Abstract
Objectives
To detect possible severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) RNA contamination of inanimate surfaces in areas at high risk of aerosol formation by patients with coronavirus disease 2019 (COVID-19).
Methods
Sampling was performed in the emergency unit and the sub-intensive care ward. SARS-CoV-2 RNA was extracted from swabbed surfaces and objects and subjected to real-time RT-PCR targeting RNA-dependent RNA polymerase and E genes. Virus isolation from positive samples was attempted in vitro on Vero E6 cells.
Results
Twenty-six samples were collected and only two were positive for low-level SARS-CoV-2 RNA, both collected on the external surface of continuous positive airway pressure helmets. All transport media were inoculated onto susceptible cells, but none induced a cytopathic effect on day 7 of culture.
Conclusions
Even though daily contact with inanimate surfaces and patient fomites in contaminated areas may be a medium of infection, our data obtained in real-life conditions suggest that it might be less extensive than hitherto recognized.
Keywords: Coronavirus disease 2019, Environmental contamination, Severe acute respiratory syndrome coronavirus-2, Surfaces, Vero E6 cells
Introduction
Current data evaluating transmission routes for severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) are scanty and controversial. Our knowledge is essentially based on the evaluation of various routes of transmission for severe acute respiratory syndrome coronavirus (SARS-CoV-1), Middle East respiratory syndrome coronavirus and influenza virus [1]. Although direct contact transmission occurs most likely through droplets, the ability of these viruses to survive on inanimate surfaces has been reported by several studies [2,3]. For instance, it has been shown that SARS-CoV-1 can survive for at least 96 hours in sputum, serum and stools [4], which may facilitate transmission if any contact with eyes, nose or mouth occurs following touching of infected material. Similarly, droplets are thought to be the major driver of SARS-CoV-2 transmission, even though the virus may potentially spread in the environment and be transferred from inanimate surfaces to hands. Recently published data indicate that fomite transmission of SARS-CoV-2 may occur, as the virus can remain viable for days on surfaces under controlled experimental conditions, in a similar way to SARS-CoV-1 [5]. Disease amplification in health-care settings has already been documented for past SARS and Middle East respiratory syndrome outbreaks [4], and contamination of inanimate materials by infectious respiratory secretions or other body fluids (saliva, urine or stools) would plausibly contribute to SARS-CoV-2 spread in the environment. The problem is further compounded by the possibility that undocumented infection could facilitate the rapid dissemination of SARS-CoV-2 [6]. Hence, efficient disinfection measures would be the first containment barrier to avoid virus spread [7]. To this end, it has recently been shown that inanimate surfaces located outside patients' rooms were free of SARS-CoV-2 RNA, suggesting that protective measures and current decontamination procedures are effective, reducing concerns over the risk of fomite transmission in the health-care setting [8].
However, recent findings indicate that extensive environmental contamination is indeed possible, as shown by a survey on individuals with coronavirus disease 2019 (COVID-19) with mild upper respiratory tract involvement. Multiple swabs taken in the patients' rooms and on staff personal protective equipment were found to be positive for SARS-CoV-2 [9]. Interestingly, all samples taken from two of three rooms after routine cleaning were negative. Apart from the above-mentioned small case series, literature on the persistence of SARS CoV-2 on inanimate surfaces is limited to anecdotal reports. In this study, we report data on the presence of SARS-CoV-2 RNA on swabs collected from inanimate surfaces in an infectious disease emergency unit and in a sub-intensive care ward that were deemed likely to be contaminated.
