Abstract
Intestinal bacterial dysbiosis is evident in children with cystic fibrosis (CF) and intestinal viruses may be contributory, given their influence on bacterial species diversity and biochemical cycles. We performed a prospective, case-control study on children with CF and age and gender matched healthy controls (HC), to investigate the composition and function of intestinal viral communities. Stool samples were enriched for viral DNA and RNA by viral extraction, random amplification and purification before sequencing (Illumina MiSeq). Taxonomic assignment of viruses was performed using Vipie. Functional annotation was performed using Virsorter. Inflammation was measured by calprotectin and M2-pyruvate kinase (M2-PK). Eight CF and eight HC subjects were included (50% male, mean age 6.9 ± 3.0 and 6.4 ± 5.3 years, respectively, p = 0.8). All CF subjects were pancreatic insufficient. Regarding the intestinal virome, no difference in Shannon index between CF and HC was identified. Taxonomy-based beta-diversity (presence-absence Bray-Curtis dissimilarity) was significantly different between CF and HC (R2 = 0.12, p = 0.001). Myoviridae, Faecalibacterium phage FP Taranis and unclassified Gokushovirinae were significantly decreased in CF compared with HC (q<0.05). In children with CF (compared to HC), the relative abundance of genes annotated to (i) a peptidoglycan-binding domain of the peptidoglycan hydrolases (COG3409) was significantly increased (q<0.05) and (ii) capsid protein (F protein) (PF02305.16) was significantly decreased (q<0.05). Picornavirales, Picornaviridae, and Enterovirus were found to positively correlate with weight and BMI (r = 0.84, q = 0.01). Single-stranded DNA viruses negatively correlated with M2-PK (r = -0.86, q = 0.048). Children with CF have an altered intestinal virome compared to well-matched HC, with both taxonomic and predicted functional changes. Further exploration of Faecalibacterium phages, Gokushovirinae and phage lysins are warranted. Intestinal viruses and their functions may have important clinical implications for intestinal inflammation and growth in children with CF, potentially providing novel therapeutic targets.
Introduction
Dysfunction of the cystic fibrosis (CF) transmembrane conductance regulator (CFTR) results in an altered intestinal milieu with proposed mechanisms including: (i) reduced bicarbonate secretion and low intestinal pH, (ii) thick and inspissated mucus, (iii) a lack of endogenous pancreatic enzymes, (iv) delayed intestinal transit and (v) possibly impaired innate immunity [1, 2]. These mechanisms along with treatment and diet regimens [3] likely result in intestinal bacterial dysbiosis and inflammation, which have repeatedly been reported in CF [4–12]. Intestinal inflammation in CF may have significant clinical relevance due to its link with growth and nutrition [9, 13, 14]. In children with CF compared to healthy controls (HC), intestinal bacterial profiles are often characterised by an increased abundance of Proteobacteria (Escherichia coli, Shigella, Enterobacter) and a decreased abundance of Bacteroidetes (Bacteroides, Alistipes) and Firmicutes (Faecalibacterium prausnitzii) [4, 6, 8, 12]. It has been suggested that in children with CF, enteric fat abundance (increased as a result of diet regimens and a lack of endogenous pancreatic enzymes) selects for pro-inflammatory gut microbiota [6]. Additionally, the faecal microbiota of people with CF may have a higher prevalence of amoxicillin resistance [15].
Intestinal viruses also have the potential to contribute to bacterial dysbiosis and intestinal inflammation, given their influence on bacterial species diversity and biochemical cycles. To the best of our knowledge, the intestinal virome has yet to be explored in CF, both in terms of characterisation and its potential contribution to microbiota, inflammation and CF pathogenesis.
Human faecal viromes typically consist of eukaryote-infecting viruses, and bacteriophages (viruses of bacterial hosts). Predominant bacteriophages in the gut include double-strand DNA (dsDNA) Caudovirales and single-strand DNA (ssDNA) Microviridae [16–19]. The community of intestinal bacteriophages show substantial inter-individual variability, but appear temporally stable within a single individual [16, 17]. Bacteriophages can influence bacterial populations via host lysis and horizontal gene transfer, as well as playing an indirect role in regulating immune function and inflammation [19–22]. Specifically, inflammatory bowel disease studies have shown changes in the intestinal virome (compared to HC) are likely due to changes in lytic phage populations, which lyse bacterial hosts, resulting in release of pathogen-associated molecular patterns and antigens that cause inflammation [19, 23].
