Abstract
For the first time in Homo sapiens history, possibly, most of human activities is stopped by coronavirus disease 2019 (COVID-19). Nearly eight billion people of this world are facing a great challenge, maybe not “to be or not to be” yet, but unpredictable. What happens to other major pandemics in the past, and how human beings went through these hurdles? The human body is equipped with the immune system that can recognize, respond and fight against pathogens such as viruses. Following the innate response, immune system processes the adaptive response by which each pathogen is encoded and recorded in memory system. The humoral reaction containing cytokines and antibodies is expected to activate when the pathogens come back. Exploiting this nature of body protection, neutralizing antibodies have been investigated. Learning from past, in parallel to SARS-CoV-2, other coronaviruses SARS-CoV and MERS-CoV who caused previous pandemics, are recalled in this review. We here propose insights of origin and characteristics and perspective for the future of antibodies development.
Keywords: SARS-CoV, MERS-CoV, COVID-19, SARS-CoV-2, Antibodies
Graphical abstract
1. Introduction
In full social distancing crisis, far from lab work, in looking anxiously at the red dots growing every day in the graphic map of coronavirus disease 2019 (COVID-19) pandemic [1], we wonder how were the other major pandemics and how antibodies, natural or artificial, can fight the diseases.
Several pandemics have occurred throughout history, some had more effects on human's life and/or on economics than the others. As scientists, a key step in vaccines research is to learn from the past. By that mean, what we are doing now might be cornerstone preparing us for future pandemics.
In this review, we look for the diversity and variability of coronaviruses, including SARS-CoV, MERS-CoV and SARS-CoV-2, which cause pandemics from 21th century. The origins, impacts and molecular structure with insights of vulnerable sites of viruses who are targets of antibodies and its humoral responses will be described.
2. Overview of antibody and immunization
In response to pathogens including viruses, human body has evolved its immune system to protect it from invasion. Following the virus invasion, antibodies (Abs) are produced after a series of immune signaling and these Abs are able to recognize a diverse array of antigens (Ag) [2]. More specifically, the paratopes (Ag-binding sites) of Abs bind epitopes on virion-associated Ags. During an Ab response, B-cells which express Ag receptors are clonally expanded [3]. Antibodies structurally are composed of heavy (μ, α, γ, δ, ε) chains that are linked by disulfide bonds with light chains (κ, λ). In the progress of B-cell development, immunoglobulin heavy (IgH) chain gene recombination typically occurs before immunoglobulin light (IgL) chain gene recombination [4].
Neutralizing antibodies (nAbs) can inhibit the viral infection via following the viral replication cycle. Attachment is the first critical step blocked by Abs by interfering with the virion-receptor binding. Moreover, Abs may induce the aggregation of viral particles which cause a reduction of individual penetration. In post-attachment step, Abs on the virion possibly dampen virus endocytosis internalization leading to the lysosomal degradation. The Abs also block fusion of virion when they intercalate between viruses and cell membrane. The next stage of interference is to uncoat or appropriate intracellular localization of core or capsid. Lastly, Abs might bind virion surface then inhibit the metabolic events that blocks the replication of viruses even after internalization [3,5,6].
The approach to nAbs design relies on the identification of antigens; in other words, the epitopes are the central of quests. However, the variable regions of the antigen induce the largest fraction of the antibodies whereas broadly nAbs represent only a minor proportion of the response. The major challenges related to both sides were previously described: viral antigen and the generation of antibodies to these sites. In the detail, the antigen concerns are (1) epitope masking or shielding by glycans or protein loops; (2) transient exposure during the entry process or via other mechanisms such as viral “breathing;” (3) the size of conserved epitopes is small resulting in the limitation of interaction, (4) epitopes are constrainedly accessible, (5) the mutability of epitopes. Regarding antibody production, (1) the need for extensive somatic mutations and focused evolution; (2) the use of specific germline allelic variants and HCDR3s of particular length and structure; and (3) the molecular mimicry of host molecules are mentioned [6].
3. Coronaviruses
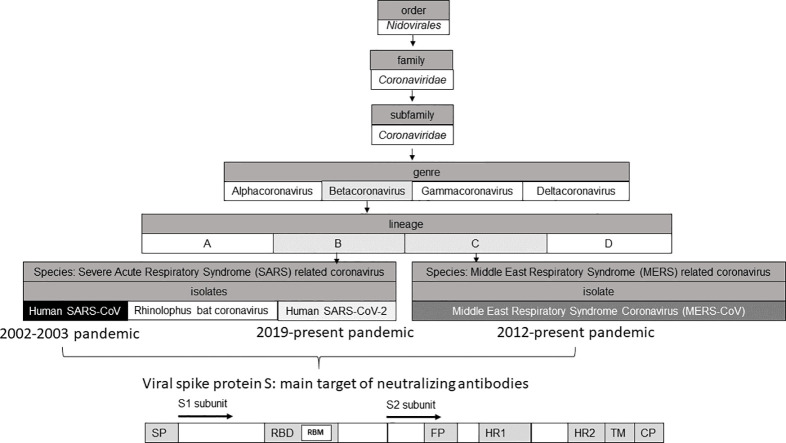
Coronaviruses are a group of related viruses in the family Coronaviridae and subfamily Coronavirinae, order Nidovirales. In the subfamily Coronavirinae, coronaviruses include 4 genres: alphacoronavirus, betacoronavirus, gammacoronavirus and deltacoronavirus (Fig. 1 ). Coronaviruses infect a wide variety of hosts including many species of birds, mammals and humans [7]. Alphacoronaviruses and betacoronaviruses circulate among mammals, gammacoronaviruses and deltacoronaviruses infect birds and mammals. Within betacoronaviruses, there are 4 lineages: lineage A contains human coronaviruses HKU1 and OC43, lineage B to which SARS-CoV (Severe Acute Respiratory Syndrome Coronavirus) belongs, lineage C belongs to MERS-CoV (Middle East Respiratory Coronavirus Syndrome) and the lineage D has the bat coronaviruses HKU4 and HKU5 who are close to MERS-CoV.
Fig. 1.
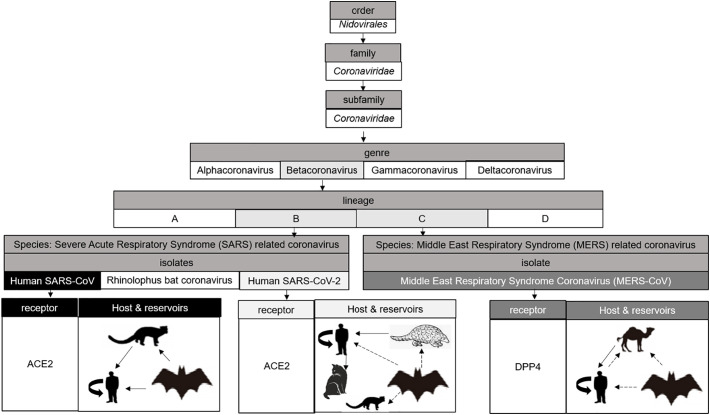
Isolates of coronaviruses discussed in this review and their receptors, host and reservoirs. SARS-CoV and SARS-CoV-2 from the lineage B use ACE2 as receptor. MERS-CoV from the lineage C enters into host cells by binding DPP4. SARS-CoV has the masked palm civets as an intermediate host in which the virus has adapted from the Chinese Horseshoe bat reservoir to ultimately infect humans [13]. SARS-CoV-2 has bats and pangolins as natural reservoir and can infect ferrets or domestics animals, with a high susceptibility in cats [14]. MERS-CoV has the origin from bats [15] but maybe this virus had an adaptation through camels before its emergence into human [16].
The first human coronaviruses 229E (HCoV-229E) and OC43 (HCoV-OC43) were isolated in the 1960s and are now classified respectively as alphacoronaviruses and betacoronaviruses. As these viruses were not very pathogenic and often associated with colds [8], this family of viruses attracted little interest from scientists until the 2000s. In November 2002, SARS-CoV, first reported in Guangdong province, China, became the first highly pathogenic coronavirus that emerged in the human population. This virus was responsible for an epidemic of severe acute respiratory syndromes that started in China before spreading rapidly over the world with around 8000 infected people and with a mortality rate of around 10%, depending on patients' age [9]. However, this coronavirus from animal origin was initially unable to use the human angiotensin 2 converting enzyme (ACE2) as receptor [[10], [11], [12]]. It has been suggested that the masked palm civet (Paguma larvata) may be an intermediate host in which the viruses have adapted to ultimately infect humans. A recent study suggests that Chinese Horseshoe bats in the family Rhinolophidae may be the natural reservoir for SARS-CoV. One of these two viral isolates in this study, WIV1, was able to recognize the human ACE2 receptor and to replicate in certain human cell lines, suggesting that this virus can directly infect humans without adaptation [13].
4. SARS-CoV: from 2002 to 2003
SARS-CoV, first reported in 2002, belongs to the SARS-related coronavirus species that also includes many bat viruses.
Coronaviruses are spherical enveloped viruses with a diameter of 80 to 120 nm [17]. The viral capsid formed by the nucleoprotein (N) and the genome is contained in the envelope and is of helical symmetry. Three structural proteins are embedded on the surface of particles, the membrane protein (M), the envelope protein (E) and the protein spike (S). They give this aspect of crown in electron microscopy that inspired the name of this viral family.
The S protein of coronaviruses (~1255 amino acids) is a highly N-glycosylated type I transmembrane protein, from 180 to 200 kDa, that plays a major role in viral entry [18]. It insures a double function in viral entry by binding the cellular receptor before conformational changes and proceeding to the fusion of the viral envelope with the membranes of the target cells. S protein has a long N-terminal domain, a short C-terminal domain and assembles into homotrimers on the surface of the viral particle [19]. S protein has a decisive role in cellular tropism and for pathogenicity [20].
S protein of SARS-CoV is composed of two functionally distinct subunits: the globular S1 subunit (~aa 12–680) allows receptor recognition, whereas the S2 subunit (~aa 681–1255) facilitates membrane fusion and anchors S into the viral membrane. S1 is organized in four distinct domains A–D. Domains S1A and S1B may be used as a receptor-binding domain (RBD, aa 318–510) containing the highly conserved receptor-binding motif (RBM, aa 424–494) [21]. Moreover, RBD contains 3 functional glycosylation sites located at amino acids 318, 330 and 357, which are necessary for S expression but do not affect ACE2 binding [22]. S1B forms an extended loop on the viral membrane-distal side and is a hypervariable region [20]. S2 contains the fusion peptides (FP1 and FP2) [23], two heptad repeat regions (HR1, aa ~889–972 and HR2, aa ~1142–1193) and the well conserved transmembrane domain [24].