Methods
Environmental samples from different surfaces were obtained using flexible nasopharyngeal nylon flocked swabs (FLOQSwabs™; Copan Italia, Brescia, Italy) dipped in 3 mL universal transport medium (UTM™; Copan Italia). In detail, flocked swabs were pre-moistened with UTM to maximize the quantity of SARS-CoV-2 RNA removed from surfaces and of any possible viable virus. Total nucleic acids (DNA/RNA) were extracted from 200 μL of UTM™ using the QIAsymphony® instrument with QIAsymphony® DSP Virus/Pathogen Midi Kit (Complex 400 protocol) according to the manufacturer's instructions (Qiagen, Hilden, Germany). Specific real-time RT-PCR targeting RNA-dependent RNA polymerase and E genes were used to detect the presence of SARS-CoV-2 according to WHO guidelines [10] and Corman et al. protocols [11]. To investigate the infectious potential of environmental samples, virus isolation from surfaces swabs was attempted with the Vero E6 (VERO C1008 (Vero 76, clone E6, Vero E6); ATCC® CRL-1586™) cell line. In detail, a 200-μL sample was inoculated onto a Vero E6 confluent 24-well microplate for virus isolation. After 1 hour of incubation at 33°C in 5% CO2 in air, the inoculum was discarded and 1 mL of medium for respiratory viruses was added (Eagle's modified minimum essential medium supplemented with 1% penicillin, streptomycin and glutamine, and 5 mg/mL trypsin) to each well. Cells were incubated at 33°C in 5% CO2 in air and observed by light microscopy every day for cytopathic effect. After a 7-day incubation, 200 μL of supernatant was used for molecular assays.
The areas subjected to environmental SARS-CoV-2 RNA sampling included the Infectious Disease Emergency Unit, where febrile patients with respiratory symptoms were evaluated, and an infectious disease sub-intensive care ward that allows advanced respiratory care. Both areas were considered contaminated areas, and staff working there wore personal protective equipment comprising liquid-repelling gowns, double gloves, class 2 filtering face-piece respirators and eye protection (goggles or face shield). Air change in our wards is typically 7 volumes per hour. Cleaning procedures were standardized. In particular, ward surfaces were routinely cleaned twice daily (morning and early afternoon) with sodium hypochlorite at a concentration of 1000 ppM of free chlorine (0.1%) on a daily basis and 5000 ppM of free chlorine (0.5%) during final sanitization after the patient(s) was(were) discharged. Swabs were performed around 12 noon, approximately 4 hours after cleaning.
Results
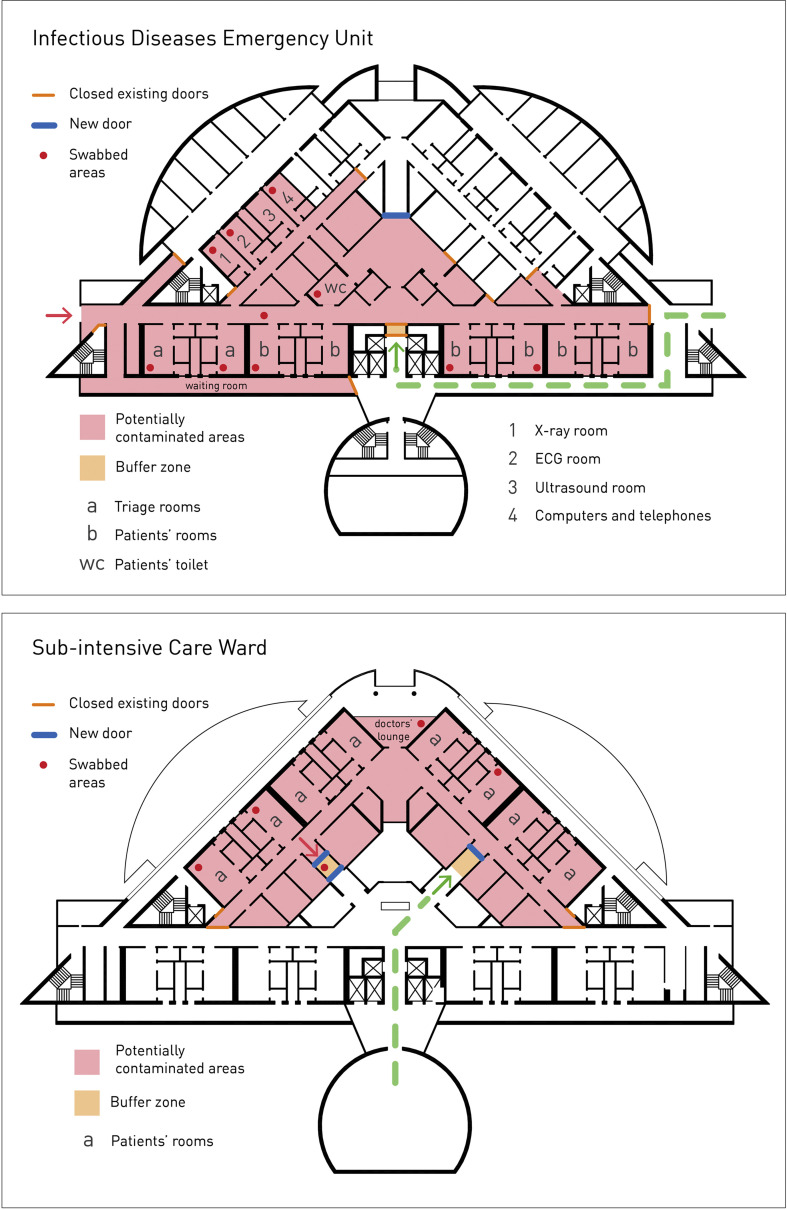
