SUMMARY
To study fidelity of RNA polymerase II (Pol II), we analyzed properties of the 6-azauracil-sensitive and TFIIS-dependent E1103G mutant of rbp1 (rpo21), the gene encoding the catalytic subunit of Pol II in Saccharomyces cerevisiae. Using an in vivo retro-transposition-based transcription fidelity assay, we observed that rpb1-E1103G causes a 3-fold increase in transcription errors. This mutant showed a 10-fold decrease in fidelity of transcription elongation in vitro. The mutation does not appear to significantly affect translocation state equilibrium of Pol II in a stalled elongation complex. Primarily, it promotes NTP sequestration in the polymerase active center. Furthermore, pre-steady-state analyses revealed that the E1103G mutation shifted the equilibrium between the closed and the open active center conformations toward the closed form. Thus, open conformation of the active center emerges as an intermediate essential for preincorporation fidelity control. Similar mechanisms may control fidelity of DNA-dependent DNA polymerases and RNA-dependent RNA polymerases.
INTRODUCTION
Accurate transcription is an essential step in utilizing genetic information, just as high fidelity of DNA replication and repair is crucial for the maintenance of genetic integrity. The mechanisms controlling the fidelity of transcription and the biological consequences of transcription errors are much less well understood. Because of the transient nature of mRNAs, the identification and characterization of defects that increase the error rate during transcription has been a particularly difficult challenge.
The consequences of increasing transcription errors have been obscured by the lack of reliable experimental systems. The “multisuppressor phenotype” caused by mutations in the two largest subunits of RNA polymerase of E. coli (Ephrati-Elizur and Luther-Davies, 1981) remains the best example of the effect of transcription errors on cellular metabolism. Mutations with pleiotropic effects, including resistance to rifampicin, in E. coli RNAP subunits were shown to have decreased fidelity of transcription in vitro (Kamzolova and Ozoline, 1982; Blank et al., 1986). None of these mutations has been mapped, and the mechanism by which they affect transcription fidelity remains unknown. Other in vivo studies of transcription fidelity in prokaryotes and eukaryotes have been focused on the effects of putative error correction factors GreA and GreB in bacteria and TFIIS in eukaryotes. None of these proteins is essential for cell viability (Christie et al., 1994; Orlova et al., 1995), and several genetic tests failed to detect significant effects of TFIIS on transcription fidelity in vivo (Shaw et al., 2002; Nesser et al., 2006). However, there is evidence that TFIIS does decrease transcription errors (Koyama et al., 2007). Obviously, new test systems and different tools, including new mutations in RNA polymerase that have an effect on the error rate in vivo and in vitro, are required to establish the physiological role of transcription accuracy.
In principle, fidelity control may occur at any step of the nucleotide addition cycle, including nucleosidetriphosphate (NTP) binding and pairing with the DNA template, phosphodiester bond formation, and pyrophosphate release after completion of the bond. The identification of these steps requires a direct observation of transient intermediates in the process by pre-equilibrium kinetic measurements. Pre-steady-state kinetic studies, combined with the structural and genetic analyses, have provided a detailed understanding of the fidelity of DNA polymerases (reviewed by [Johnson, 1993; Joyce and Benkovic, 2004]). DNA polymerases and RNA-dependent RNA polymerases discriminate between the correct and incorrect substrates by the induced-fit mechanism in which phosphodiester bond formation requires a conformational change in the enzyme active center (isomerization) induced by an accurate base pairing of the NTP to the template base. Despite significant progress in understanding the structure of RNA polymerases (Steitz, 2006; Cramer, 2006; Kornberg, 2007), some of the key functional interactions of the incoming NTP with RNA polymerase remain to be identified. A number of the residues predicted to be involved in NTP selection based on the cocrystals of bacterial and yeast ternary elongation complexes (TECs) with NTPs, failed to exhibit the expected phenotype, as noted below.
First, NTP selection has been proposed to depend on distinct binding sites for complementary and noncomplementary NTPs. The existence of “active” and “inhibitory” conformations of bound NTPs was first proposed for E. coli RNA polymerase based on the pre-steady-state kinetic analyses of misincorporation (Erie et al., 1993) and the electron and nuclear paramagnetic resonance measurement of the distance between the two metal ions in the active center (Eichhorn et al., 1994). This concept has been developed in recent crystallographic studies. The incoming NTP has been detected in the template-independent entry (E) site (Westover et al., 2004) or in the template-dependent preselection (PS) site (Temiakov et al., 2004; Vassylyev et al., 2007b). The correct NTP is believed to be subsequently transferred to the active (A) site, structurally identified in the yeast (Wang et al., 2006) and bacterial (Vassylyev et al., 2007b) TECs. Based on these studies, Arg678 and Asp814 residues in the β subunit of E. coli RNA polymerase were predicted to be crucial for the coordination of the NTP-associated metal ion in the active site (Holmes et al., 2006). However, mutation of these residues did not affect transcription. Unexpectedly, a substitution of a semi-conserved Asp675 to tyrosine dramatically increased misincorporation (Holmes et al., 2006). Asp675 is located far away from the E and A sites, and, while it has been proposed to affect the transfer of the NTP from the E site to the A site, this mechanism requires further experimental proof.
Second, NTP selection has been proposed to be mediated by the movement of the “trigger loop,” a mobile element of the largest subunit of RNA polymerase, which closes on the active site in the presence of a cognate NTP (Wang et al., 2006; Vassylyev et al., 2007a). While the role of the trigger loop in catalysis and pausing has been well supported by functional studies (Toulokhonov et al., 2007), its possible role in fidelity remains to be demonstrated biochemically. The two-pawl ratchet mechanism of elongation, which proposed the role of the trigger loop in fidelity based on the effects of two mutations in the trigger loop of E. coli RNA polymerase on misincorporation and transcription pausing (Bar-Nahum et al., 2005), has been challenged by the more recent studies of these and other mutant polymerase variants (Toulokhonov et al., 2007). The structural data suggest that the trigger loop might be involved in discrimination against dNTPs (Wang et al., 2006). It is unclear whether other parts of the active site are involved in discrimination against dNTPs. The mutation of Asn458 residue in the β′ subunit of E. coli RNA polymerase dramatically increases the efficiency of dNTP incorporation (Svetlov et al., 2004), but the mutation of the homologous residue in S. cerevisiae RNA polymerase does not affect transcription (Wang et al., 2006).
In summary, the complexity of the Pol II structure appears to limit the capacity of the structure-driven site-directed mutagenesis for identification of functionally meaningful mutations. We reasoned that if a mutation in a Pol II subunit has a synthetic phenotype with deletion of DST1 (PPR2) gene, which encodes putative transcription error-correction factor TFIIS (Jeon and Agarwal, 1996), this mutation might decrease transcription fidelity. In this work, we characterize the rpb1-E1103G mutation, rendering the cells dependent on TFIIS (Malagon et al., 2006). The in vivo phenotype of this mutant and its location in the downstream DNA binding cleft at the base of the trigger loop and in vicinity of bridge helix suggested its potential impact on fidelity of Pol II.
