Abstract
In Neurofibromatosis type 1, NF1 gene mutations in Schwann cells (SC) drive benign plexiform neurofibroma (PNF), and no additional SC changes explain patient-to-patient variability in tumor number. Evidence from twin studies suggests that variable expressivity might be caused by unidentified modifier genes. Whole exome sequencing of SC and fibroblast DNA from the same resected PNFs confirmed biallelic SC NF1 mutations; non-NF1 somatic SC variants were variable and present at low read number. We identified frequent germline variants as possible neurofibroma modifier genes. Genes harboring variants were validated in two additional cohorts of NF1 patients and by variant burden test. Genes including CUBN, CELSR2, COL14A1, ATR and ATM also showed decreased gene expression in some neurofibromas. ATM-relevant DNA repair defects were also present in a subset of neurofibromas with ATM variants, and in some neurofibroma SC. Heterozygous ATM G2023R or homozygous S707P variants reduced ATM protein expression in heterologous cells. In mice, genetic Atm heterozygosity promoted Schwann cell precursor self-renewal and increased tumor formation in vivo, suggesting that ATM variants contribute to neurofibroma initiation. We identify germline variants, rare in the general population, overrepresented in NF1 patients with neurofibromas. ATM and other identified genes are candidate modifiers of PNF pathogenesis.
Keywords: Neurofibromatosis type 1, neurofibroma, genomics, mutation, ATM, DNA damage
INTRODUCTION
Neurofibromatosis type 1 (NF1) is one of the most common human monogenic disorders, with a frequency of 1:3000 in the human population [22, 50]. Loss-of-function NF1 germline mutations are identified in up to 95% of NF1 patients, with no evidence for other NF1causing genes [37]. NF1 is a Rasopathy, one of a group of disorders caused by RAS-MAPK signaling pathway gene mutations [51]. The NF1 gene (17q11.2, 283kb) encodes a RAS-GAP protein, so that inactivating NF1 gene mutations increase levels of cellular RAS-GTP, thereby affecting cellular proliferation, migration, and differentiation [60]. NF1 patients often show cognitive and motor deficits, and can develop neoplasms including astrocytoma or myeloid leukemia (JMML), or vascular and bone anomalies [51]. Neurofibromas are benign peripheral nerve sheath tumors and the characteristic lesions found in individuals with NF1 [36]. More than 95% of NF1 patients develop neurofibromas associated with cutaneous nerves called dermal neurofibroma (DNF) [2, 50], and up to half of NF1 patients develop plexiform neurofibromas (PNF), largely associated with deep nerves [36, 47]. PNF can cause significant morbidity and can transform to malignant peripheral nerve sheath tumors (MPNST), highlighting their clinical importance [15, 31]. In mice, loss of Nf1 in Schwann cells and their precursors is sufficient to generate plexiform and dermal neurofibromas, consistent with the major role of Nf1 in driving neurofibroma formation [49, 73].
Cancer is a genetic disease, and many mutations are found in most adult cancers [66]. However, high-density SNP arrays failed to identify recurrent genetic changes in PNF (apart from alterations at the NF1 locus itself) supporting the idea that few genetic changes occur in these benign tumors [5, 42, 50]. Atypical neurofibromas (ANF), thought to be precursor lesions for MPNST, can show deletion at the CDKN2A locus, with no other consistent genomic changes; histologic criteria define some ANF as atypical neurofibroma of uncertain biologic potential (ANNUBP) [5, 38]. Also, using whole-exome sequencing (WES) in PNF samples, recurrent somatic mutations were not identified [42]. Indeed, a relatively silent genome (somatic changes 0 – 30) characterizes many types of pediatric tumors [12, 63]. Analysis of germline mutations in sarcoma, including 28 MPNST, identified an excess of damaging variants in cancer predisposition genes. Sarcoma patients were more likely than controls to have multiple pathogenic variants, especially in DNA repair-related genes [4, 10], but germline modifiers of neurofibroma have not been studied. Any sequence variation that alters a gene’s interaction with other genes/pathways may act as a modifier allele in a specific disease context.
Neurofibromas are complex peripheral nerve tumors composed of 20 – 80% Schwann cells (SCs), abundant macrophages (~30%), and variable numbers of mast cells, endothelial cells, and fibroblasts (FBs), including endoneurial and perineurial fibroblasts [62]. SC NF1 mutations drive neurofibroma formation, as biallelic NF1 loss of function mutations occur in neurofibroma SCs, but not FBs [33]. Also, biallelic loss of Nf1 in the SC lineage is sufficient to generate mouse neurofibromas, in the presence or absence of hemizygous loss of Nf1 in non-neoplastic cells [51].
We searched for germline variants that might modify neurofibroma formation. Evidence from comparisons of monozygotic twins to more distant relatives strongly supports effects of genetic modifier genes on dermal neurofibroma number [18, 52, 61]. Scoring plexiform neurofibromas by number in twin pairs, sibling pairs and more distant relatives, demonstrated a correlation between higher numbers of plexiform neurofibromas and closer relatives [56], [18]. This suggests that as-yet-unidentified genetic modifier genes, distinct from NF1 itself, exist for PNF, and that variants in specific genes may influence DNF and/or PNF number.
Any type of sequence variation can produce modifier effects: modifier variants can occur in exons, intragenic sequences, or regulatory regions. Modifier alleles in exons can include missense, frameshift, deletion, and gain-of-function alleles [53]. Even small differences in gene expression levels can cause phenotype-modifying effects [68]. Modifier alleles are not necessarily clinically pathogenic, as they alone are not sufficient for disease.
Next-generation sequencing (NGS) technologies have enabled whole exome sequencing on increasingly smaller DNA samples [3, 19, 21, 24]. We found that increasing the sensitivity of WES by independently analyzing neurofibroma cell types allowed detection of most NF1 mutations, and of rare non-NF1 somatic variants in neurofibroma cells. We identified germline variants predicted to have altered protein function (deleterious/damaging) and which are rare or absent in the general population. Variants in these genes were confirmed in validation DNF and PNF datasets. Gene-based burden tests confirmed that many of these genes are more likely to have genetic changes in neurofibroma patients than in general population. We confirmed some ATM germline variants by Sanger sequencing. DNA damage repair defects occurred in neurofibromas, correlating with ATM variants. Two ATM germline variant, heterozygous G2023R and homozygous S707P, decreased ATM protein expression in mK4 cells. In model systems, reduced Atm gene expression promoted the self-renewal of Schwann cell precursor cells (SCP), a cell-of-origin for neurofibroma, and tumorigenesis, and Atm haploinsufficiency increased tumor number in mice. Taken together, the data implicate the ATM tumor suppressor in PNF initiation and identify other candidates for investigation.
METHODS
Sample Collection
Fresh plexiform neurofibromas (n=9) were obtained after medically mandated surgeries. All samples were obtained with patient consent under IRB approval.
Cell Sorting and Flow Cytometry
Surgical PNF neurofibroma specimens were enzymatically dissociated as described [70]. We incubated cell suspensions anti-p75/NGFR (C40–1457, Becton-Dickinson) bound to phycoerythrin (PE), and anti-EGFR (cat # 61R-E109BAF, Fitzgerald, Acton, MA) bound to FITC on ice in PBS/ 0.2%BSA/0.01% NaN3 for 30 min. We removed cells with macrophage (CD11b) and endothelial cell (CD31) markers. After washing, we re-suspended cells in PBS/0.2%BSA/0.01% NaN3/ 2 μg/mL 7-aminoactinomycin D (7-AAD, Invitrogen). We carried out isotopic controls with irrelevant mouse IgG1-PE and mouse-IgG1-FITC in parallel. Cells were FACS-sorted using a four-laser FACSDiva (Becton-Dickinson) to acquire live Schwann cells (P75+/CD11b-/CD31-/7-AAD-) and fibroblasts (P75-/EGFR+/CD31-/CD11b-/7-AAD-). Three primary human PNF tumors (P7–8) were dissociated, and SC and FB separated by incubation in medium with forskolin and heregulin (for SC), or DMEM and 10% FBS (for fibroblasts) for 3 – 7 serial passages, as described [55].
Library Preparation and Whole Exome Sequencing
Double stranded DNA (1ug) determined by Invitrogen Qbit spectrofluorometric measurement was sheared by sonication to an average size of 200bp on a Covaris S2. Library construction was performed on a Wafergen Apollo324. Each library was fitted with one of 48 adapters for multiplexing. After 9 cycles of PCR amplification using the Clontech Advantage II kit, 1ug of genomic library was recovered for exome enrichment using the NimbleGen EZ Exome V2 kit (Roche Nimblegen, Inc, Madison, WI), targeting ~30,000 coding genes (~300,000 exons, total size 36.5 Mbp) from RefSeq (January 2010), CCDS (September 2009) and miRBase (September 2009). Libraries were enriched per manufacturer’s recommendations and sequenced on an Illumina HiSeq2500, generating ~30 million paired end reads 125 bases long each equivalent to 7.5GB of usable high-quality sequence per sample.
Variant Calls
The Genome Analysis Toolkit (GATK) pipeline (https://www.broadinstitute.org/gatk/ was used, with modifications in the “Best Practices” document on their website. Briefly, after applying Illumina’s Chastity filter, raw sequenced reads were aligned using the Burrows Wheeler Aligner (BWA, http://bio-bwa.sourceforge.net) against reference human genome (GRCh37.v4). For each sample, reads appearing to be PCR artifacts were flagged, reads that overlap known or putative insertions/deletions (INDELs) realigned, then all base quality scores were recalibrated to the empirical error rate from non-polymorphic sites. The GATK HaplotypeCaller module was used to create a gVCF file for each patient sample containing confidence values for every position in the exome (variant and/or reference). A set of variant calls with the GenotypeGVCFs module was generated using every compatible sample sequenced by the sequencing facility to date. The Variant Quality Score Recalibration (VQSR) method was applied to filter variant calls. Finally, variant calls specific to the samples were extracted. Variants marked “PASS” were considered for further analysis. Variant calling was performed with GATK v2.8.
For INDELs, samples were individually preprocessed by realigning reads around putative INDELs using GATK’s IndelRealigner tool, marking putative polymerase chain reaction duplicate reads with Picard’s MarkDuplicates tool and by recalibrating base quality scores and calculating Base Alignment Quality scores with GATK’s CountCovariates and TableRecalibration tools. After preprocessing, samples were jointly processed with HaplotypeCaller to generate initial variant calls. Variants were then filtered using GATK Variant Quality Score Recalibration.
Gene-Based Burden Test
The Test Rare vAriants with Public Data (TRAPD, https://github.com/mhguo1/TRAPD) workflow generates a burden score for each subject by taking a weighted linear combination of the mutation counts within a gene. TRAPD performs a one-sided Fisher’s exact test to determine if there is a greater burden of qualifying variants in cases as compared to controls for each gene. After annotating variants using Variant Effect Predictor (VEP v96), non-common (allele frequency < 0.05) and non-synonymous variants found in protein coding regions were analyzed against the ExAC database (v1.0, http://exac.broadinstitute.org) admixed American (AMR) sub-population as a control group (n = 5789).
Copy-Number Variation in Coding Regions
ExomeCNV [58] (v1.4) was used to detect copy-number variations from matched samples. It calculates log coverage-ratio between case (i.e. SC) and control (i.e. matched FB) to calls CNV/LOH events for each exon, then combines exonic CNV/LOH into segments using the Circular Binary Segmentation (CBS) algorithm.
