Abstract
Liver is the place where cholesterol is synthesized, transported, secreted, and transformed, thus liver takes an irreplaceable role in cholesterol homeostasis. Hepatic cholesterol metabolism differs between breeds, yet the molecular mechanism is unclear. In this study Large White (LW) and Erhualian (EHL) piglets (at birth and 25-day-old) were used, 6 each time point per breed. Erhualian piglets had significantly lower body and liver weight compared with Large White at birth and weaning, but the liver/ body weight ratio was higher at weaning, associated with increased serum and liver cholesterol and triglyceride content. The mRNA expression of Cholesterol-7alpha-hydroxylase (CYP7a1) and Recombinant Acetyl Coenzyme Acetyltransferase 2 (ACAT2) were down-regulated in Erhualian piglets at birth, while hepatic Sterol-regulatory element binding protein 2 (SREBP2) mRNA expression was up-regulated in Erhualian piglets at weaning, as well as SREBP2 protein content, compared with Large White piglets. At birth, the depressed CYP7a1 transcription in Erhualian piglets was associated with decreased Histone H3 (H3) and increased Histone H3 lysine 27 trimethylation (H3K27me3). While the results revealed significant promoter hypermethylation of 3-Hydroxy-3-methylglutaryl-CoA reductase (HMGCR) promoter in Erhualian piglets at weaning, together with increased Histone H3 lysine 9 monomethylation (H3K9me1) and Histone H3 lysine 4 trimethylation (H3K4me3). These results suggest that epigenetic modification may be an important mechanism in hepatic cholesterol metabolism among different species, which is vital for maintaining cholesterol homeostasis and decreasing risk of cardiovascular disease.
Keywords: piglets, cholesterol metabolism, liver, epigenetic regulation, CYP7a1, HMGCR
Background
Cholesterol participates in the formation of cell membrane (1) and can synthesis bile acid and vitamin D as a precursor, which is of great significance to the life activities of the body (2, 3). Liver cholesterol content is determined by the net balance of cholesterol synthesis, transport and catabolism, i.e., cholesterol can be converted into bile acids (4). HMGCR is the key enzymes in cholesterol biosynthesis (5). HMGCR transcription is regulated by Sterol-regulatory element binding proteins (SREBPs), of which SREBP2 is the major subtype that regulates liver cholesterol (6, 7). Meanwhile, the content of cholesterol in the cell can affect the activity of HMGCR and SREBP2, thereby forming a feedback loop that forms the homeostasis of cholesterol (8). The transport of cholesterol is that into and out of the liver. Low-density lipoprotein (LDL) molecules are the main operator of cholesterol in the blood which are responsible for transporting liver cholesterol to other peripheral tissues, while High-density lipoprotein (HDL) transports cholesterol from extrahepatic tissues back to the liver. Low-density lipoprotein receptor (LDL-R) mediates the uptake of low-density lipoprotein cholesterol (LDL-C), which is very important in regulating serum concentrations of total cholesterol (Tch) and LDL-C. The key enzyme of cholesterol synthesis of bile acid in liver through the predominant pathway is CYP7a1 (9), however Cholesterol-27alpha-hydroxylase (CYP27a1) is the rate-limiting enzyme in the alternative pathway of bile acid synthesis.
The content of cholesterol in liver and serum is different vary from species to species and even within the same species. Studies have shown that the serum cholesterol content of different breeds of pigs was different (10, 11). Serum cholesterol concentration was lower in Full blood Chianina compared with crossbreds (Hereford × Angus crossbred), which was affected by sex class and dietary (12). There are significant differences in the sensitivity of different breeds of rabbits to hypercholesterolemia caused by a high cholesterol diet. Polymorphisms at genetic loci may be associated with significant heterogeneity in sensitivity to dietary cholesterol. These polymorphisms include absorption of dietary cholesterol, conversion of liver cholesterol to bile acids, feedback inhibition of endogenous cholesterol synthesis, or regulation of the LDL-R pathway (13–15). The expression level of LDL-R mRNA was different in muscle of pigs from different genetic groups, which was correlated with Intramuscular fat of longus dorsi (IMF) content in the Longissimus dorsi muscle of pigs (16). Indeed, while there are many reports about the difference of liver cholesterol content among varieties, molecular mechanisms of the difference were seldom studied.
HMGCR is essential for cholesterol synthesis. CYP7a1 is the rate-limiting enzymes in the classical pathway of bile acids synthesis in liver (17). Epigenetic regulation including DNA methylation and histone modification on fetal gene expression during cholesterol metabolism have been well-demonstrated (18). In angiocardiopathy and cancer researches, CYP7a1 and HMGCR gene are described to be vulnerable to epigenetic regulation including DNA methylation and histone modification (18). However, whether interspecific differences affects gene expression via epigenetics has not been well-explored. Meaney provide a clue that DNA acylation has a key role in the overall coordination of cholesterol homeostasis (19). Different species modifies hepatic cholesterol metabolic gene transcription by DNA methylation and histones modification like H3K4me3, H3K27me3, and H3K9me1 (19–21). Activation of the CYP7a1 gene is associated with an increase in H3 acetylation and H3K4me3 and a decrease in H3K9me1 and H3K27me3. Meanwhile, the activation of HMGCR transcription is accompanied by an increase in H3 acetylation and a decrease in histones H3, H3K9me1, and H3K27me3 (21–24). Based on these studies, we hypothesized that epigenetic events have a crucial function in modifying liver cholesterol metabolism in Large White and Erhualian piglets.