In recent years there has been a focus on the role of viruses in CF lung disease [24]. In CF airways, environmental stress and frequent antibiotic treatment has been suggested to enhance bacteriophage mobility and promote bacteriophage-mediated spread of antibiotic resistance genes [25, 26]. The respiratory viromes of adults with CF have previously been explored in several small studies using a metagenomics approach, [27–30] however only Willner et al. (2009) [27] included healthy controls in their study. To the best of our knowledge, the intestinal virome in CF has not been explored, particularly in comparison to matched-healthy controls.
The aim of this study was to investigate the composition and function of viral communities inhabiting the intestines of children with CF. We hypothesized that the composition and functional capacity of viral communities in the intestines of children with CF are different when compared to HC. Furthermore, we hypothesized that these alterations have clinical implications in children with CF.
Results
Population characteristics
There were eight children with CF and eight paired HC matched for age and gender included in this study. The mean age of CF and HC subjects was 6.9 ± 3.0 and 6.4 ± 3.2 years, respectively (p = 0.8), with a range of 3 to 12 years for both cohorts. Each group consisted of four males (50%). All CF children were pancreatic insufficient (PI) and five were homozygous for the F508del mutation in the CFTR gene (a disease-causing CF mutation) (63%), whilst the remaining three were heterozygous for the F508del mutation and a second disease-causing mutation (37%). No CF participants were on CFTR modulator therapy. Clinical characteristics are presented in Table 1.
Table 1. Clinical characteristics.
| ID | Gender | Group | Pancreas | Age (years) | Genotype | Calprotectin(mg/kg) | M2-PK (U/ml) |
|---|---|---|---|---|---|---|---|
| CF1 | M | CF | PI | 3.0 | F508del / F508del | 204.7 | 142.5 |
| HC1 | M | HC | HC | 3.0 | - / - | - | - |
| CF2 | M | CF | PI | 3.1 | F508del / F508del | 56.8 | 35.0 |
| HC2 | M | HC | HC | 3.2 | - / - | 17.6 | < 1 |
| CF3 | F | CF | PI | 5.8 | F508del / F508del | 171.4 | 6.1 |
| HC3 | F | HC | HC | 3.1 | - / - | 97.3 | < 1 |
| CF4 | F | CF | PI | 6.5 | F508del / G551D | 45.2 | 7.4 |
| HC4 | F | HC | HC | 5.8 | - / - | 30.5 | < 1 |
| CF5 | M | CF | PI | 8.2 | F508del / F508del | 51.7 | 10.1 |
| HC5 | M | HC | HC | 8.6 | - / - | - | - |
| CF6 | M | CF | PI | 8.4 | F508del / 394delTT | 141.1 | 5.2 |
| HC6 | M | HC | HC | 11.7 | - / - | < 19.5 | 3.0 |
| CF7 | F | CF | PI | 8.8 | F508del / F508del | < 19.5 | 6.4 |
| HC7 | F | HC | HC | 7.6 | - / - | - | - |
| CF8 | F | CF | PI | 11.8 | F508del / G551D | 149.2 | - |
| HC8 | F | HC | HC | 8.5 | - / - | < 19.5 | < 1 |
Demographics and fecal inflammatory marker results of participants. No CF participants were on CFTR modulator therapy. M2-PK, M2-pyruvate kinase; PI, pancreatic insufficient; -, not tested or data not available.
Virome characteristics
Viral community structures were successfully assessed using Illumina MiSeq sequencing of a viral DNA and RNA enrichment of stool samples. Sequencing data were processed using the Vipie pipeline and contained an average of 3,909,006 ± 1,283,246 reads per sample, which after quality filtering and de novo assembly resulted in a median (IQR; range) of 863 (625–2946; 227–7737) contigs per sample. On average, samples contained 568,964 ± 201,042 reads that were mapped to viruses (75.6% ± 27.5%), bacterial ribosomes (23.7% ± 27.2%) and humans (0.63% ± 0.88%) (S3 Data). Bacterial contamination rates (based on reads mapping to bacterial ribosomes) ranged from 0.1% to 82.2% and both bacterial and human reads were filtered out from the analysis. Rarefaction curves of viral taxa suggest the number of samples included in this study did not reach virome saturation, suggesting that a larger number of samples per cohort would be needed to capture the entire human virome (S1A and S1B Fig). However, saturation of rarefaction curves for individual samples was achieved, suggesting that all viruses in a given sample were recovered (S1C and S1D Fig).