Human Gene Expression Data
We analyzed gene expression levels in human (GEO accession: GSE14038, human) microarray data. The heatmaps represent log2 fold change to the average expression levels of 10 normal cultured human SC samples (NHSC).
Comparison with Dermal Neurofibroma WGS data
We parsed 40 matched VCF files (tumor and matched blood samples n=13 patients) from Synapse (http://www.synapse.org; Synapse ID: syn4984604), then applied the filtering strategy above to detect germline and somatic variants.
Comparison with Plexiform Neurofibroma WES data
From dbGaP (Study Accession: phs001403.v1.p1), we obtained 44 whole-exome sequencing alignment files (hg18 as reference genome) from 21 NF1 patients (blood samples and matched plexiform neurofibroma). SNP/INDEL/LOH events were called using VarScan2 (v2.3.9), hg18/NCBI36 reference genome, and the parameter set described above. Genomic coordinates of called variants were converted into their hg19/GRCh37 counterparts, using CrossMap (v0.2.1) and the NCBI36-to-GRCh37 chain file [74]. Once variants were annotated using Variant Effect Predictor (VEP, v83) and the annotation databases described, we analyzed variants as described in supplemental method.
DNA Comet Assay
Neurofibroma-derived Schwann cells were exposed to 1.0 uM doxorubicin, and single-cell gel electrophoresis (comet assay) performed under alkaline conditions (Trevigen), at designated repair time points [41]. Comet images (75–100) were captured and analyzed using Comet Assay IV software (Perceptive Instruments). The percentage of nuclei with DNA comet tail moment value 0–9.9, 10–19.9, 20–29.9, 30–39.9, 40–49.9, >=50 were calculated to determine the DNA damage repair capacity; higher tail moment values = unrepaired DNA damage.
CRISPR/Cas9 to construct missense variants in mK4 cells
We constructed guide RNA (gRNA) sequence to target specific regions of the ATM gene (gRNA sequences are shown in Suppl. Fig 8a) into the pX458 plasmid, which also encodes GFP. Single strand donor DNAs were from IDT (~100nt length; see supplemental material for full sequences of donor DNA). We delivered the plasmid containing sequence encoding gRNA and donor DNA oligo into mK4 cells by FuGene (Promega, E2311) transfection. 72 hours after transfection, we sorted GFP positive cells by FACS, and seeded single cell into 96 well plates in tissue culture medium. After 3 weeks, we extracted DNA from single cell colonies and did PCR and Sanger sequencing to confirm missense variants.
SCP Culture and Sphere Counting
We dissociated DRG from E12.5 embryos with 0.25% Trypsin (Life Technologies) for 20 min at 37°C and obtained single-cell suspensions with narrow-bore pipettes and a 40 μm strainer (BD-Falcon), subsequently plating the cells in 24 well low attachment plates (Corning). The free-floating cells were cultured in serum-free medium with EGF and FGF as described [70]. For passage, we dissociated spheres with 0.05% Trypsin (Life Technologies) at 37° for 3–5 minutes. For shRNA treatment and sphere counts, we plated SCP cells at low density (250/cm2) to avoid sphere fusion (500 cells/well in 24 well plates). We treated cells with lentiviral particles at MOI=10, 24 hours after plating. We performed two independent experiments, each with 4 replicates, and counted sphere number with an inverted phase contrast microscope after 4 days.
Tumorigenesis Assay in Nude Mice
We anaesthetized athymic nu/nu mice in isoflurane and subcutaneously injected 4.5 × 105 mouse sphere cells/injection in 33% matrigel into both left and right flanks. Tumors were monitored weekly. At 11 weeks after injection, we dissected visible tumors and fixed them in 4% paraformaldehyde overnight, then embedded them in paraffin.
Immunohistochemistry Staining
Paraffin blocks were sectioned at 6μm. de-paraffinized in xylene and rehydrated, boiled in citrate buffer for antigen retrieval, blocked 1hr in 10% normal goat/horse serum in 1xPBS and 0.3% Triton-X100, then incubated with primary antibody S100B (Dako, Z0311, rabbit, 1:10,000), SOX10 (Santa Cruz,sc-17342, goat, 1:200), CD31 (Abcam, ab28364, rabbit, 1:100) and Ki67 (Cell Signaling, 12202S, rabbit, 1:250) overnight and secondary antibody-biotinylated goat anti-rabbit or horse anti-goat (Vector lab BA-1000 or BA-9500, 1:200) for 1hr after rinsing with PBS. Sections were further processed with the Elite ABC kit and DAB and mounted slides in histomount after dehydrating and clearing. Images were captured on a Nikon Eclipse 80i microscope using a Nikon DS-Fi1 camera. γH2AX staining used anti-phospho-Histone H2A.X (Ser139) (EMD Millipore, clone JBW301, cat # 05636) and was processed as above except that (Vector lab, A-2011, 1:500) for 20 mins and DAPI (0.1ug/ml) for 5 mins were used after incubating with secondary fluorescent Avidin DCS antibody, sections mounted in Fluoromount G (Electron Microscopy Sciences) and imaged on a Nikon C2 Confocal, using a 60X oil objective and optimized Zoom (4.78). Foci in ≥ 120 nuclei were blindly counted by 3 investigators/ sample. For 53BP1 focus staining, each dermal neurofibroma generated 6–8 primary Schwann cell cultures (plated on coverslips), and 250–500 nuclei were scored for 53BP1 foci. In each of 3 independent experiments cells were immunestained with anti-53BP1, and Alexa Fluor 594 donkey anti-rabbit (Molecular Probes) to visualize foci under epifluorescence microscopy. Schwann cell nuclei were scored for number of 53BP1 foci at 400X magnification.
MRI and Tumor Burden Analysis
MRI data were collected on a 7-T Bruker BioSpec system equipped with 400G/cm gradients and tumor burden was calculated as described previously [71]. Un-paired t-test was applied to do statistical comparison between Nf1 fl/fl; DhhCre and Atm+/−; Nf1 fl/fl; DhhCre two groups of mouse.
Mouse Dissection and Tumor Number and Size Calculation
To quantify tumor numbers and size, we perfused mice and used a Leica dissecting microscope to dissect the spinal cord with attached DRG and nerve roots at the age of 7 months, as described previously [72]. A tumor was defined as a mass surrounding the DRG or nerve roots, with a diameter greater than 1 mm, measured perpendicular to DRG/nerve roots. Tumor numbers within each mouse and diameter for each tumor were measured with Image J.
RESULTS
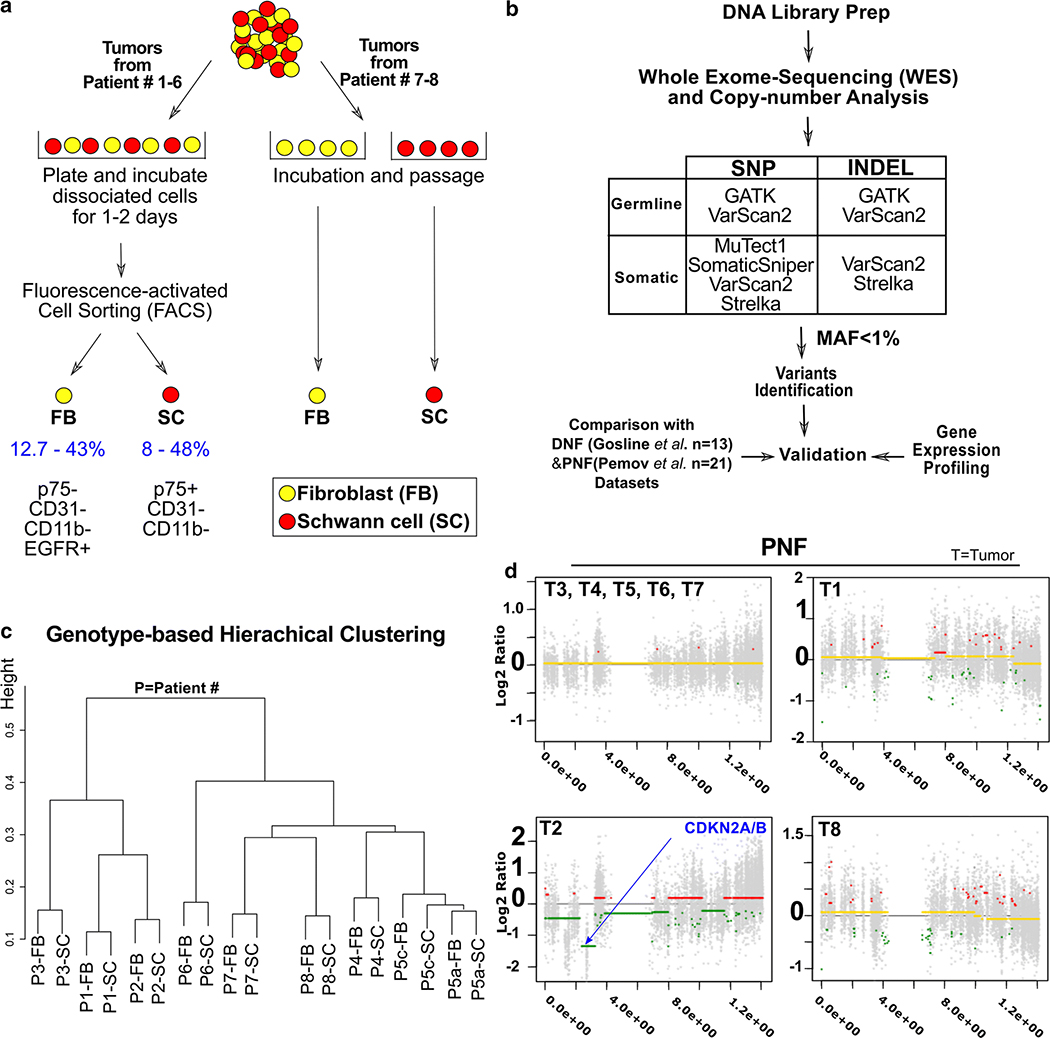
To identify variants in genes that might modify neurofibroma pathogenesis, we separated SCs and matched FBs from 9 PNFs (T1-T9) from 8 NF1 patients (P1-P8); T5a and T5c are from the same patient. Patient blood was not available, so we used FB DNA to identify individual germline alterations using WES (Fig. 1a and b). SC and FB samples from P1-P6 tumors were prepared by FACS, while samples from P7-P8 were selectively cultured (Fig. 1a). We used WES to detect variants in coding regions and canonical splice sites. The average depth of coverage was 101x and 107x for FB and SC DNA, respectively (Suppl. Table 1). Clustering analysis demonstrated more variance among patients than between cell types (SC and FB), consistent with the low mutational burden previously documented in neurofibroma [5, 42](Fig. 1c). Rare variants [minor allele frequency (MAF) of < 1%] present both in FB and SC from a tumor were denoted germline variants. We hypothesized that modifier gene variants would be present in both SC and FB.
Figure 1. Workflow for variant calling.
(a) Patient samples were separated into Schwann cells (SC) and fibroblasts (FB) by acute cell sorting or differential primary cell culture. (b) SNP and INDEL variant calling used 5 different methods and downstream analyses. MAF=minor allele frequency. (c) Genotype similarity among 9 matched cell samples. (d) Copy-number patterns in chromosome 9; P3-P7 show normal allele ratios. P1 and P8 showed frequent small regions of altered allele rations. Only P2 showed a CDKN2A deletion typical of atypical neurofibroma,a whole-gene deletion of CDKN2A, present in a fraction of SC (logRatio = −9.94 at chr9:21968184–21968284, logRatio = −5.12 at chr9:21970900–21971207, logRatio = -inf at chr9:21974475–21974826, logRatio = -inf at chr9:21994137–21994453) and a partial deletion of CDNK2B (logRatio = −4.21 at chr9:22005985–22006246, logRatio= −1.45 at chr9:2200871522008952).