Materials and Methods
Animal and Sampling
The experimental animals include Large White and Erhualian pig, and the animal experiments were performed in the Conservation and Breeding Farm at Jiangsu Polytechnic College of Agriculture and Forestry, Jurong, Jiangsu Province, China. LW and EHL piglets (at birth and 25-day-old) were used in the study, 6 each time point per breed. All piglets were killed and sampled after weighing. Serum specimens were prepared and stored at −20°C. Liver specimens were collected within 20 min of death, quickly frozen in liquid nitrogen and stored at −80°C until further analysis. Animal experiments were conducted under the guidance of the Animal Ethics Committee of Northwest A&F University, China.
Cholesterol in Serum and Liver
Total cholesterol in serum and liver was measured using a commercial cholesterol test kit (AO10027, Jinma Biotechnology Co., Ltd., Wenzhou, China). Respective assay kits (006340 and 006328, respectively, Beijing BHKT Clinical Reagent Co., Ltd., Beijing, China) were used to detect the concentrations of LDL-C and High-density lipoprotein cholesterol (HDL-C) in the serum. Liver cholesterol was extracted according to previously reported methods (Gibney and Nolan 2010). Briefly, 200 mg frozen liver sample was pipette in 1 ml of lysis buffer (18 mmol/L Tris, pH 7.5, 300 mmol/L mannitol, 50 mmol/L Ethylene Diamine Tetraacetic Acid(EDTA), 0.1 mmol/L PMSF) by a Polytron homogenizer (PT1200E, Brinkman Instruments, Littau, Switzerland). Two hundred microliter of homogenate were vigorously mixed with 800 μl chloroform/methanol (2:1, vol/vol) and centrifuged at 3,000 g for 5 min.
Cholesterol was extracted from the organic phase, air-dried and reconstituted in 30 μl of a mixture of tert-butanol and methanol (13:2, vol/vol). The cholesterol content was determined by cholesterol assay kit (Jinma Biotechnology Co., Ltd., Wenzhou, China).
Total RNA Isolation and mRNA Quantification
Total RNA was isolated from liver samples using Trizol Reagent (Tiangen Biotech Co., Ltd., Beijing, China). Each sample was synthesized from total RNA using iScript cDNA Synthesis Toolkit (Promega, Madison, WI, USA) according to the manufacturer's instructions. Diluted 2 μl of cDNA (1:50) for real-time polymerase chain reaction (PCR). Primer sequences are shown in Table 1 and were synthesized by Invitrogen (Shanghai, China). Real-time PCR was performed in Mx3000P (Stratagene, USA). Simulated reverse transcription (RT) and no template controls were set to monitor the possible contamination of genomic and exogenous DNA both at RT and PCR. The specificity of amplification was determined by melting curve analysis and PCR product sequence.
Table 1.
Primer sequences of the target genes.
| Target | Primer sequence (5′-3′) | Product (bp) | GenBank No. |
|---|---|---|---|
| SREBP1 | F: GCGACGGAGCCTCTGGTAGT | 217 | NM214157.1 |
| R: GCAAGACGGCGGATTTATTCA | |||
| SREBP2 | F: GCCTACCGCAAGGTGTTTC | 305 | DQ020476.1 |
| R: GTCATTTGCTGGCAGTCGTT | |||
| HMGCR | F: CAGGCTGAAGTAAGGGAGA | 174 | DQ432054.1 |
| R: CACGAAGTAGGTGGCGAGA | |||
| LDL-R | F: ACTGCTCATCCTCCTCTT | 109 | AF065990.1 |
| R: TTCCGTGGTCTTCTGGTA | |||
| LXR | F: ATTTCCAGGAGTGCCGTCTT | 102 | AB254406.1 |
| R: CTTGCCGCTTCAGTTTCTT | |||
| FXR | F: CGGAGAAGCATTACCA | 137 | XM 001928800.2 |
| R: AAGCATTCAGCCAACA | |||
| CYP7a1 | F: TATTCCTTCCGTTACCGAGTG | 262 | AK230868.1 |
| R: ACCTGACCAGTTCCGAGAT | |||
| CYP27a1 | F: TGTGGCTCGCATCGTTC | 153 | EF625352.1 |
| R: TCACCTGGCAGCTCCTT | |||
| GAPDH | F: TACATGGTCTACATGTTCCAGTATG | 285 | DQ403065 |
| R: CAGGAGGCATTGCTGACAATCTTG |
Tissue Protein Extraction and Western Blot Analysis
Extract total and nuclear proteins from 100 mg of frozen liver tissue, as previously described (30). Detection of protein concentration with Pierce BCA Protein Assay Kit (Thermo Scientific, USA). Western blot analysis for SREBP2 (ab30682, Abcam, UK, diluted 1:200) and HMGCR (sc-33827, Santa Cruz, USA, diluted 1:100) were follow the manufacturer's instructions. HMGCR and SREBP2 was normalized with Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (KC-5G4, Kangcheng, China, diluted 1:10,000).
Chromatin Immunoprecipitation (ChIP)
ChIP experiment was performed according to the modified based on previous reports (37, 34). Briefly, 200 mg frozen liver sample was taken in liquid nitrogen, ground, and then resuspended in phosphate buffered saline (PBS) containing a mixture of protease inhibitors cocktail (Roche Diagnostics GmbH, Mannheim, Germany) and cross-linked in 1% formaldehyde for 10 min at room temperature. Then use 2.5 M glycine to stop the cross-linking reaction.