The mechanism of interactions with peptidases (aminopeptidase APN, ACE2, DPP4) as a cellular receptor for most coronaviruses is not known. Indeed, the binding of coronaviruses to their receptor is not enough and S protein on the surface of the virus must undergo proteolytic maturation. Coronaviruses do not use the catalytic activity of peptidases serving as receptors for this maturation but enter after the action of proteases located close to the receptors. The binding of SARS-CoV to its ACE2 receptor is followed by internalization and decrease in ACE2 enzyme activity on the cell surface, which may partly explain the severity of SARS-CoV infections [25].
4.1. Anti-S1 & RBD antibodies
Neutralizing Abs can fight against viral infections by blocking binding to cellular receptors or by interfering with viral fusion. Besides, in the case of enveloped viruses, the Abs can recruit effector cells or the complement, thus allowing the destruction of the infected cells or the lysis of the viral particles [6]. The S1 domain contains most of the epitopes recognized by nAbs during infection. The RBD located in this S1 domain would be the most important target for nAbs against SARS-CoV, MERS-CoV and the novel coronavirus SARS-CoV-2 [[26], [27], [28], [29]]. More specifically, certain secondary structures such as extended loops seem to be particularly immunogenic.
RBD of SARS-CoV is composed of 193 amino acids (N318-V510) within S protein. Five regions on the S glycoprotein of SARS-CoV (residues 274–306, 510–586, 587–628, 784–803 and 870–893), in which three first regions belong to S1 subunit in the CTD2 and CTD3 (C-terminal domain) and two later belong to HR1 domain of the S2 subunit, were predicted to be associated with a robust immune response to SARS-CoV [30]. Several specific-nAbs for SARS-CoV were discovered; unfortunately none of them are under clinical trial [31] (Fig. 2 ).
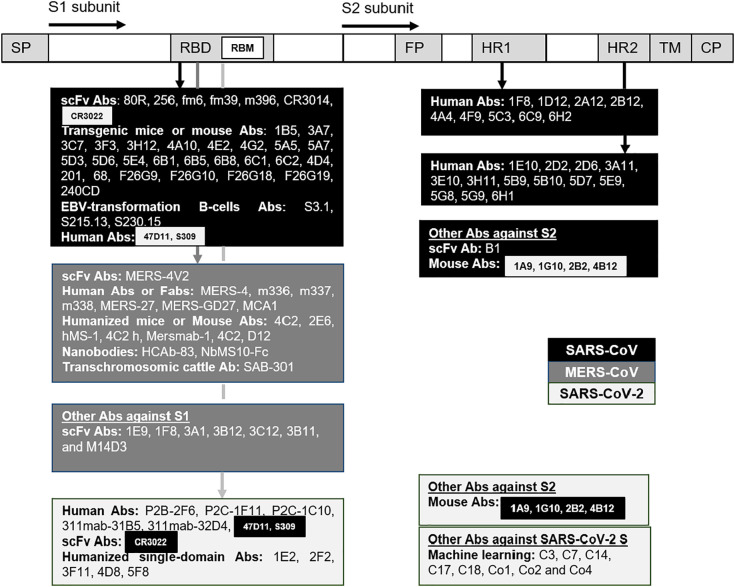
Fig. 2.
S protein of SARS-CoV, MERS-CoV, SARS-CoV-2 with its subdomains are the target of antibodies. The antibodies cited in this review have different origins or techniques, and some of them have specific targets such as the receptor binding domain (RBD) containing the receptor binding motif (RBM), the heptad repeat regions (HR1 and HR2). Some antibodies could bind SARS-CoV and SARS-CoV-2. Background color: Black for SARS-CoV, dark grey for MERS-CoV, grey for SARS-CoV-2. SP: Signal peptide, FP: Fusion peptide, TM: Transmembrane domain, CP: Cytoplasm domain.
The human single-chain variable region fragment (scFv) antibody 80R blocked ACE-RBD interaction (epitope aa 324–503) [32] but some 80R-escape variants were found with the mutations mostly locating at lysine D480 [33]. The target epitope of 80R is not conserved in SARS-CoV-2 then it does not affect this novel virus [34]. Another nAb generated from a non-immune scFv library, named 256, could bind to an epitope of RBD but did not inhibit RBD binding. 256 is weak but specific to D480A-muted strains of 80R-escape variants. Some engineered broad nAbs, fm6 and fm39, also showed a high affinity to D480A-muted strains [33]. m396 (epitope aa 482–491) from human antibody fab library was cross-reactive [35] and used the D95 of m396 to form a salt bridge with R395 or an electrostatic interaction with D408 of SARS-CoV RBD [34]. m396 potently neutralized GD03 strain isolated from the second outbreak which resisted neutralization by 80R and S3.1. m396 also neutralized isolates from the first SARS-CoV outbreak (Urbani, Tor2) and from palm civets (SZ3, SZ16) [36]. Another human monoclonal antibody from scFv libraries CR3014 (epitope aa 318–510) showed potent effects on SARS-CoV neutralization; however, this virus can escape CR3014 upon P426L mutation in the S glycoprotein [37]. Same as 80R, m396 and CR3014 RBD-specific SARS-CoV antibodies failed to bind the S protein of SARS-CoV-2 [34]. CR3022, always from scFv libraries, could bind noncompetitively the SARS-CoV RBD (epitope aa 318–510) and had a synergistic neutralizing effect with CR3014 on SARS-CoV, even with the escaped P426L-muted variants [37].
By using Xenomouse in which mouse immunoglobulin genes were replaced by human immunoglobulin genes, 19 neutralizing mAbs bound S1 were found. 18 of them, 1B5 [38], 3A7, 3C7, 3F3, 3H12, 4A10, 4E2, 4G2, 5A5, 5A7, 5D3, 5D6, 5E4, 6B1, 6B5, 6B8, 6C1 and 6C2 bound RBD (aa 318–510) to avoid virus binding to the ACE2 receptor. The last one, 4D4, bound an epitope (aa 12–261) located on the N-terminal of RBD and inhibited post-binding event but not the RBD binding. Truncation of the first 300 amino acids of S1 blocked the trimerization and the fusion of S protein [39]. Synergistic effects in some SARS-CoV strains of 4D4 with other mAbs targeting S1 or S2 proteins such as 3C7 (S1), 1F8 (HR1) or 5E9 (HR2) were also reported [38,40]. The tri-combination of 3C7, 3H12 and 4D4 could effectively neutralize escape variants.
Other neutralizing human monoclonal Abs from transgenic mice were also reported. Ab 201 interfered with ACE2 binding by targeting S1 protein at the epitope aa 490–510. In contrast to 201, Ab 68 bound epitope aa 130–150 at the N-terminal of RBD but did not affect ACE2 binding [41].
F26 family of monoclonal Abs generated from mice (F26G9, F26G10, F26G18 and F26G19) showed neutralizing effect against SARS-CoV [42]. F26G18 binding RBD at the epitope aa 460–476 showed the most potent effect [43]. F26G19 (epitope aa 486–492 on RBD [44]) or 80R could also bind SARS-CoV by forming salt bridge R426 (RBD)-D56 or D480 (RBD)-R162, respectively [34].
SARS-CoV mouse antibody 240CD had a nanomolar affinity for the SARS-CoV-2 RBD but did not significantly block ACE-2 receptor binding [45]. As 240CD, CR3022 also has high affinity to SARS-CoV-2 and moreover, CR3022 had cross-neutralizing activity with this novel coronavirus [34].
The effects of neutralizing human monoclonal antibodies, S3.1, S215.13 [46] and S230.15, from Epstein-Bar virus transformation of human B cells were observed. As m396, S230.15 had potent inhibitory activity against isolates from the first, second SARS-CoV outbreaks and from palm civets (SZ3, SZ16) [36].
4.2. Anti-S2 antibodies
In contrast to RBD, the fusion domains are more difficult to access due to the tight folding of viral glycoproteins or the excessively transient exposure during the fusion stage. This is why few epitopes are described in these regions [6]. Interestingly, the S2 specific mAbs can neutralize pseudotyped viruses which expressing different S proteins containing RBD sequences of various clinical isolates [47]. The S2 protein is highly conserved. No mutation in HR1 was reported in an analysis of the amino acid sequences of the S protein from 94 SARS-CoV clinical isolates. Only few mutations in HR2, at amino acids K1163 or Q1183 for example, were observed in this study [47].
Some S2 epitopes inducing nAbs were reported. A peptide containing aa 1055–1192 can elicit neutralizing activity [48]. Two other proteins Trx-F3 and Trx-F9 containing linear antigenic determinants (Leu 803 to Ala 828 and Pro 1061 to Ser 1093, respectively) on the S2 domain were identified by using sera from convalescent SARS-CoV patients. Trx-F3 was capable of inducing nAbs in some animals [49].
Some human mAbs anti-HR1 (1F8, 1D12, 2A12, 2B12, 4A4, 4F9, 5C3, 6C9, 6H2) and anti-HR2 (1E10, 2D2, 2D6, 3A11, 3E10, 3H11, 5B9, 5B10, 5D7, 5E9, 5G8, 5G9, 6H1) were reported. With these Abs, the authors showed that the combination of HmAbs targeting different regions of the S protein would likely increase the broad neutralization against different isolates [47].
A human scFv antibody, named B1, showed a high affinity to an epitope (aa 1023–1189) on S2 protein. This antibody also showed potent neutralizing activities against SARS-CoV in vitro [50]. B1, 1F8 and 5E9 nAbs against epitopes on SARS-CoV S2 also showed effectiveness in neutralization [51].
The protective immunity by the time in patients after SARS-CoV natural infection was observed. After 6 years, the humoral immunity continuously decreased and eventually disappeared in most infected individuals. The IgG Ab could be an indicator of neutralizing Ab for the humoral response to SARS-CoV infection [52].
5. MERS-CoV: from 2012 to present
MERS-CoV, a zoonotic virus, belonging to lineage C in the genre betacoronavirus of the family Coronaviridae, caused the Middle East respiratory syndrome, in 2012. As of August 11, 2016, the virus had infected 1791 patients, with a mortality rate of 35.6% [53]. The natural reservoir of MERS-CoV is assumedly bats whereas intermediate host is possibly dromedary camels [[54], [55], [56], [57]]. BtCoV-HKU4 and BtCoV-HKU5 bat viruses have been shown to be the closest phylogenetically even if these viruses are not direct ancestors [15]. The first transmission of a bat virus to camels for an adaptation before its emergence into human was suggested. nAbs anti-MERS-CoV could accordingly be found in camels. Moreover, the viruses circulating in dromedaries and in humans are very close suggesting that the dromedary is a reservoir of the virus [58,59]. The genomic structures of bat, human and camel MERS-CoVs are similar but their genomic sequences are different [16].