Twenty-six samples were collected in the locations shown in Fig. 1 and from various surfaces and objects as shown in Table 1 . In the sub-intensive care ward, swabs were performed in a double room where two patients with CPAP helmets were allocated. In the emergency room, samples were collected from two different rooms. Each room accommodated three patients, one of them with a CPAP helmet. Samples were collected using identical procedures.
Fig. 1.
Areas in the potentially contaminated infectious diseases emergency unit (a) and pre-intensive care (b) ward where swabbing was performed. Red dots indicate the precise locations where swabbing was carried out. Abbreviations: ID, infectious diseases; ECG, electrocardiography.
Table 1.
Inanimate surfaces on which swabbing was performed
| Potentially contaminated areas | Inanimate surfaces (n of samples) |
|---|---|
| Pre-intensive care ward | |
| Ward and buffer zone | Exit buffer zone (1) |
| Computer keyboards in staff lounge (1) | |
| Rooms of patients A and B with CPAP helmet | Bed rails (1) |
| Infusion pump (1) | |
| Multi-parameter monitor (1) | |
| Nurse buzzer (1) | |
| CPAP helmet, exterior surface (1) | |
| Table (1) | |
| Room of patient C in high-flow oxygen therapy | Bed rails (1) |
| Multi-parameter monitor (1) | |
| Infusion pump (1) | |
| Table (1) | |
| Staff personal protective equipment | Liquid-repelling gowns after 1 hour of use (1) |
| Face shield and eye goggles (1) | |
| Woven gown worn over liquid-repelling gown after 1 hour of use (1) | |
| Staff gloves, internal pair (1) | |
| Infectious Diseases Emergency Unit | |
| Ward | Computer keyboards in triage and examination rooms (1) |
| Telephones (1) | |
| Door knobs and water taps in patient toilets (1) | |
| Staff equipment | Portable X-ray machine (1) |
| Electrocardiograph machine (1) | |
| Medication cart (1) | |
| Patient rooms | Beds (1) |
| Bed of patient with CPAP helmet (1) | |
| Staff equipment | Liquid-repelling gowns, after 7 hours of use (1) |
| Face shields (1) | |
| Nurse gloves, internal pair gloves (1) | |
| Staff mobile phone (1) | |
Abbreviation: CPAP, continuous positive airway pressure.
In swabbing the patients' rooms, we followed a specific order, working our way down from objects and surfaces with the highest contamination risk to those with the lowest risk. Considering the patient as the contamination source, we first examined the external surface of the CPAP helmet, the fomite closer to the face. The two samples taken there were positive for SARS-CoV-2 RNA. Of note, SARS-CoV-2 RNA quantified by real-time RT-PCR in these two samples showed a very low RNA level. All the remaining 24 samples, taken farther from the patient, were SARS-CoV-2 RNA negative.