RESULTS
The rpb1-E1103G Mutation Increases the Frequency of Retrotransposition Errors
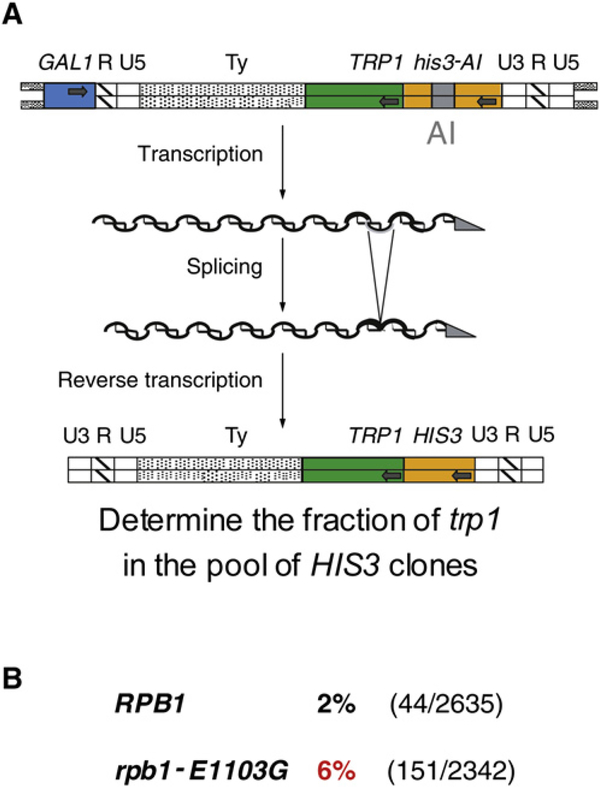
In order to identify rpb1 mutations that cause decreased fidelity, we developed an assay to monitor the fidelity of retrotransposition. This approach differs in two key ways from previous attempts to identify RNA polymerase mutations with reduced fidelity, which are based on suppression of nonsense mutations (Blank et al., 1986; Shaw et al., 2002; Nesser et al., 2006). First, the nonsense suppression assays detect only base substitution events in the three base codon target, whereas the retrotransposition assay can detect substitution, deletions, or additions throughout the reporter gene. Second, during retrotransposition, transcription errors are preserved as permanent by reverse transcription and integration of the retroelement into the genome. Our assay is based on the galactose-inducible Ty1 retrotransposon developed by (Curcio and Garfinkel, 1991), which generates a functional HIS3 gene by a pathway that requires transcription, splicing, and reverse transcription. To that system, we added the TRP1 gene as a passenger during retrotransposition and then monitored whether it had been copied faithfully (Figure 1A). In wild-type cells, 2% of the retrotransposition events resulted in a defective trp1 gene reflecting basal level errors made by RNA polymerase and reverse transcription (Figure 1B). Which polymerase made those errors cannot be distinguished by this approach. Notably, when the wild-type copy of RPB1 was replaced with the rpb1-E1103 allele, the frequency of defective trp1 genes among the retrotransposition events increased to 6%, which suggests an at least 2.5-fold rpb1-E1103-dependent increase in the error rate (p = 0.04). Combined with the demonstration that rpb1-E1103G shows a synthetic lethal interaction with the TFIIS defective mutation dst1 (Malagon et al., 2006), these results made rpb1-E1103G a strong candidate for a mutation causing a direct effect on the fidelity of transcription. To test that hypothesis, we assessed the fidelity of transcription in a variety of in vitro assays.
Figure 1. The E1103G Substitution in Rpb1 Confers a Transcription Fidelity Defect In Vivo.
(A) The experimental setup was designed to determine the frequency of errors in a TRP1 reporter gene during retrotransposition and is shown at the top of the figure.
(B) Frequency of TRP1 inactivation in the strain his3-Δ200 trp1-Δ361 ura3–167 leu2Δ::G418 spt3–101 rpb1-Δ::NAT [pJS366] expressing wild-type Rpb1 [pJS757 RPB1] or Rpb1-E1103G [pJS781 /pb1-E1103G]. The numbers of the clones with trp1/HIS3 along with the total number of HIS3 clones are shown in parentheses.
E1103G Mutation in Rpb1 Compromises Fidelity of Transcription
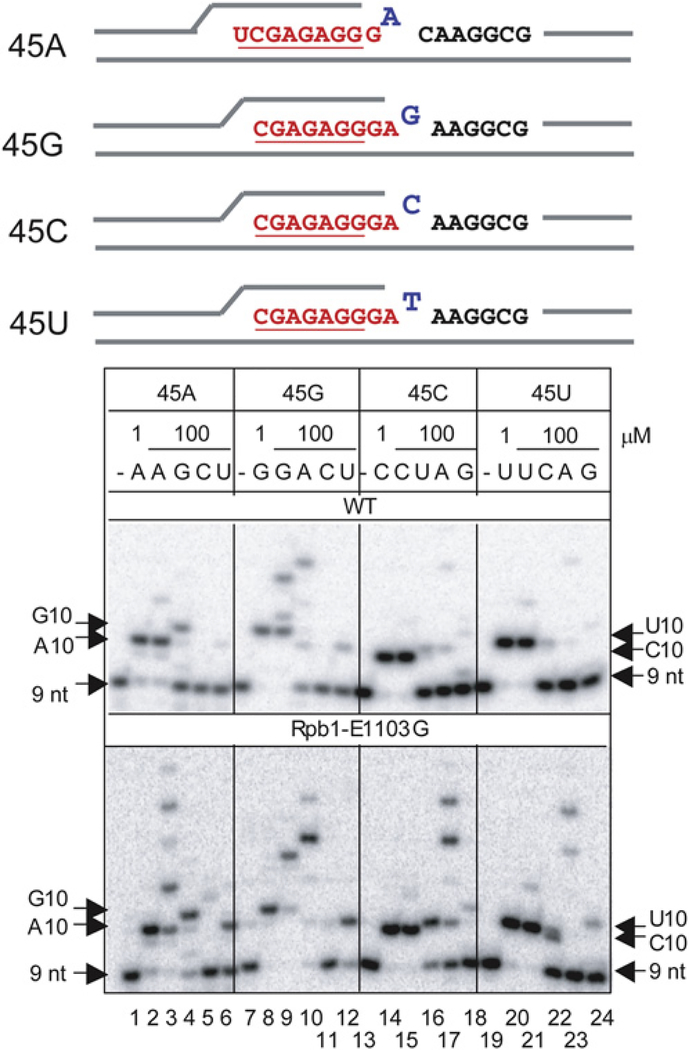
Reported mutations in various DNA and RNA polymerases unequally affect different types of misincorporation, including transitions, transversions, and the ability to discriminate between dNTP and rNTP substrates (Gohara et al., 2004; Svetlov et al., 2004; Holmes et al., 2006). We, therefore, evaluated different types of misincorporation by E1103G Pol II. Incorporation of each of the four NTPs has been tested on templates encoding either A, C, G, or U at the tenth base in the transcript (Figure 2). The product RNAs of the same lengths but with different 3′ ends are distinguished by distinct electrophoretic mobility (compare the 10 nt RNAs in lanes 2 and 4 ending in A or G, respectively, or lanes 15 and 16 ending in C or U, respectively), eliminating the possibility that the RNA extension resulted from incorporation of trace amounts of the complementary NTP. The result shown in Figure 2 confirms that the E1103G mutation dramatically enhances various types of misincorporation. First, there is a strong increase in the “transition-type” misincorporation, which results in purine-to-purine or pyrimidine-to-pyrimidine substitutions (lanes 4, 10, 16, and 22, compare within lanes between the upper panel and the lower panel). Second, the Rpb1-E1103G has a dramatic effect on “transversion-type” substitutions as shown in lane 6 where rU is incorporated opposite dT (rU-dT). Similarly, rU-dC misincorporation (lane 12) is elevated in the mutant, as is the more bulky rA-dG misincorporation (lane 17). We also generated “mature” elongation complexes (Ujvari et al., 2002) with RNA longer than 50 nt and observed a similar effect of Rpb1-E1103G on misincorporation (Figure S1 available online).
Figure 2. The E1103G Substitution in Rpb1 Promotes Misincorporation and Mismatch Extension In Vitro.
TECs were assembled with fully complementary 45 nt template and nontemplate DNA strands and 7 nt or 8 nt RNA oligonucleotides. The assembled TECs were walked to position +9 with 1 μM each of ATP and GTP or only GTP (45A), washed, and incubated with purified NTPs for 5 min. The resulting 10 nt products are named according to the base at the 3’ end (A10 incorporated AMP at the 3′ end, etc.). Positions of the RNAs are indicated by arrows.
The relatively efficient rA-dG transversion observed on template 45C (Figure 2, lane 17), where the encoded CMP is followed by AMP, might suggest that this event occurs by template misalignment (Kashkina et al., 2006). However, misincorporation patterns within other sequence contexts are not consistent with the template misalignment mechanism. First, transitions are always faster than transversions (Figure 2 and Figure S2B), as would be expected when the mispairing occurs in the active site. Second, the efficiency of transitions does not depend on the downstream base (Figure S2A). Third, the rA-dG transversion remains much more efficient than the rG-dG transversion even when the encoded CMP is followed by GMP (Figure S2B). Fourth, the rC-dT transversion is less efficient than the rU-dT transversion, even in the “AC” sequence context (Figure 2, lanes 5 and 6). In summary, most of the misincorporation events analyzed in this work are likely to occur by mispairing rather than by template misalignment.