Washing the pellets with PBS and sodium dodecyl sulfate (SDS) lysis buffer (50 M Tris-HCl pH 8.1, 10 mM EDTA, 1% SDS containing protease inhibitors. The samples after cross-linked were sonicated for 10 min on ice with 10 s on/off intervals (Sonics Vibra, USA). To remove cell debris from the crude chromatin preparations the samples were then centrifuged at 12,000 rpm for 10 min at 4°C. According to the results of 1% agarose gel measurement, the average length of ultrasonic chromatin was about 500 bp. Prepare salmon sperm DNA/G protein agarose beads (60 μl, 50% slurry, Biyuntian, Biotechnology, China) in advance, dilute the Protein-DNA complex with ChIP dilution solution, and incubate 2 μg of the corresponding antibody at 4°C overnight [histone H3 antibody, ab1791, Abcam; anti-acetyl-histone H3, 06-599, Millipore; monomethyl-Histone H3K9 (Lys9) 17-680, Millipore; trimethyl-histone H3K27 (Lys27), 17-622, Millipore; trimethyl-histone H3K4 antibody, ab1012, Abcam]. Negative control group did not add antibodies. Add G protein agarose beads (120 μl slurry 50%) to capture chromatin immunoprecipitation complex. The pellets containing the immunoprecipitation complex were washed sequentially, and the antibody/protein/DNA complex were eluted from the protein G agarose beads. Finally, reversing cross-linking DNA fragments at 65°C for 5 h to purify the DNA that released from the immunoprecipitation complex. Immunoprecipitated DNA was used as a specific primer for the RT-PCR template to amplify the genomic sequence of the promoter regions of the HMGCR and CYP7a1 genes.SREBP2 has no genomic DNA sequence, so this study did not use ChIP-qPCR to detect the SREBP2 promoter.
Statistical Analysis
All data are expressed as mean ± SEM and were analyzed using independent-samples test with SPSS 13.0 for windows. The method of 2−ΔΔCt was used to analyze the real-time PCR data. P < 0.05 is considered statistically significant.
Result
Body Weight and Cholesterol in Serum and Liver
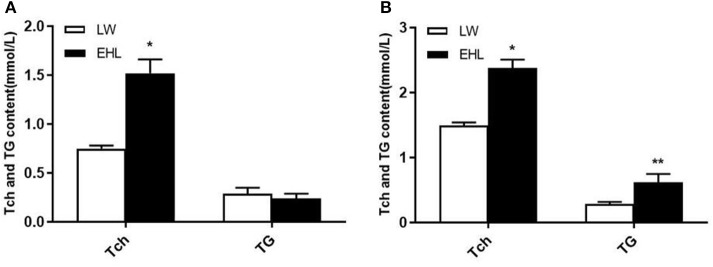
As shown in Table 2, the piglets born from EHL sows had significantly lower body weight and liver weight at birth and weaning, while the ratio of liver weight to body weight (liver index) did not change at birth. The piglets born from EHL significantly increased liver index at weaning. This growth retardation is associated with significantly increased serum concentration and liver content of Tch (P < 0.05; Figure 1), HDL-C (P < 0.01), and LDL-C (P < 0.01) at birth and weaning.
Table 2.
Body weight, liver weight, liver index, and the content of cholesterol in serum and liver of the offspring piglets at birth and weaning (n = 6 piglets each time point per breed).
| Parameters | LW | EHL | ||
|---|---|---|---|---|
| Birth (0 d) | Weaning (25 d) | Birth (0 d) | Weaning (25 d) | |
| Liver weight (g) | 31.76 ± 1.72 | 178.74 ± 15.40 | 17.16 ± 1.21** | 119.84 ± 6.72** |
| Body weight (kg) | 1.31 ± 0.04 | 8.10 ± 0.34 | 0.75 ± 0.03** | 4.08 ± 0.25** |
| Liver index (g/kg) | 24.27 ± 0.91 | 21.92 ± 1.15 | 23.04 ± 2.01 | 29.51 ± 1.10** |
| TG (mmol/L) | 0.29 ± 0.06 | 0.29 ± 0.03 | 0.24 ± 0.05 | 0.62 ± 0.128** |
| Tch (mmol/L) | 0.75 ± 0.03 | 1.49 ± 0.05 | 1.52 ± 0.14* | 2.38 ± 0.13* |
| HDL-C (mmol/L) | 0.30 ± 0.02 | 0.47 ± 0.02 | 0.43 ± 0.03** | 1.16 ± 0.08** |
| LDL-C (mmol/L) | 0.23 ± 0.02 | 0.47 ± 0.01 | 0.52 ± 0.07** | 0.76 ± 0.05** |
P < 0.05 and
P < 0.01. Data are presented as mean ± SEM.
Figure 1.
Total cholesterol and triglyceride content of the offspring piglets at birth (A) and weaning (B) (n = 6 piglets each time point per breed). *P < 0.05 and **P < 0.01. Data are presented as mean ± SEM.