Genomic Changes in the neurofibroma samples
We used ExomeCNV [58], to confirm a quiet neurofibroma genome. Six of 9 samples (T3, T4, T5a, T5c, T6 and T7) showed no significant copy number alterations (CNA), consistent with diagnosis of benign neurofibroma, as predicted (Fig. 1d). CNA events found in SC versus FB were detected in 3/9 paired samples (T1, T2, T8), although the histology of all evaluable tumors was consistent with the diagnosis of neurofibroma. In T2 a deletion was present on chromosome 9 including the CDKN2A locus (chr9:19049657–22451910, log2 ratio = −0.46), and CNA in SC across the genome were present (Fig. 1d). In T2, from IHC analysis, a slight increase in cellularity was observed, but mitoses were absent, and atypia remained mild (not shown), defining T2 as a neurofibroma. T1 and T8 did not show copy-number alterations in the CDKN2A locus, but showed short segments of gain or loss on many chromosomes (Fig. 1d). In T1, no features of atypical neurofibroma were present; mitoses were absent, atypia was mild, and collagen was retained throughout. T8 sections were unavailable.
NF1 Mutations
Consistent with previous research [33], we identified bi-allelic NF1 mutations in 8/9 of tumors (Supp. Fig1a). Many have been reported as pathogenic in HGMD or ClinVar. Others were predicted to be deleterious/damaging by bioinformatics prediction tools (SIFT, Meta-SVM, and/or MetaLR). As expected, the alternative reads ratio of NF1 germline mutations (present in both FB and SC) was ~50%. Most NF1 mutations caused premature termination or frameshift predicting loss of protein function (Supp. Fig1b and Supp. Table2). We identified somatic NF1 mutations (alternative reads ratio 5–50%) present in SCs but not fibroblasts in 9/9 samples. Implying the possible existence of sub-clonal SC populations harboring different NF1 somatic variants in single tumors, we identified >1 somatic variant in some tumor SC preparations. We did not have DNA after WES to confirm NF1 somatic variants; however, many have been annotated in HGMD or ClinVar as having clinical impact; others may not be pathogenic.
Identification of non-NF1 genetic variants in neurofibroma cells
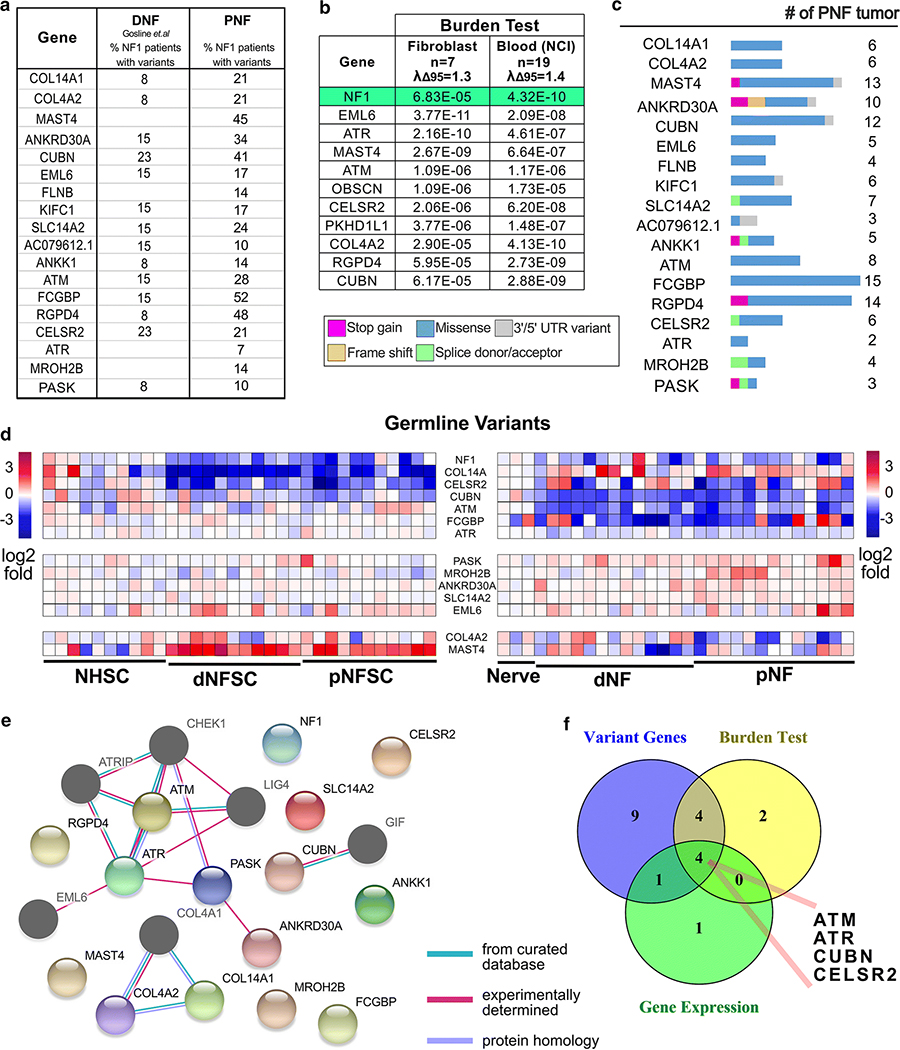
We identified 18 genes that showed germline variants only, with minor allele frequency (MAF) <1% in the general population, in at least 3 individuals (group II, Fig2a). We also identified genes in group I (genes with bi-allelic variants, one allele germline variant showed both in FB and SC, and second allele SC specific somatic variant, including NF1, PKHD1L1 and OBSCN, see Suppl. Fig4) and 25 genes with only somatic variants (group III, variants in SC only, Suppl. Fig 3) in ≥2 patients, or variants in genes relevant to cancer (Suppl. Tables 3–4). We ignored recurrent variants in repetitive regions, which when tested, failed to confirm with Sanger sequencing or digital droplet PCR (not shown).
Figure 2. Genes harboring germline variants.
(a) A list of non-synonymous rare (MAF < 0.01) germline variants. (b) Genes with significant burdens (p < 1E-5) predicted by burden tests using non-common (allele frequency < 0.05) non-synonymous variants found in protein coding regions, against ExAC’s ad mixed American (AMR) sub-population. The level of artifact inflation/deflation were measured using the λΔ95 metric implemented in TRAPD. We detected the small levels of inflation (i.e. above the expected λΔ95 > 1.00) and could not eliminate entire artifacts probably due to the small case sample size and incomplete ancestry information. (c) The percent of PNF (average of cell data set from Figure 1 and Pemov et al. data) and DNF containing variants in each designated gene are shown. (d) Heatmap of RNA expression of those genes with rare germline variants in tumor SC vs. normal SC and/or tumor tissue vs. nerve. (e) STRING analysis showing the predicted interactions (confidence level >= 0.15, Maximum interaction number of the first shell <= 5) of the candidate genes, and identifies DNA damage and repair as a candidate pathway. Nodes shown in color are genes with variants identified in PNF tumors. The dark-gray nodes are genes are cross-link genes. (f) A Venny diagram showing the candidate gene filtering strategy.
Whole genome sequencing (WGS) data from patient DNF and their 13 matched blood samples [24] (http://www.synapse.org, Synapse ID: syn4984604), and whole exome sequencing (WES) data from 21 paired blood and plexiform neurofibroma samples [42] verified over-representation of variants in identified candidate genes. Of 21 genes with germline variants (including 3 genes with bi-allelic variants) identified in PNF cells, 16 also showed germline variants in the DNF and PNF validation cohorts (Fig 2a). A single SNP variant is usually underpowered to detect statistically meaningful signals between case and control samples. However, by aggregating SNP variants across a candidate gene, a detection power can be significantly improved, with reduced bias. To test the association of candidate genes harboring the germline variants with NF1 disease, we performed burden tests using cases from each PNF cohort versus control samples from the ExAC dataset. We analyzed the 26 samples of mixed American ancestry; 7 in one cohort, and 19 in the other. The burden tests implemented in the TRAPD package revealed significant association between the burden of rare or low-frequency variants in 11 genes in both plexiform neurofibroma cohorts, as compared to the average number of SNPs in each of these genes in a large population of similar ethnicity; those genes with significantly different burdens (p < 1E-5) are shown in Fig. 2b. The level of artifact inflation/deflation measured using the λΔ95 metric implemented in TRAPD was similar to those of other small sample sets compared to population databases (1.3; 1.4) (Suppl. Fig. 2a).
Most identified variants occurred at private sites in each tumor. ANKRD30A, ATM, CELSR2, COL14A1, COL4A2, CUBN, KIFC1, PKHD1L1, RGPD4 and SLC14A2 showed an identical variant in 2 datasets (Suppl. Tables 3, 5, 7 highlighted in yellow or green). CUBN, SLC14A2 and RGPD4 showed an identical variant in all 3 datasets (Suppl. Table 3, 5, 7 highlighted in orange). Additional genes harboring multiple germline variants were COL14A1, COL4A2 and MAST4.
Most germline variants identified were missense. Many were predicted as deleterious/damaging to the protein by multiple bioinformatics tools. Loss of function ‘splicing donor/acceptor’ or ‘stop gained’ variants were found in genes including MROH2B, PASK, ANKRD30A and ANKK1 (Fig. 2c). Supplemental Fig 2b shows the position of the variants with respect to known domains; predicted deleterious/damaging alterations were present in COL14A1, CELSR2, CUBN and FCGBP. Rare changes in PASK and MROH2B were also predicted to be deleterious/damaging. The consequences of these variants is unclear, with most not having clinical interpretations listed in ClinVar. For variants with clinical interpretation which achieved consensus, there were 7 variants considered likely benign and 5 variants considered unlikely inheritance, with only 1 pathogenic variant. While this would be problematic if we were searching for the primary cause of disease, for modifier variants, such results are not unexpected. Given that reduced protein levels can result from reduced mRNA expression, we analyzed available mRNA expression data from neurofibroma compared to normal peripheral nerve [39]. Like NF1, significant down-regulation of CELSR2, KIFC1, COL14A1, and CUBN was found in tumors compared to nerve (Fig. 2d).
Somatic variants were rarely shared among the 3 cohorts, and most were missense variants with very low alternative reads ratio. However, rare somatic variants were predicted to induce stop gains (Suppl. Fig 3a), and the mRNA levels of some candidate genes were reduced in tumors versus normal tissue (Suppl. Fig 3b). Bioinformatics tools predicted effects of some identified variants on protein function (Suppl. Tables 4; Supplemental Fig3c).
Of note, two large genes, OBSCN and PKHD1L1, like NF1, showed bi-allelic variants in one and two tumor samples each (Group I), and in the DNF and PNF validation cohorts (Suppl. Fig 4a and 4b). Other PNF tumors harbored either one allele germline or somatic variant in OBSCN and/or PKHD1L1. PKHD1L1 gene variants at Y884 and G1223 were identified in both a PNF and a DNF sample; one variant, T3104I, occurred in 3/30 PNF (Suppl. Tables 3, 5 and 7). Supplemental Figs 4b and 4c show the type and position of the variants. Many of these variants were predicted to be deleterious/damaging to the protein by bioinformatics tools.