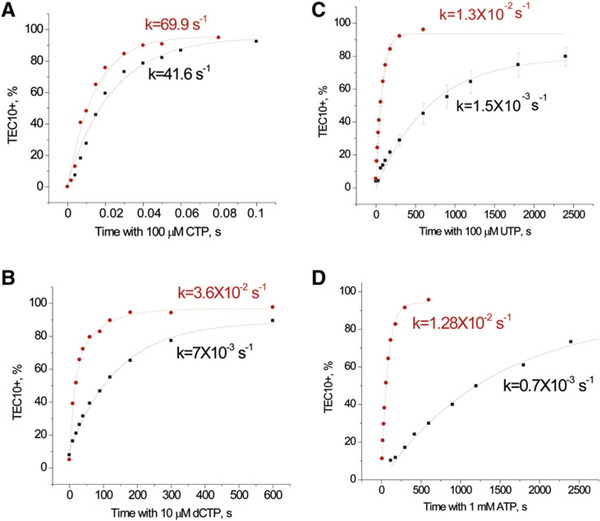
The accuracy of RNA synthesis can only be determined by comparing the difference between the correct and incorrect NTP incorporation for a given template position. In Figure 3, we compare the rates of CTP incorporation and dCTP, UTP, and ATP misincorporation by the elongation complex containing a 9 nt transcript (TEC9) formed by the WT or E1103G Pol II. The E1103G mutation in Rpb1 confers a 1.7-fold increase in the correct CTP incorporation rate (Figure 3A), but its effect on misincorporation is much more pronounced. dCTP is incorporated by the mutant TEC9 5-fold faster (Figure 3B), and UTP and ATP are misincorporated 9- and 18-fold faster, respectively (Figures 3C and 3D), than the same nucleotides by the WT Pol II. As a result, the accuracy of NTP incorporation by E1103G Pol II in this particular position is substantially decreased compared to the WT Pol II.
Figure 3. The E1103G Mutation Promotes Misincorporation of Various Substrates.
TEC8 obtained on the template 45C (Figure 2) was incubated with 10 μM ATP for 30 s, after which 100 μM of CTP (A), 10 μM dCTP (B), 100 μM UTP (C), or 1 μM ATP (D) was added. The reaction with CTP was stopped with 1 M HCl. Black symbols and lines show the data set for WT Pol II, and red symbols and lines represent E1103G Pol II. The data shown are an average of three parallels, and the error bars show the standard deviation. The curves represent single-exponential fits of the data, and the apparent rates (k) are shown.
The mutant relatively successfully selects against dCTP, but it would be wrong to conclude that the E1103G mutation primarily affects recognition of the base complementarity and not the ribose moiety. Apparently, dCTP is a special case, because it is a relatively good substrate for the WT Pol II. At a 10 μM concentration, dCTP incorporation is completed by WT Pol II in 10 min, whereas the comparably efficient incorporation of other dNTPs on their corresponding complementary templates occurred only at 10- to 100-fold higher dNTP concentrations. On all of these templates, E1103G Pol II incorporated the complementary dNTP 10–15 times faster than the WT Pol II (Figure S3 and data not shown). Thus, the effect of the mutation on ribose recognition is comparable with its effect on transition-type misincorporation, indicating that the mutant enzyme is impaired in the recognition of the Watson-Crick base pairing, and the chemical structure of the sugar ring.
Comparisons of incorporation and misincorporation rates at physiological concentrations of NTPs by the WT and mutant Pol II variants provide only an estimation of the impact of the mutation on transcription fidelity. Because the mutation may affect the polymerization rates and the affinity to the nucleotide substrate, fidelity is calculated as the ratio of kpol/KD for the correct NTP to kpol/KD for the incorrect substrate (Wong et al., 1991). The apparent maximal polymerization rate, kpol, and the apparent dissociation constant, KD, were obtained for the WT and E1103G Pol II from the hyperbolic dependence of the reaction rates on the NTP concentration as described in the Supplemental Experimental Procedures. The resulting parameters are shown in Table 1. It is clear that the most prominent effect of the E1103G mutation is an increase in the kpol for UTP, from 0.008 to 0.16 s−1. The subtle increase in the correct NTP incorporation rate and a significant increase of misincorporation rate suggest disruption of a mechanism that maintains nucleotide addition fidelity. We first addressed the effect of the E1103G mutation on translocation of Pol II because a shift of the translocation equilibrium toward the posttranslocated state has been proposed earlier to decrease transcription fidelity of E. coli RNA polymerase (Bar-Nahum et al., 2005).
Table 1.
Rpb1-E1103G Mutation Decreases Transcription Fidelity In Vitro
| Pol II Variant | NTP | kpol, s−1 | KD, μM | kpol/Kd, s−1μM−1 | Fidelity |
|---|---|---|---|---|---|
| WT | CTP (correct) | 75 ± 4 | 68 ± 7 | 1.1 ± 0.17 | |
| UTP (incorrect) | 0.0078 ± 0.0009 | 3531 ± 556 | (2.2 ± 0.6) × 10−6 | (5 ± 2.1) × 105 | |
| Rpb1-E1103G | CTP (correct) | 97 ± 7.3 | 42 ± 9 | 2.3 ± 0.66 | |
| UTP (incorrect) | 0.164 ± 0.03 | 4326 ± 1063 | (3.8 ± 1.63) × 10−5 | (6 ± 4.3) × 104 | |
TEC9 was obtained on the template 45C and chased with 5–500 μM CTP or 0.1–5 μM UTP. The error values for kpol and KD estimates were obtained from the hyperbolic fit as described in the Supplemental Experimental Procedures
Substrate-Independent and NTP-Induced Translocation of Pol II
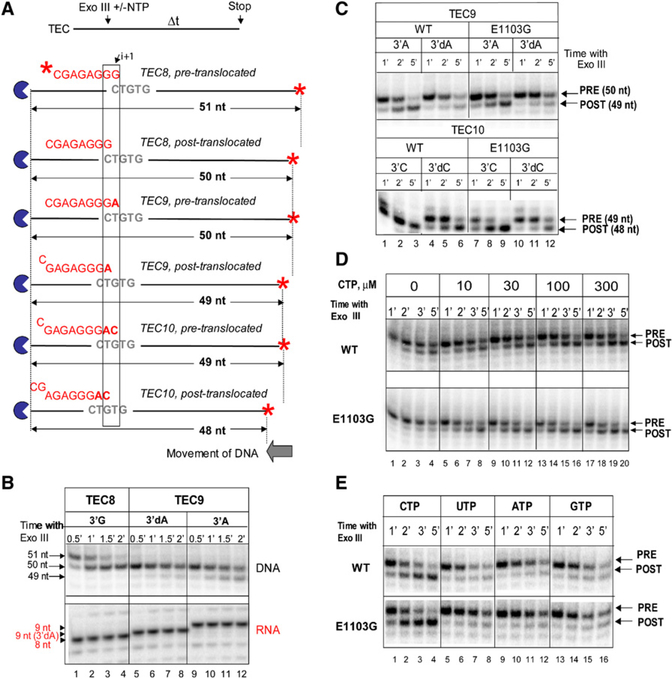
The pre- and posttranslocated states of the Pol II TEC can be distinguished using DNA footprinting with exonuclease III (Exo III). The detailed description of the method is provided in the Supplemental Experimental Procedures. Figure 4A illustrates that the size of the template DNA fragment protected by Pol II from degradation by Exo III should depend on translocation state of the TEC. Figure 4B demonstrates that in TEC8, the upstream boundary of the WT Pol II detected after 30 s incubation with Exo III is located 51 nt from the downstream (labeled) end of the template (lane 1); in TEC9, the protected fragment is 50 nt in the first time point (lane 9). Upon longer incubation with Exo III, the footprint slowly shifts one base pair downstream in both TEC8 and TEC9, but further exonuclease degradation does not occur (lanes 2–4 and 10–12). The longer protected fragment defines the position of pretranslocated TEC. The rate of the conversion of the upstream pretranslocated boundary to the downstream posttranslocated boundary reflects the equilibrium between the pre- and posttranslocated states. It is important to emphasize that the apparent rate of the conversion between the states is affected by Exo III affinity for the DNA in the TEC and the subsequent rate of DNA degradation by Exo III. Therefore, the dynamics of the posttranslocated boundary appearance does not provide quantitative information about the translocation rate. Short time points in the footprinting experiment give the best approximation of the pre-/posttranslocation equilibrium. The detailed Exo III mapping, taken together with the results of pyrophosphorolysis (Figures S5 and S6 and data not shown), suggest that 60%−70% of TEC9 formed by WT Pol II resides in the pretranslocated state.