Expression of Hepatic Genes Involved in Cholesterol Metabolism
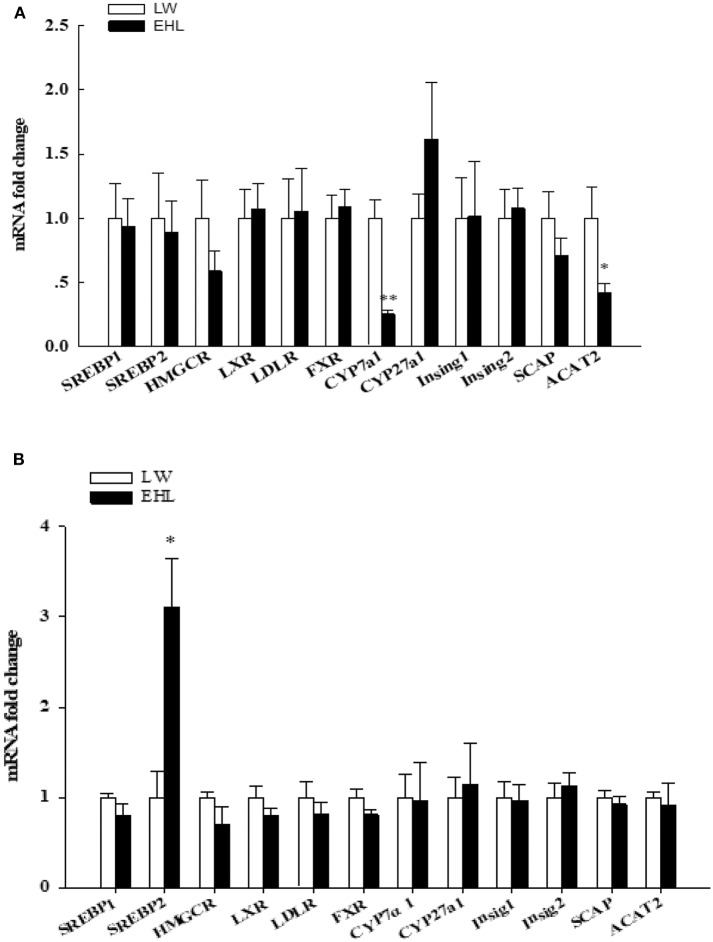
Using RT-PCR to quantitative detection mRNA abundance of 12 genes that involved in liver cholesterol metabolism. Among these genes, ACAT2 (P < 0.05) and CYP7a1 (P < 0.01) were found to be significantly down-regulated in the liver of EHL piglets at birth (Figure 2A). The levels of hepatic SREBP2 mRNA in EHL piglets significantly higher than LW piglets at weaning (P < 0.05; Figure 2B).
Figure 2.
Hepatic expression of genes involved in cholesterol metabolism of the offspring piglets at birth (A) and weaning (B) (n = 6 piglets each time point per breed). *P < 0.05, **P < 0.01. Data are presented as mean ± SEM.
Hepatic SREBP2 and HMGCR Protein Content
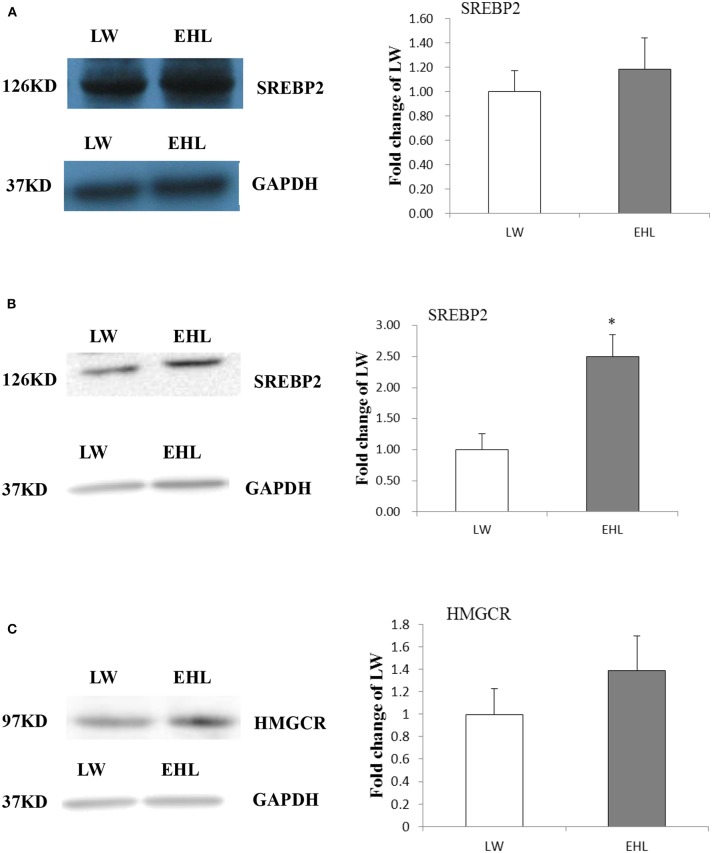
Western blot was used to detect the protein content of SREBP2 and HMGCR in the liver. The content of SREBP2 protein (Figure 3A, P > 0.05) at birth and HMGCR protein (Figure 3C, P > 0.05) at weaning was not affected by variety and age. However, the SREBP2 content in nuclear lysate was significantly increased (Figure 3B, P < 0.05) in the liver of EHL piglets at weaning, which was consistent with liver mRNA abundance.
Figure 3.
Hepatic SREBP2 and HMGCR protein content. (A) Expressed SREBP2 protein content at birth; (B) expressed SREBP2 protein content at weaning; (C) expressed HMGCR protein content at weaning (n = 6 piglets each time point per breed). *P < 0.05. Data are presented as mean ± SEM.
DNA Methylation
MeDIP analysis showed that the HMGCR promoter was significantly hypermethylated (P < 0.05) in liver of EHL piglets at weaning (Figure 4). Because no CpG island was expected to exist in the CYP7a1 promoter, MeDIP analysis of the CYP7a1 promoter was excluded from this study.
Figure 4.
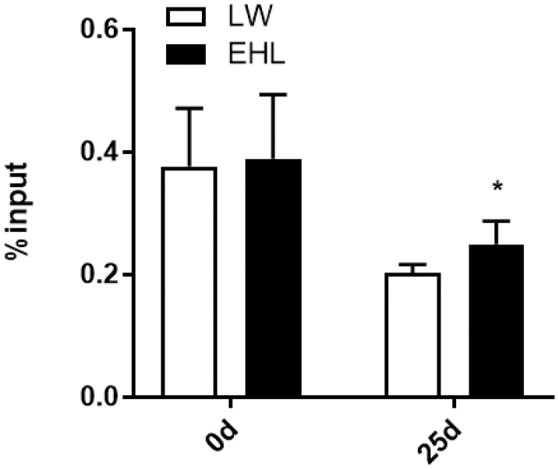
DNA methylation at HMGCR promoter in the liver of weaning piglets (n = 6 piglets each time point per breed). *P < 0.05. Data are presented as mean ± SEM.