The first patient treated with CPAP had been admitted to the Emergency Room 24 hours before swabbing and had a 10-day history of fever and cough. The second patient was admitted to the sub-intensive care unit for 3 days and had become symptomatic 12 days earlier. Both patients had a positive nasal swab for SARS-CoV-2 RNA on admission and both had pneumonia. The former had a cycle threshold (Ct) value of 23.0 and the latter a Ct value of 26.3.
All 26 samples were inoculated onto susceptible Vero E6 cells. None of the inoculated samples induced a cytopathic effect on day 7 of culture. To further confirm the negative data, supernatants collected on day 7 were tested by real-time RT-PCR and were all negative.
Discussion
Our findings are at variance with the recently published case report analysis that showed that most surfaces were contaminated by SARS-CoV-2 RNA [9], although no evidence of viable virus with consequent infectious potential was provided. Importantly, staff personal protective equipment and equipment, as well as patient fomites, in our potentially contaminated areas were SARS-CoV-2 RNA free. This is relevant to daily practice, because contact with patient fomites in contaminated areas is thought to be a risk factor for infection. These results obtained in real-life conditions are also somehow in contrast with those recreated in a laboratory environment aerosolized with a SARS-CoV-2 strain grown in vitro [5]. In that study, several materials were examined for their ability to maintain virus viability, and it was found that SARS-CoV-2 was more stable on plastic and stainless steel than on copper and cardboard, and viable virus was detected for up to 72 hours. In line with the above interpretation and not unexpectedly, our findings cannot be compared with those reported under strictly controlled experimental conditions, with constant room temperature and humidity [5]. Indeed, surfaces are cleaned twice daily and temperature and humidity may change over time. Another notable issue is that, for patients on Venturi masks or CPAP helmets, oxygen support was set up with specific filters, which are likely to reduce virus spread. Such differences may account for the discrepancies observed in the two studies.
In our study, the only two positive samples were taken from the plastic of the CPAP helmet, a few centimetres from the patient's airways, whereas all other surfaces, including plastic bed rails, only 60 cm from the patient's face and in continuous contact with his/her hands, were SARS-CoV-2 RNA free. Hence, our data suggest that although environmental contamination may occur in real-life conditions, it might be less extensive than hitherto recognized. Moreover, the inability of the SARS-CoV-2 RNA collected from the CPAP helmet to infect susceptible cell monolayers suggests that recent contamination of plastic surfaces, which apparently maintain SARS-CoV-2 infectivity for several hours [5], is unlikely to contribute to nosocomial spread, which is most likely occurring either via aerosol or droplets, and/or direct physical contact by patients and staff.
A possible limitation of our study may be the timing of swabbing which was relatively close to the cleaning procedures and the effectiveness of flocked swabbing for environmental sampling. Sample collection from the environment was set-up on the basis of a precise workflow and it cannot apply to surfaces that are not systematically cleaned, eg computer keyboards, telephones, multi-parameter monitors, infusion pumps, etc. to cite but a few. The use of flocked swabs is also reported by others in COVID-19 surveillance [12], and their use has been previously validated [13].
Transparency declaration
The authors declare that they have no conflicts of interest.
Funding statement
This work was supported by the Italian Ministry of Health to Fondazione IRCCS Policlinico San Matteo (Ricerca Corrente) for consumables and research staff salaries. Resident salaries were funded by the Italian Ministry of Education, University and Research.