Structurally, MERS-CoV is a spherical, enveloped, single-stranded, positive sense RNA beta-coronavirus [60]. MERS-CoV utilizes its S protein to mediates cell internalization via binding with the receptor dipeptidyl peptidase 4 (DPP4) on the surface of cells instead of the receptor ACE2 of SARS-CoV and SARS-CoV-2. S protein is therefore the most exposed and immunogenic viral protein [61]. The association of MERS-CoV S protein is similar to that of SARS-CoVs including: the distal subunit S1 containing the RBD and the membrane-anchored subunit S2 containing a putative fusion peptide, transmembrane domain and two heptad repeat regions HR1 and HR2. This S protein is also the target to develop nAbs, particularly the RBD [62].
Using the fragment containing residues 358–588 of S protein, the neutralization against MERS-CoV of induced Abs were observed [63]. Other studies, also approaching RBD, reported the generated Abs with the epitopes aa 377–662 or aa 377–588 of MERS-CoV RBD. The latter elicited the strongest effect which effectively neutralized MERS-CoV infection [64,65]. The epitope aa 736–761 also induced nAbs [66].
Using a novel panning strategy, seven anti-S1 scFvs Abs, named 1E9, 1F8, 3A1, 3B12, 3C12, 3B11, and M14D3, which bind one or several of these three different epitopes (aa 21–358, 349–751 and 349–590) were identified. They neutralized MERS-CoV infection at nanomolar of concentration [67].
Other Abs, such as MERS-4 and its variant MERS-4 V2, were intriguingly discovered to bind RBD and compete with DPP4 but from outside. MERS-4 Fab and MERS-4V2 scFv interact with β5-β6, β6-β7, and β7-β8 loops of RBD resulting in the inhibition of MERS-CoV infection [68,69].
Three human monoclonal Abs, m336, m337, and m338 bind RBD of MERS-CoV at extremely low concentration, 4.2, 9.3, and 15 nM, respectively. m336, that neutralized 50% of both pseudotyped and live MERS-CoV at 0.005 and 0.07 μg/mL, respectively, suggested the prophylaxis and therapy of MERS-CoV infection [70].
In another study, using MERS-RBD to immunize mice, two monoclonal nAbs 4C2 and 2E6 recognizing an epitope that partially overlaps the receptor-binding footprint in MERS-CoV RBD were identified. The 4C2 could further reduce the number of viral particles in MERS-CoV infected mice [71].
Recently, other nAbs, such as human mAbs or Fabs (MERS-27, MERS-GD27, or MCA1), humanized mAbs (hMS-1, 4C2 h), mouse mAbs (Mersmab1, 4C2, or D12), single-domain antibodies (nanobodies Nbs) HCAb-83 or NbMS10-Fc and transchromosomic cattle antibody SAB-301 recognizing epitopes on the RBD have been demonstrated to neutralize pseudotyped and/or live MERS-CoVs [31]. Only SAB-301 is under phase I clinical trial [72].
Despite the efforts of scientists to find anti-MERS-CoV antibodies, no vaccine has been found yet and the virus continues to circulate in human beings and other species.
6. SARS-CoV-2: from 2019 to present
The novel SARS-CoV-2 coronavirus, first appeared in Wuhan, China, in December 2019, is creating a pandemic over the world with the number of confirmed cases reached 3,529,408, of which 248,025 were dead up to the 4th May 2020 [1]. Phylogenetic analysis of SARS-CoV-2 demonstrated similarity with SARS-CoV and bat-derived SARS-like coronaviruses (SL-CoVs) with 79.6% and 88% sequence identity, respectively. They belong to lineage B of the beta coronavirus genus [73,74]. SARS-CoV-2 seems to be more contagious but less pathogenic than SARS-CoV [75]. COVID-19 is a self-limiting disease in >80% of patients. Same as Spanish influenza viruses, SARS-CoV and SARS-CoV-2 induce a “cytokine storm” but to different degrees. The difference of some conserved interferon antagonists and of inflammasome activators explains their abilities to modulate antiviral and proinflammatory responses.
Along with the race of finding therapeutic treatment, nAbs and vaccine development are also important to control the spread in the long run. SARS-CoV-2 entries the host via the binding of its spike S protein to the ACE2 receptor - sharing receptor, but with higher affinity than SARS-CoV S [76], suggesting a basis for the greater human-to-human transmission of SARS-CoV-2 [51,77].
S protein of SARS-CoV-2 composed of 1273 amino acids [76] uses its N-terminal S1 subunit to bind ACE2 receptor with a better affinity than SARS-CoV S glycoprotein for entry [78]. Effectively, S1 subunit divides into an N-terminal domain (NTD) and a receptor-binding domain (RBD). The latter is necessary for viral binding and a potential target for nAbs. During infection, SARS-CoV-2 first binds the host cell through interaction between its S1-RBD and ACE2, triggering conformational changes in the S2 subunit that is indispensable for virus fusion and entry into the target cell [79,80]. Some recent studies also confirmed that RBD is a conformational epitope [78]. Antibodies binding RBD may sterically hinder binding to the nearby peptide S14P5 of ACE2 receptor, thereby abolishing virus infection [34].
SARS-CoV-2 nAbs could be detected in patients from 10 to 15 days after symptoms onset and the positive rate for IgG reached close to 100% around 20 days [81,82] with the highest level during day 31–40 since onset. Some patients (5.7%) had neutralizing Abs titers under the detectable level (ID50: <40) [83]. The level of IgG antibodies was different between gender, age and clinical classification. The average IgG antibody level in female patients was higher than in male patients [84]. Patient over 40 years old developed higher levels of SARS-CoV-2 specific nAbs than the younger persons. Patients with a worse clinical classification had a higher antibody titer [83]. This remark is useful to select a research candidate and to save research time. The passive antibody therapy, such as plasma fusion containing polyclonal antibodies from COVID-19 neutralized patients has been tested. This method was tested as an option to treat other viruses such as influenza, Ebola or SARS-CoV [[85], [86], [87], [88], [89]]. The lack of human sera, and the possibility of contamination with other infectious agents limit this strategy. However, several groups have reported some positive results demonstrating the potential of this approach. After one dose of 200 mL of convalescent plasma derived from recently recovered donors with the neutralizing antibody titers above 1:640, the patients with SARS-CoV-2 positive revealed an improvement. Among ten patients, seven patients were virus-negative post transfusion [90]. Whereas in another study, among 5 patients received transfusion with convalescent plasma with a neutralization titer >40, 3 have been discharged from the hospital (length of stay: 53, 51, and 55 days), and 2 are in stable condition at 37 days after transfusion [91]. More studies might brighter this approach but evaluation in clinical trials are also still far from a bold conclusion.
6.1. Effect of cross-reactive antibodies on SARS-CoV-2 pandemic
Due to the high similarity of S proteins from SARS-CoV and MERS-CoV [73], their specific cross-nAbs were tested against SARS-CoV-2 infection in the COVID-19 outbreak. Serum Abs from recovered SARS-CoV patients could efficiently cross-neutralize SARS-CoV-2 but with lower efficiency as compared to SARS-CoV [92]. Cross-reactive Abs against SARS-CoV-2 S protein mostly target non-RBD regions [93]. Using simulation technique, the binding of five Abs against SARS-CoV, six Abs anti-MERS-CoV to RBD of SARS-CoV-2 was predicted with Rosetta antibody-antigen docking protocols. The amino acid position 445–449 (VGGNY) and 470–486 (TEIYQAGSTPCNGVEGF) were found to be conserved in SARS-CoV-2. Moreover, in addition to the amino acid positions 71–77 (GTNGTKR) in the NTD region of the S protein, aa 445–449 and 470–486 are potential for further development [94].
The difference between RBD of SARS-CoV and SARS-CoV-2 is located at the C-terminus residues. This change has an important impact on the cross-reactivity of nAbs. This difference was observed using bioinformatic approaches of epitope analysis. The antibody epitope score of SARS-CoV-2 is higher than SARS-CoV. Moreover, compared with the conserved regions, the non-conserved regions had a significantly higher antibody epitope score indicating that non-conserved regions of spike proteins are much more antigenic. The non-conserved regions also showed significantly higher surface epitope accessibility scores suggesting an easier accessibility for antibody recognition of non-conserved regions. The divergence of spike proteins is considered as a major change in the antibody epitopes. The search for SARS-CoV-2 requires more effort than simply screening SARS-CoV antibodies [95].
Antibody response to RBD is viral species-specific. Effectively, none of the found SARS-CoV-2 antibodies nor the infected plasma cross-reacted with RBDs from either SARS-CoV or MERS-CoV. In a study, 206 monoclonal antibodies specific to the RBD SARS-CoV-2 were identified in eight patients. These mAbs are different in: antibody heavy and light chains, antibody clones, CDR3 length… which lead to different binding and neutralizing capacities. ACE2 is out-competed with almost 100% efficacity by some mAbs such as P2B-2F6 and P2C-1F11. Interestingly the latter and a moderate antibody P2C-1C10 seems to target the different epitopes, and they could be combined for synergistic antiviral effect [96]. CR3022, a SARS-CoV RBD-specific antibody, can bind strongly with a kd of 6.3 nM to an epitope on RBD that does not overlap with the SARS-Cov-2 ACE-2 binding site [34]. Despite its strong binding, CR3022 could not neutralize SARS-CoV-2 [97].
S1 is a specific antigen for SARS-CoV-2 diagnostics [98]. The S1 subunit of SARS-CoV or SARS-CoV-2 has four core domains S1A through S1D. The human 47D11 antibody binds the S1B of both viruses, without competing with S1B binding to ACE2 receptor expressed at the cell surface, and showed cross-neutralizing activity by an unknown mechanism that is different from receptor binding interference [99]. An immunogenic domain in the S2 subunit of SARS-CoV S (aa 1029–1192) was highly conserved in several strains of SARS-CoV-2. Four murine monoclonal Abs, 1A9, 1G10, 2B2 and 4B12, against this S2 subunit of SARS-CoV can also cross-reactive with the S protein of SARS-CoV-2. Interestingly, 1A9 can strongly bind the S2 subunit of SARS-CoV-2 through a novel epitope (aa 1111–1130) and can detect S protein in SARS-CoV-2 during infection [100]. This epitope also overlaps with one of two cytotoxic T-lymphocyte epitopes (aa 884–891 and 1116–1123) of SARS-CoV S2 subunit [101]. 1A9 is therefore suggested to induce both humoral and cellular immune responses against SARS-CoV and SARS-CoV-2.