To determine if some germline or somatic variants were consistently changed in tandem we used cBioPortal. We generated an Oncoprint of combined germline and somatic variants in PNF, but did not identify patterns of alterations (Suppl. Fig. 5). Notably, T1 (with a CDNKN2A deletion) and T2 (with small genome-level alterations) did not show increased numbers of variants in these genes, consistent with the histological diagnosis of neurofibroma.
Pathway and signaling analysis of genes showing variants
Identification of multiple genes in a similar pathway support the relevance of the identified pathway to disease. STRING analysis of the genes showing germline variants in the PNF datasets identified DNA damage and repair as a candidate pathway. (Fig. 2e). “Biological Process” term-based GO enrichment analysis confirmed the enriched functional category of telomerase catalytic core complex assembly (GO:1904882 (p=0.00000489) and related terms--ATM and ATR (data not shown). Fig. 2f shows the genes identified as showing germline variant, verified by variant burden test; and showing reduced gene expression. Four common genes were identified: ATM, ATR, CUBN, and CELSR2.
ATM germline variants correlate with DNA damage in neurofibromas
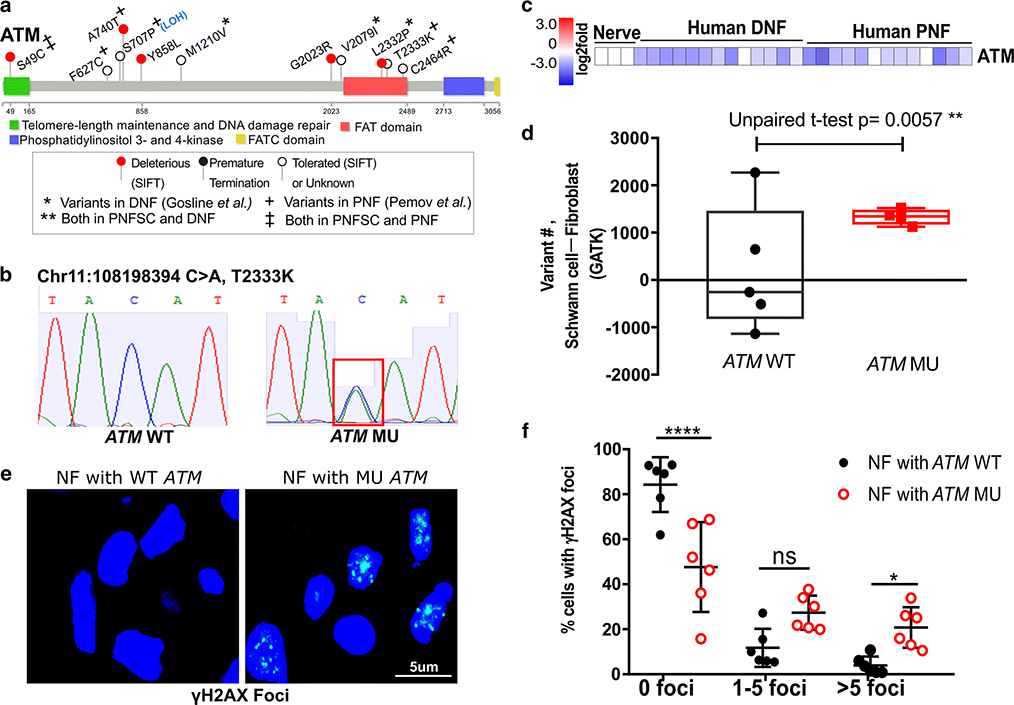
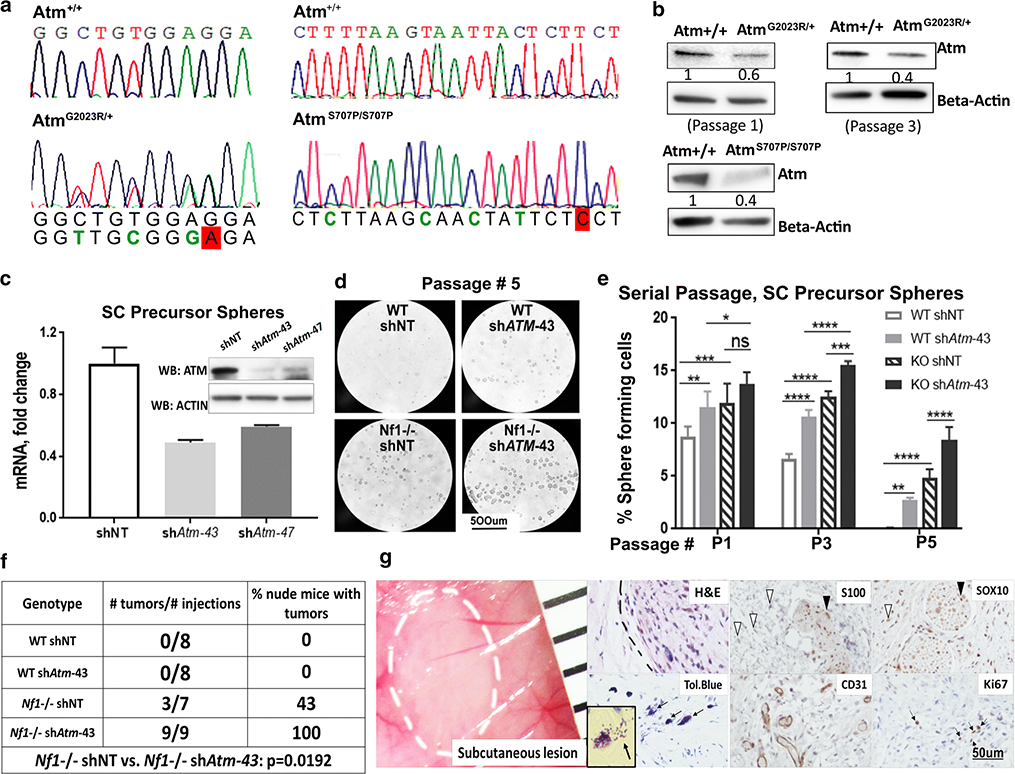
We focused on ATM, a DNA repair-related tumor suppressor gene, which showed variants in the germline of 27% of individuals in PNF cohorts (44% in our test cohort and 24% in the validation PNF cohort), and in the DNF validation cohort at lower incidence (15%) (Fig. 2a). The ATM germline variants are described (Fig. 3a, Suppl. Fig 6, Suppl. Tables 3, 5 and 7). Clinically, none of the identified ATM variants have been deemed pathogenic in Ataxia Telangiectasia (A-T) patients. Some are of uncertain significance while other are designated benign. Instead, consistent with the fact that modifiers need not be pathogenic on their own [25], they may have modifier functions in the context of NF1 mutant Schwann cells. The role of individual ATM variants on the function of the very large ATM protein will require further analysis in neurofibroma cells. Indicating possible disease relevance in cancer predisposition, however, the ATM variants S49C and F858L were reported in A-T patients who developed breast cancer [57, 67]. Notably, a breast cancer patient with an ATM F858L variant showed excess loss of heterozygosity (LOH) in breast cancer tissue [27], consistent with a modifier role for this variant. S49C, present in 2/30 PNF, was associated with risk for melanoma, prostate cancer and oropharyngeal cancer [16]. Western blot analyses of a T-cell line derived from a patient with G2023R (T5a/c) allele did not show expression of full length ATM protein, suggesting that this variant may be not stable [9]. S707P showed LOH in one PNF sample. Although C2464R was reported to not interfere with ATM kinase activation [59], ATML2332P, T2333K and C2464R are located in the FAT domain of the ATM protein, which is important for regulating ATM activity [34] (Fig. 3a, Suppl. Fig 6).
Figure 3. Germline ATM variants and related DNA damage in neurofibroma cells.
(a) Lollipop plots show ATM germline variants and their predicted functional effect on protein structure in all analyzed PNF and DNF. (b) Sanger sequencing confirmation of an ATM germline variant. (c) ATM mRNA is reduced in neurofibroma. Expression is shown as Log2 expression change, normalized to the average expression level of ATM in controls (normal human nerve). (d) Variants are increased in SC versus FB in neurofibromas from patients with ATM variants. The change in number of total somatic (including synonymous and nonsynonymous) variants between SC and FB per PNF tumor (ATM wildtype or ATM variant) are indicated (un-paired t-test). (e) Fluorescent detection of γH2AX foci in neurofibroma sections with ATM wildtype (ATM WT) or ATM variant (ATM MU). Blue: DAPI (nuclei); Green: γH2AX foci. (f) Analysis of γH2AX foci in nuclei of neurofibroma tissue sections with ATM WT or ATM variant. The percent of nuclei with 0, 1–5, or >5 γH2AX foci within a nucleus was calculated. (2Way ANOVA, *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001.)
We verified ATM variants in 3 PNF specimens identified by WES using Sanger sequencing. Results are shown for a missense variant (T2223K) (Fig3b), and two others (Suppl. Fig 7a and 7b). DNA was unavailable to verify other variants. In addition, ATM gene expression was down-regulated in DNF and PNF tumors as compared to normal nerve (Fig. 2d and 3c), although not in cultured Schwann cells (Fig. 2d). We hypothesized that NF1 patient PNF cells with ATM germline variants might have compromised ability to repair DNA damage, because ATM acts upstream in the repair of DNA double-strand breaks via homologous recombination [8, 11].
We compared numbers of synonymous and non-synonymous DNA variants in SC (with biallelic NF1 loss) versus FB, comparing cells from PNF with (n=4) and without (n=5) ATM variants. Importantly, there were significantly more variants in SC than in FB in all 4 PNF with variant ATM compared to those without ATM variants (p=0.0057; Fig. 3d). In this context, it should be noted that deficiency for various DNA repair pathways, including downstream HR proteins, crosslink repair and mismatch repair can lead to increased levels of point mutations [46, 75]. The above effect (Fig. 3d) is likely to result from oncogenic stress in SC with bi-allelic NF1 mutations (with elevated Ras-GTP) versus stress in FB, which retain one functional NF1 allele. Oncogenic stress activates the RAS/MAPK pathway, which can itself compromise the DNA damage response [1, 32, 69]. ATM variant in some neurofibroma samples additively increase the defect of DNA damage repair.
To further test whether NF1 patient neurofibroma cells with ATM variants are compromised in DNA damage repair, we monitored foci assembled by γH2AX, a marker for DNA strand breaks [45, 54], in PNF tissue sections (Fig 3e). ATM WT PNF contained few cells with more than 5 γH2AX foci, and most cells harbored none, while in PNF sections with ATM variants significantly more γH2AX foci are present (Fig 3f). There was no significant difference in Ki67 staining between neurofibromas with and without ATM variants (Suppl. Fig 7c), excluding the possibility that γH2AX foci are increased due to increased cell proliferation. Senescent cells are rare in neurofibromas [13], and thus unlikely to account for the increase in foci. Thus, increased levels of γH2AX foci suggest increased levels of unrepaired DNA damage in neurofibromas with ATM germline variants.