Editor: S.J. Cutler
Contributor Information
The COVID19 IRCCS San Matteo Pavia Task Force:
R. Bruno, M.U. Mondelli, E. Brunetti, A. Di Matteo, E. Seminari, L. Maiocchi, V. Zuccaro, L. Pagnucco, S. Ludovisi, R. Lissandrin, A. Parisi, P. Sacchi, S.F.A. Patruno, G. Michelone, R. Gulminetti, D. Zanaboni, S. Novati, R. Maserati, P. Orsolini, and M. Vecchia
Appendix 1. COVID19 IRCCS San Matteo Pavia Task Force
ID Staff
Raffaele Bruno, Mario U Mondelli, Enrico Brunetti, Angela Di Matteo, Elena Seminari, Laura Maiocchi, Valentina Zuccaro, Layla Pagnucco, Serena Ludovisi, Raffaella Lissandrin, Aldo Parisi, Paolo Sacchi, Savino FA Patruno, Giuseppe Michelone, Roberto Gulminetti, Domenico Zanaboni, Stefano Novati, Renato Maserati, Paolo Orsolini, Marco Vecchia.
ID Residents
Erika Asperges, Marta Colaneri, Alessandro Di Filippo, Margherita Sambo, Simona Biscarini, Matteo Lupi, Silvia Roda, Teresa Chiara Pieri, Ilaria Gallazzi, Michele Sachs, Pietro Valsecchi.
Emergency Care Unit
ECU Staff
Stefano Perlini, Claudia Alfano, Marco Bonzano, Federica Briganti, Giuseppe Crescenzi, Anna Giulia Falchi, Roberta Guarnone, Barbara Guglielmana, Elena Maggi, Ilaria Martino, Pietro Pettenazza, Serena Pioli di Marco, Federica Quaglia, Anna Sabena, Francesco Salinaro, Francesco Speciale, Ilaria Zunino.
ECU Residents
Marzia De Lorenzo, Gianmarco Secco, Lorenzo Dimitry, Giovanni Cappa, Igor Maisak, Benedetta Chiodi, Massimiliano Sciarrini, Bruno Barcella, Flavia Resta, Luca Moroni, Giulia Vezzoni, Lorenzo Scattaglia, Elisa Boscolo, Caterina Zattera, Tassi Michele Fidel, Capozza Vincenzo, Damiano Vignaroli, Marco Bazzini.
Intensive Care Unit
Giorgio Iotti, Francesco Mojoli, Mirko Belliato, Luciano Perotti, Silvia Mongodi, Guido Tavazzi.
Paediatric Unit
Gianluigi Marseglia, Amelia Licari, Ilaria Brambilla.
Virology Staff
Daniela Barbarini, Antonella Bruno, Patrizia Cambieri, Giulia Campanini, Caterina Cavanna, Giuditta Comolli, Marta Corbella, Rossana Daturi, Milena Furione, Bianca Mariani, Piero Marone, Roberta Maserati, Vincenzina Monzillo, Stefania Paolucci, Maurizio Parea, Elena Percivalle, Antonio Piralla, Francesca Rovida, Antonella Sarasini, Maurizio Zavattoni.
Virology Technical Staff
Guy Adzasehoun, Marco Ardizzone, Laura Bellotti, Vincenzo Brunco, Ermanna Cabano, Giuliana Casali, Laura Capella, Debora Devitis, Luca Dossena, Gabriella Frisco, Gabriella Garbagnoli, Federica Gardellini, Alessia Girello, Andrea Guerrizio, Viviana Landini, Claudia Lucchelli, Valentina Maliardi, Pasquale Piemontese, Simona Pezzaia, Marta Premoli, Chiara Rebuffa.
Virology Residents
Jessica Bagnarino, Federica Bergami, Alice Bonetti, Giacomo Caneva, Irene Cassaniti, Alfonso Corcione, Raffella Di Martino, Annapia Di Napoli, Alessandro Ferrari, Guglielmo Ferrari, Loretta Fiorina, Arianna Gallone, Federica Giardina, Assunta Girardi, Alessandra Mercato, Federica Novazzi, Giacomo Ratano, Beatrice Rossi, Graziella Saverimpilla, Irene Maria Sciabica, Monica Tallarita, Edoardo Vecchio Nepita, Jessica Vitali.
Research Laboratories, Division of Infectious Diseases and Immunology
Antonella Cerino, Stefania Varchetta, Barbara Oliviero, Stefania Mantovani, Dalila Mele.