In a serologic cross-reactivity test, Khan et al. found out that 4 out of 5 showed high IgG seroreactivity across the 4 common human coronaviruses but all showed low IgG seroreactivity to SARS-CoV-2, SARS-CoV, and MERS-CoV [102]. The weak cross-immunity against SARS-CoV-2 from others betacoronaviruses, such as HCoV-OC43 and HCoV-HKU1, could restraint the transmission of SARS-CoV-2 but a resurgence is possible in the future [103]. Moreover, spike- and non-spike specific CD4+ T cell responses were detectable not only in SARS-CoV-2 infected patients but also in uninfected individuals. If there is an absence of antibody cross-reactivity, T lymphocyte cross-reactivity present in 50% of cases will be responsible for the epidemiological evolution of SARS-CoV-2 infection [104].
6.2. Anti-SARS-CoV-2 specific antibodies
Up to this moment, only few tests of specific Abs against SARS-CoV-2 have been reported. 311mab-31B5 and 311mab-32D4 human monoclonal Abs could strongly and specifically bind the RBD protein. These mAbs could efficiently block SARS-CoV-2-ACE2 interaction and neutralize pseudovirus entry into host cells ectopically expressing ACE2 [105].
Peptides S14P5 and S21P2 in the two distinct peptide pools S14 and S21 from SARS-CoV-2 S library were strongly detected in COVID-19 patients but not in SARS-CoV patients by using pools of overlapping linear peptides and functional assays [78]. With the data from antibodies depletion assays, researchers indicated that S14P5 and S21P2 were necessary for SARS-CoV-2 neutralization. Moreover, pool S51 contains very conserved fusion peptide in coronavirus [106,107] and is partially overlapped in the sera of SARS-CoV and SARS-CoV-2 patients. These results suggested that S51 may be a potential pan-coronavirus epitope. Sera from recalled SARS-CoV patients could neutralize SARS-CoV, but not the SARS-CoV-2 pseudotyped lentiviruses [78].
In an effort to screen a set of B cell and T cell epitopes of SARS-CoV toward to the spike S and nucleocapsid (N) proteins of SARS-CoV-2, 27 epitope-sequences were identical within SARS-CoV-2 proteins among 115 T cell epitopes. However, 19 out of 27 epitopes are associated with five distinct MHC alleles (at 4-digit resolution): HLA-A*02:01, HLA-B*40:01, HLA-DRA*01:01, HLA-DRB1*07:01, and HLA-DRB1*04:01. For B cell epitopes, they found 49 identical match epitope-sequences that have potential for developing effective vaccines to combat the SARS-CoV-2 [108]. Based on the sequence of the spike glycoprotein, seven epitope residue/regions (491–505, 558–562, 703–704, 793–794, 810, 914, and 1140–1146) in the surface glycoprotein were predicted to be associated with a robust immune response to SARS-CoV-2 [30]. Other candidate epitopes need to be confirmed [95,108].
Using the memory B cells from a survivor who was SARS-CoV infected in 2003, one nAb anti-RBD named S309 was found to bind to SARS-CoV-2 without interfering ACE2 binding. Besides, S309 could recognize a N343-glycan epitope that is distant from the RBM of SARS-CoV-2. Interestingly, N343-glycan of SARS-CoV-2 corresponds to SARS-CoV N330 and they are highly conserved. S309 potently neutralized both pseudotyped SARS-CoV and SARS-CoV-2 and also the authentic SARS-CoV-2 [109].
Using machine learning approaches with the data from other virus outbreaks, some synthetic nAbs named C3, C7, C14, C17, C18, Co1, Co2 and Co4 showed a potential to against SARS-CoV-2. The authors also confirmed that the mutations of Methionine and Tyrosine could increase the affinity of antibody-target binding [110].
19 potential immunogenicity B-cell epitopes, including 2 epitopes located within the RBD region were reported using in silico analysis. 17 of them have >14 amino acids. The B-cell epitopes which had highest score in this study is the 1052-FPQSAPH-1058 located at position 1052aa of S protein. 499 T-cell epitopes bound 34 most popular HLA alleles in the Chinese population were also found. Around 30 candidate vaccine peptides in which 5 peptides located within the RBD region and 17 of them contained both B- and T-cell epitopes, were designed [111]. These vaccine candidates are theoretically able to induce either specific humoral or cellular immune against SARS-CoV-2.
A panel of five humanized single domain antibodies (sdAbs) or nanobodies, 1E2, 2F2, 3F11, 4D8 and 5F8, was recently discovered. These sdAbs bound SARS-CoV-2 tightly but not SARS-CoV, except for 5F8 could bind both viruses but with weaker affinity to SARS-CoV. They also showed neutralization activity against both pseudotyped and authentic SARS-CoV-2. 1E2, 3F11 and 4D8 completely prevented SARS-CoV-2 RBD-ACE2 binding but this effect of 2F2 and 5F8 was only partial. Interestingly, the fusion of the human IgG1 Fc to these sdAbs improved their neutralization activity by 10- to 80-fold [112].
Due to the lack of repairing mechanism of RNA virus replicase complex, SARS-CoV-2 mutations frequently occur during viral replication [111]. The genetic drifts of SARS-CoV-2 are a selective evolution toward less immunogenicity for host immune surveillance by T- or B-cells. The latter appearing strains are less immunogenic than earlier ones [113]. Antigenic drift is also reported in the COVID-19 pandemic. The highly prevalent 23403A > G (p.D614G) variant in the European population may result in vaccine mismatches with little protection to that group of patients [114,115].
Though SARS-COV-2 genome has a much lower mutation rate and genetic diversity than SARS, some of its mutations attract the special attention of scientists. Single amino acid mutation R408I in RBD can reduce the affinity of ACE2 receptor binding [115] that leads to a low or ineffective vaccine for the future epidemic. Effectively, sequence alignment showed that this 408R is strictly conserved in SARS-CoV-2, SARS-CoV. 408R located at the interface between RBD and ACE2, but positioned relatively far away from ACE2, does not have direct interaction with ACE2. 408R can form a hydrogen bond with the 90 N of ACE2. This hydrogen bond is suggested to contribute to the high binding affinity of ACE2 binding [115].
7. Discussion
Science, with new advances, somehow might find the therapy to protect human beings from COVID-19. Among these, plasma therapy composing of antibodies and humoral immune components has been doing great and being one of the first solutions. Because of that, the quest of an antibody always becomes a “must-do-first” when human population facing new pandemic. However, only one lesson could be obvious is we can never get the answer for every pandemic at ones.
Some coronaviruses can infect birds, bats and other species, some are phylogenetically similar to known pathogenic human coronaviruses. The search for the reservoir has resulted in the vast expansion of the library of known coronaviruses which suggests that additional emergence events are possible.
Pandemics will create the urgent need for vaccines around the world simultaneously. But it is not because of this urgency that we can license a vaccine when its benefits and side effects are not clear. Researching a vaccine for influenza viruses, HIV or SARS-CoV-2 is always challenging. Firstly, although the immunogen for protection of a virus, glycoproteins gp120 or gp41 of HIV or S protein of SARS-CoV-2 for example, can be quickly detected, but the immune response needs to be optimized with a good antigen design. Secondly, any drug has side effects immediately or in long-term, directly by the composition of the medication or indirectly by the response of the body to the medication. Pre-clinical experiences with vaccine candidates for SARS-CoV and MERS-CoV are typical examples in aggravating lung disease, either directly or by antibody-dependent enhancement [[116], [117], [118]]. Thirdly, in natural acquired infection, the time point of the detection of nAbs can be easily observed, from 10 to 14 days post-infection in SARS-CoV-2 case for example [119,120], but the potential duration of immunity response is not clear. Therefore, the use of singe-dose or several doses of vaccines needs to be confirmed. Moreover, once a vaccine has been approved, that does not mean that virus research and monitoring can stop. Indeed, influenza and HIV viruses have been reported to have high mutations, making it difficult to find broadly neutralizing vaccines. In the actual pandemic of SARS-CoV-2, drift variants have been reported and that can affect COVID-19 vaccine development [114,115]. No vaccine is available against any coronavirus [121]. Monoclonal antibodies cocktails including multiple epitopes targeting Abs could be taken into account to broaden spectrum of therapy.
Vaccine development is a long and expensive process. From identifying a virus to producing vaccines to market, it takes us a few years. If pandemic gets end before vaccines are approved, the research of vaccine candidates under development need to be continued and ready for clinical trials, in order to get emergency authorization when an outbreak recurs. This statement draws on experience from Ebola pandemic in which vaccine was still under development when the Ebola outbreak ended in 2016. Ebola vaccine is recently approved [[122], [123], [124]] and already used in the recent outbreak in the Democratic Republic of Congo [125].
Studying and understanding the antigen-antibody mechanism of a virus can be used as a precondition to accelerate the studying another virus during an outbreak. In the COVID-19 pandemic, some nAbs studies were based on the research of previous viruses such as SARS, MERS, Ebola and HIV. Through machine learning approaches with the data composed of HIV gp41-antibodies complexes and of 13 more different virus types, some potential nAbs against SARS-CoV-19 were found [110]. This case shows the usefulness of this review of host-antibody interactions from coronaviruses pandemics for young or mature scientists working on vaccine research.
The key findings in coronavirus antibodies investigation could reveal S protein and its subunits as major frame for antibody generation in which RBD seems to be the most efficient peptides. A lot of effort has been tried but still plenty of gaps to fill up. Other approaches and therapies are also needed to protect us from coronavirus infection.
Acknowledgments
Acknowledgments
We would like to thank Dr. Thi-Ngoc-Anh Nguyen and Dr. Bruno Beaumelle for their advice.