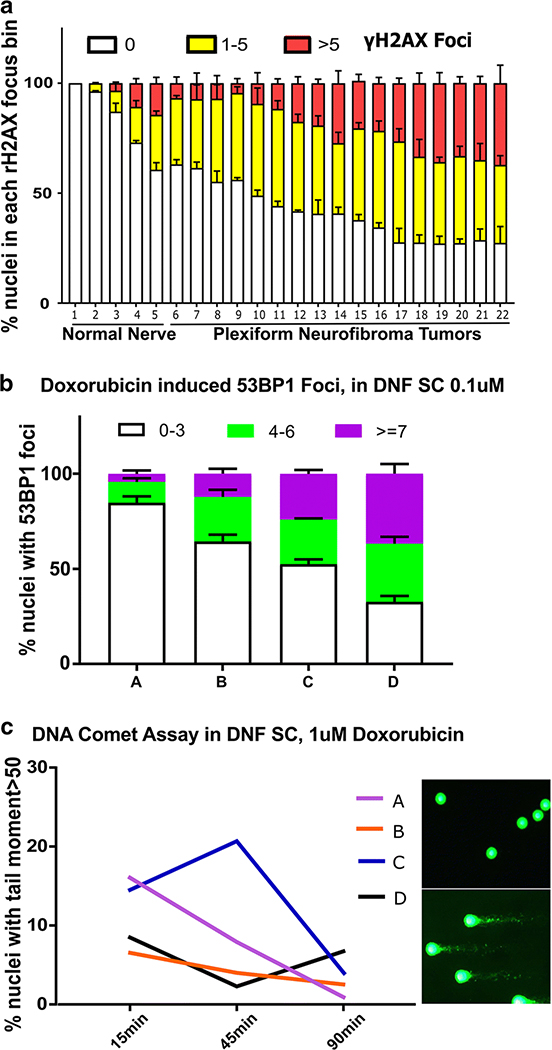
These results predicted that subsets of unselected neurofibromas might show increased numbers of γH2AX foci. Indeed, quantification of γH2AX foci in neurofibromas of unknown ATM genomic status (n=17) versus normal peripheral nerve (n=5) identified a subset of tumors with increased γH2AX foci (Fig. 4a). We also cultured DNF-derived Schwann cells from 4 individuals (with unknown ATM genomic status), and exposed them to the DNA damaging agent doxorubicin. Cells from each individual showed different levels of nuclear focus formation, as marked by 53BP1 (TP53-binding protein 1) which is recruited to sites of DNA damage (Fig. 4b), and variable timing of DNA damage level repair as monitored by comet assay, a single cell gel electrophoresis assay which indicates residual (unrepaired) DNA strand breaks (Fig. 4c). The variability in DNA damage repair capacity in Schwann cells from different DNFs, indicated by assembly of γH2AX and 53BP1 foci that correspond to DNA double-strand breaks, and as measured using a Comet assay, is consistent with the idea that variants in DNA damage related genes are relevant in some but not all neurofibromas.
Fig 4. Variable DNA damage repair in neurofibroma cells from patients with unknown ATM status.
(a) Analysis of γH2AX foci in nuclei of peripheral nerve and neurofibroma tissue sections. The percent of nuclei with 0, 1–5, or >5 γH2AX foci within a nucleus was calculated. (b) 53BP1 foci in Schwann cells cultured from DNFs. The percent of nuclei with 0–3, 4–7 or >7 53BP1 foci was counted 1 recovery after exposure to 0.1uM Doxorubicin (2-way ANOVA). (c) Comet assay. Time course shows the percent of nuclei with DNA comet tail moment >50 in each of 4 individual DNF samples exposed to 1uM doxorubicin. Repair was delayed in 2 of the samples. Images of DNA comet with DNA damage repaired (upper right) and unrepaired (lower right).
ATM missense alleles decrease ATM protein expression
Analysis of individual ATM missense variants is important, but expression of the very large ATM cDNA, especially stable expression, has proven challenging [48]. We used the CRISPR/Cas9 system to introduce the ATM missense variant G2023R and S707P into immortalized mouse kidney (MK4) cells (Suppl. Fig 8a), and verified the presence of a heterozygous G2023R missense variant and a homozygous S707P missense variant in single colonies by Sanger sequencing (Fig. 5a). We chose these variants because western blot analyses of a T-cell line derived from a A-T patient with G2023R (T5a/c) allele did not show expression of full length ATM protein, suggesting that this variant might not be not stable [9] and because S707P showed LOH in one PNF sample. Fig. 5b show western blots of protein lysates from parental cells and cells expressing one allele ATM G2023R and two alleles S707P. Both changes decreased protein expression by about half. We also generated clones without ATM alterations after CRISPR/CAS9; in those clones, ATM protein expression level was equal to that of parental cells (not shown).
Fig 5. Atm missense variants effect on protein expression and ATM effects on SCP self-renewal and tumorigenesis.
(a) Sanger sequencing identified a heterozygous Atm G2023R variant and a homozygous S707P variant in colonies of mK4 cells. (b) Western blot indicated that heterozygous Atm G2023R allele and homozygous Atm S707P change reduce of protein expression (relative ATM protein quantification by ImageJ). (c)Atm mRNA and protein are reduced in E12.5 mouse SCP treated with shAtm versus non-targeting (NT) control. (d) Photomicrographs of SCP spheres formed by wild type or Nf1−/− SCP with non-targeting control or shAtm, 4 days after plating at passage 5. (e) shAtm increases self-renewal of SCP spheres (2-way ANOVA, *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001). (f) shAtm increases SCP tumor number. A Fisher Exact test was performed. (g) Left, gross image of a xenograft tumor under the skin of a mouse grafted with Nf1−/−; shATM SCPs. The ruler shows 1 mm markings. Right, histological analysis of paraffin sections. The inset shows a higher magnification image of a de-granulated metachromatic (purple) mast cell in a toluidine (Tol.) blue stained section.
shAtm promotes Schwann cell precursor self-renewal and neurofibroma formation
To directly test if decreased ATM function affects SC tumorigenesis, we used embryonic Schwann cell precursor cells (SCPs), a cell-of-origin for PNF51. SCPs grow as self-renewing spheres that can be passaged in vitro, and form small neurofibromas on transplantation [29, 70]. Two shRNAs (shAtm-43 and shAtm-47) showed similar decreases of Atm in extent of mRNA reduction (Fig. 5c); the average percent of reduction over 4 experiments was 49% vs. 55%. Reduction in protein was also observed. Genetically reducing Atm significantly increased the number of floating spheres formed by wild type and Nf1−/− SCPs (Fig. 5d, 5e). Representative experiments are shown for one shRNA, but both gave similar results. Reduced ATM significantly increased SCP self-renewal, especially when SCP were deficient in Nf1 and with increasing numbers of passages, consistent with an increase in self-renewal (Fig. 5e). In contrast to these effects on SCPs, there was no effect of shAtm on proliferation of wild type or NF1 deficient immortalized human SC (Suppl. Fig. 8b and 8c), or mouse Schwann cells (eSC) (Suppl. Fig. 8d and 8e).
To test if reducing Atm in SCP increases tumorigenic potential, we injected SCPs subcutaneously into nude mice. Wild type SCP (with or without shAtm) did not form tumors. Half of the mice grafted with Nf1−/− SCP formed lesions, confirming previous results [70]. Consistent with increased tumor initiation, all mice grafted with Nf1−/−; shAtm SCP formed neurofibromas (Fig. 5f). Histology of the neurofibroma-like lesions formed by SCPs of both genotypes were similar; the benign lesions contained SOX10+ cells (SCPs and SCs) and S100+ SCs (Fig. 5g). Blood vessels and nerves infiltrated the lesions. Cell proliferation was low, with occasional Ki67 positive cells (Fig. 5g). Each lesion showed mast cell accumulation, a feature of neurofibromas; numbers of mast cells were similar between Nf1 null neurofibromas with shAtm or non-targeting controls (not shown). Thus, reducing Atm in Nf1 mutant SCPs increases self-renewal in vitro and increases neurofibroma initiation in vivo.
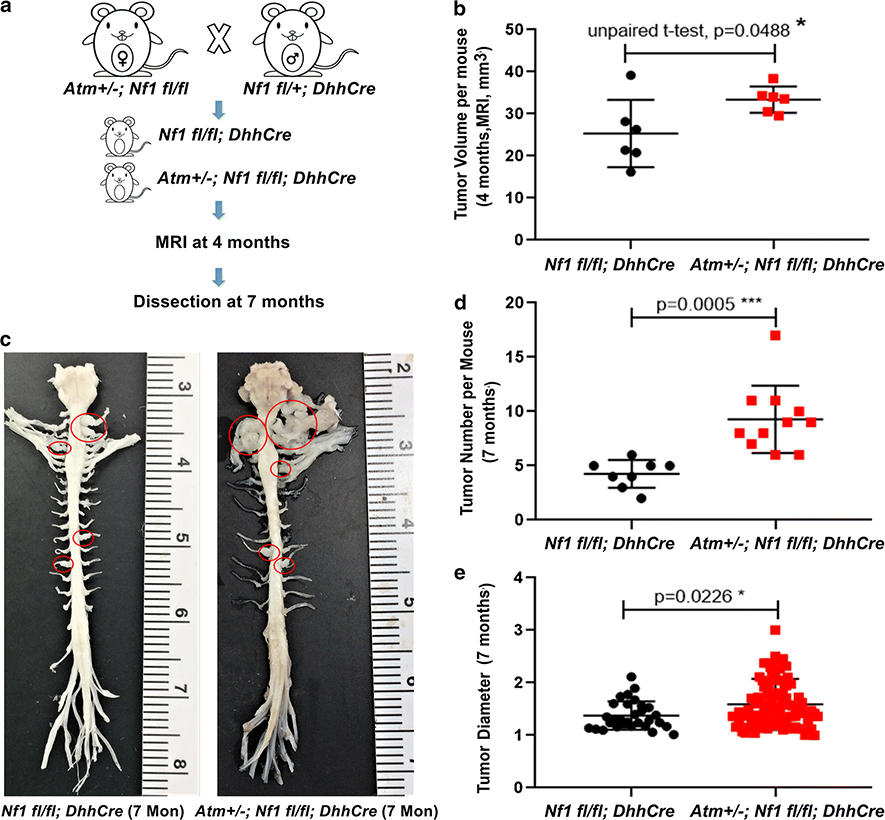
Atm heterozygosity increases neurofibroma number
These shRNA experiments left open the possibility of off-target effects of shAtm, and the possibility that Atm was more than 50% reduced in the SCPs, a situation unlikely in ATM variants. To test if a 50% reduction in Atm increases neurofibroma formation in vivo we turned to a mouse model. In the DhhCre;Nf1fl/fl model all mice form paraspinal neurofibromas, which are visible on magnetic resonance imaging (MRI) in 4 month old mice [73]. We generated Atm+/−; DhhCre;Nf1fl/fl mice and aged them to 4 months of age (Fig. 6a). At this early time point, Atm+/−; DhhCre;Nf1fl/fl mice showed increased tumor burden versus littermate controls (Fig. 6b). This could result from increased tumor number and/or tumor size. To discriminate these alternatives, we aged mice to 7 mo. and performed gross dissections, and histology of tumors. Consistent with shAtm increasing SCP self-renewal and tumor formation on grafting (Fig. 5), on dissecting mice at 7 months old we observed increased tumor number in Atm+/−; DhhCre;Nf1/fl/fl mice versus littermate controls (DhhCre;Nf1/fl/fl); tumor size only slightly changed (Fig6c, d, e). Although Atm+/−; DhhCre;Nf1/fl/fl sections showed increased numbers of mast cells per high powered field (Suppl. Fig 9b), there was no change in tumor grade between the groups; mitoses and atypia were absent, and cell proliferation (Ki67+ cells) was similar (Suppl. Fig 9a). The data are consistent with Atm heterozygosity modifying neurofibroma number.
Fig 6. Atm heterozygosity increases neurofibroma number.