Figure 4. Translocation Properties of the WT and E1103G Pol II.
(A) Experimental setup.
(B) Dynamics of DNA degradation by Exo III reveals equilibrium between pre- and posttranslocated states of the TEC. TEC9 variants were obtained by 5 min incubation of TEC8 with 10 μM ATP or 3′dATP. The RNAs are shown in the lower panel. Note the aberrantly high mobility of the 9 nt RNA containing 3′dAMP.
(C) The E1103G mutation confers an equilibrium shift toward the pretranslocated state in the stalled TEC. TEC10 variants were obtained by 5 min incubation of TEC9 with 10 μM of CTP or 3′dCTP.
(D) Forward translocation of Pol II in TEC9 containing terminating 3′dAMP is induced by the incoming substrate. CTP was added at the final concentrations indicated on top of the gel.
(E) NTP-stabilized forward translocation of Pol II in TEC9 containing terminating 3′dAMP requires complementary substrate. CTP, UTP, GTP, and ATP were added at 1 μM.
The sequence specificity of Exo III (Linxweiler and Horz, 1982) precludes direct comparison of TECs stalled at different template positions. In TECs stalled at the same DNA position, the difference in Exo III affinity to different DNA sequences can be disregarded because the digestion occurs in the same local sequence context. Thus, Exo III can be used to detect shifts in the equilibrium between the pre- and posttranslocated states in the same TEC caused either by modifications of the nucleic acid scaffold or by mutations in Pol II itself. For example, substitution of AMP with the terminating 3′-dAMP in RNA9 of TEC9 (Figure 4B, lanes 5–8) results in an equilibrium shift toward the pretranslocated state as compared with the TEC9 carrying regular rAMP at the 3′ end of the RNA (lanes 9–12). The E1103G mutation in Rpb1 also affects the translocation equilibrium. Figure 4C illustrates that TEC9 or TEC10 formed by E1103G Pol II (lanes 7–9) are less inclined to reside in the posttranslocated state than the same TECs formed by the WT Pol II (lanes 1–3). The difference in translocation equilibrium of WT and mutant Pol II variants is also observed in the TECs with a 3′-dNMP (Figure 4C, compare lanes 4–6 and 10–12). The decrease of the posttranslocated fraction in E1103G Pol II has been unexpected because E1103G Pol II is faster than WT Pol II (Malagon et al., 2006; also Figure 3A and Table 1). The increased catalytic activity of RNA polymerases is traditionally associated with a shift of the translocation equilibrium toward the posttranslocated state (Bar-Nahum et al., 2005; Nedialkov et al., 2003; Toulokhonov et al., 2007). To resolve this apparent paradox, we analyzed the translocation equilibria in the WT and E1103G Pol II in the presence of the incoming substrate NTP.
Figure 4D shows that the addition of 300 μM CTP to TEC9 (3′-dAMP) shifts the equilibrium toward the posttranslocated state (compare lanes 1–4 and 17–20 in the top [WT] and bottom [E1103G] panels). Lower concentrations of CTP enhance translocation more in the mutant than in the WT TEC9 (Figure 4D, compare lanes 5–8 for the top and bottom panels). The enhanced NTP-dependent translocation equilibrium shift in the mutant is also evident at higher NTP concentrations (compare the upper and lower panels in the lanes 9–12 and 13–16; see also Figure 4E, lanes 1–4). Noncomplementary NTPs do not affect translocation equilibrium even at 1 mM concentration (Figure 4E, lanes 5–16). Thus, the E1103G mutation shifts the equilibrium toward the pretranslocated state when the TECs are deprived of a substrate NTP (Figures 4B and 4C) but induces the equilibrium shift toward the posttranslocated state when substrate NTP is present (Figures 4D and 4E).
The better stabilization of the mutant TEC in posttranslocated state by incoming NTP could be due to a better binding (higher affinity) of the substrate NTP to E1103G Pol II. However, the apparent affinity (KD) to either CTP or UTP is similar in the WT and mutant Pol II (Table 1). Therefore, the equilibrium shift toward the posttranslocated state, observed in the presence of the incoming NTP, is likely to be caused by NTP-dependent conformational change, which stabilizes a posttranslocated TEC-NTP reaction intermediate. By this reasoning, E1103G Pol II should undergo the conformational change faster, or would be more stable in this specific conformation, than the WT Pol II. However, kinetics of the correct and incorrect NTP incorporation analyzed so far (Figure 2 and 3) are consistent with a simple mechanism of NTP addition, in which the initial substrate binding is immediately followed by a phosphoryl transfer reaction (Figure S8), and do not provide information about the reaction intermediates. We modified the pre-steady-state experimental setup to analyze the TEC-NTP intermediates that precede the bond formation.
Sequestration of the Incoming NTP by Pol II
It was likely that the conformational change that stabilizes the posttranslocation state converts the TEC to a conformation in which the bound substrate NTP is positioned for the catalysis. To distinguish between this isomerization and the subsequent phosphoryl transfer, we used a “pulse-chase” experimental setup (Dahlberg and Benkovic, 1991; Patel et al., 1991; Arnold et al., 2004; Anand and Patel, 2006). The pulse-chase method involves exposure of the TEC to radioactively labeled NTP followed by addition of an excess of the unlabeled substrate. The rapid dilution of the labeled substrate does not prevent completion of incorporation of labeled NTP, which has been already bound to the TEC. On the contrary, instant quench of the reaction with HCl reveals only the fraction of TEC that has completed incorporation of the labeled nucleotide at the moment of HCl addition (Dahlberg and Benkovic, 1991).
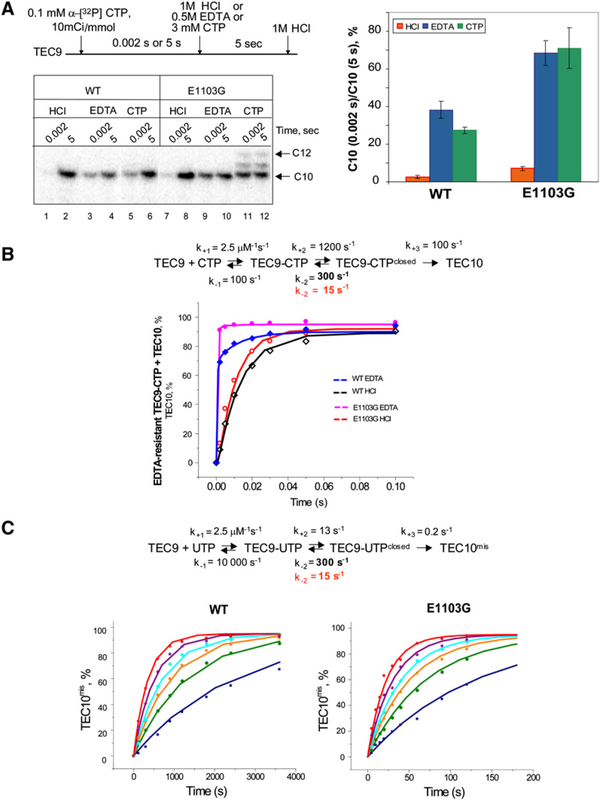
WT TEC9 carrying an unlabeled transcript has been incubated with α-[P32] CTP for 0.002 s, followed by addition of a 30-fold molar excess of unlabeled CTP or HCl. The amount of the 10 nt RNA synthesized in the pulse-chase setup is significantly higher than the amount of the RNA obtained in the acid quench setup (Figure 5A, lane 1 and red bars in the graph). The comparison of the 10 nt bands obtained after 0.002 and 5 s pulses (lanes 5 and 6 and green bars in the graph) suggests that about one-third of the WT TEC9 sequesters the labeled CTP within 0.002 s. The E1103G TEC9 is markedly different from the WT TEC9 in the pulse-chase test. While only a small amount of CTP is incorporated into the RNA in 0.002 s (lane 7), a greater fraction of the mutant TEC successfully sequesters the labeled CTP and generates labeled RNA in the presence of excess unlabeled CTP (compare WT in lane 5 with the mutant in lane 11; also, compare the green bars in the graph).