Funding
This review did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
References
- 1.University J.H. COVID-19 Dashboard by the Center for Systems Science and Engineering (CSSE) 2020. https://gisanddata.maps.arcgis.com/apps/opsdashboa
- 2.Vanblargan L.A., Goo L., Pierson T.C. Deconstructing the antiviral neutralizing-antibody response: implications for vaccine development and immunity. Microbiol. Mol. Biol. Rev. 2016;80:989–1010. doi: 10.1128/MMBR.00024-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Klasse P.J., Sattentau Q.J. Occupancy and mechanism in antibody-mediated neutralization of animal viruses. J. Gen. Virol. 2002;83:2091–2108. doi: 10.1099/0022-1317-83-9-2091. [DOI] [PubMed] [Google Scholar]
- 4.Georgiou G., Ippolito G.C., Beausang J., Busse C.E., Wardemann H., Quake S.R. The promise and challenge of high-throughput sequencing of the antibody repertoire. Nat. Biotechnol. 2014;32:158–168. doi: 10.1038/nbt.2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Klasse P.J. Neutralization of virus infectivity by antibodies: old problems in new perspectives. Adv. Biol. 2014;2014:1–24. doi: 10.1155/2014/157895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Corti D., Lanzavecchia A. 2013. Broadly Neutralizing Antiviral Antibodies. [DOI] [PubMed] [Google Scholar]
- 7.Monto A.S. Coronaviruses. Yale J. Biol. Med. 1974;47:234–251. [PMC free article] [PubMed] [Google Scholar]
- 8.Hamre D., Procknow J.J. A new virus isolated from the human respiratory tract. Exp. Biol. Med. 1966;121:190–193. doi: 10.3181/00379727-121-30734. [DOI] [PubMed] [Google Scholar]
- 9.Peiris J.S.M., Guan Y., Yuen K.Y. Severe acute respiratory syndrome. Nat. Med. 2004;10:S88–S97. doi: 10.1038/nm1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lau S.K.P., Woo P.C.Y., Li K.S.M., Huang Y., Tsoi H.W., Wong B.H.L., Wong S.S.Y., Leung S.Y., Chan K.H., Yuen K.Y. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc. Natl. Acad. Sci. U. S. A. 2005;102:14040–14045. doi: 10.1073/pnas.0506735102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li W., Shi Z., Yu M., Ren W., Smith C., Epstein J.H., Wang H., Crameri G., Hu Z., Zhang H., Zhang J., McEachern J., Field H., Daszak P., Eaton B.T., Zhang S., Wang L.F. Bats are natural reservoirs of SARS-like coronaviruses. Science (80-.) 2005;310:676–679. doi: 10.1126/science.1118391. [DOI] [PubMed] [Google Scholar]
- 12.Pfefferle S., Oppong S., Drexler J.F., Gloza-Rausch F., Ipsen A., Seebens A., Müller M.A., Annan A., Vallo P., Adu-Sarkodie Y., Kruppa T.F., Drosten C. Distant relatives of severe acute respiratory syndrome coronavirus and close relatives of human coronavirus 229E in bats, Ghana. Emerg. Infect. Dis. 2009;15:1377–1384. doi: 10.3201/eid1509.090224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ge X.Y., Li J.L., Lou Yang X., Chmura A.A., Zhu G., Epstein J.H., Mazet J.K., Hu B., Zhang W., Peng C., Zhang Y.J., Luo C.M., Tan B., Wang N., Zhu Y., Crameri G., Zhang S.Y., Wang L.F., Daszak P., Shi Z.L. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature. 2013;503:535–538. doi: 10.1038/nature12711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shi J., Wen Z., Zhong G., Yang H., Wang C., Huang B., Liu R., He X., Shuai L., Sun Z., Zhao Y., Liu P., Liang L., Cui P., Wang J., Zhang X., Guan Y., Tan W., Wu G., Chen H., Bu Z. Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS–coronavirus 2. Science (80-.) 2020;7015 doi: 10.1126/science.abb7015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Q., Qi J., Yuan Y., Xuan Y., Han P., Wan Y., Ji W., Li Y., Wu Y., Wang J., Iwamoto A., Woo P.C.Y., Yuen K.Y., Yan J., Lu G., Gao G.F. Bat origins of MERS-CoV supported by bat coronavirus HKU4 usage of human receptor CD26. Cell Host Microbe. 2014;16:328–337. doi: 10.1016/j.chom.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cui J., Li F., Shi Z.L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019;17:181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Masters P.S. The molecular biology of coronaviruses. Adv. Virus Res. 2006;65:193–292. doi: 10.1016/S0065-3527(06)66005-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Belouzard S., Chu V.C., Whittaker G.R. Activation of the SARS coronavirus spike protein via sequential proteolytic cleavage at two distinct sites. Proc. Natl. Acad. Sci. U. S. A. 2009;106:5871–5876. doi: 10.1073/pnas.0809524106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Delmas B., Laude H. Assembly of coronavirus spike protein into trimers and its role in epitope expression. J. Virol. 1990;64:5367–5375. doi: 10.1128/jvi.64.11.5367-5375.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hulswit R.J.G., de Haan C.A.M., Bosch B.J. 1st ed. Elsevier Inc; 2016. Coronavirus Spike Protein and Tropism Changes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li F., Li W., Farzan M., Harrison S.C. Structural biology: structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science (80-.) 2005;309:1864–1868. doi: 10.1126/science.1116480. [DOI] [PubMed] [Google Scholar]
- 22.Chakraborti S., Prabakaran P., Xiao X., Dimitrov D.S. The SARS coronavirus S glycoprotein receptor binding domain: fine mapping and functional characterization. Virol. J. 2005;2:1–10. doi: 10.1186/1743-422X-2-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.B.S. glycoproteins, identification of the fusion peptide-containing region in. J. Virol. 2016;90:5586–5600. doi: 10.1128/JVI.00015-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ingallinella P., Bianchi E., Finotto M., Cantoni G., Eckert D.M., Supekar V.M., Bruckmann C., Carfi A., Pessi A. Structural characterization of the fusion-active complex of severe acute respiratory syndrome (SARS) coronavirus. Proc. Natl. Acad. Sci. U. S. A. 2004;101:8709–8714. doi: 10.1073/pnas.0402753101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bosch B.J., Smits S.L., Haagmans B.L. Membrane ectopeptidases targeted by human coronaviruses. Curr. Opin. Virol. 2014;6:55–60. doi: 10.1016/j.coviro.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hofmann H., Hattermann K., Marzi A., Gramberg T., Geier M., Krumbiegel M., Kuate S., Uberla K., Niedrig M., Pohlmann S. S protein of severe acute respiratory syndrome-associated coronavirus mediates entry into hepatoma cell lines and is targeted by neutralizing antibodies in infected patients. J. Virol. 2004;78:6134–6142. doi: 10.1128/jvi.78.12.6134-6142.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dua L., Yang Y., Zhou Y., Lu L., Li F., Jiang S. MERS-CoV spike protein: a key target for antivirals. Expert Opin. Ther. Targets. 2017;21:131–143. doi: 10.1016/j.physbeh.2017.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen Y., Lu S., Jia H., Deng Y., Zhou J., Huang B., Yu Y., Lan J., Wang W., Lou Y., Qin K., Tan W. A novel neutralizing monoclonal antibody targeting the N-terminal domain of the MERS-CoV spike protein. Emerg. Microbes Infect. 2017;6 doi: 10.1038/emi.2017.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang L., Shi W., Chappell J.D., Joyce M.G., Zhang Y., Kanekiyo M., Becker M.M., van Doremalen N., Fischer R., Wang N., Corbett K.S., Choe M., Mason R.D., Van Galen J.G., Zhou T., Saunders K.O., Tatti K.M., Haynes L.M., Kwong P.D., Modjarrad K., Kong W.-P., McLellan J.S., Denison M.R., Munster V.J., Mascola J.R., Graham B.S. Importance of neutralizing monoclonal antibodies targeting multiple antigenic sites on the Middle East Respiratory Syndrome coronavirus spike glycoprotein to avoid neutralization escape. J. Virol. 2018;92:1–21. doi: 10.1128/jvi.02002-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grifoni A., Sidney J., Zhang Y., Scheuermann R.H., Peters B., Sette A. A sequence homology and bioinformatic approach can predict candidate targets for immune responses to SARS-CoV-2. Cell Host Microbe. 2020;27:671–680.e2. doi: 10.1016/j.chom.2020.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jiang S., Hillyer C., Du L. Neutralizing antibodies against SARS-CoV-2 and other human coronaviruses. Trends Immunol. Xx. 2020:1–5. doi: 10.1016/j.it.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sui J., Li W., Roberts A., Matthews L.J., Murakami A., Vogel L., Wong S.K., Subbarao K., Farzan M., Marasco W.A. Evaluation of human monoclonal antibody 80R for immunoprophylaxis of severe acute respiratory syndrome by an animal study, epitope mapping, and analysis of spike variants. J. Virol. 2005;79:5900–5906. doi: 10.1128/jvi.79.10.5900-5906.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sui J., Aird D.R., Tamin A., Murakami A., Yan M., Yammanuru A., Jing H., Kan B., Liu X., Zhu Q., Yuan Q.A., Adams G.P., Bellini W.J., Xu J., Anderson L.J., Marasco W.A. Broadening of neutralization activity to directly block a dominant antibody-driven SARS-coronavirus evolution pathway. PLoS Pathog. 2008;4 doi: 10.1371/journal.ppat.1000197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tian X., Li C., Huang A., Xia S., Lu S., Shi Z., Lu L., Jiang S., Yang Z., Wu Y., Ying T. Potent binding of 2019 novel coronavirus spike protein by a SARS coronavirus-specific human monoclonal antibody. Emerg. Microbes Infect. 