(a) Schematic of generation of neurofibroma mice breeding with or without Atm heterozygosity. (b) Volumetric measurements of MRI scans of mice at 4 months of age. (n=6 Nf1 fl/fl; DhhCre, n=6 Atm+/−; Nf1 fl/fl; DhhCre). Tumor volume was increased in Atm+/−; Nf1 fl/fl; DhhCre mice. Un-paired t-test, *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001 (c) Representative images of gross dissections of spinal cords attached to peripheral nerves and tumors from mice at 7 months of age. (d, e) Statistical analysis (n=8 mice, Nf1 fl/fl; DhhCre, n=11 mice, Atm+/−; Nf1 fl/fl; DhhCre, at 7 months of age. (d) Tumor number/mouse, unpaired t-test. (e)Tumor diameter: Nf1 fl/fl; DhhCre mouse total of 30 tumors and Atm+/−; Nf1 fl/fl; DhhCre total of 102 tumors, unpaired t-test.
DISCUSSION
NF1 is a monogenic disorder, and NF1 mutation alone accounts for NF1 disease. Modifier genes can affect disease expressivity. We conclude that plexiform neurofibroma initiation can be modified by germline variants in the DNA repair-related gene ATM. This conclusion is based on our identification of ATM germline variants in 8/30 (27%) of resected PNF from NF1 patients and on data in cell culture models and a genetically engineered mouse model system showing that reduced Atm increases neurofibroma initiation. There is also an increase in germline variants in other identified genes in these samples, all from patients with neurofibromas. Our results are in line with studies of other tumor predisposition cohorts, including large scale studies on families with predisposition to sarcoma, which show increased number of variants in specific genes [4, 10]. Whether ATM also plays a role in DNF initiation remains unclear; fewer (2/13; 15.4%) DNF showed ATM variants. Other recurrent germline variants, predicted to be deleterious/damaging, also occur in PNF and in DNF (e.g. COL14A1, CELSR2, CUBN, OBSCN and PKHD1L1) and require investigation for roles in NF1. Much larger cohorts will define the relative percentages of dermal and plexiform neurofibromas with ATM variants, and variants in other genes.
We inferred germline sequence by comparing genotypes of tumor FB and matched SC from each PNF. Identification of variants in the same genes - in addition to NF1- in our analysis of cells and in 2 other cohorts, for which blood was used as a control, supports our use of FB as a germline control. However, it remains formally possible that in some tumors these alterations are not germline, but occur in development prior to somatic NF1 loss in SCs. During embryological development Schwann cells develop from neural crest cells, as do endoneurial fibroblasts [30]. Perineurial fibroblasts are not crest-derived [20]. Therefore, if somatic loss of function in NF1 occurred prior to divergence of perineurial and SC lineages during development, or if endoneurial FB were sufficiently abundant, we might have identified some FB with biallelic NF1 mutations. Our data are consistent with findings that SCs are the only cell type within a neurofibroma harboring a ‘second hit’ in the NF1 gene [33].
Separating neurofibroma FBs and SCs enabled the detection of germline and somatic NF1 variants in PNF. In individual NF1 patients, somatic NF1 mutation in each DNF tumor results from an independent mutational event in Schwann cells [33, 64], and we show that this also occurs in PNF (e.g. the two PNFs from P5). Also, in some PNF, a second, rare, somatic NF1 mutant can be detected. A second heterozygous SC may undergo a somatic NF1 mutation in a neurofibroma-bearing peripheral nerve, or an adjacent fascicle contains Schwann cells with a different somatic mutation. Whether the low alternative reads ratio somatic variants are drivers or passengers in neurofibroma development, or sequencing artifacts, remains to be determined.
We detected low frequency somatic variants in a set of genes. Recurrent somatic changes were not reported in a previous study, because the authors only studied variants with >30% alternative reads ratio [42]. Interestingly, an HMCN1 somatic mutation was reported in 1 of 7 DNFs from one NF1 patient [19]; our analysis detected HMCN1 variants predicted to be deleterious/damaging to protein in 3 of 9 PNFs, so analysis of additional tumors may identify recurrent alterations. The significance of these remains unclear.
We used changes in gene expression as an additional criterion to ensure that a gene flagged as variant is expressed in the cells/tissue of interest, as recommended [28]. CELSR2 expression was down-regulated in dermal and PNF Schwann cells and tumors, and showed germline variants in PNF (47%) and DNF (23%), some of which were potentially deleterious/damaging. CELSR2 encodes a Ciliopathy gene implicated in Joubert syndrome, and plays roles in the planar cell polarity pathway [7].
OBSCN and PDKHL1 each showed germline alterations in PNF and DNF. PKHD1L1 is a poorly studied homolog of the autosomal recessive polycystic kidney disease gene, containing 78 exons and spanning 168-kb [26]. OBSCN haploinsufficiency predisposes to dilated cardiomyopathy [35], so alterations in one allele might be disease relevant. OBSCN contains a Rho-GEF domain and a kinase domain (Suppl. Fig. 4c); reduced levels of OBSCN decreased Rho-GTP signaling [44]. OBSCN encodes numerous cell type specific protein isoforms (50kd - 900kd) generated through alternative splicing. Variants in OBSCN have been reported in breast and colon cancers; intriguingly, reduced levels of OBSCN increased breast cancer cell viability after DNA damage [43]. OBSCN is also a very large gene with many variants reported in ExAC, so more work will be needed to prove functional effects in NF1 tumors.
Most germline variants we defined in NF1 patients in the DNA repair genes ATR and ATM were heterozygous, consistent with roles for their haploinsufficiency in predisposition to cancer [40, 65]. Our studies in cell culture and in mouse models strongly support a role for Atm heterozygosity in modifying neurofibroma number in mice. Haploinsufficiency of ATM in a RAS-driven model of pancreatic cancer increased numbers of tumors and numbers of metastases [17]. Furthermore, we demonstrated that ATM variants correlate with increased numbers of γH2AX foci in human samples, similar to increased percentage of cells with γH2AX foci in precancerous epithelial cells [17]. The DNA comet assay supports the hypothesis that DNF SC (with increased Ras-GTP) from some individuals have reduced DNA damage repair capacity. We found that patients harboring ATM variants, but not those with WT ATM, also display low but increased levels of somatic mutations, demonstrated by increased levels of variants in SCs versus fibroblasts (Fig. 3d). While neurofibromas are characterized by a stable genome, ATM variants could result in rare downstream mutations, some of which could contribute to the development of neurofibroma growth or progression. It should be noted that a small increase in the level of somatic mutations associated with ATM variants could be in any gene or in other areas in the genome, and not necessarily lead to readily detectible recurrent mutations. Also, while ATM has canonical roles in repair of DNA double strand breaks (DSBs), it also has non-canonical roles in cell survival and genome stability, and as a Redox sensor that could contribute to its role in self-renewal [6, 14].
Our identification of ATM missense variants in NF1 patients with plexiform neurofibroma is consistent with missense variants functioning as modifier variants. Of note, the ATM S49C variant is reported in the ExAC dataset in the control European population at 1.1%. This frequency would preclude this variant from consideration as a primary, highly penetrant, cause of a Mendelian condition, and this variant would be considered benign for the primary NF1 phenotype as reported in ClinVar. However, it is not necessary that a modifier is rare in the general population, because modifiers alter a primary gene’s impact. Thus, without NF1 mutations, these modifier alleles might have a lesser effect.
While to date we have been unable to generate Atm missense mutants in primary cells to test the functional significance of identified variants on DNA repair, mRNA stability, or protein kinase activity in NF1 mutant cells, the increased mutational burden in ATM mutant human neurofibroma SC suggests that in SC with biallelic NF1 mutations and elevated RAS-GTP, effects of ATM variants are enhanced. Importantly, ATM expression was decreased in PNF and DNF compared to normal nerve in our previous microarray analysis and G2023R, identified in one patient (T5a/c) is an A-T patient mutation [9]. In immortalized mK4 cells, cells with the heterozygous missense G2023R allele and homozygous S707P alleles, each decreased protein expression. Potentially, other missense variants identified by sequencing may also decrease ATM expression, and so mimic the heterozygous loss of Nf1 in our murine study (Fig. 6). As new technologies enable missense mutant generation without requiring clonal selection, it will be of importance to study the properties of each identified variant to identify their effect in NF1.
Recent studies identified ATM and ATR as potential germline risk alleles for human sarcoma, including MPNST. The ATM and ATR kinases are activated by different types of DNA-damage and regulate DNA damage response. Their functions are largely not redundant [34], and ATM and ATR proteins can work together to co-regulate downstream processes [8]. In our study, ATM -- but not ATR -- variants correlated with increased DNA variant burden in SC versus FB. Our finding of increased γH2AX foci in tumors harboring identified ATM variants supports a role for these variants in DNA repair, and P1 and P2, harboring germline sequence variants in ATM and ATR, respectively, showed mild genome-wide genomic CNA (Fig. 1d). Also, Nf1;p53 mice develop MPNST that show a weak mutator phenotype [23]. However, we cannot exclude the possibility that some ATM variants are silent polymorphisms or have effects outside DNA repair functions. In addition to modifying NF1 initiation, reduced levels of ATM might facilitate loss of the previously normal NF1 allele in neurofibroma, or facilitate progression to malignancy, possibilities we have not yet tested. Consistent with Atm heterozygosity modifying neurofibroma number, our in vivo experiments show that Atm loss increases neurofibroma number without changing tumor size or grade. Future studies will be needed to assess whether at later time points Atm increases transformation to malignancy.
In conclusion, we identified predicted deleterious/damaging damgermline variants in 21 genes, in addition to the known driver mutation NF1, in neurofibroma cells. None of these have previously been evaluated in NF1 tumorigenesis. Genes, including ATM, showing germline variants in neurofibroma are potential modifiers of tumor formation and/or transformation to malignancy, and sets of variants occurring in different neurofibromas may contribute to clinical variability.
Supplementary Material
ACKNOWLEDGEMENTS
This work was funded by NIH-R37-NS083580 (NINDS Javits Neuroscience Investigator Award to NR) and an Innovation Award from Cincinnati Children’s Hospital. We thank Dr. Carlos Prada for review of the manuscript, Sage Bioinformatics (Sara J.C. Gosline) for access to the dermal neurofibroma dataset prior to publication, A. Pemov and D. Stewart (NCI) for assistance with data transfer, and Doug Marchuk (Duke) for suggesting use of the Burden test. We thank the CHTN Midwestern Division for some samples used in this study. All procedures involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Footnotes
CONFLICTS OF INTEREST
The authors declare no conflicts of interest.
Publisher's Disclaimer: This Author Accepted Manuscript is a PDF file of an unedited peer-reviewed manuscript that has been accepted for publication but has not been copyedited or corrected. The official version of record that is published in the journal is kept up to date and so may therefore differ from this version.