Figure 5. The E1103G Mutation in Rpb1 Enhances Sequestration of the Incoming NTP and Suppresses Isomerization Reversal of the Elongation Complex.
(A) E1103G Pol II sequesters the incoming NTP more efficiently than the WT Pol II. TEC9 was chased with α-[P32] CTP for 0.002 or 5 s, and the reaction was quenched with 1 M HCl or 0.25 M EDTA or diluted with 3 mM unlabeled CTP and quenched with HCl after 5 s incubation. The longer RNA products (lanes 11 and 12) result from misincorporation of the unlabeled CMP in place of AMP and extension of the mismatch with the next CTP. The plot shows an average of the results of three experiments, and the error bars indicate standard deviation.
(B) EDTA quench reveals the equilibrium between the open and closed conformations of the active center. The experiment was done as in Figure 3A, but the reaction was stopped with HCl (open symbols) or EDTA (solid symbols). The solid lines represent kinetic simulation according to the reaction scheme showed on top. The details of kinetic simulation are described in the Supplemental Experimental Procedures and Figure S8.
(C) The reverse isomerization rate defines the efficiency of misincorporation. UTP concentrations in the reactions were 200 μM (blue), 400 μM (green), 600 μM (orange), 800 μM (cyan), 1200 μM (purple), and 2000 μM (red). The solid lines show kinetic simulation of the data according to the mechanism for misincorporation.
The more efficient “capture” of CTP by the mutant enzyme was further confirmed by EDTA quench. The active center of RNA polymerase contains a stably bound Mg2+ ion, and incoming NTP is brought to the active center of Pol II in a complex with the second Mg2+ ion. If EDTA chelates even one of the Mg2+ ions, catalysis cannot occur. Quenching by EDTA occurs instantly in solution, unless Mg2+ ions are sequestered within the polymerase. The EDTA quench and pulse chase approaches yield similar results for both the WT (lanes 3 and 5) and mutant (lanes 9 and 11) polymerases, suggesting that Pol II sequesters the entire NTP-Mg+2 complex rather than NTP or Mg+2 alone, and that EDTA inactivates only the free pool of NTPs without accessing NTP bound to Pol II.
Importantly, not all polymerases easily sequester the incoming NTP-Mg+2 from EDTA. Poliovirus 3Dpol has no difference in a single NTP incorporation kinetics quenched with HCl or EDTA in the presence of Mg2+ (Arnold and Cameron, 2004). Consequently, presence of an EDTA-resistant step prior to bond formation observed for Pol II in this work and for human Pol II in the previous works (Nedialkov et al., 2003; Zhang and Burton, 2004) is not caused by a slower diffusion rate and reactivity of EDTA compared to HCl.
NTP Sequestration, TEC Isomerization, and Transcription Fidelity
The pulse-chase experiment convincingly demonstrated that we could monitor formation of the NTP-TEC reaction intermediate by EDTA quench. The kinetics of CTP incorporation by the WT TEC9 in this setup is profoundly biphasic (Figure 5B, closed sym-bols), similar to the kinetics observed in human Pol II (Nedialkov et al., 2003; Zhang and Burton, 2004). In the E1103G Pol II, the difference between the accumulation of the 10 nt product in the EDTA quench and the acid quench is amplified. This trend could not be modeled by the increase of phosphoryl transfer rate in the mutant Pol II (Figure S8B). To explain the biphasic ki-netic of the EDTA-resistant intermediate accumulation, we intro-duced a reversible conformational change step into the reaction scheme (Figure 5B). The rate of the fast phase in the EDTA-quenched reaction allowed us to estimate the forward rates for NTP binding (k+1) and isomerization (k+2) as at least 2.5 mM−1s−1 and 1200 s−1, respectively. The biphasic time course of the inter-mediate formation by the WT Pol II could be modeled with a 300 s−1 reverse isomerization rate k−2. In the mutant TEC, k−2 was estimated to be not higher than 15 s−1.
To test whether the decreased rate of isomerization reversal could explain the impaired fidelity of E1103G Pol II, we used the same reaction mechanism to perform kinetic simulation of UTP misincorporation (Figure 5C). We reasoned that the reverse isomerization rate k−2 for incorrect NTP should be at least the same or higher than k−2 for the correct substrate. We accepted reverse isomerization rates of 300 and 15 s−1 for the WT and mutant TEC for misincorporation modeling and obtained the best fit with 13 s−1 direct isomerization rate and 0.2 s−1 chemistry rate (Figure 5C). Thus, the difference between the WT and mutant Pol II misincorporation kinetics could be attributed to the change of a single parameter, the isomerization reversal rate.
DISCUSSION
E1103G substitution in the catalytic subunit of Pol II was selected from a library of randomly mutagenized rpbl clones for sensitivity to 6-azauracil and for strict dependence on TFIIS for viability. A retrotransposition-based in vivo fidelity test suggested that E1103G confers a transcription fidelity defect. In vitro, E1103G Pol II makes a stunningly broad range of transcription errors. The E1103G mutation promotes transition-type and transversion-type misincorporation and decreases selectivity against dNTPs.
To identify the fidelity checkpoint affected by the mutation, we performed pre-steady-state kinetic analysis of transcription by the WT and E1103G Pol II. The analysis revealed that, before proceeding to bond formation, Pol II rapidly tightens the incoming NTP in the active center. A conformational change (isomerization) positioning the substrate for catalysis was proposed based on structural analyses of the core Pol II elongation complex (Gnatt et al., 2001), the position of the NTP in the Pol II TEC structure (Kettenberger et al., 2004), and on transient kinetic analyses of transcription (Anand and Patel, 2006; Zhang and Burton, 2004). Isomerization has been directly demonstrated in the T7 RNA polymerase TEC (Temiakov et al., 2004), Pol II TEC (Wang et al., 2006), and bacterial RNA polymerase TEC (Vassylyev et al., 2007b).
Our observations demonstrate that in yeast Pol II, the sequestration of the substrate in the active site is significantly faster than the bond formation rate, similar to isomerization of human Pol II (Zhang and Burton, 2004) and in contrast to isomerization of T7 RNA polymerase (Anand and Patel, 2006). Allosteric isomerization has been previously suggested as a mechanism of the NTP preselection in RNA polymerases (Kettenberger et al., 2004; Temiakov et al., 2004). The reversible nature of isomerization and the role of the open conformation of the active site in fidelity control have been hypothesized based on localization of mutations that increase misincorporation by T7 RNA polymerase (Huang et al., 2000). We propose a kinetic model that employs a fast isomerization rate for the correct NTP and a slow isomerization rate for the incorrect NTP. Our data obtained in the EDTA quench and pulse-chase experimental setups (Figure 5) indicate that in the WT Pol II, isomerization is reversible. The equilibrium between the closed and open conformations of the WT TEC with a noncomplementary NTP is dramatically shifted toward the open conformation (Figure 5C), making release of the incorrect NTP from the TEC much more likely than its incorporation. The high rate of the cognate substrate tightening (1200 s−1) ensures that, even if the TEC goes through several cycles of reversible isomerization before the chemistry occurs, the correct NTP is not necessarily rejected (Figure 5B). Thus, the slowing of the isomerization reversal in E1103G Pol II promotes misincorporation to a higher extent than the correct NTP incorporation. Consequently, isomerization reversal is likely to represent the transcription fidelity checkpoint that is impaired in E1103G Pol II. Our findings do not exclude the possibility of other fidelity checkpoints, which may take place before or after the NTP sequestration step. For instance, slowing down of phosphoryl transfer itself may significantly reduce misincorporation (Wong et al., 1991; Tsai and Johnson, 2006). Inhibition of pyrophosphate release may cause a dynamic bond reversal in human Pol II (Gong et al., 2005; Xiong and Burton, 2007).