Histone Modifications
The enrichment of four histone modification marks, namely histone H3 acetylation (H3AC), histone H3 lysine 4 trimethylation (H3K4me3), histone H3 lysine 27 trimethylation (H3K27me3), and histone H3 lysine 9 monomethylation (H3K9me1), as well as histone H3 on the promoter of HMGCR and CYP7a1, analyzed by ChIP with specific antibodies. As shown in Figure 5A, when expressed as a percentage, the activation of hepatic HMGCR gene transcription in EHL piglets at birth was associated with a 685.1% decrease in H3AC (P < 0.01). When expressed as a ratio relative to H3, the transcription of HMGCR gene was related to the decrease of H3AC (P < 0.05) by 593.8%, and increase of H3K9me1 (P < 0.05) by 77.5%. Figure 5B showed significant decrease in histone H3 (P < 0.05; −2.9%), K27H3 (P < 0.05; −4.13%), and H3AC (P < 0.01; −9.83%) at weaning, expressed as percentage of the input, yet significant increase in H3K9me1 (P < 0.05; 102.1%) and H3K4me3 (P < 0.05; 8.57%) in EHL piglets at weaning, when expressed as the ratio relative to H3.
Figure 5.
Histone modification at HMGCR and CYP7a1 promoter in the liver of weaning piglets (n = 6 piglets each time point per breeds). (A) Expressed HMGCR as the percentage of the input and the ratio relative to H3 at birth; (B) expressed HMGCR as the percentage of the input and the ratio relative to H3 at weaning; (C) expressed CYP7a1 as the percentage of the input and the ratio relative to H3 at birth; (D) expressed CYP7a1 as the percentage of the input and the ratio relative to H3 at weaning. *P < 0.05 and **P < 0.01. Data are presented as mean ± SEM.
Figure 5C shows the enrichment and the modified forms of histone H3 in the CYP7a1 promoter. The decrease in CYP7a1 transcription in EHL piglets at birth was related to the downward trend (P = 0.07) in H3K4 (−33.5%) and uptrend (P = 0.09) in H3K27 (75.3%) and significantly decreased (P < 0.01) total H3 (−52.2%), expressed as percentage of input. A significant increase (P < 0.01) of total H3AC (85.6%) and H3K27me3 (172.8%) and a trend of increase (P = 0.06) of H3K9me1 (172.4%) were seen at the CYP7a1 promoter in EHL piglets at birth when expressed as the ratio to total H3. While a trend of increase in H3AC (P = 0.07; 6.55%) and H3K4 (P = 0.06; 3.53%) and significantly decreased H3K27me3 (P < 0.05; −6.48%) were seen at the CYP7a1 promoter in EHL piglets at weaning, expressed as percentage of input. But when expressed as the ratio relative to H3 (Figure 5D), H3K27me3 (−62.6%) significantly (P < 0.01) decreased but H3K4me3 (P = 0.09; 49.1%) had the trend of increasement at weaning. There was no significant change in H3K4me3 of the promoter HMGCR or CYP7a1 when expressed as a percentage.
Discussion
EHL piglets had lower body and liver weight (P < 0.01) compared with LW piglets in both ages, while the liver/body weight ratio was higher at weaning, associated with the variety of pigs. Large White pig belongs to the breed with excellent growth rate, large body shape and high fecundity, but Erhualian pig is contrary. We have presented evidences that serum concentration and liver content of Tch and HDL-C were higher in EHL piglets, which was in agreement with early findings. Pond et al. found that the different content of serum cholesterol and HDL in variety was passed on from generation to generation when the serum of four-breed swine population was measured (11). We found that serum concentration of LDL-C was lower at birth but higher at weaning in EHL, but the reasons for these changes were not fully understood.
Cholesterol synthesis in pigs predominantly occurs in the liver. Among the ileum, cerebrum, kidney, heart, liver, semitendinosus muscle, longissimus muscle, and subcutaneous fat, the liver was the only tissue showing significant difference in cholesterol content among pigs with high or low serum cholesterol concentrations, indicating that liver is important to regulate cholesterol balance in pigs (25). There are four main pathways for cholesterol metabolism in liver: the first is that the de novo cholesterol biosynthesis mediated by HMGCR pathway; the second is that CYP7a1 and CYP27a1 catalyze the synthesis of bile acids; the third is that HDL mediate reverse cholesterol transport and LDL-R mediate endocytosis; the last is that cholesterol secrete into bloodstream via LDL-C. It has been reported that ACAT2 participate in cholesterol absorption, esterify dissociative cholesterol in cell, and reduce the toxic effect of high free cholesterol on cell (26–28).
Similarly, we demonstrated that the expression of CYP7a1 and ACAT2 were decreased in EHL at birth. Serao at al. found that different pig breeds and lines present different content of intramuscular fat (16). Because no significant change in mRNA expression in liver tissue of Isigs, Sterol-regulatory element binding protein cleavage-activating protein (SCAP), Sterol-regulatory element binding protein 1 (SREBP1), SREBP2, HMGCR, or liver content of SREBP1 protein and HMGCR protein, we demonstrated that the liver cholesterol biosynthesis was not increased in both pig breeds at birth. While the cholesterol esterification via ACAT2 and transformation via CYP7a1 were lower in EHL at birth, which is further clarified the liver cholesterol transformation to bile acids, cholesterol absorption, and dissociative cholesterol esterification were reduced. At 25-day-old, the increase of liver cholesterol content in EHL piglets is related to the significant increase of liver SREBP2 mRNA expression and the increase of SREBP2 protein in liver nuclear lysate, but no difference in CYP7a1. Thus, the liver cholesterol metabolism was balanced by reversed cholesterol transport in EHL, but the liver cholesterol content was higher in EHL compared with LW. This might be explained by higher cholesterol biosynthesis and lower cholesterol transformation in liver.