Pharmacy Unit
Monica Calvi, Michela Tizzoni.
Hospital Management
Carlo Nicola, Antonio Triarico, Vincenzo Petronella, Carlo Marena, Alba Muzzi, Paolo Lago.
Data Unit
Francesco Comandatore, Gherard Batisti Biffignandi, Stefano Gaiarsa, Marco Rettani, Claudio Bandi, Alessandra Ferrari.
References
- 1.Otter J.A., Donskey C., Yezli S., Douthwaite S., Goldenberg S.D., Weber D.J. Transmission of SARS and MERS coronaviruses and influenza virus in healthcare settings: the possible role of dry surface contamination. J Hosp Infect. 2016;92:235–250. doi: 10.1016/j.jhin.2015.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shiu E.Y.C., Leung N.H.L., Cowling B.J. Controversy around airborne versus droplet transmission of respiratory viruses: implication for infection prevention. Curr Opin Infect Dis. 2019;32:372–379. doi: 10.1097/QCO.0000000000000563. [DOI] [PubMed] [Google Scholar]
- 3.Bean B., Moore B.M., Sterner B., Peterson L.R., Gerding D.N., Balfour H.H. Survival of influenza viruses on environmental surfaces. J Infect Dis. 1982;146:47–51. doi: 10.1093/infdis/146.1.47. [DOI] [PubMed] [Google Scholar]
- 4.Chowell G., Abdirizak F., Lee S., Lee J., Jung E., Nishiura H. Transmission characteristics of MERS and SARS in the healthcare setting: a comparative study. BMC Med. 2015;13:210. doi: 10.1186/s12916-015-0450-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van Doremalen N., Bushmaker T., Morris D.H., Holbrook M.G., Gamble A., Williamson B.N. Aerosol and surface stability of SARS-CoV-2 as compared with SARS-CoV-1. N Engl J Med. 2020 doi: 10.1056/NEJMc2004973. Mar 17. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li R., Pei S., Chen B., Song Y., Zhang T., Yang W. Substantial undocumented infection facilitates the rapid dissemination of novel coronavirus (SARS-CoV2) Science. 2020 doi: 10.1126/science.abb3221. Mar 16; pii: eabb3221. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kampf G., Todt D., Pfaender S., Steinmann E. Persistence of coronaviruses on inanimate surfaces and their inactivation with biocidal agents. J Hosp Infect. 2020;104:246–251. doi: 10.1016/j.jhin.2020.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Colaneri M., Seminari E., Piralla A., Zuccaro V., Di Filippo A., Baldanti F. Lack of SARS-CoV-2 RNA environmental contamination in a tertiary referral hospital for infectious diseases in Northern Italy. J Hosp Infect. 2020 doi: 10.1016/j.jhin.2020.03.018. Mar 19. pii: S0195-6701, 30117-1. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ong S.W.X., Tan Y.K., Chia P.Y., Lee T.H., Ng O.T., Wong M.S.Y. Surface environmental, and personal protective equipment contamination by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) from a symptomatic patient. JAMA. 2020 doi: 10.1001/jama.2020.3227. Mar 4. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Diagnostic detection of 2019-nCoV by real-time RT-PCR. https://www.who.int/docs/default-source/coronaviruse/protocol-v2-1.pdf Available from:
- 11.Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K.W. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25 doi: 10.2807/1560-7917.ES.2020.25.3.2000045. Jan. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang J., Feng H., Zhang S., Ni Z., Ni L., Chen Y. SARS-CoV-2 RNA detection of hospital isolation wards hygiene monitoring during the Coronavirus Disease 2019 outbreak in a Chinese hospital. Int J Infect Dis. 2020;94:103–106. doi: 10.1016/j.ijid.2020.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dalmaso G., Bini M., Paroni R., Ferrari M. Qualification of high-recovery, flocked swabs as compared to traditional rayon swabs for microbiological environmental monitoring of surfaces. PDA J Pharm Sci Technol. 2008;62:191–199. [PubMed] [Google Scholar]