Alpha diversity of virome
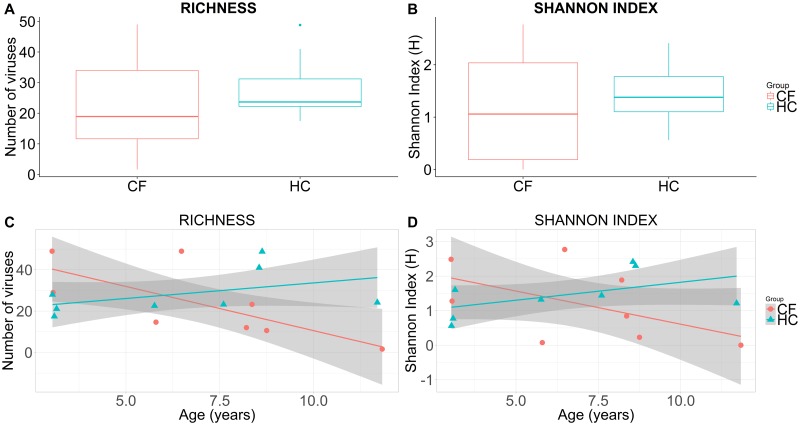
Regarding the virome alpha diversity, children with CF and HC had no significant difference in richness (number of unique viruses) (mean difference (95% CI) of -4.7 (-24.9 to 15.6), p = 0.6) and Shannon index (mean difference (95% CI) of -0.3 (-1.4 to 0.9), p = 0.6) (Fig 1A and 1B). Controlling for age, there was also no difference between children with CF and HC for richness (estimate (SE) -4.1 (7.4), p = 0.6) and Shannon index (estimate (SE) -0.2 (0.5), p = 0.6). However, with advancing age, alpha diversity indices (richness and Shannon index) decreased in CF and increased in HC cohorts (Fig 1C and 1D). The intercepts for the two generalised linear models that fit the trends were significantly different (richness model p = 0.008; Shannon index model p = 0.045) suggesting the effect of age should be considered.
Fig 1.
Boxplots of sample richness (number of unique viruses) (A) and Shannon index (B) in CF and HC cohorts. Scatterplots of sample richness (number of unique viruses) (C) and Shannon index (D) against age in CF and HC cohorts. Cohort mean and 95% confidence intervals are constructed from generalised linear models and presented as lines and shaded regions, respectively (C, D).
Beta diversity of virome
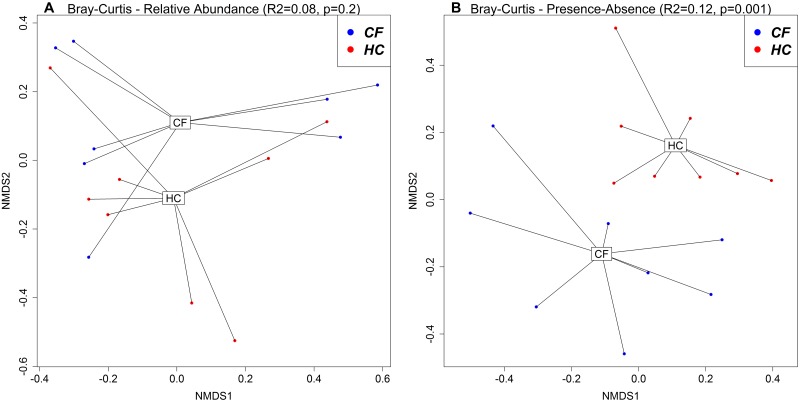
Visualization of taxonomy-based beta diversity (Fig 2) showed a significant difference in viral communities between CF and HC cohorts based on presence-absence data using Bray-Curtis dissimilarity (BC) (R2 = 0.12, p = 0.001), but not on relative abundance data using BC (R2 = 0.08, p = 0.2). PERMANOVA showed no significant difference in viral communities between males and females (relative abundance BC R2 = 0.09, p = 0.09; presence-absence BC R2 = 0.07, p = 0.2). PERMANOVA showed a significant difference in viral communities across ages (relative abundance BC R2 = 0.11, p = 0.02; presence-absence BC R2 = 0.09, p = 0.04).
