and adverse health effects. Occup. Environ. Med. 60 (8), 612–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).MacNee W (2001) Oxidative stress and lung inflammation in airways disease. Eur. J. Pharmacol. 429 (1–3), 195–207 Epub 2001/11/08. PubMed PMID: 11698041. [DOI] [PubMed] [Google Scholar]
- (25).Rahman I (2002) Oxidative stress and gene transcription in asthma and chronic obstructive pulmonary disease: antioxidant therapeutic targets. Curr. Drug Targets: Inflammation Allergy 1 (3), 291–315 Epub 2003/10/17. PubMed PMID: 14561194. [DOI] [PubMed] [Google Scholar]
- (26).Arashiro M, Lin Y-H, Sexton KG, Zhang Z, Jaspers I, Fry RC, Vizuete WG, Gold A, and Surratt JD (2016) In vitro exposure to isoprene-derived secondary organic aerosol by direct deposition and its effects on COX-2 and IL-8 gene expression. Atmos. Chem. Phys. 16, 14079–90. [Google Scholar]
- (27).Lin Y-H, Arashiro M, Clapp PW, Cui T, Sexton KG, Vizuete W, Gold A, Jaspers I, Fry RC, and Surratt JD (2017) Gene Expression Profiling in Human Lung Cells Exposed to Isoprene-Derived Secondary Organic Aerosol. Environ. Sci. Technol. 51 (14), 8166–75 Epub 2017/06/22. PubMed PMID: 28636383; PMCID: PMC5610912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Bartel DP (2004) MicroRNAs: Genomics, Biogenesis, Mechanism, and Function. Cell 116 (2), 281–97. [DOI] [PubMed] [Google Scholar]
- (29).O’Brien J, Hayder H, Zayed Y, and Peng C (2018) Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. (Lausanne, Switz.) 9, 402 Epub 2018/08/21. PubMed PMID: 30123182; PMCID: PMC6085463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Lin Y-H, Zhang H, Pye HOT, Zhang Z, Marth WJ, Park S, Arashiro M, Cui T, Budisulistiorini SH, Sexton KG, Vizuete W, Xie Y, Luecken DJ, Piletic IR, Edney EO, Bartolotti LJ, Gold A, and Surratt JD (2013) Epoxide as a precursor to secondary organic aerosol formation from isoprene photooxidation in the presence of nitrogen oxides. Proc. Natl. Acad. Sci. U. S. A. 110 (17), 6718–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Zhang Z, Lin Y-H, Zhang H, Ball LM, and Gold A (2012) Technical Note: Synthesis of isoprene atmospheric oxidation products: isomeric epoxydiols and the rearrangement products. Atmos. Chem. Phys. 12 (18), 8529–35. [Google Scholar]
- (32).Lin Y-H, Zhang Z, Docherty KS, Zhang H, Budisulistiorini SH, Rubitschun CL, Shaw SL, Knipping EM, Edgerton ES, Kleindienst TE, Gold A, and Surratt JD (2012) Isoprene Epoxydiols as Precursors to Secondary Organic Aerosol Formation: Acid-Catalyzed Reactive Uptake Studies with Authentic Compounds. Environ. Sci. Technol. 46 (1), 250–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Zhang H, Surratt JD, Lin Y-H, Bapat J, and Kamens RM (2011) Effect of relative humidity on SOA formation from isoprene/NO photooxidation: enhancement of 2-methylglyceric acid and its corresponding oligoesters under dry conditions. Atmos. Chem. Phys. 11 (13), 6411–24. [Google Scholar]
- (34).Zhang H, Surratt JD, Lin YH, Bapat J, and Kamens RM (2011) Effect of relative humidity on SOA formation from isoprene/NO photooxidation: enhancement of 2-methylglyceric acid and its corresponding oligoesters under dry conditions. Atmos. Chem. Phys. 11 (13), 6411–24. [Google Scholar]
- (35).ATCC. BEAS-2B (ATCC CRL-9609) Culture Method [cited 2019 October 31]. Available from: https://atcc.org/Products/All/CRL-9609.aspx#culturemethod.
- (36).Malm S, Amouzougan E, and Klimecki W (2018) Fetal bovine serum induces sustained, but reversible, epithelial-mesenchymal transition in the BEAS-2B cell line. Toxicol. In Vitro 50, 383–90 PubMed PMID: 29678786; PMCID: 6084805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Cui T, Zeng Z, Dos Santos EO, Zhang Z, Chen Y, Zhang Y, Rose CA, Budisulistiorini SH, Collins LB, Bodnar WM, de Souza RAF, Martin ST, Machado CMD, Turpin BJ, Gold A, Ault AP, and Surratt JD (2018) Development of a hydrophilic interaction liquid chromatography (HILIC) method for the chemical characterization of water-soluble isoprene epoxydiol (IEPOX)-derived secondary organic aerosol. Environ. Sci. Process Impacts. 20 (11), 1524–36 Epub 2018/09/28. PubMed PMID: 30259953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Nanostring. Gene Expression Data Analysis Guidelines 2019. [cited 2019 October 31]. Available from: https://www.nanostring.com/support/data-analysis/nsolver-data-analysis-support.