Based on our and other precious studies, liver modulates cholesterol transport, biosynthesis and transformation in pigs predominantly via epigenetic regulation (20, 21). We chose CYP7a1 and HMGCR for epigenetic because the promoter sequences of these two genes can be used in pigs and both of them are key enzymes in the cholesterol metabolism (20, 29, 30). CpG island cytosine methylation located in promoter genes is associated with gene suppression (31, 32), while histone acetylation is related to the activation of transcription (20, 29, 30). According to the type of histone, the position of amino acid residues and the number of methyl groups (mono-, di- and trimethylation), histone methylation can inhibit or activate gene transcription. H3K4me3 is generally considered an activation marker, while H3K9me1 and H3K27me3 have repressive effects on transcriptional (22–24). These views are confirmed in a representative study of transcriptional suppression of CYP7a1 in EHL is associated with increase in activation markers, H3K9me1 and H3K27me3 at birth, but the increased H3 acetylation was due to decreased histone H3, which content varies with species (33–37). Since increased histone H3K9me1 and H3K4me3, together with up-regulated SREBP2 mRNA expression, HMGCR gene should be activated, but no significant alteration was observed for the mRNA expression and protein content of HMGCR in EHL and LW at weaning. While the liver content of cholesterol was higher in EHL, we speculate that HMGCR differ in its susceptibility to cholesterol (38–42).
These results showed that different species or breeds have different cholesterol levels and metabolic. Besides, diet and environment have influence on cholesterol metabolism. Serao et al. studied the relationship between lipid content and gene expression in the muscle of three breeds of pigs, and found that the expression of LDL-R mRNA was different in different breeds of pigs. It suggests that endocytosis of cholesterol in muscle tissue was different (16). The study showed that yolk cholesterol content was not only related to the breed of laying hens, but also to the age of the hen (43). Literature indicates that fatty acid composition varies between breeds. Due to the higher oleic acid concentration, the proportion of monounsaturated fatty acids (MUFA) in marbled Wagyu and Hanwoo beef was higher. They can lower LDL-C while increasing HDL-C, and may reduce risk for cardiovascular diseases (44). Clinical studies have shown that, low concentrations of SFA C12: 0, C14: 0 are benefit and important for longissimus lumborum muscle of Boer crossbreed goats compared with Santa Inês breed sheep. They are promoting the accumulation of low-density lipoprotein, which increases the risk factors of cardiovascular disease in humans (45). A large number of experiments indicated that the cholesterol content had variety specificity. Environmental affects cholesterol metabolism, and this effect is different in specie and tissue. Recently, methylation is very important in the development of human cardiovascular diseases. With the rapid development of high-throughput methylation technology, it has made a great breakthrough in epigenomic research, other biological and clinical fields. Studies have shown that epigenetic regulation involved in cholesterol synthesis, absorption, elimination, and storage so related to total cholesterol levels. Even be involved in the regulation of lipid concentration variability, and leading to cardiovascular disease. As methylation level increasing, the risk of Coronary heart disease (CHD), and higher TG and LDL-C levels and lower HDL-C level result in the change of DNA methylation.
In conclusion, we showed that the liver content of cholesterol was higher, and the ability of cholesterol biosynthesis was stronger in EHL compared with LW, and the molecular mechanism for this difference is enzymatic regulatory and age-dependent. These results provide a new idea for the study of lipid metabolism and meat quality difference mechanism between Erhualian pigs and large white pigs, and provide a reference for the study of the cardiovascular diseases.
Data Availability Statement
All datasets generated for this study are included in the article/supplementary material.
Ethics Statement
This animal study was reviewed and approved by NWAFU.
Author Contributions
XL and RC contributed to the conception of the study. HX and XZ contributed significantly to analysis and manuscript preparation. HX, XJ, and HZ performed the data analyses and wrote the manuscript. HW, YM, and SC helped perform the analysis with constructive discussions.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank the agricultural science and technology innovation and research (2015NY173), study and application of welfare breeding technology for sows (K3380216087), we also thank the study and application of welfare breeding technology for broilers and Shaanxi agricultural finance special fund project.