Fig 2. NMDS plots based on Bray-Curtis dissimilarities using relative abundance (A) and presence-absence (B) data between CF and HC cohorts.
Differences in viral communities between CF and HC populations
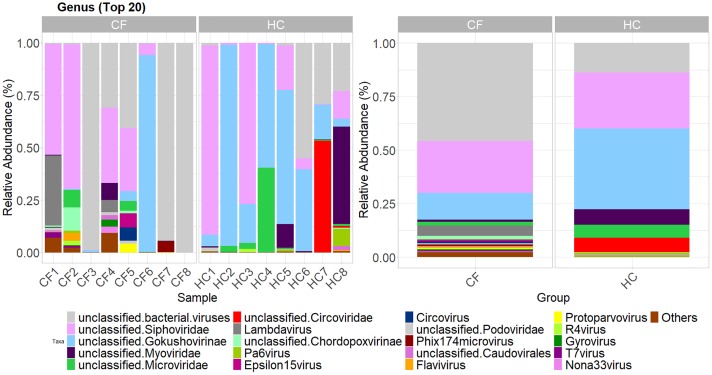
The relative abundance of all viruses at each of the taxonomic ranks in CF and HC subjects are presented in S2A–S2E Fig. Relative abundances of the top 20 most abundant viral genera in CF and HC are presented in Fig 3. The relative abundance of Myoviridae (family) and one of its species, Faecalibacterium phage FP Taranis were significantly decreased in children with CF compared with HC (q<0.05) (S3A and S3C Fig). Unclassified Gokushovirinae was also significantly decreased in children with CF compared with HC (q<0.05) (S3B Fig). Unclassified Gokushovirinae consisted of two species of Gokushovirus (WZ-2015a and human gut gokushovirus isolate SH).
Fig 3. Relative abundance of top 20 most abundant viral genera for CF and HC subjects.
Samples ordered in increasing age (from left to right).
Porcine circovirus (PCV) and porcine parvovirus (PPV) are known to be in porcine-derived pancreatic enzyme replacement therapy (PERT) products prescribed to patients with CF [31, 32]. We identified PCV type 2 (PCV2) and PPV in 4/8 (50%) and 3/8 (38%) of CF subjects respectively, and not in any HC subjects.
Functional annotations
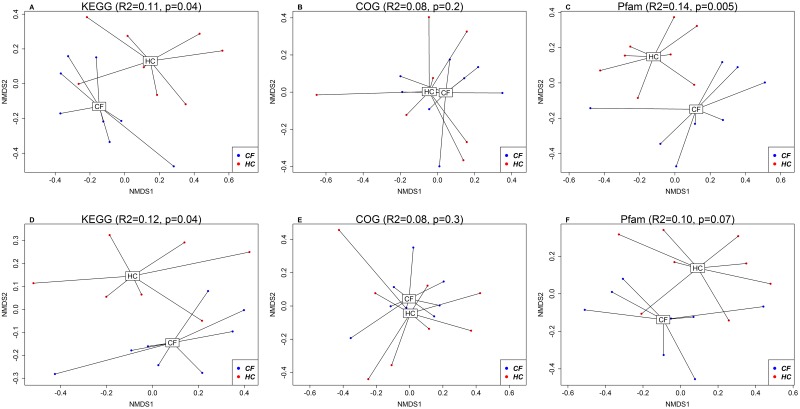
The functional annotations were performed using Virsorter [33] to search against the Kyoto Encyclopedia of Genes and Genomes (KEGG) [34], Clusters of Orthologous Groups (COG) [35] and Pfam [36] databases. On average, 225 (138) predictions were identified per sample. Predictions were mapped to: (i) 71 unique KEGG terms (4.8% predictions mapped); (ii) 146 unique COG terms (10.4% predictions mapped); and (iii) 223 unique Pfam terms (16.6% predictions mapped). Visualization of beta diversity based on the functional annotation (Fig 4) showed a significant difference in KEGG terms between CF and HC cohorts based on relative abundance data using BC dissimilarity (R2 = 0.11, p = 0.04) and presence-absence data using BC dissimilarity (R2 = 0.12, p = 0.04). There was no significant difference in COG terms between CF and HC cohorts based on relative abundance BC (R2 = 0.08, p = 0.2) and presence-absence BC (R2 = 0.08, p = 0.3). There was a significant difference in Pfam terms between CF and HC cohorts based on relative abundance BC (R2 = 0.14, p = 0.005), but not on presence-absence BC (R2 = 0.10, p = 0.07).