References
- 1.Abulaiti A, Fikaris AJ, Tsygankova OM, Meinkoth JL (2006) Ras induces chromosome instability and abrogation of the DNA damage response. Cancer Res 66: 10505–10512 Doi 10.1158/0008-5472.CAN-06-2351 [DOI] [PubMed] [Google Scholar]
- 2.Allaway RJ, Gosline SJC, La Rosa S, Knight P, Bakker A, Guinney J, Le LQ (2018) Cutaneous neurofibromas in the genomics era: current understanding and open questions. Br J Cancer 118: 1539–1548 Doi 10.1038/s41416-018-0073-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anastasaki C, Dahiya S, Gutmann DH (2017) KIR2DL5 mutation and loss underlies sporadic dermal neurofibroma pathogenesis and growth. Oncotarget 8: 47574–47585 Doi 10.18632/oncotarget.17736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ballinger ML, Goode DL, Ray-Coquard I, James PA, Mitchell G, Niedermayr E, Puri A, Schiffman JD, Dite GS, Cipponi A et al. (2016) Monogenic and polygenic determinants of sarcoma risk: an international genetic study. Lancet Oncol 17: 1261–1271 Doi 10.1016/S1470-2045(16)30147-4 [DOI] [PubMed] [Google Scholar]
- 5.Beert E, Brems H, Daniels B, De Wever I, Van Calenbergh F, Schoenaers J, DebiecRychter M, Gevaert O, De Raedt T, Van Den Bruel A et al. (2011) Atypical neurofibromas in neurofibromatosis type 1 are premalignant tumors. Genes Chromosomes Cancer 50: 1021–1032 Doi 10.1002/gcc.20921 [DOI] [PubMed] [Google Scholar]
- 6.Berger ND, Stanley FKT, Moore S, Goodarzi AA (2017) ATM-dependent pathways of chromatin remodelling and oxidative DNA damage responses. Philos Trans R Soc Lond B Biol Sci 372: Doi 10.1098/rstb.2016.0283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boutin C, Labedan P, Dimidschstein J, Richard F, Cremer H, Andre P, Yang Y, Montcouquiol M, Goffinet AM, Tissir F (2014) A dual role for planar cell polarity genes in ciliated cells. Proc Natl Acad Sci U S A 111: E3129–3138 Doi 10.1073/pnas.1404988111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caron P, Choudjaye J, Clouaire T, Bugler B, Daburon V, Aguirrebengoa M, Mangeat T, Iacovoni JS, Alvarez-Quilon A, Cortes-Ledesma F et al. (2015) Non-redundant Functions of ATM and DNA-PKcs in Response to DNA Double-Strand Breaks. Cell Rep 13: 15981609 Doi 10.1016/j.celrep.2015.10.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carranza D, Vega AK, Torres-Rusillo S, Montero E, Martinez LJ, Santamaria M, Santos JL, Molina IJ (2017) Molecular and Functional Characterization of a Cohort of Spanish Patients with Ataxia-Telangiectasia. Neuromolecular Med 19: 161–174 Doi 10.1007/s12017-016-8440-8 [DOI] [PubMed] [Google Scholar]
- 10.Chan SH, Lim WK, Ishak NDB, Li ST, Goh WL, Tan GS, Lim KH, Teo M, Young CNC, Malik S et al. (2017) Germline Mutations in Cancer Predisposition Genes are Frequent in Sporadic Sarcomas. Sci Rep 7: 10660 Doi 10.1038/s41598-017-10333-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen CC, Kass EM, Yen WF, Ludwig T, Moynahan ME, Chaudhuri J, Jasin M (2017) ATM loss leads to synthetic lethality in BRCA1 BRCT mutant mice associated with exacerbated defects in homology-directed repair. Proc Natl Acad Sci U S A 114: 76657670 Doi 10.1073/pnas.1706392114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chmielecki J, Bailey M, He J, Elvin J, Vergilio JA, Ramkissoon S, Suh J, Frampton GM, Sun JX, Morley S et al. (2017) Genomic Profiling of a Large Set of Diverse Pediatric Cancers Identifies Known and Novel Mutations across Tumor Spectra. Cancer Res 77: 509–519 Doi 10.1158/0008-5472.CAN-16-1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Courtois-Cox S, Genther Williams SM, Reczek EE, Johnson BW, McGillicuddy LT, Johannessen CM, Hollstein PE, MacCollin M, Cichowski K (2006) A negative feedback signaling network underlies oncogene-induced senescence. Cancer Cell 10: 459–472 Doi 10.1016/j.ccr.2006.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cremona CA, Behrens A (2014) ATM signalling and cancer. Oncogene 33: 3351–3360 Doi 10.1038/onc.2013.275 [DOI] [PubMed] [Google Scholar]
- 15.De Raedt T, Brems H, Wolkenstein P, Vidaud D, Pilotti S, Perrone F, Mautner V, Frahm S, Sciot R, Legius E (2003) Elevated risk for MPNST in NF1 microdeletion patients. Am J Hum Genet 72: 1288–1292 Doi 10.1086/374821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dombernowsky SL, Weischer M, Allin KH, Bojesen SE, Tybjaerg-Hansen A, Nordestgaard BG (2008) Risk of cancer by ATM missense mutations in the general population. J Clin Oncol 26: 3057–3062 Doi 10.1200/JCO.2007.14.6613 [DOI] [PubMed] [Google Scholar]
- 17.Drosos Y, Escobar D, Chiang MY, Roys K, Valentine V, Valentine MB, Rehg JE, Sahai V, Begley LA, Ye J et al. (2017) ATM-deficiency increases genomic instability and metastatic potential in a mouse model of pancreatic cancer. Sci Rep 7: 11144 Doi 10.1038/s41598-017-11661-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Easton DF, Ponder MA, Huson SM, Ponder BA (1993) An analysis of variation in expression of neurofibromatosis (NF) type 1 (NF1): evidence for modifying genes. Am J Hum Genet 53: 305–313 [PMC free article] [PubMed] [Google Scholar]
- 19.Emmerich D, Zemojtel T, Hecht J, Krawitz P, Spielmann M, Kuhnisch J, Kobus K, Osswald M, Heinrich V, Berlien P et al. (2015) Somatic neurofibromatosis type 1 (NF1) inactivation events in cutaneous neurofibromas of a single NF1 patient. Eur J Hum Genet 23: 870–873 Doi 10.1038/ejhg.2014.210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Erlandson RA, Woodruff JM (1982) Peripheral nerve sheath tumors: an electron microscopic study of 43 cases. Cancer 49: 273–287 [DOI] [PubMed] [Google Scholar]
- 21.Faden DL, Asthana S, Tihan T, DeRisi J, Kliot M (2017) Whole Exome Sequencing of Growing and Non-Growing Cutaneous Neurofibromas from a Single Patient with Neurofibromatosis Type 1. PLoS One 12: e0170348 Doi 10.1371/journal.pone.0170348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ferner RE, Huson SM, Thomas N, Moss C, Willshaw H, Evans DG, Upadhyaya M, Towers R, Gleeson M, Steiger C et al. (2007) Guidelines for the diagnosis and management of individuals with neurofibromatosis 1. J Med Genet 44: 81–88 Doi 10.1136/jmg.2006.045906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garza R, Hudson RA 3rd, McMahan CA, Walter CA, Vogel KS (2007) A mild mutator phenotype arises in a mouse model for malignancies associated with neurofibromatosis type 1. Mutat Res 615: 98–110 Doi 10.1016/j.mrfmmm.2006.11.031 [DOI] [PubMed] [Google Scholar]
- 24.Gosline SJ, Weinberg H, Knight P, Yu T, Guo X, Prasad N, Jones A, Shrestha S, Boone B, Levy SE et al. (2017) A high-throughput molecular data resource for cutaneous neurofibromas. Sci Data 4: 170045 Doi 10.1038/sdata.2017.45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hamilton BA, Yu BD (2012) Modifier genes and the plasticity of genetic networks in mice. PLoS Genet 8: e1002644 Doi 10.1371/journal.pgen.1002644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hogan MC, Griffin MD, Rossetti S, Torres VE, Ward CJ, Harris PC (2003) PKHDL1, a homolog of the autosomal recessive polycystic kidney disease gene, encodes a receptor with inducible T lymphocyte expression. Hum Mol Genet 12: 685–698 [PubMed] [Google Scholar]
- 27.Izatt L, Greenman J, Hodgson S, Ellis D, Watts S, Scott G, Jacobs C, Liebmann R, Zvelebil MJ, Mathew C et al. (1999) Identification of germline missense mutations and rare allelic variants in the ATM gene in early-onset breast cancer. Genes Chromosomes Cancer 26: 286–294 [PubMed] [Google Scholar]
- 28.Jalali Sefid Dashti M, Gamieldien J (2017) A practical guide to filtering and prioritizing genetic variants. Biotechniques 62: 18–30 Doi 10.2144/000114492 [DOI] [PubMed] [Google Scholar]
- 29.Joseph NM, Mosher JT, Buchstaller J, Snider P, McKeever PE, Lim M, Conway SJ, Parada LF, Zhu Y, Morrison SJ (2008) The loss of Nf1 transiently promotes self-renewal but not tumorigenesis by neural crest stem cells. Cancer Cell 13: 129–140 Doi 10.1016/j.ccr.2008.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Joseph NM, Mukouyama YS, Mosher JT, Jaegle M, Crone SA, Dormand EL, Lee KF, Meijer D, Anderson DJ, Morrison SJ (2004) Neural crest stem cells undergo multilineage differentiation in developing peripheral nerves to generate endoneurial fibroblasts in addition to Schwann cells. Development 131: 5599–5612 Doi 10.1242/dev.01429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim A, Stewart DR, Reilly KM, Viskochil D, Miettinen MM, Widemann BC (2017) Malignant Peripheral Nerve Sheath Tumors State of the Science: Leveraging Clinical and Biological Insights into Effective Therapies. Sarcoma 2017: 7429697 Doi 10.1155/2017/7429697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Knauf JA, Ouyang B, Knudsen ES, Fukasawa K, Babcock G, Fagin JA (2006) Oncogenic RAS induces accelerated transition through G2/M and promotes defects in the G2 DNA damage and mitotic spindle checkpoints. J Biol Chem 281: 3800–3809 Doi 10.1074/jbc.M511690200 [DOI] [PubMed] [Google Scholar]
- 33.Maertens O, Brems H, Vandesompele J, De Raedt T, Heyns I, Rosenbaum T, De Schepper S, De Paepe A, Mortier G, Janssens S et al. (2006) Comprehensive NF1 screening on cultured Schwann cells from neurofibromas. Hum Mutat 27: 1030–1040 Doi 10.1002/humu.20389 [DOI] [PubMed] [Google Scholar]
- 34.Marechal A, Zou L (2013) DNA damage sensing by the ATM and ATR kinases. Cold Spring Harb Perspect Biol 5: Doi 10.1101/cshperspect.a012716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marston S, Montgiraud C, Munster AB, Copeland O, Choi O, Dos Remedios C, Messer AE, Ehler E, Knoll R (2015) OBSCN Mutations Associated with Dilated Cardiomyopathy and Haploinsufficiency. PLoS One 10: e0138568 Doi 10.1371/journal.pone.0138568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mautner VF, Asuagbor FA, Dombi E, Funsterer C, Kluwe L, Wenzel R, Widemann BC, Friedman JM (2008) Assessment of benign tumor burden by whole-body MRI in patients with neurofibromatosis 1. Neuro Oncol 10: 593–598 Doi 10.1215/15228517-2008-011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Messiaen LM, Callens T, Mortier G, Beysen D, Vandenbroucke I, Van Roy N, Speleman F, Paepe AD (2000) Exhaustive mutation analysis of the NF1 gene allows identification of 95% of mutations and reveals a high frequency of unusual splicing defects. Hum Mutat 15: 541–555 Doi [DOI] [PubMed] [Google Scholar]