References
- 1.Goedeke L, Fernandez-Hernando C. Regulation of cholesterol homeostasis. Cell Mol Life Sci. (2012) 69:915–30. 10.1007/s00018-011-0857-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ikonen E. Cellular cholesterol trafficking and compartmentalization. Nat Rev Mol Cell Biol. (2008) 9:125–38. 10.1038/nrm2336 [DOI] [PubMed] [Google Scholar]
- 3.Ouimet M, Franklin V, Mak E, Liao X, Tabas I, Marcel YL. Autophagy regulates cholesterol efflux from macrophage foam cells via lysosomal acid lipase. Cell Metab. (2011) 13:655–67. 10.1016/j.cmet.2011.03.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Khovidhunkit W, Kim MS, Memon RA, Shigenaga JK, Moser AH, Feingold K, et al. Effects of infection and inflammation on lipid and lipoprotein metabolism: mechanisms and consequences to the host. J Lipid Res. (2004) 45:1169–96. 10.1194/jlr.R300019-JLR200 [DOI] [PubMed] [Google Scholar]
- 5.Brown MS, Goldstein JL. Multivalent feedback regulation of HMG CoA reductase, a control mechanism coordinating isoprenoid synthesis and cell growth. J Lipid Res. (1980) 21:505–17. [PubMed] [Google Scholar]
- 6.Brown MS, Goldstein JL. The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell. (1997) 89:331–40. 10.1016/S0092-8674(00)80213-5 [DOI] [PubMed] [Google Scholar]
- 7.Luskey KL. Structure and expression of 3-hydroxy-3-methylglutaryl coenzyme a reductase. Ann N Y Acad Sci. (1986) 478:249–54. 10.1111/j.1749-6632.1986.tb15535.x [DOI] [PubMed] [Google Scholar]
- 8.deBose-Boyd RA. Feedback regulation of cholesterol synthesis: sterol-accelerated ubiquitination and degradation of HMG CoA reductase. Cell Res. (2008) 18:609–21. 10.1038/cr.2008.61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chiang JY. Regulation of bile acid synthesis: pathways, nuclear receptors, and mechanisms. J Hepatol. (2004) 40:539–51. 10.1016/j.jhep.2003.11.006 [DOI] [PubMed] [Google Scholar]
- 10.Pond WG, Mersmann HJ, Young LD. Heritability of plasma cholesterol and triglyceride concentrations in swine. Proc Soc Exp Biol Med. (1986) 182:221–4. 10.3181/00379727-182-42331 [DOI] [PubMed] [Google Scholar]
- 11.Pond WG, Su DR, Mersmann HJ. Divergent concentrations of plasma metabolites in swine selected for seven generations for high or low plasma total cholesterol. J Anim Sci. (1997) 75:311–6. 10.2527/1997.752311x [DOI] [PubMed] [Google Scholar]
- 12.Wheeler TL, Davis GW, Stoecker BJ, Harmon CJ. Cholesterol concentration of longissimus muscle, subcutaneous fat and serum of two beef cattle breed types. J Anim Sci. (1987) 65:1531–7. 10.2527/jas1987.6561531x [DOI] [PubMed] [Google Scholar]
- 13.Beynen AC, Katan MB, van Zutphen LF. Hypo- and hyperresponders: Individual differences in the response of serum cholesterol concentration to changes in diet. Adv Lipid Res. (1987) 22:115–71. 10.1016/B978-0-12-024922-0.50008-4 [DOI] [PubMed] [Google Scholar]
- 14.Adams WC, Gaman EM, Feigenbaum AS. Breed differences in the responses of rabbits to atherogenic diets. Atherosclerosis. (1972) 16:405–11. 10.1016/0021-9150(72)90087-1 [DOI] [PubMed] [Google Scholar]
- 15.Lacombe C, Nibbelink M. Breed differences in nutritionally induced hyperlipoproteinemia in the rabbit. Artery. (1980) 7:419–27. [PubMed] [Google Scholar]
- 16.Serao NLV, Veroneze R, Ribeiro AM, Verardo LL, Braccini Neto J, Gasparino E, et al. Candidate gene expression and intramuscular fat content in pigs. J Anim Breed Genet. (2011) 128:28–34. 10.1111/j.1439-0388.2010.00887.x [DOI] [PubMed] [Google Scholar]
- 17.Gao X, Fan JG. Study Group of Liver and Metabolism Chinese Society of Endocrinology. Diagnosis and management of non-alcoholic fatty liver disease and related metabolic disorders: consensus statement from the study group of liver and metabolism, Chinese society of endocrinology. J Diabetes. (2013) 5:406–15. 10.1111/1753-0407.12056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grimaldi V, Vietri MT, Schiano C, Picascia A, de Pascale MR, Fiorito C, et al. Epigenetic reprogramming in atherosclerosis. Curr Atheroscler Rep. (2015) 17:476. 10.1007/s11883-014-0476-3 [DOI] [PubMed] [Google Scholar]
- 19.Meaney S. Epigenetic regulation of cholesterol homeostasis. Front Genet. (2014) 5:311. 10.3389/fgene.2014.00311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chittur SV, Sangster-Guity N, McCormick PJ. Histone deacetylase inhibitors: a new mode for inhibition of cholesterol metabolism. BMC Genomics. (2008) 9:507. 10.1186/1471-2164-9-507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meaney S. Epigenetic regulation of oxysterol formation. Biochimie. (2013) 95:531–7. 10.1016/j.biochi.2012.08.020 [DOI] [PubMed] [Google Scholar]