Fig 4. NMDS plots based on Bray-Curtis dissimilarities using relative abundance (A-C; top row) and presence-absence (D-F; bottom row) data on functional annotation (using KEGG, COG and Pfam databases) between CF and HC cohorts.
The relative abundance of peptidoglycan-binding (PGRP) domain of the peptidoglycan hydrolases (COG3409 [COG]) was significantly elevated in children with CF compared with HC (q<0.05) (S4A Fig). The relative abundance of capsid protein (F protein) (PF02305.16 [Pfam]) was significantly decreased in children with CF compared with HC (q<0.05) (S4B Fig). There was no significant difference in the relative abundance of any KEGG terms between CF and HC cohorts (q<0.05), however zinc D-Ala-D-Ala carboxypeptidase (K08640 [KEGG]) came close to significance (q<0.1) (S4C Fig).
Age-related changes in the intestinal virome
Exploring the effect of age, linear models of the differentially abundant viruses and protein sequences were constructed and presented in S5 Fig. Comparing children with CF and HC whilst controlling for age, the relative abundance of: (i) Myoviridae was significantly decreased with age in CF (estimate (SE) -1.1×10−4 (3.7×10−5), p = 0.01); (ii) unclassified Gokushovirinae was not significantly different (estimate (SE) -0.02 (0.01), p = 0.2); (iii) Faecalibacterium phage FP Taranis was significantly decreased in CF (estimate (SE) -4.5×10−5 (1.6×10−5), p = 0.02); (iv) a peptidoglycan-binding (PGRP) domain of peptidoglycan hydrolases (COG3409) was significantly increased in CF (estimate (SE) 2.0×10−3 (4.5×10−4), p = 0.0006); (v) capsid protein (F protein) (PF02305.16) was significantly decreased in CF (estimate (SE) -0.008 (0.002), p = 0.003); and (vi) zinc D-Ala-D-Ala carboxypeptidase (K08640) was significantly decreased in CF (estimate (SE) -0.005 (0.001), p = 0.009) and significantly increased with age in HC (estimate (SE) 0.0007 (0.0003), p = 0.02);
Associations with anthropometrics and inflammatory biomarkers in children with CF
In children with CF, there were no significant correlations between alpha diversity indices (richness and Shannon diversity index) and anthropometric (height, weight and body mass index (BMI)) z-scores or intestinal inflammatory makers (calprotectin and M2-PK). Correlations between the relative abundances of viruses with anthropometric z-scores and intestinal inflammatory makers in children with CF are presented in S1 Table. Picornavirales, Picornaviridae and Enterovirus positively correlated with weight and BMI (r = 0.84, q = 0.01). Anelloviridae also positively correlated with weight and BMI (r = 0.79, q = 0.03). Parvoviridae and Protoparvovirus negatively correlated with weight (r = -0.87, q = 0.03).
Regarding inflammatory markers, children with CF had significantly elevated median (IQR) faecal calprotectin levels compared to HC (98.9 mg/kg (50.1–104.9) vs. 19.5 mg/kg (19.5–30.5), respectively, p = 0.047; lower limit of detection was 19.5 mg/kg) (S6A Fig). Median (IQR) faecal M2-PK levels were also significantly elevated in CF compared with HC (7.4 U/ml (6.3–22.6) vs. 1.0 U/ml (1.0–1.0), respectively, p = 0.005; lower limit of detection was 1.0 U/ml) (S6B Fig). Podoviridae positively correlated with M2-PK (r = 0.85, q = 0.03). Single-stranded DNA viruses and unclassified ssDNA viruses (order) negatively correlated with M2-PK (r = -0.86, q = 0.048).
Correlations between the relative abundances of KEGG, COG and Pfam terms with anthropometric z-scores and intestinal inflammatory makers in children with CF are presented in S2 Table. N-acetylmuramoyl-L-alanine amidase (K01447) positively correlated with height (r = 0.85, q = 0.02). No correlations with q<0.05 were identified between COG or Pfam terms and: (i) anthropometric z-scores, or (ii) intestinal inflammatory markers.