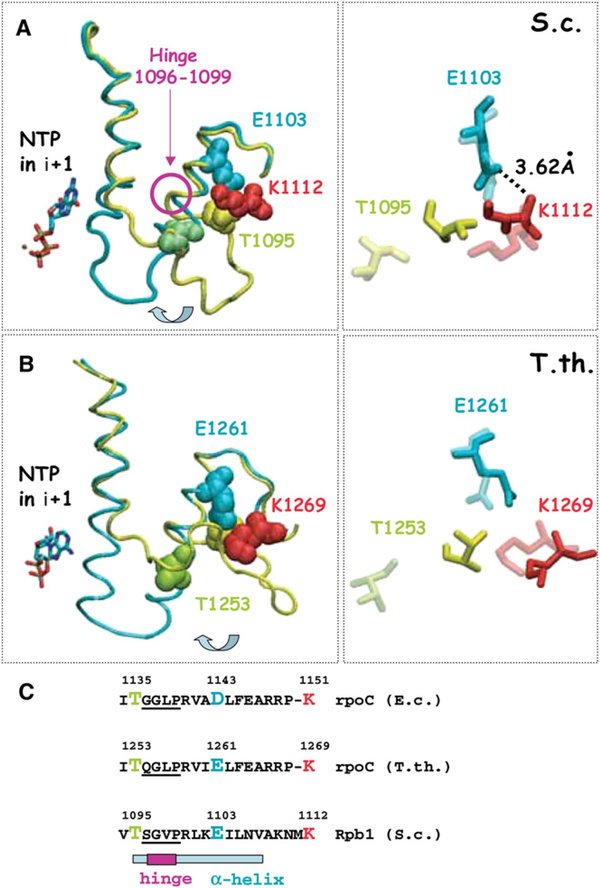
The mechanism for fidelity control involving a reversal of TEC isomerization is consistent with the recently revealed dynamic structural features of the bacterial and yeast elongation complexes. Glu1103 (and its homologs in bacterial RNA polymerases) is located in the “cleft” at the base of the trigger loop, a mobile Rpb1 element tightly closed on the active site in the TECs containing the correct NTP substrates (Vassylyev et al., 2007b; Wang et al., 2006). The trigger loop is unresolved (Cramer et al., 2001; Gnatt et al., 2001) or turned away from the active site in the free polymerase (Tuske et al., 2005) and in the TECs containing incorrect NTPs (Wang et al., 2006; Vassylyev et al., 2007b). The trigger loop consists of two short alpha helices connected by a flexible loop, which performs a long distance movement during the NTP-induced isomerization. In the closed position, the loop and the NTP-proximal α-helix form multiple hydrogen bonds with the NTP (Wang et al., 2006; Vassylyev et al., 2007b). The Glu1103 residue is located at the far end of the α-helix furthest from the NTP-substrate site, raising a question about indirect involvement of this residue in trigger loop closure. We noted that in the open loop conformation, Thr1095 of the flexible part of the trigger loop is positioned in close proximity to Lys1112, which forms a hydrogen bond with Glu1103. In addition, proximity of Glu1103 to Thr1095, and a highly variable positioning of Thr1095 in different crystal forms of Pol II, suggests a possibility of direct interaction of these two residues. Upon the trigger loop closure, Thr1095 moves at a large distance toward the active center, and the Glu1103-Lys1112 interaction becomes broken. The structural alignment of these three residues and their spatial rearrangement during the trigger loop closure is conserved in bacterial polymerase and yeast Pol II (Figures 6A and 6B). The direct or Lys1112-mediated interaction of Glu1103 with the trigger loop might stabilize the open state of the active center (Figure 6). This model for the action of Glu1103 is further supported by the presence of a typical Gly-X-Pro hinge motif (Tieleman et al., 2001) between Glu1103 and Lys1112 on one side and Thr1095 on the other side, which may enable the loop rotation toward or away from the NTP. This model is fully consistent with a dramatic increase in NTP sequestration by E1103G Pol II TEC as compared with the WT TEC (Figure 5). It also might explain the suppression of transient pauses during transcription by E1103G Pol II (Malagon et al., 2006) because a specific positioning of the trigger loop, distinct from its closed position, contributes to the paused conformation of E. coli RNA polymerase (Toulokhonov et al., 2007). Other mutations in this position obtained by site-directed mutagenesis, such as E1103A, also suppress the pausing and increase sequestration of the incoming NTP (Figure S9).
Figure 6. Glu1103 May Stabilize an Open Conformation of the Trigger Loop by Interaction with Lys1112 and Thr1095.
(A) TEC structures of S. cerevisiae Pol II with the closed trigger loop (cyan) and open trigger loop (yellow) (PDB 2E2H [Wang et al., 2006] and 1Y1V [Kettenberger et al., 2004]) aligned using the VMD software (Humphrey et al., 1996). Positions of Glu1103, Lys1112, and Thr1095 are shown for the open state (solid) and closed state (semitransparent) on the right panel. A hydrogen bond between the side chain of Glu1103 and the backbone amide of Lys1112 is shown.
(B) Alignment of T. thermophilus RNA polymerase TEC structures (PDB 2O5J [Vassylyev et al., 2007b] and 2CW0 [Tuske et al., 2005]).
(C) Sequence alignment of the regions undergoing a major shift during the trigger loop closing in yeast (S.c., Saccharomyces cerevisiae) Pol II and bacterial (E.c., E. coli; T. th., T. thermophilus) RNA polymerases.
The predominance of the pretranslocated conformation of TEC9 (the key complex in this study) in the absence of incoming substrate, taken together with a robust sequestration of NTP within the first 0.002 s, deserves separate discussion. Indeed, the posttranslocated state has been considered ready for catalysis based on an assumption that an NTP can bind to the empty active site by thermally driven diffusion through a special funnelshaped secondary pore connecting the active site with the exterior of the enzyme (Batada et al., 2004). All preinsertion sites for the incoming NTP are occluded by the 3′ end of the RNA in the pretranslocated TEC, suggesting that translocation is a prerequisite for the NTP binding. How does NTP enter the predominantly pretranslocated TEC in such a short time? The elongation complexes that predominantly reside in the pretranslocation state might translocate back and forth with a very high frequency, permitting the NTP entry through the pore and sequestration within 2 milliseconds. However, the estimated NTP diffusion rate through the highly negatively charged pore estimated from collision theory is ~200 nts per s, and the number of binding events will be ~20 per s (Batada et al., 2004), which is too low to explain the rapid NTP sequestration observed in our work. Alternatively, the incoming NTP might bind to the pretranslocated complex and subsequently transfer to the active center. The possibility of a sequence-specific preloading of NTP to the pretranslocated complex has been proposed based on the influence of the NTPs complementary to i+2, i+3, and i+4 positions on phosphodiester bond formation by human Pol II (Gong et al., 2005; Xiong and Burton, 2007). NTP loading may precede translocation in bacteriophage T7 RNA polymerase, where the TEC containing an incoming NTP has been crystallized in a conformation best described as a transition between the pre- and posttranslocated states (Yin and Steitz, 2004; Temiakov et al., 2004).
Our findings establish a mechanism for transcription fidelity control, which is surprisingly similar to the mechanism of substrate selection proposed for a number of single-subunit DNA polymerases and RNA-dependent RNA polymerases. The specificity of T7 DNA polymerase is determined by the relative rates of chemistry and the reverse of the enzyme-NTP complex isomerization: in a complex with the correct NTP, the chemistry is much faster; in a complex with the incorrect NTP, the reversed isomerization is much faster (Johnson, 1993; Arnold and Cameron, 2004; Joyce and Benkovic, 2004; Tsai and Johnson, 2006). Thus, mechanisms of substrate selection may be conserved in different classes of polymerases.