2020;9:382–385. doi: 10.1080/22221751.2020.1729069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Prabakaran P., Gan J., Feng Y., Zhu Z., Choudhry V., Xiao X., Ji X., Dimitrov D.S. Structure of severe acute respiratory syndrome coronavirus receptor-binding domain complexed with neutralizing antibody. J. Biol. Chem. 2006;281:15829–15836. doi: 10.1074/jbc.M600697200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhu Z., Chakraborti S., He Y., Roberts A., Sheahan T., Xiao X., Hensley L.E., Prabakaran P., Rockx B., Sidorov I.A., Corti D., Vogel L., Feng Y., Kim J., Wang L., Baric R., Lanzavecchia A., Curtis K.M., Nabel G.J., Subbarao K., Jiang S., Dimitrov D.S. Potent cross-reactive neutralization of SARS coronavirus isolates by human monoclonal antibodies. PNAS. 2007;104:12123–12128. doi: 10.1073/pnas.0701000104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ter Meulen J., Van Den Brink E.N., Poon L.L.M., Marissen W.E., Leung C.S.W., Cox F., Cheung C.Y., Bakker A.Q., Bogaards J.A., Van Deventer E., Preiser W., Doerr H.W., Chow V.T., De Kruif J., Peiris J.S.M., Goudsmit J. Human monoclonal antibody combination against SARS coronavirus: synergy and coverage of escape mutants. PLoS Med. 2006;(3):1071–1079. doi: 10.1371/journal.pmed.0030237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Coughlin M.M., Babcook J., Prabhakar B.S. Human monoclonal antibodies to SARS-coronavirus inhibit infection by different mechanisms. Virology. 2009;394:39–46. doi: 10.1016/j.virol.2009.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xiao X., Feng Y., Chakraborti S., Dimitrov D.S. Oligomerization of the SARS-CoV S glycoprotein: dimerization of the N-terminus and trimerization of the ectodomain. Biochem. Biophys. Res. Commun. 2004;322:93–99. doi: 10.1016/j.bbrc.2004.07.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Coughlin M., Lou G., Martinez O., Masterman S.K., Olsen O.A., Moksa A.A., Farzan M., Babcook J.S., Prabhakar B.S. Generation and characterization of human monoclonal neutralizing antibodies with distinct binding and sequence features against SARS coronavirus using XenoMouse®. Virology. 2007;361:93–102. doi: 10.1016/j.virol.2006.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Greenough T.C., Babcock G.J., Roberts A., Hernandez H.J., Thomas W.D., Jr., Coccia J.A., Graziano R.F., Srinivasan M., Lowy I., Finberg R.W., Subbarao K., Vogel L., Somasundaran M., Luzuriaga K., Sullivan J.L., Ambrosino D.M. Development and characterization of a severe acute respiratory syndrome–associated coronavirus–neutralizing human monoclonal antibody that provides effective immunoprophylaxis in mice. J. Infect. Dis. 2005;191:507–514. doi: 10.1086/427242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Berry J.D., Jones S., Drebot M.A., Andonov A., Sabara M., Yuan X.Y., Weingartl H., Fernando L., Marszal P., Gren J., Nicolas B., Andonova M., Ranada F., Gubbins M.J., Ball T.B., Kitching P., Li Y., Kabani A., Plummer F. Development and characterisation of neutralising monoclonal antibody to the SARS-coronavirus. J. Virol. Methods. 2004;120:87–96. doi: 10.1016/j.jviromet.2004.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Berry J.D., Hay K., Rini J.M., Yu M., Wang L., Plummer F.A., Corbett C.R., Andonov A. Neutralizing epitopes of the SARS-CoV S-protein cluster independent of repertoire, antigen structure or mAb technology. MAbs. 2010;2:53–66. doi: 10.4161/mabs.2.1.10788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pak J.E., Sharon C., Satkunarajah M., Auperin T.C., Cameron C.M., Kelvin D.J., Seetharaman J., Cochrane A., Plummer F.A., Berry J.D., Rini J.M. Structural insights into immune recognition of the severe acute respiratory syndrome coronavirus S protein receptor binding domain. J. Mol. Biol. 2009;388:815–823. doi: 10.1016/j.jmb.2009.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Joyce M.G., Sankhala R.S., Chen W.-H., Choe M., Bai H., Hajduczki A., Yan L., Sterling S.L., Peterson C.E., Green E.C., Smith C., de Val N., Amare M., Scott P., Laing E.D., Broder C.C., Rolland M., Michael N.L., Modjarrad K. BioRxiv; 2020. A Cryptic Site of Vulnerability on the Receptor Binding Domain of the SARS-CoV-2 Spike Glycoprotein. [DOI] [Google Scholar]
- 46.Traggiai E., Becker S., Subbarao K., Kolesnikova L., Uematsu Y., Gismondo M.R., Murphy B.R., Rappuoli R., Lanzavecchia A. An efficient method to make human monoclonal antibodies from memory B cells: potent neutralization of SARS coronavirus. Nat. Med. 2004;10:871–875. doi: 10.1038/nm1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Elshabrawy H.A., Coughlin M.M., Baker S.C., Prabhakar B.S. Human monoclonal antibodies against highly conserved HR1 and HR2 domains of the SARS-CoV spike protein are more broadly neutralizing. PLoS One. 2012;7:1–9. doi: 10.1371/journal.pone.0050366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Keng C.-T., Zhang A., Shen S., Lip K.-M., Fielding B.C., Tan T.H.P., Chou C.-F., Loh C.B., Wang S., Fu J., Yang X., Lim S.G., Hong W., Tan Y.-J. Amino acids 1055 to 1192 in the S2 region of severe acute respiratory syndrome coronavirus S protein induce neutralizing antibodies: implications for the development of vaccines and antiviral agents. J. Virol. 2005;79:3289–3296. doi: 10.1128/jvi.79.6.3289-3296.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang H., Wang G., Li J., Nie Y., Shi X., Lian G., Wang W., Yin X., Zhao Y., Qu X., Ding M., Deng H. Identification of an antigenic determinant on the S2 domain of the severe acute respiratory syndrome coronavirus spike glycoprotein capable of inducing neutralizing antibodies. J. Virol. 2004;78:6938–6945. doi: 10.1128/jvi.78.13.6938-6945.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Duan J., Yan X., Guo X., Cao W., Han W., Qi C., Feng J., Yang D., Gao G., Jin G. A human SARS-CoV neutralizing antibody against epitope on S2 protein. Biochem. Biophys. Res. Commun. 2005;333:186–193. doi: 10.1016/j.bbrc.2005.05.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhou G., Zhao Q. Perspectives on therapeutic neutralizing antibodies against the Novel Coronavirus SARS-CoV-2. Int. J. Biol. Sci. 2020;16:1718–1723. doi: 10.7150/ijbs.45123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tang F., Quan Y., Xin Z., Wrammert J., Ma M., Lv H., Wang T., Yang H., Jan H. Lack of peripheral memory B cell responses in recovered patients with severe acute respiratory syndrome: a six-year follow-up study. J. Immunol. 2011;186:7264–7268. doi: 10.4049/jimmunol.0903490. [DOI] [PubMed] [Google Scholar]
- 53.Fehr A.R., Channappanavar R., Perlman S. Middle East respiratory syndrome: emergence of a pathogenic human coronavirus. Annu. Rev. Med. 2017;68:387–399. doi: 10.1146/annurev-med-051215-031152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Du L., Jiang S. Middle East respiratory syndrome: current status and future prospects for vaccine development. Expert. Opin. Biol. Ther. 2015;15:1647–1651. doi: 10.1517/14712598.2015.1092518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Annan A., Baldwin H.J., Corman V.M., Klose S.M., Owusu M., Nkrumah E.E., Badu E.K., Anti P., Agbenyega O., Meyer B., Oppong S., Sarkodie Y.A., Kalko E.K.V., Lina P.H.C., Godlevska E.V., Reusken C., Seebens A., Gloza-Rausch F., Vallo P., Tschapka M., Drosten C., Drexler J.F. Human betacoronavirus 2c EMC/2012-related viruses in bats, Ghana and Europe. Emerg. Infect. Dis. 2013;19:456–459. doi: 10.3201/eid1903.121503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Memish Z.A., Mishra N., Olival K.J., Fagbo S.F., Kapoor V., Epstein J.H., AlHakeem R., Al Asmari M., Islam A., Kapoor A., Briese T., Daszak P., Al Rabeeah A.A., Lipkin W.I. Middle East respiratory syndrome coronavirus in bats, Saudi Arabia. Emerg. Infect. Dis. 2013;19:1819–1823. doi: 10.3201/eid1911.131172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lu G., Hu Y., Wang Q., Qi J., Gao F., Li Y., Zhang Y., Zhang W., Yuan Y., Bao J., Zhang B., Shi Y., Yan J., Gao G.F. Molecular basis of binding between novel human coronavirus MERS-CoV and its receptor CD26. Nature. 2013;500:227–231. doi: 10.1038/nature12328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Reusken C.B.E.M., Haagmans B.L., Müller M.A., Gutierrez C., Godeke G.J., Meyer B., Muth D., Raj V.S., De Vries L.S., Corman V.M., Drexler J.F., Smits S.L., El Tahir Y.E., De Sousa R., van Beek J., Nowotny N., van Maanen K., Hidalgo-Hermoso E., Bosch B.J., Rottier P., Osterhaus A., Gortázar-Schmidt C., Drosten C., Koopmans M.P.G. Middle East respiratory syndrome coronavirus neutralising serum antibodies in dromedary camels: a comparative serological study. Lancet Infect. Dis. 2013;13:859–866. doi: 10.1016/S1473-3099(13)70164-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chan J.F.W., Lau S.K.P., To K.K.W., Cheng V.C.C., Woo P.C.Y., Yue K.Y. Middle East respiratory syndrome coronavirus: another zoonotic betacoronavirus causing SARS-like disease. Clin. Microbiol. Rev. 2015;28:465–522. doi: 10.1128/CMR.00102-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zaki A.M., Van Boheemen S., Bestebroer T.M., Osterhaus A.D.M.E., Fouchier R.A.M. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 2012;367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 61.Modjarrad K. MERS-CoV vaccine candidates in development: the current landscape. Vaccine. 2016;34:2982–2987. doi: 10.1016/j.vaccine.2016.03.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang N., Shi X., Jiang L., Zhang S., Wang D., Tong P., Guo D., Fu L., Cui Y., Liu X., Arledge K.C., Chen Y.H., Zhang L., Wang X. Structure of MERS-CoV spike receptor-binding domain complexed with human receptor DPP4. Cell Res. 2013;23:986–993. doi: 10.1038/cr.2013.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mou H., Raj V.S., van Kuppeveld F.J.M., Rottier P.J.M., Haagmans B.L., Bosch B.J. The receptor binding domain of the new Middle East respiratory syndrome coronavirus maps to a 231-residue region in the spike protein that efficiently elicits neutralizing antibodies. J. Virol. 2013;87:9379–9383. doi: 10.1128/jvi.01277-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Du L., Zhao G., Kou Z., Ma C., Sun S., Poon V.K.M., Lu L., Wang L., Debnath A.K., Zheng B.-J., Zhou Y., Jiang S. Identification of a receptor-binding domain in the S protein of the novel human coronavirus Middle East respiratory syndrome coronavirus as an essential target for vaccine development. J. Virol. 