- 38.Miettinen MM, Antonescu CR, Fletcher CDM, Kim A, Lazar AJ, Quezado MM, Reilly KM, Stemmer-Rachamimov A, Stewart DR, Viskochil D et al. (2017) Histopathologic evaluation of atypical neurofibromatous tumors and their transformation into malignant peripheral nerve sheath tumor in patients with neurofibromatosis 1-a consensus overview. Hum Pathol 67: 1–10 Doi 10.1016/j.humpath.2017.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miller SJ, Jessen WJ, Mehta T, Hardiman A, Sites E, Kaiser S, Jegga AG, Li H, Upadhyaya M, Giovannini M et al. (2009) Integrative genomic analyses of neurofibromatosis tumours identify SOX9 as a biomarker and survival gene. EMBO Mol Med 1: 236–248 Doi 10.1002/emmm.200900027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.O’Driscoll M (2008) Haploinsufficiency of DNA Damage Response Genes and their Potential Influence in Human Genomic Disorders. Curr Genomics 9: 137–146 Doi 10.2174/138920208784340795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Olive PL, Wlodek D, Banath JP (1991) DNA double-strand breaks measured in individual cells subjected to gel electrophoresis. Cancer Res 51: 4671–4676 [PubMed] [Google Scholar]
- 42.Pemov A, Li H, Patidar R, Hansen NF, Sindiri S, Hartley SW, Wei JS, Elkahloun A, Chandrasekharappa SC, Program NCS et al. (2017) The primacy of NF1 loss as the driver of tumorigenesis in neurofibromatosis type 1-associated plexiform neurofibromas. Oncogene 36: 3168–3177 Doi 10.1038/onc.2016.464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Perry NA, Shriver M, Mameza MG, Grabias B, Balzer E, Kontrogianni-Konstantopoulos A (2012) Loss of giant obscurins promotes breast epithelial cell survival through apoptotic resistance. FASEB J 26: 2764–2775 Doi 10.1096/fj.12-205419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Perry NA, Vitolo MI, Martin SS, Kontrogianni-Konstantopoulos A (2014) Loss of the obscurin-RhoGEF downregulates RhoA signaling and increases microtentacle formation and attachment of breast epithelial cells. Oncotarget 5: 8558–8568 Doi 10.18632/oncotarget.2338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Podhorecka M, Skladanowski A, Bozko P (2010) H2AX Phosphorylation: Its Role in DNA Damage Response and Cancer Therapy. J Nucleic Acids 2010: Doi 10.4061/2010/920161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Polak P, Kim J, Braunstein LZ, Karlic R, Haradhavala NJ, Tiao G, Rosebrock D, Livitz D, Kubler K, Mouw KW et al. (2017) A mutational signature reveals alterations underlying deficient homologous recombination repair in breast cancer. Nat Genet 49: 1476–1486 Doi 10.1038/ng.3934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Prada CE, Rangwala FA, Martin LJ, Lovell AM, Saal HM, Schorry EK, Hopkin RJ (2012) Pediatric plexiform neurofibromas: impact on morbidity and mortality in neurofibromatosis type 1. J Pediatr 160: 461–467 Doi 10.1016/j.jpeds.2011.08.051 [DOI] [PubMed] [Google Scholar]
- 48.Qi J, Shackelford R, Manuszak R, Cheng D, Smith M, Link CJ, Wang S (2004) Functional expression of ATM gene carried by HSV amplicon vector in vitro and in vivo. Gene Ther 11: 25–33 Doi 10.1038/sj.gt.3302140 [DOI] [PubMed] [Google Scholar]
- 49.Radomska KJ, Coulpier F, Gresset A, Schmitt A, Debbiche A, Lemoine S, Wolkenstein P, Vallat JM, Charnay P, Topilko P (2019) Cellular Origin, Tumor Progression, and Pathogenic Mechanisms of Cutaneous Neurofibromas Revealed by Mice with Nf1 Knockout in Boundary Cap Cells. Cancer Discov 9: 130–147 Doi 10.1158/21598290.CD-18-0156 [DOI] [PubMed] [Google Scholar]
- 50.Rasmussen SA, Friedman JM (2000) NF1 gene and neurofibromatosis 1. Am J Epidemiol 151: 33–40 [DOI] [PubMed] [Google Scholar]
- 51.Ratner N, Miller SJ (2015) A RASopathy gene commonly mutated in cancer: the neurofibromatosis type 1 tumour suppressor. Nat Rev Cancer 15: 290–301 Doi 10.1038/nrc3911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rieley MB, Stevenson DA, Viskochil DH, Tinkle BT, Martin LJ, Schorry EK (2011) Variable expression of neurofibromatosis 1 in monozygotic twins. Am J Med Genet A 155A: 478–485 Doi 10.1002/ajmg.a.33851 [DOI] [PubMed] [Google Scholar]
- 53.Riordan JD, Nadeau JH (2017) From Peas to Disease: Modifier Genes, Network Resilience, and the Genetics of Health. Am J Hum Genet 101: 177–191 Doi 10.1016/j.ajhg.2017.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM (1998) DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem 273: 58585868 Doi 10.1074/jbc.273.10.5858 [DOI] [PubMed] [Google Scholar]
- 55.Rosenbaum T, Rosenbaum C, Winner U, Muller HW, Lenard HG, Hanemann CO (2000) Long-term culture and characterization of human neurofibroma-derived Schwann cells. J Neurosci Res 61: 524–532 Doi [DOI] [PubMed] [Google Scholar]
- 56.Sabbagh A, Pasmant E, Laurendeau I, Parfait B, Barbarot S, Guillot B, Combemale P, Ferkal S, Vidaud M, Aubourg P et al. (2009) Unravelling the genetic basis of variable clinical expression in neurofibromatosis 1. Hum Mol Genet 18: 2768–2778 Doi 10.1093/hmg/ddp212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sandoval N, Platzer M, Rosenthal A, Dork T, Bendix R, Skawran B, Stuhrmann M, Wegner RD, Sperling K, Banin S et al. (1999) Characterization of ATM gene mutations in 66 ataxia telangiectasia families. Hum Mol Genet 8: 69–79 [DOI] [PubMed] [Google Scholar]
- 58.Sathirapongsasuti JF, Lee H, Horst BA, Brunner G, Cochran AJ, Binder S, Quackenbush J, Nelson SF (2011) Exome sequencing-based copy-number variation and loss of heterozygosity detection: ExomeCNV. Bioinformatics 27: 2648–2654 Doi 10.1093/bioinformatics/btr462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Scott SP, Bendix R, Chen P, Clark R, Dork T, Lavin MF (2002) Missense mutations but not allelic variants alter the function of ATM by dominant interference in patients with breast cancer. Proc Natl Acad Sci U S A 99: 925–930 Doi 10.1073/pnas.012329699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Simanshu DK, Nissley DV, McCormick F (2017) RAS Proteins and Their Regulators in Human Disease. Cell 170: 17–33 Doi 10.1016/j.cell.2017.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sites ER, Smolarek TA, Martin LJ, Viskochil DH, Stevenson DA, Ullrich NJ, Messiaen LM, Schorry EK (2017) Analysis of copy number variants in 11 pairs of monozygotic twins with neurofibromatosis type 1. Am J Med Genet A 173: 647–653 Doi 10.1002/ajmg.a.38058 [DOI] [PubMed] [Google Scholar]
- 62.Staser K, Yang FC, Clapp DW (2012) Pathogenesis of plexiform neurofibroma: tumorstromal/hematopoietic interactions in tumor progression. Annu Rev Pathol 7: 469–495 Doi 10.1146/annurev-pathol-011811-132441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sweet-Cordero EA, Biegel JA (2019) The genomic landscape of pediatric cancers: Implications for diagnosis and treatment. Science 363: 1170–1175 Doi 10.1126/science.aaw3535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Thomas L, Kluwe L, Chuzhanova N, Mautner V, Upadhyaya M (2010) Analysis of NF1 somatic mutations in cutaneous neurofibromas from patients with high tumor burden. Neurogenetics 11: 391–400 Doi 10.1007/s10048-010-0240-y [DOI] [PubMed] [Google Scholar]
- 65.van Os NJ, Roeleveld N, Weemaes CM, Jongmans MC, Janssens GO, Taylor AM, Hoogerbrugge N, Willemsen MA (2016) Health risks for ataxia-telangiectasia mutated heterozygotes: a systematic review, meta-analysis and evidence-based guideline. Clin Genet 90: 105–117 Doi 10.1111/cge.12710 [DOI] [PubMed] [Google Scholar]
- 66.Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA Jr., Kinzler KW (2013) Cancer genome landscapes. Science 339: 1546–1558 Doi 10.1126/science.1235122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vorechovsky I, Luo L, Lindblom A, Negrini M, Webster AD, Croce CM, Hammarstrom L (1996) ATM mutations in cancer families. Cancer Res 56: 4130–4133 [PubMed] [Google Scholar]
- 68.Vu V, Verster AJ, Schertzberg M, Chuluunbaatar T, Spensley M, Pajkic D, Hart GT, Moffat J, Fraser AG (2015) Natural Variation in Gene Expression Modulates the Severity of Mutant Phenotypes. Cell 162: 391–402 Doi 10.1016/j.cell.2015.06.037 [DOI] [PubMed] [Google Scholar]
- 69.Weyemi U, Lagente-Chevallier O, Boufraqech M, Prenois F, Courtin F, Caillou B, Talbot M, Dardalhon M, Al Ghuzlan A, Bidart JM et al. (2012) ROS-generating NADPH oxidase NOX4 is a critical mediator in oncogenic H-Ras-induced DNA damage and subsequent senescence. Oncogene 31: 1117–1129 Doi 10.1038/onc.2011.327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Williams JP, Wu J, Johansson G, Rizvi TA, Miller SC, Geiger H, Malik P, Li W, Mukouyama YS, Cancelas JA et al. (2008) Nf1 mutation expands an EGFR-dependent peripheral nerve progenitor that confers neurofibroma tumorigenic potential. Cell Stem Cell 3: 658–669 Doi 10.1016/j.stem.2008.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wu J, Dombi E, Jousma E, Scott Dunn R, Lindquist D, Schnell BM, Kim MO, Kim A, Widemann BC, Cripe TPet al (2012) Preclincial testing of sorafenib and RAD001 in the Nf(flox/flox) ;DhhCre mouse model of plexiform neurofibroma using magnetic resonance imaging. Pediatr Blood Cancer 58: 173–180 Doi 10.1002/pbc.23015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wu J, Keng VW, Patmore DM, Kendall JJ, Patel AV, Jousma E, Jessen WJ, Choi K, Tschida BR, Silverstein KA et al. (2016) Insertional Mutagenesis Identifies a STAT3/Arid1b/beta-catenin Pathway Driving Neurofibroma Initiation. Cell Rep 14: 1979–1990 Doi 10.1016/j.celrep.2016.01.074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wu J, Williams JP, Rizvi TA, Kordich JJ, Witte D, Meijer D, Stemmer-Rachamimov AO, Cancelas JA, Ratner N (2008) Plexiform and dermal neurofibromas and pigmentation are caused by Nf1 loss in desert hedgehog-expressing cells. Cancer Cell 13: 105–116 Doi 10.1016/j.ccr.2007.12.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhao H, Sun Z, Wang J, Huang H, Kocher JP, Wang L (2014) CrossMap: a versatile tool for coordinate conversion between genome assemblies. Bioinformatics 30: 1006–1007 Doi 10.1093/bioinformatics/btt730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zou X, Owusu M, Harris R, Jackson SP, Loizou JI, Nik-Zainal S (2018) Validating the concept of mutational signatures with isogenic cell models. Nat Commun 9: 1744 Doi 10.1038/s41467-018-04052-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.