EXPERIMENTAL PROCEDURES
Reagents
NTPs were purchased from Amersham Pharmacia Biotech (Piscataway, NJ). The NTPs used for misincorporation assays were additionally purified by chromatography on Dowex 1X2–400 using a 0 to 1 M LiCl gradient in 3 mM HCl. Pol II containing a histidine-tagged Rpb3 subunit was purified on HisTrap HP cartridges (Amersham Pharmacia Biotech, Piscataway, NJ) with a 10–100 mM imidazole gradient elution, followed by chromatography on a MonoQ HR10/10 column (Amersham Pharmacia Biotech, Piscataway, NJ) (Kireeva et al., 2003). E1103G Pol II was additionally purified on hepa-rin-Sepharose and on a MonoQ HR5/5 column (Amersham Pharmacia Bio-tech, Piscataway, NJ).
In Vitro Transcription
Elongation complexes were assembled and immobilized on Ni-nitrilotriacetate (Ni-NTA) agarose (QIAGEN, Valencia, CA) as described previously (Kireeva et al., 2003). Before the chase, the TECs were eluted from Ni-NTA agarose by 100 mM imidazole. The eluate was filtered using Ultrafree-MC 0.45 μm cen-trifugal filters (Millipore, Billerica, MA) to remove the agarose beads and diluted with transcription buffer (TB; 20 mM Tris-HCl, pH 7.9, 5 mM MgCl2, 1 mM 2-mercaptoethanol, 40 mM KCl, 0.1 mg/ml BSA, 12% glycerol) to bring the imidazole concentration below 10 mM. The time courses for the correct NTP incorporation were obtained using RQF3 and RQF4 rapid quench flow instruments (KinTek Corporation, Austin, TX). The TECs were incubated with NTPs for 0.002–0.5 s at 25°C. The reactions were stopped by addition of HCl to 1 M or EDTA to 0.25 M final concentration. The reactions with the incorrect substrates were stopped manually by gel-loading buffer (5 M urea; 25 mM EDTA final concentrations). The RNA products were resolved in 20% denatur-ing polyacrylamide gels, visualized with a Typhoon 8600 phosphorimager (Amersham Pharmacia Biotech, Piscataway, NJ), and quantified using Image-Quant software (Molecular Dynamics, part of Amersham Pharmacia Biotech, Piscataway, NJ).
Data Analyses
The percentage of TECs that incorporated the given NTP was quantified as a fraction of the RNAs of that length and longer from the total radioactivity in the lane and plotted versus time. The data sets were fit with a single-exponential equation using the OriginPro 7.5 software (OriginLab Corporation, Northhampton, MA). Kinetic simulations were performed using KinTekSim and KinTek Global Kinetic Explorer (Kintek Corp.)
Exonuclease III Footprinting
For Exo III footprinting, the TECs were assembled with RNA7 on TDS65 with the fully complementary NDS65 (Table S1). The RNA7 and TDS65 were labeled at the 5′ end (Kireeva et al., 2003). The TECs were eluted from the beads as described above. The footprinting reactions were initiated by mixing 15 μl of the TEC with 15 μl TB containing 100 units of Exo III (New England Biolabs, Bedford, MA) and NTP substrates. The reaction was stopped by mixing with the gel-loading buffer.
Supplementary Material
ACKNOWLEDGMENTS
We thank Mikhail Bubunenko, Donald Court, and Eric Galburt for helpful discussions, and we thank Vladimir Tchernaenko for advice in NTP purification, Deanna Gotte and Brenda Shafer for technical assistance, and Donald Court for critical reading of the manuscript. The contents of this publication do not necessarily reveal the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Footnotes
SUPPLEMENTAL DATA
The Supplemental Data include Supplemental Experimental Procedures, one table, and nine figures and can be found with this article online at http://www.molecule.org/cgi/content/full/30/5/557/DC1/.
REFERENCES
- Anand VS, and Patel SS (2006). Transient state kinetics of transcription elongation by T7 RNA polymerase. J. Biol. Chem. 281, 35677–35685. [DOI] [PubMed] [Google Scholar]
- Arnold JJ, and Cameron CE (2004). Poliovirus RNA-dependent RNA polymerase (3Dpol): pre-steady-state kinetic analysis of ribonucleotide incorporation in the presence of Mg2+. Biochemistry 43, 5126–5137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold JJ, Gohara DW, and Cameron CE (2004). Poliovirus RNA-dependent RNA polymerase (3Dpol): pre-steady-state kinetic analysis of ribonucleotide incorporation in the presence of Mn2+. Biochemistry 43, 5138–5148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Nahum G, Epshtein V, Ruckenstein AE, Rafikov R, Mustaev A, and Nudler E (2005). A ratchet mechanism of transcription elongation and its control. Cell 120, 183–193. [DOI] [PubMed] [Google Scholar]
- Batada NN, Westover KD, Bushnell DA, Levitt M, and Kornberg RD (2004). Diffusion of nucleoside triphosphates and role of the entry site to the RNA polymerase II active center. Proc. Natl. Acad. Sci. USA 101, 17361–17364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blank A, Gallant JA, Burgess RR, and Loeb LA (1986). An RNA polymerase mutant with reduced accuracy of chain elongation. Biochemistry 25, 5920–5928. [DOI] [PubMed] [Google Scholar]
- Christie KR, Awrey DE, Edwards AM, and Kane CM (1994). Purified yeast RNA polymerase II reads through intrinsic blocks to elongation in response to the yeast TFIIS analogue, P37. J. Biol. Chem. 269, 936–943. [PubMed] [Google Scholar]
- Cramer P (2006). Mechanistic studies of the mRNA transcription cycle. Bio-chem. Soc. Symp. 73, 41–47. [DOI] [PubMed] [Google Scholar]
- Cramer P, Bushnell DA, and Kornberg RD (2001). Structural basis of transcription: RNA polymerase II at 2.8 angstrom resolution. Science 292, 1863–1876. [DOI] [PubMed] [Google Scholar]
- Curcio MJ, and Garfinkel DJ (1991). Regulation of retrotransposition in Saccharomyces cerevisiae. Mol. Microbiol. 5, 1823–1829. [DOI] [PubMed] [Google Scholar]
- Dahlberg ME, and Benkovic SJ (1991). Kinetic mechanism of DNA polymerase I (Klenow fragment): identification of a second conformational change and evaluation of the internal equilibrium constant. Biochemistry 30, 4835–4843. [DOI] [PubMed] [Google Scholar]
- Eichhorn GL, Chuknyisky PP, Butzow JJ, Beal RB, Garland C, Janzen CP, Clark P, and Tarien E (1994). A structural model for fidelity in transcription. Proc. Natl. Acad. Sci. USA 91, 7613–7617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ephrati-Elizur E, and Luther-Davies S (1981). A novel form of suppression due to an altered RNA polymerase. Mol. Gen. Genet. 181, 390–394. [DOI] [PubMed] [Google Scholar]
- Erie DA, Hajiseyedjavadi O, Young MC, and von Hippel PH (1993). Multiple RNA polymerase conformations and GreA: control of the fidelity of transcription. Science 262, 867–873. [DOI] [PubMed] [Google Scholar]
- Gnatt AL, Cramer P, Fu J, Bushnell DA, and Kornberg RD (2001). Structural basis of transcription: an RNA polymerase II elongation complex at 3.3 A resolution. Science 292, 1876–1882. [DOI] [PubMed] [Google Scholar]
- Gohara DW, Arnold JJ, and Cameron CE (2004). Poliovirus RNA-dependent RNA polymerase (3Dpol): kinetic, thermodynamic, and structural analysis of ribonucleotide selection. Biochemistry 43, 5149–5158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong XQ, Zhang C, Feig M, and Burton ZF (2005). Dynamic error correction and regulation of downstream bubble opening by human RNA polymerase II. Mol. Cell 18, 461–70. [DOI] [PubMed] [Google Scholar]
- Holmes SF, Santangelo TJ, Cunningham CK, Roberts JW, and Erie DA (2006). Kinetic investigation of Escherichia coli RNA polymerase mutants that influence nucleotide discrimination and transcription fidelity. J. Biol. Chem. 281, 18677–18683. [DOI] [PubMed] [Google Scholar]