- 22.Huang Y, Greene E, Murray Stewart T, Goodwin AC, Baylin SB, Woster PM, et al. Inhibition of lysine-specific demethylase 1 by polyamine analogues results in reexpression of aberrantly silenced genes. Proc Natl Acad Sci USA. (2007) 104:8023–8. 10.1073/pnas.0700720104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kalushkova A, Fryknas M, Lemaire M, Fristedt C, Agarwal P, Eriksson M, et al. Polycomb target genes are silenced in multiple myeloma. PLoS ONE. (2010) 5:e11483. 10.1371/journal.pone.0011483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sohi G, Marchand K, Revesz A, Arany E, Hardy DB. Maternal protein restriction elevates cholesterol in adult rat offspring due to repressive changes in histone modifications at the cholesterol 7alpha-hydroxylase promoter. Mol Endocrinol. (2011) 25:785–98. 10.1210/me.2010-0395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harris KB, Cross HR, Pond WG, Mersmann HJ. Effect of dietary fat and cholesterol level on tissue cholesterol concentrations of growing pigs selected for high or low serum cholesterol. J Anim Sci. (1993) 71:807–10. 10.2527/1993.714807x [DOI] [PubMed] [Google Scholar]
- 26.Chang TY, Chang CC, Lin S, Yu C, Li B, Miyazaki LA. Roles of acyl-coenzyme A: cholesterol acyltransferase-1 and−2. Curr Opin Lipidol. (2001) 12:289–96. 10.1097/00041433-200106000-00008 [DOI] [PubMed] [Google Scholar]
- 27.Mukherjee S, Kunitake GR, Alfinslater B. The esterification of cholesterol with palmitic acid by rat liver homogenates. J Biol Chem. (1958) 230:91–6. [PubMed] [Google Scholar]
- 28.Goodman DS, Deykin D, Shiratori T. The formation of cholesterol esters with rat liver enzymes. J Biol Chem. (1964) 239:1335–45. [PubMed] [Google Scholar]
- 29.Gibney ER, Nolan CM. Epigenetics and gene expression. Heredity. (2010) 105:4–13. 10.1038/hdy.2010.54 [DOI] [PubMed] [Google Scholar]
- 30.Xu D, Luo HW, Hu W, Hu SW, Yuan C, Wang GH, et al. Intrauterine programming mechanism for hypercholesterolemia in prenatal caffeine-exposed female adult rat offspring. FASEB J. (2018) 32:5563–76. 10.1096/fj.201701557R [DOI] [PubMed] [Google Scholar]
- 31.Miranda TB, Jones PA. DNA methylation: the nuts and bolts of repression. J Cell Physiol. (2007) 213:384–90. 10.1002/jcp.21224 [DOI] [PubMed] [Google Scholar]
- 32.Weber M, Schubeler D. Genomic patterns of DNA methylation: targets and function of an epigenetic mark. Curr Opin Cell Biol. (2007) 19:273–80. 10.1016/j.ceb.2007.04.011 [DOI] [PubMed] [Google Scholar]
- 33.Perkins PS, Young RW. Histone proteins in fetal and adult human retinas. Jpn J Ophthalmol. (1987) 31:590–7. [PubMed] [Google Scholar]
- 34.Perkins PS, Young RW. Comparisons of histones in retinal and brain nuclei from newborn and adult mice. Brain Res. (1987) 430:161–8. 10.1016/0165-3806(87)90150-7 [DOI] [PubMed] [Google Scholar]
- 35.Lindner H, Sarg B, Grunicke H, Helliger W. Age-dependent deamidation of h1(0) histones in chromatin of mammalian tissues. J Cancer Res Clin Oncol. (1999) 125:182–6. 10.1007/s004320050261 [DOI] [PubMed] [Google Scholar]
- 36.Hake SB, Garcia BA, Duncan EM, Kauer M, Dellaire G, Shabanowitz J, et al. Expression patterns and post-translational modifications associated with mammalian histone h3 variants. J Biol Chem. (2006) 281:559–68. 10.1074/jbc.M509266200 [DOI] [PubMed] [Google Scholar]
- 37.Sellos D, van Wormhoudt A. [Characterization of histones and chromatin of the hepatopancreas in Palaemon serratus (Crustacea Natantia)]. Biochimie. (1979) 61:393–404. 10.1016/S0300-9084(79)80133-9 [DOI] [PubMed] [Google Scholar]
- 38.Bosron WF, Lumeng L, Li TK. Genetic polymorphism of enzymes of alcohol metabolism and susceptibility to alcoholic liver disease. Mol Aspects Med. (1988) 10:147–58. 10.1016/0098-2997(88)90019-2 [DOI] [PubMed] [Google Scholar]
- 39.Hukkanen J, Pelkonen O, Raunio H. Expression of xenobiotic-metabolizing enzymes in human pulmonary tissue: possible role in susceptibility for ild. Eur Respir J Suppl. (2001) 32:122–26s. [PubMed] [Google Scholar]
- 40.Wang HR, Li XB, Yang QN, Tang MH, Xiao CB, Wan L. [Metabolic stability and metabolic enzyme phaenotypes of lanceolatin b in liver microsomes of different species by UPLC-MS/MS]. Zhongguo Zhong Yao Za Zhi. (2016) 41:124–28. 10.4268/cjcmm20160124 [DOI] [PubMed] [Google Scholar]
- 41.Allegri G, Bertazzo A, Biasiolo M, Costa CV, Ragazzi E. Kynurenine pathway enzymes in different species of animals. Adv Exp Med Biol. (2003) 527:455–63. 10.1007/978-1-4615-0135-0_53 [DOI] [PubMed] [Google Scholar]
- 42.Rippa M, Giovannini PP, Barrett MP, Dallocchio F, Hanau S. 6-phosphogluconate dehydrogenase: the mechanism of action investigated by a comparison of the enzyme from different species. Biochim Biophys Acta. (1998) 1429:83–92. 10.1016/S0167-4838(98)00222-2 [DOI] [PubMed] [Google Scholar]
- 43.Yang PK, Tian YD, Sun GR, Jiang RR, Han RL, Kang XT. Deposition rule of yolk cholesterol in two different breeds of laying hens. Genet Mol Res. (2013) 12:5786–92. 10.4238/2013.November.22.5 [DOI] [PubMed] [Google Scholar]
- 44.Gotoh T, Joo ST. Characteristics and health benefit of highly marbled Wagyu and Hanwoo beef. Korean J Food Sci Anim Resour. (2016) 36:709–18. 10.5851/kosfa.2016.36.6.709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gama K, Pereira Filho JM, Soares RF, Cordao MA, Cezar MF, Batista AS, et al. Fatty acid, chemical, and tissue composition of meat comparing Santa Ines breed sheep and Boer crossbreed goats submitted to different supplementation strategies. Trop Anim Health Prod. (2019) 52:601–10. 10.1007/s11250-019-02047-1 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All datasets generated for this study are included in the article/supplementary material.