Discussion
We have demonstrated the intestines of children with CF exhibit an altered intestinal virome, which includes both taxonomic and functional changes, when compared to matched healthy controls. To our knowledge, this is the first study to explore the intestinal virome in children with CF using a metagenomics approach on an enriched virome preparation. We observed high inter-individual variability in intestinal viruses and distinct clustering (based on presence-absence data for the BC analysis) between CF and HC cohorts. The results of this study suggest that the virome has a potential role in intestinal inflammation and growth impairment in children with CF. Whether this is a direct influence or indirectly through bacterial population changes is unknown and will require further investigation. This could be one explanation for why therapies directed at specifically modulating microbiota (e.g. probiotics [37]) may not be sufficient. A holistic approach that incorporates other microbes (i.e. viruses) may be necessary to restore intestinal homeostasis in children with CF.
The intestinal inflammation present in children with CF is clinically relevant given its negative correlation with growth (9). We have identified distinct mechanisms which may promote intestinal inflammation in children with CF, including: (i) excess production of bacteriophage endolysins (e.g. peptidoglycan-binding (PGRP) domain of peptidoglycan hydrolases) which have potential pro inflammatory effects [19, 36], (ii) under expression of viruses which lyse obligate intra-cellular parasitic bacteria (e.g. Gokushovirinae) [38] and bacteriophages associated with beneficial bacteria, Faecalibacterium prausnitzii (e.g. Faecalibacterium phage FP Taranis) [39, 40], and (iii) over expression of bacteriophages associated with potentially detrimental Proteobacteria (e.g. Podoviridae) [41]. Additionally, we identified several viruses which positively (e.g. Anelloviridae and Enterovirus) and negatively (e.g. Protoparvovirus) correlated with growth measures in children with CF. Functional annotation of intestinal viruses revealed distinct clusters between CF and HC when referencing the KEGG (Fig 4A and 4D) and Pfam (based on relative abundance BC) databases (Fig 4C). A peptidoglycan-binding (PGRP) domain of peptidoglycan hydrolases (COG3409) was significantly elevated in children with CF compared to HC. These endolysins are synthesised by bacteriophages at the end of their lytic cycle and result in the degradation of their host’s peptidoglycan-based cell wall to allow for the release of viral progeny [36]. This finding may provide one hypothesis for the reduced bacterial richness and Shannon index previously reported in CF children [4, 6]. Furthermore, lysis of bacteria may release nucleic acids, proteins and lipids that serve as inflammatory response triggers, inducing cellular infiltration, cytokine production and even tissue damage [19], a possible contributory factor to the elevated M2-PK in children with CF.
Consistent with current literature [16–19], intestinal viruses were predominantly bacteriophages, namely dsDNA Caudovirales and ssDNA Microviridae. Gokushovirinae were significantly decreased in children with CF. Gokushovirinae are generally considered lytic viruses and typically infect obligate intra-cellular parasitic bacteria, e.g. Chlamydia and Spiroplasma [38]. Furthermore, Gokushovirinae peptidases resemble peptidases from Firmicutes (e.g. Faecalibacterium) and Proteobacteria (e.g. Ahrensia, Providencia) suggesting a horizontal acquisition of genes during a co-infection or infection of the prophage-bearing host cell [42]. The importance of ssDNA viruses in CF intestinal dysbiosis, which were predominantly Gokushovirinae in our cohort, are further emphasised by the negative correlation with M2-PK. M2-PK is expressed by rapidly proliferating intestinal cells and elevated in the setting of intestinal inflammation and colorectal cancer [43], however the mechanism for a negative correlation with ssDNA viruses is unclear. The decreased relative abundance of Gokushovirinae in our CF cohort is also consistent with the reduction of capsid protein (F protein), which is a major capsid component of ssDNA bacteriophages [36].
The relevance of intestinal viruses and inflammation is highlighted by Faecalibacterium phage FP Taranis, which was significantly decreased in children with CF. The bacterial host for this phage is F. prausnitzii [39], which has previously been reported to be decreased in children and adolescence with CF [44], as well as in other inflammatory conditions including inflammatory bowel disease and psoriasis [45]. F. prausnitzii is considered a beneficial bacteria as it is an important butyrate producer with anti-inflammatory activity through the production of MAM protein [40]. Therefore, reduced Faecalibacterium phage FP Taranis may be reflective of reduced F. prausnitzii and reduced anti-inflammatory capacity i.e. resulting in elevated inflammatory biomarkers (calprotectin and M2-PK) in children with CF. Additionally, Podoviridae (a family of dsDNA viruses) positively correlated with M2-PK and several phage from this family are associated with potentially detrimental Proteobacteria (Enterobacteria, Escherichia, Pseudomonas and Salmonella Phage) [41].