2013;87:9939–9942. doi: 10.1128/jvi.01048-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Du L., Kou Z., Ma C., Tao X., Wang L., Zhao G., Chen Y., Yu F., Tseng C.-T.K., Zhou Y. A truncated receptor-binding domain of MERS-CoV spike protein potently inhibits MERS-CoV infection and induces strong neutralizing antibody responses: implication for developing therapeutics and vaccines. PLoS One. 2013;8 doi: 10.1371/journal.pone.0081587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yang Y., Deng Y., Wen B., Wang H., Meng X., Lan J., Gao G.F., Tan W. The amino acids 736–761 of the MERS-CoV spike protein induce neutralizing antibodies: implications for the development of vaccines and antiviral agents. Viral Immunol. 2014;27:543–550. doi: 10.1089/vim.2014.0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tang X.C., Agnihothram S.S., Jiao Y., Stanhope J., Graham R.L., Peterson E.C., Avnir Y., Tallarico A.S.C., Sheehan J., Zhu Q., Baric R.S., Marasco W.A. Identification of human neutralizing antibodies against MERS-CoV and their role in virus adaptive evolution. Proc. Natl. Acad. Sci. U. S. A. 2014;111:1–9. doi: 10.1073/pnas.1402074111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jiang L., Wang N., Zuo T., Shi X., Poon K.M.V., Wu Y., Gao F., Li D., Wang R., Guo J., Fu L., Yuen K.Y., Zheng B.J., Wang X., Zhang L. Potent neutralization of MERS-CoV by human neutralizing monoclonal antibodies to the viral spike glycoprotein. Sci. Transl. Med. 2014;6 doi: 10.1126/scitranslmed.3008140. [DOI] [PubMed] [Google Scholar]
- 69.Zhang S., Zhou P., Wang P., Li Y., Jiang L., Jia W., Wang H., Fan A., Wang D., Shi X., Fang X., Hammel M., Wang S., Wang X., Zhang L. Structural definition of a unique neutralization epitope on the receptor-binding domain of MERS-CoV spike glycoprotein. Cell Rep. 2018;24:441–452. doi: 10.1016/j.celrep.2018.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ying T., Du L., Ju T.W., Prabakaran P., Lau C.C.Y., Lu L., Liu Q., Wang L., Feng Y., Wang Y., Zheng B.-J., Yuen K.-Y., Jiang S., Dimitrov D.S. Exceptionally potent neutralization of Middle East respiratory syndrome coronavirus by human monoclonal antibodies. J. Virol. 2014;88:7796–7805. doi: 10.1128/jvi.00912-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li Y., Wan Y., Liu P., Zhao J., Lu G., Qi J., Wang Q., Lu X., Wu Y., Liu W., Zhang B., Yuen K.Y., Perlman S., Gao G.F., Yan J. A humanized neutralizing antibody against MERS-CoV targeting the receptor-binding domain of the spike protein. Cell Res. 2015;25:1237–1249. doi: 10.1038/cr.2015.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhou Y., Yang Y., Huang J., Jiang S., Du L. Advances in MERS-CoV vaccines and therapeutics based on the receptor-binding domain. Viruses. 2019;11:1–18. doi: 10.3390/v11010060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhou P., Lou Yang X., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., Chen H.D., Chen J., Luo Y., Guo H., Di Jiang R., Liu M.Q., Chen Y., Shen X.R., Wang X., Zheng X.S., Zhao K., Chen Q.J., Deng F., Liu L.L., Yan B., Zhan F.X., Wang Y.Y., Xiao G.F., Shi Z.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N., Bi Y., Ma X., Zhan F., Wang L., Hu T., Zhou H., Hu Z., Zhou W., Zhao L., Chen J., Meng Y., Wang J., Lin Y., Yuan J., Xie Z., Ma J., Liu W.J., Wang D., Xu W., Holmes E.C., Gao G.F., Wu G., Chen W., Shi W., Tan W. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yuen K.S., Ye Z.W., Fung S.Y., Chan C.P., Jin D.Y. SARS-CoV-2 and COVID-19: the most important research questions. Cell Biosci. 2020;10:1–5. doi: 10.1186/s13578-020-00404-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science (80-.) 2020;367:1260–1263. doi: 10.1126/science.aax0902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhou Peng, Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L., Chen H.-D., Chen J., Luo Y., Guo H., Jiang R.-D., Liu M.-Q., Chen Y., Shen X.-R., Wang X., Zheng X.-S., Zhao K., Chen Q.-J., Deng F., Liu L.-L., Yan B., Zhan F.-X., Wang Y.-Y., Xiao G.-F., Shi Z.-L. In: Nature. Skipper, editor. Vol. 579. Nature Research; 2020. A pneumonia outbreak associated with a new coronavirus of probable bat origin; pp. 270–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Poh C.M., Carissimo G., Bei W., Amrun S.N., Lee C.Y.-P., Chee R.S.-L., Yeo N.K.-W., Lee W.-H., Leo Y.-S., Chen M.I.-C., Tan S.-Y., Chai L.Y.A., Kalimuddin S., Thien S.-Y., Young B.E., Lye D.C., Wang C.-I., Renia L., Ng L.F. Potent neutralizing antibodies in the sera of convalescent COVID-19 patients are directed against conserved linear epitopes on the SARS-CoV-2 spike protein. BioRxiv. 2020 doi: 10.1101/2020.03.30.015461. [DOI] [Google Scholar]
- 79.Du L., He Y., Zhou Y., Liu S., Zheng B.J., Jiang S. The spike protein of SARS-CoV - a target for vaccine and therapeutic development. Nat. Rev. Microbiol. 2009;7:226–236. doi: 10.1038/nrmicro2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Du L., Yang Y., Zhou Y., Lu L., Li F., Jiang S. MERS-CoV spike protein: a key target for antivirals. Expert Opin. Ther. Targets. 2017;21:131–143. doi: 10.1080/14728222.2017.1271415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Long Q., Deng H., Chen J., Hu J., Liu B., Liao P., Lin Y., Yu L., Mo Z., Xu Y., Gong F., Wu G., Zhang X., Chen Y., Li Z., Wang K., Zhang X., Tian W., Niu C., Yang Q., Xiang J., Du H., Liu H., Lang C., Luo X., Wu S., Cui X., Zhou Z., Wang J., Xue C., Li X., Wang L., Tang X., Zhang Y., Qiu J., Liu X., Li J., Zhang D., Zhang F., Cai X., Wang D., Hu Y., Ren J., Tang N., Liu P., Li Q., Huang A. MedRxiv; 2020. Antibody Responses to SARS-CoV-2 in COVID-19 Patients: The Perspective Application of Serological Tests in Clinical Practice. [Google Scholar]
- 82.Zhao J., Yuan Q., Wang H., Liu W., Liao X., Su Y., Wang X., Yuan J., Li T., Li J., Qian S., Hong C., Wang F., Liu Y., Wang Z., He Q., He B., Zhang T., Ge S., Liu L., Zhang J., Xia N., Zhang Z. Antibody Responses to SARS-CoV-2 in Patients of Novel Coronavirus Disease 2019. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa344. Oxford Academic. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wu F., Wang A., Liu M., Wang Q., Chen J., Xia S., Ling Y., Zhang Y., Xun J., Lu L., Jiang S., Lu H., Wen Y., Huang J. Neutralizing antibody responses to SARS-CoV-2 in a COVID-19 recovered patient cohort and their implications. MedRxiv. 2020 doi: 10.1101/2020.03.30.20047365. [DOI] [Google Scholar]
- 84.Zeng Fanfan, Dai C., Cai P., Wang J., Xu L., Li J., Hu G., Wang L. A comparison study of SARS-CoV-2 IgG antibody between male and female COVID-19 patients: a possible reason underlying different outcome between gender. J. Med. Virol. 2020 doi: 10.1002/jmv.25989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wong V., Dai D., Wu A., Sung J. Treatment of severe acute respiratory syndrome. Hong Kong Med J. 2003;9:199–201. doi: 10.1378/chest.126.3.670. [DOI] [PubMed] [Google Scholar]
- 86.Zhou B., Zhong N., Guan Y. Treatment with convalescent plasma for influenza A (H5N1) infection [9] N. Engl. J. Med. 2007;357:1450–1451. doi: 10.1056/NEJMc070359. [DOI] [PubMed] [Google Scholar]
- 87.Van Griensven J., Edwards T., De Lamballerie X., Semple M.G., Gallian P., Baize S., Horby P.W., Raoul H., Magassouba N., Antierens A., Lomas C., Faye O., Sall A.A., Fransen K., Buyze J., Ravinetto R., Tiberghien P., Claeys Y., De Crop M., Lynen L., Bah E.I., Smith P.G., Delamou A., De Weggheleire A., Haba N. Evaluation of convalescent plasma for Ebola virus disease in Guinea. N. Engl. J. Med. 2016;374:33–42. doi: 10.1056/NEJMoa1511812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Soo Y.O.Y., Cheng Y., Wong R., Hui D.S., Lee C.K., Tsang K.K.S., Ng M.H.L., Chan P., Cheng G., Sung J.J.Y. Retrospective comparison of convalescent plasma with continuing high-dose methylprednisolone treatment in SARS patients. Clin. Microbiol. Infect. 2004;10:676–678. doi: 10.1017/CBO9781107415324.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Cheng Y., Wong R., Soo Y.O.Y., Wong W.S., Lee C.K., Ng M.H.L., Chan P., Wong K.C., Leung C.B., Cheng G. Use of convalescent plasma therapy in SARS patients in Hong Kong. Eur. J. Clin. Microbiol. Infect. Dis. 2005;24:44–46. doi: 10.1007/s10096-004-1271-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Duan K., Liu B., Li C., Zhang H., Yu T., Qu J., Zhou M. Effectiveness of convalescent plasma therapy in severe COVID-19 patients. PNAS. 2020;117:9490–9496. doi: 10.1073/pnas.2004168117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Shen C., Wang Z., Zhao F., Yang Y., Li J., Yuan J., Wang F., Li D., Yang M., Xing L., Wei J., Xiao H., Yang Y., Qu J., Qing L., Chen L., Xu Z., Peng L., Li Y., Chen F., Zheng H., Huang K., Jiang Y., Liu D., Zhang Z., Liu Y., Liu L. Treatment of 5 critically ill patients with COVID-19 with convalescent plasma. JAMA. 2020;323:1582–1589. doi: 10.1001/jama.2020.4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhu Y., Yu D., Han Y., Yan H., Chong H., Ren L., Wang J., Li T., He Y. Cross-reactive neutralization of SARS-CoV-2 by serum antibodies from recovered SARS patients and immunized animals. BioRxiv. 2020 doi: 10.1126/sciadv.abc9999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lv H., Wu N.C., Tsang O.T.-Y., Yuan M., Perera R.A.P.M., Leung W.S., So R.T.Y., Chan J.M.C., Yip G.K., Chik T.S.H., Wang Y., Choi C.Y.C., Lin Y., Ng W.W., Zhao J., Poon L.L.M., Peiris J.S.M., Wilson I.A., Mok C.K.P. Cross-reactive antibody response between SARS-CoV-2 and SARS-CoV infections. Cell Rep. 2020 doi: 10.1016/j.celrep.2020.107725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Park T., Lee S.-Y., Kim S., Kim M.J., Kim H.G., Jun S., Il Kim S., Kim B.T., Park E.C., Park D. Spike protein binding prediction with neutralizing antibodies of SARS-CoV-2. BioRxiv. 2020 doi: 10.1101/2020.02.22.951178. [DOI] [Google Scholar]