- Huang J, Brieba LG, and Sousa R (2000). Misincorporation by wild-type and mutant T7 RNA polymerases: identification of interactions that reduce misincorporation rates by stabilizing the catalytically incompetent open conformation. Biochemistry 39, 11571–11580. [DOI] [PubMed] [Google Scholar]
- Humphrey W, Dalke A, and Schulten K (1996). VMD: visual molecular dynamics. J. Mol. Graph. 14, 33–38. [DOI] [PubMed] [Google Scholar]
- Jeon C, and Agarwal K (1996). Fidelity of RNA polymerase II transcription controlled by elongation factor TFIIS. Proc. Natl. Acad. Sci. USA 93, 13677–13682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson KA (1993). Conformational coupling in DNA polymerase fidelity. Annu. Rev. Biochem. 62, 685–713. [DOI] [PubMed] [Google Scholar]
- Joyce CM, and Benkovic SJ (2004). DNA polymerase fidelity: kinetics, structure, and checkpoints. Biochemistry 43, 14317–14324. [DOI] [PubMed] [Google Scholar]
- Kamzolova SG, and Ozoline ON (1982). An RNA polymerase with reduced fidelity of RNA synthesis from an E. coli mutant suggests the existence of a correction system of non-complementary nucleotide incorporation during transcription. Mol. Biol. Rep. 8, 133–135. [DOI] [PubMed] [Google Scholar]
- Kashkina E, Anikin M, Brueckner F, Pomerantz RT, McAllister WT, Cramer P, and Temiakov D (2006). Template misalignment in multisubunit RNA polymerases and transcription fidelity. Mol. Cell 24, 257–266. [DOI] [PubMed] [Google Scholar]
- Kettenberger H, Armache KJ, and Cramer P (2004). Complete RNA polymerase II elongation complex structure and its interactions with NTP and TFIIS. Mol. Cell 16, 955–965. [DOI] [PubMed] [Google Scholar]
- Kireeva ML, Lubkowska L, Komissarova N, and Kashlev M (2003). Assays and affinity purification of biotinylated and nonbiotinylated forms of double-tagged core RNA polymerase II from Saccharomyces cerevisiae. Methods Enzymol. 370, 138–155. [DOI] [PubMed] [Google Scholar]
- Kornberg RD (2007). The molecular basis of eucaryotic transcription. Cell Death Differ. 14, 1989–1997. [DOI] [PubMed] [Google Scholar]
- Koyama H, Ito T, Nakanishi T, and Sekimizu K (2007). Stimulation of RNA polymerase II transcript cleavage activity contributes to maintain transcriptional fidelity in yeast. Genes Cells 12, 547–559. [DOI] [PubMed] [Google Scholar]
- Linxweiler W, and Horz W (1982). Sequence specificity of exonuclease III from E. coli. Nucleic Acids Res. 10, 4845–4859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malagon F, Kireeva ML, Shafer BK, Lubkowska L, Kashlev M, and Strathern JN (2006). Mutations in the Saccharomyces cerevisiae RPB1 gene conferring hypersensitivity to 6-azauracil. Genetics 172, 2201–2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nedialkov YA, Gong XQ, Hovde SL, Yamaguchi Y, Handa H, Geiger JH, Yan H, and Burton ZF (2003). NTP-driven translocation by human RNA polymerase II. J. Biol. Chem. 278, 18303–18312. [DOI] [PubMed] [Google Scholar]
- Nesser NK, Peterson DO, and Hawley DK (2006). RNA polymerase II subunit Rpb9 is important for transcriptional fidelity in vivo. Proc. Natl. Acad. Sci. USA 103, 3268–3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlova M, Newlands J, Das A, Goldfarb A, and Borukhov S (1995). Intrinsic transcript cleavage activity of RNA polymerase. Proc. Natl. Acad. Sci. USA 92, 4596–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel SS, Wong I, and Johnson KA (1991). Pre-steady-state kinetic analysis of processive DNA replication including complete characterization of an exonuclease-deficient mutant. Biochemistry 30, 511–525. [DOI] [PubMed] [Google Scholar]
- Shaw RJ, Bonawitz ND, and Reines D (2002). Use of an in vivo reporter assay to test for transcriptional and translational fidelity in yeast. J. Biol. Chem. 277, 24420–24426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steitz TA (2006). Visualizing polynucleotide polymerase machines at work. EMBO J. 25, 3458–3468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svetlov V, Vassylyev DG, and Artsimovitch I (2004). Discrimination against deoxyribonucleotide substrates by bacterial RNA polymerase. J. Biol. Chem. 279, 38087–38090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temiakov D, Patlan V, Anikin M, McAllister WT, Yokoyama S, and Vassylyev DG (2004). Structural basis for substrate selection by t7 RNA polymerase. Cell 116, 381–391. [DOI] [PubMed] [Google Scholar]
- Tieleman DP, Shrivastava IH, Ulmschneider MR, and Sansom MS (2001). Proline-induced hinges in transmembrane helices: possible roles in ion channel gating. Proteins 44, 63–72. [DOI] [PubMed] [Google Scholar]
- Toulokhonov I, Zhang J, Palangat M, and Landick R (2007). A central role of the RNA polymerase trigger loop in active-site rearrangement during transcriptional pausing. Mol. Cell 27, 406–419. [DOI] [PubMed] [Google Scholar]
- Tsai YC, and Johnson KA (2006). A new paradigm for DNA polymerase specificity. Biochemistry 45, 9675–9687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuske S, Sarafianos SG, Wang X, Hudson B, Sineva E, Mukhopadhyay J, Birktoft JJ, Leroy O, Ismail S, Clark AD Jr., et al. (2005). Inhibition of bacterial RNA polymerase by streptolydigin: stabilization of a straight-bridge-helix active-center conformation. Cell 722, 541–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ujvari A, Pal M, and Luse DS (2002). RNA polymerase II transcription complexes may become arrested if the nascent RNA is shortened to less than 50 nucleotides. J. Biol. Chem. 277, 32527–32537. [DOI] [PubMed] [Google Scholar]
- Vassylyev DG, Vassylyeva MN, Perederina A, Tahirov TH, and Artsimovitch I (2007a). Structural basis for transcription elongation by bacterial RNA polymerase. Nature 448, 157–162. [DOI] [PubMed] [Google Scholar]
- Vassylyev DG, Vassylyeva MN, Zhang J, Palangat M, Artsimovitch I, and Landick R (2007b). Structural basis for substrate loading in bacterial RNA polymerase. Nature 448, 163–168. [DOI] [PubMed] [Google Scholar]
- Wang D, Bushnell DA, Westover KD, Kaplan CD, and Kornberg RD (2006). Structural basis of transcription: role of the trigger loop in substrate specificity and catalysis. Cell 127, 941–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westover KD, Bushnell DA, and Kornberg RD (2004). Structural basis of transcription: nucleotide selection by rotation in the RNA polymerase II active center. Cell 119, 481–89. [DOI] [PubMed] [Google Scholar]
- Wong I, Patel SS, and Johnson KA (1991). An induced-fit kinetic mechanism for DNA replication fidelity: direct measurement by single-turnover kinetics. Biochemistry 30, 526–537. [DOI] [PubMed] [Google Scholar]
- Xiong Y, and Burton ZF (2007). A tunable ratchet driving human RNA polymerase II translocation adjusted by accurately templated nucleoside triphosphates loaded at downstream sites and by elongation factors. J. Biol. Chem. 282, 36582–36592. [DOI] [PubMed] [Google Scholar]
- Yin YW, and Steitz TA (2004). The structural mechanism of translocation and helicase activity in T7 RNA polymerase. Cell 116, 393–404. [DOI] [PubMed] [Google Scholar]
- Zhang C, and Burton ZF (2004). Transcription factors IIF and IIS and nucleoside triphosphate substrates as dynamic probes of the human RNA polymerase II mechanism. J. Mol. Biol. 342, 1085–1099. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.