- (39).Zerbino DR, Achuthan P, Akanni W, Barrell D, Bhai J, Billis K, Cummins C, Gall A, Girón CG, Gil L, Gordon L, Haggerty L, Haskell E, Hourlier T, Izuogu OG, Janacek SH, Juettemann T, Kiang J, Laird MR, Lavidas I, Liu Z, Loveland JE, Maurel T, McLaren W, Moore B, Mudge J, Murphy DN, Newman V, Nuhn M, Ogeh D, Ong CK, Parker A, Patricio M, Riat HS, Schuilenburg H, Sheppard D, Sparrow H, Taylor K, Thormann A, Vullo A, Walts B, Zadissa A, Frankish A, Hunt SE, Kostadima M, Langridge N, Martin FJ, Muffato M, Perry E, Ruffier M, Staines DM, Trevanion SJ, Aken BL, Cunningham F, Yates A, and Flicek P (2018) Ensembl 2018. Nucleic Acids Res. 46 (D1), D754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Rager JE, Bailey KA, Smeester L, Miller SK, Parker JS, Laine JE, Drobná Z, Currier J, Douillet C, Olshan AF, Rubio-Andrade M, Stýblo M, García-Vargas G, and Fry R C. (2014) Prenatal arsenic exposure and the epigenome: altered microRNAs associated with innate and adaptive immune signaling in newborn cord blood. Environ. Mol. Mutagen. 55 (3), 196–208 PubMed PMID: 24327377; PMCID: 4023469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Lin Y-H, Knipping EM, Edgerton ES, Shaw SL, and Surratt J (2013) Investigating the influences of SO2 and NH3 Levels on Isoprene-Derived Secondary Organic Aerosol Formation Using Conditional Sampling Approaches. Atmos. Chem. Phys. 13 (16), 8457–70. [Google Scholar]
- (42).Budisulistiorini SH, Baumann K, Edgerton ES, Bairai ST, Mueller S, Shaw SL, Knipping EM, Gold A, and Surratt JD (2016) Seasonal characterization of submicron aerosol chemical composition and organic aerosol sources in the southeastern United States: Atlanta, Georgia, and Look Rock, Tennessee. Atmos. Chem. Phys. 16 (8), 5171–89. [Google Scholar]
- (43).Budisulistiorini SH, Li X, Bairai ST, Renfro J, Liu Y, Liu YJ, McKinney KA, Martin T, McNeill VF, Pye HOT, Nenes A, Neff ME, Stone EA, Mueller S, Knote C, Shaw SL, Zhang Z, Gold A, and Surratt JD (2015) Examining the effects of anthropogenic emissions on isoprene-derived secondary organic aerosol formation during the 2013 Southern Oxidant and Aerosol Study (SOAS) at the Look Rock, Tennessee ground site. Atmos. Chem. Phys. 15 (15), 8871–88. [Google Scholar]
- (44).Santerre M, Bagashev A, Gorecki L, Lysek KZ, Wang Y, Shrestha J, Del Carpio-Cano F, Mukerjee R, and Sawaya BE (2019) HIV-1 Tat protein promotes neuronal dysregulation by inhibiting E2F transcription factor 3 (E2F3). J. Biol. Chem. 294 (10), 3618–33 Epub 2018/12/29. PubMed PMID: 30591585; PMCID: PMC6416426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Liang R, Li Y, Wang M, Tang SC, Xiao G, Sun X, Li G, Du N, Liu D, and Ren H (2018) MiR-146a promotes the asymmetric division and inhibits the self-renewal ability of breast cancer stem-like cells via indirect upregulation of Let-7. Cell Cycle 17 (12), 1445–56 Epub 2018/06/3. PubMed PMID: 29954239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Dang J, Yang M, Zhang X, Ruan H, Qin G, Fu J, Shen Z, Tan A, Li R, and Moore J (2018) Associations of Exposure to Air Pollution with Insulin Resistance: A Systematic Review and Meta-Analysis. Int. J. Environ. Res. Public Health 15 (11), 2593 Epub 2018/11/23. PubMed PMID: 30463387; PMCID: PMC6266153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Liu C, Xu X, Bai Y, Wang TY, Rao X, Wang A, Sun L, Ying Z, Gushchina L, Maiseyeu A, Morishita M, Sun Q, Harkema JR, and Rajagopalan S (2014) Air pollution-mediated susceptibility to inflammation and insulin resistance: influence of CCR2 pathways in mice. Environ. Health Perspect. 