- 95.Zheng M., Song L. Novel antibody epitopes dominate the antigenicity of spike glycoprotein in SARS-CoV-2 compared to SARS-CoV. Cell. Mol. Immunol. 2020;2:9–11. doi: 10.1038/s41423-020-0385-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ju B., Zhang Q., Ge X., Wang R., Yu J., Shan S., Zhou B., Song S., Tang X., Yu I., Ge J., Lan U., Yuan J., Wang H., Zhao J., Zhang S., Wang Y., Xuanling S., Liu L., Wang X., Zhang Z., Zhang L. BioRxiv; 2020. Potent human neutralizing antibodies elicited by SARS-CoV-2 infection. [DOI] [PubMed] [Google Scholar]
- 97.Yuan M., Wu N.C., Zhu X., Lee C.-C.D., So R.T.Y., Lv H., Mok C.K.P., Wilson I.A. A highly conserved cryptic epitope in the receptor-binding domains of SARS-CoV-2 and SARS-CoV. Science (80-.) 2020;7269 doi: 10.1126/science.abb7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.OKBA N.M.A., Muller M.A., Li W., Wang C., GeurtsvanKessel C.H., Corman V.M., Lamers M.M., Sikkema R.S., de Bruin E., Chandler F.D., Yazdanpanah Y., Le Hingrat Q., Descamps D., Houhou-Fidouh N., Reusken C.B.E.M., Bosch B.-J., Drosten C., Koopmans M.P.G., Haagmans B.L. Severe Acute Respiratory Syndrome Coronavirus 2-Specific Antibody Responses in Coronavirus Disease 2019 Patients. Emerg. Infect. Dis. 2020 doi: 10.3201/eid2607.200841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wang C., Li W., Drabek D., Okba N.M.A., van Haperen R., Osterhaus A.D.M.E., van Kuppeveld F.J.M., Haagmans B.L., Grosveld F., Bosch B.-J. A human monoclonal antibody blocking SARS-CoV-2 infection. Nat. Commun. 2020 doi: 10.1038/s41467-020-16256-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zheng Z., Monteil V.M., Maurer-Stroh S., Yew C.W., Leong C., Mohd-Ismail N.K., Arularasu S.C., Chow V.T.K., Lin R.T.P., Mirazimi A., Hong W., Tan Y.-J. Monoclonal antibodies for the S2 subunit of spike of SARS-CoV cross-react with the newly-emerged SARS-CoV-2. BioRxiv. 2020 doi: 10.1101/2020.03.06.980037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Poh W.P., Narasaraju T., Pereira N.A., Zhong F., Phoon M.C., Macary P.A., Wong S.H., Lu J., Koh D.R., Chow V.T.K. Characterization of cytotoxic T-lymphocyte epitopes and immune responses to SARS coronavirus spike DNA vaccine expressing the RGD-integrin-binding motif. J. Med. Virol. 2009;81:1131–1139. doi: 10.1002/jmv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Khan S., Nakajima R., Jain A., Ramiro de Assis R., Jasinskas A., Obiero J.M., Adenaiye O., Tai Sheldon, Hong F., Milton D.K., Davies H., Felgner P.L. Analysis of serologic cross-reactivity between common human coronaviruses and SARS-CoV-2 using coronavirus antigen microarray. BioRxiv. 2020 doi: 10.1101/2020.03.24.00654. [DOI] [Google Scholar]
- 103.Kissler S.M., Tedijanto C., Goldstein E., Grad Y.H., Lipsitch M. Projecting the transmission dynamics of SARS-CoV-2 through the postpandemic period. Science (80-.) 2020;5793 doi: 10.1126/science.abb5793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Grifoni A., Weiskopf D., Ramirez S.I., Mateus J., Jennifer M., Moderbacher C.R., Rawlings S.A., Sutherland A., Premkumar L., Jadi R.S., Marrama D., De Silva A.M., Frazier A., Carlin A., Greenbaum J.A., Peters B., Krammer F., Smith D.M., Crotty S., Sette A. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell. 2020 doi: 10.1016/j.cell.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Chen X., Li R., Pan Z., Qian C., Yang Y., You R., Zhao J., Liu P., Gao L., Li Z., Huang Q., Xu L., Tang J., Tian Q., Yao W., Hu L., Yan X., Zhou X., Wu Y., Deng K., Zhang Z., Qian Z., Chen Y., Ye L. Human Monoclonal Antibodies Block the Binding of SARS-CoV-2 Spike Protein to Angiotensin Converting Enzyme 2 Receptor. Cell. Mol. Immunol. 2020 doi: 10.1038/s41423-020-0426-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Madu I.G., Roth S.L., Belouzard S., Whittaker G.R. Characterization of a highly conserved domain within the severe acute respiratory syndrome coronavirus spike protein S2 domain with characteristics of a viral fusion peptide. J. Virol. 2009;83:7411–7421. doi: 10.1128/jvi.00079-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Alsaadi E.A.J., Neuman B.W., Jones I.M. 2019. A Fusion Peptide in the Spike Protein of, Viruses; p. 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ahmed S.F., Quadeer A.A., McKay M.R. Preliminary identification of potential vaccine targets for the COVID-19 Coronavirus (SARS-CoV-2) based on SARS-CoV immunological studies. Viruses. 2020;12 doi: 10.3390/v12030254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Pinto D., Park Y.-J., Beltramello M., Walls A.C., Tortorici M.A., Bianchi S., Jaconi S., Culap K., Zatta F., DeMarco A., Peter A., Guarino B., Spreafico R., Cameroni E., Case J.B., Chen R.E., Havenar-Daughton C., Snell G., Telenti A., Virgin H., Lanzavecchia A., Diamond M.S., Fink K., Veesler D., Corti D. Structural and functional analysis of a potent sarbecovirus neutralizing antibody. BioRxiv. 2020 doi: 10.1101/2020.04.07.023903. [DOI] [Google Scholar]
- 110.Magar R., Yadav P., Farimani A.B. Potential neutralizing antibodies discovered for novel corona virus using machine learning. ArXiv. 2020 doi: 10.1038/s41598-021-84637-4. http://arxiv.org/abs/2003.08447 arXiv:2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Feng Y., Qiu M., Zou S., Li Y., Luo K., Chen R., Sun Y., Wang K., Zhuang X., Zhang S., Chen S., Mo F. BioRxiv; 2020. Multi-epitope Vaccine Design Using an Immunoinformatics Approach for 2019 Novel Coronavirus in China (SARS-CoV-2) [DOI] [Google Scholar]
- 112.Xiaojing Chi, X. Liu, C. Wang, X. Zhang, L. Ren, Q. Jin, J. Wang, W. Yang, Humanized Single Domain Antibodies Neutralize SARS-CoV-2 by Targeting Spike Receptor Binding Domain, BioRxiv. (2020) 2020.04.14.042010. [DOI] [PMC free article] [PubMed]
- 113.Zhu J., Kim J., Xiao X., Wang Y., Luo D., Chen R., Xu L., Zhang H., Xiao G., Zhan X., Wang T., Xie Y. The immune vulnerability landscape of the 2019 Novel Coronavirus. BioRxiv. 2020 doi: 10.1101/2020.02.08.939553. [DOI] [Google Scholar]
- 114.Koyama T., Weeraratne D., Snowdon J.L., Parida L., Heights Y., Heights Y. Emergence of Drift Variants That May Affect COVID-19 Vaccine Development and Antibody Treatment. Pathogens. 2020 doi: 10.3390/pathogens9050324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Jia Y., Shen G., Zhang Y., Huang K., Ho H., Hor W., Yang C. Analysis of the mutation dynamics of SARS-CoV-2 reveals the spread history and emergence of RBD mutant with lower ACE2 binding affinity. BioRxiv. 2020 doi: 10.1101/2020.04.09.034942. [DOI] [Google Scholar]
- 116.Ricke D., Malone R.W. Medical countermeasures analysis of 2019-nCoV and vaccine risks for antibody-dependent enhancement (ADE) Lancet. 2020 doi: 10.2139/ssrn.3546070. [DOI] [Google Scholar]
- 117.Yasui F., Kai C., Kitabatake M., Inoue S., Yoneda M., Yokochi S., Kase R., Sekiguchi S., Morita K., Hishima T., Suzuki H., Karamatsu K., Yasutomi Y., Shida H., Kidokoro M., Mizuno K., Matsushima K., Kohara M. Prior immunization with severe acute respiratory syndrome (SARS)-associated coronavirus (SARS-CoV) nucleocapsid protein causes severe pneumonia in mice infected with SARS-CoV. J. Immunol. 2008;181:6337–6348. doi: 10.4049/jimmunol.181.9.6337. [DOI] [PubMed] [Google Scholar]
- 118.Liu L., Wei Q., Lin Q., Fang J., Wang H., Kwok H., Tang H., Nishiura K., Peng J., Tan Z., Wu T., Cheung K., Chan K., Alvarez X., Qin C., Lackner A., Perlman S., Yuen K., Chen Z. Anti – spike IgG causes severe acute lung injury by skewing macrophage responses during acute SARS-CoV infection. JCI Insight. 2019;4 doi: 10.1172/jci.insight.123158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wu F., Wang Aojie, Liu M., Wang Q., Chen J., Xia S., Ling Y., Zhang Y., Xun J., Lu L., Jiang S., Lu H., Wen Y., Huang J. Neutralizing Antibody Responses to SARS-CoV-2 in a COVID-19 Recovered Patient Cohort and Their Implications. 2020. https//Ssrn.Com/Abstract=3566211 Available SSRN. [DOI]
- 120.Lin L., Lu L., Cao W., Li T. Hypothesis for potential pathogenesis of SARS-CoV-2 infection-a review of immune changes in patients with viral pneumonia. Emerg. Microbes Infect. 2020;9:727–732. doi: 10.1080/22221751.2020.1746199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Padron-regalado E. Vaccines for SARS-CoV-2: lessons from other coronavirus strains. Infect. Dis. Ther. 2020 doi: 10.1007/s40121-020-00300-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Henao-restrepo A.M., Camacho A., Longini I.M., Watson C.H., Edmunds W.J., Egger M., Carroll M.W., Dean N.E. Efficacy and effectiveness of an rVSV-vectored vaccine in preventing Ebola virus disease: final results from the Guinea ring vaccination, open-label, cluster-randomised trial (Ebola Ça Suffit!) Lancet. 2017;389:505–518. doi: 10.1016/S0140-6736(16)32621-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Jendrossek M., Edmunds W.J., Rohan H., Clifford S., Mooney T.A., Eggo R.M. Health care worker vaccination against Ebola: vaccine acceptance and employment duration in Sierra Leone. Vaccine. 2019;37:1101–1108. doi: 10.1016/j.vaccine.2018.12.060. [DOI] [PubMed] [Google Scholar]
- 124.Keshwara R., Johnson R.F., Schnell M.J. Toward an effective Ebola virus vaccine. Annu. Rev. Med. 2017;68:371–386. doi: 10.1146/annurev-med-051215-030919. [DOI] [PubMed] [Google Scholar]
- 125.Kalenga O.I., Moeti M., Sparrow A., Nguyen V.K., Lucey D., Ghebreyesus T.A. The ongoing Ebola epidemic in the Democratic Republic of Congo, 2018–2019. N. Engl. J. Med. 2019;381:373–383. doi: 10.1056/NEJMsr1904253. [DOI] [PubMed] [Google Scholar]