122 (1), 17–26 Epub 2013/10/24. PubMed PMID: 24149114; PMCID: PMC3888572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Singh S, Prakash YS, Linneberg A, and Agrawal A (2013) Insulin and the lung: connecting asthma and metabolic syndrome. J. Allergy 2013, 627384 Epub 2013/11/10. PubMed PMID: 24204385; PMCID: PMC3800560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Soberanes S, Panduri V, Mutlu GM, Ghio A, Bundinger GR, and Kamp DW (2006) p53 mediates particulate matter-induced alveolar epithelial cell mitochondria-regulated apoptosis. Am. J. Respir. Crit Care Med. 174 (11), 1229–38 Epub 2006/09/02. PubMed PMID: 16946128; PMCID: PMC2648105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Deng X, Rui W, Zhang F, and Ding W (2013) PM2.5 induces Nrf2-mediated defense mechanisms against oxidative stress by activating PIK3/AKT signaling pathway in human lung alveolar epithelial A549 cells. Cell Biol. Toxicol. 29 (3), 143–57 Epub 2013/03/26. PubMed PMID: 23525690. [DOI] [PubMed] [Google Scholar]
- (51).Pardo M, Xu F, Shemesh M, Qiu X, Barak Y, Zhu T, and Rudich Y (2019) Nrf2 protects against diverse PM2.5 components-induced mitochondrial oxidative damage in lung cells. Sci. Total Environ. 669, 303–13 Epub 2019/03/18. PubMed PMID: 30878937. [DOI] [PubMed] [Google Scholar]
- (52).Kang KW, Lee SJ, and Kim SG (2005) Molecular mechanism of nrf2 activation by oxidative stress. Antioxid. Redox Signaling 7 (11–12), 1664–73 Epub 2005/12/17. PubMed PMID: 16356128. [DOI] [PubMed] [Google Scholar]
- (53).Wittkopp S, Staimer N, Tjoa T, Stinchcombe T, Daher N, Schauer JJ, Shafer MM, Sioutas C, Gillen DL, and Delfino RJ (2016) Nrf2-related gene expression and exposure to traffic-related air pollution in elderly subjects with cardiovascular disease: An exploratory panel study. J. Exposure Sci. Environ. Epidemiol. 26 (2), 141–9 Epub 2015/01/08. PubMed PMID: 25564368; PMCID: PMC4495007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Hu WW, Campuzano-Jost P, Palm BB, Day DA, Ortega AM, Hayes PL, Krechmer JE, Chen Q, Kuwata M, Liu YJ, Sá SSd, McKinney K, Martin ST, Hu M, Budisulistiorini SH, Riva M, Surratt JD, Clair JMS, Wertz G-V, Yee LD, Goldstein AH, Carbone S, Brito J, Artaxo P, deGouw JA, Koss A, Wisthaler A, Mikoviny T, Karl T, Kaser L, Jud W, Hansel A, Docherty KS, Alexander ML, Robinson NH, Coe H, Allan JD, Canagaratna MR, Paulot F, and Jimenez JL (2015) Characterization of a real-time tracer for isoprene epoxydiols-derived secondary organic aerosol (IEPOX-SOA) from aerosol mass spectrometer measurements. Atmos. Chem. Phys. 15 (20), 11807–33. [Google Scholar]
- (55).Henze DK, and Seinfeld JH (2006) Global secondary organic aerosol from isoprene oxidation. Geophys. Res. Lett. 33, 33. [Google Scholar]
- (56).Henze DK, Seinfeld JH, Ng NL, Kroll JH, Fu T-M, Jacob DJ, and Heald CL (2008) Global modeling of secondary organic aerosol formation from aromatic hydrocarbons: high- vs. low-yield pathways. Atmos. Chem. Phys. 8 (9), 2405–20. [Google Scholar]
- (57).Rattanavaraha W, Chu K, Budisulistiorini SH, Riva M, Lin Y-H, Edgerton ES, Baumann K, Shaw SL, King L, Weber RJ, Neff ME, Stone EA, Offenberg JH, Zhang Z, Gold A, and Surratt JD (2016) Assessing the impact of anthropogenic pollution on isoprene-derived secondary organic aerosol formation in PM2.5 collected from the Birmingham, Alabama, ground site during the 2013 Southern Oxidant and Aerosol Study. Atmospheric Chemistry and Physics. 16 (8), 4897–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1: IEPOX-responsive miRNA–mRNA pairings from the target filter analysis
Table S2: MAE-responsive miRNA–mRNA pairings from the target filter analysis (XLSX)