The associations between intestinal viruses and anthropometrics in children with CF require further exploration as the potential mechanisms are unclear. We identified that in children with CF, the family Anelloviridae and the genus Enterovirus positively correlated with weight and BMI z-scores (S1 Table). A phage lysin, N-acetylmuramoyl-L-alanine amidase (K01447) also positively correlated with height z-scores in children with CF (S2 Table). Protoparvovirus negatively correlated with weight z-scores and the only identified species of this genus was PPV. Porcine parvovirus is currently not thought to infect humans [46] and the negative correlation of Protoparvovirus with weight is more than likely reflective of an increase in PERT dosing to improve weight gain in our CF clinic. The presence of the ssDNA viruses PPV and PCV2 only in CF subjects may have unintentionally provided quality assurance of our workflow, given that they are both known to be in porcine-derived PERT products prescribed to patients with CF [31, 32]. Porcine circovirus 2 is ubiquitous among swine populations and not thought to infect immunocompetent humans [47].
The trends in alpha diversity indices between CF and HC cohorts with advancing age appeared divergent (Fig 1C and 1D) and there was a significant difference in viral communities across ages in all children. Additionally, children with CF clustered separately from HC (Fig 2B), despite the high degree of inter-individual variability (Fig 2A and 2B). These findings highlight the need to further explore and/or control for age in future studies.
Our study is limited by the developing nature of virus reference databases. Unknown viruses may influence our estimates of alpha and beta diversity. Our sequencing effort appeared adequate for individual samples (saturation of rarefaction curves S1C and S1D Fig), and similar to Shkoporov et al. (2019) [48], with 75% of reads mapped to viruses. A larger sample size is required for future studies to account for the high degree of inter-individual variability in the intestinal virobiota, to get a complete picture of the virome of CF patients, and minimise the influence of outliers (which is controlled for using the ANCOM analysis [49]). Although the percentage of predictions mapped to functional terms is small, a recent review of human gut virome literature identified 18 studies, none of which performed a functional annotation [50]. Bacterial contamination rates in our sequencing data were quite varied and could require optimisation, however the level is on par with other current studies [27, 28]. Bacterial community data (i.e. 16S rRNA sequencing data) was only available for two CF and five HC participants, therefore we were unable to assess the relationships between intestinal bacteria and viruses. Paired 16S rRNA and viral sequencing data would likely provide important insights into bacteriophage-host relationships. Given our study only investigated children aged 3 to 12 years, further studies on infants, young children and adolescents are warranted, particularly in light of the age-related differences. Additionally, potential confounding factors, including antibiotic usage and altered dietary regimens between CF and HC were not controlled for with this study. Future longitudinal studies across a wider age range with a multi-omics approach will likely provide greater insights including phage-host associations, patient immune response effects and potentially unravel therapeutic targets in CF gastrointestinal disease. Mechanisms promoting intestinal inflammation in children with CF also warrant further investigation, particularly the excess production of bacteriophage endolysins and their downstream effects. Future in-vitro studies using CF-intestinal epithelial cell lines or patient derived models with intestinal organoids should be considered for future research.
In conclusion, there exists an altered intestinal virome in children with CF compared with HC, which provides novel insights into paediatric CF gastrointestinal disease. Faecalibacterium phage, Gokushovirinae and phage lysins are considerations for future areas of research, specifically in the context of intestinal inflammation in CF. A high degree of inter-individual variation in the intestinal virome, which changes across age, suggests that larger, longitudinal studies are warranted. Intestinal viruses and their functions may have important clinical implications for intestinal inflammation and growth in children with CF, potentially providing novel therapeutic targets.
Materials and methods
Study population
We performed a prospective, cross-sectional, case-control study in children with CF and HC. The subjects and samples for this analysis were collected as part of three prior studies evaluating the progression of intestinal microbiota and inflammation in children with CF [4, 7, 11]. Children with CF and HC were prospectively recruited from the outpatient clinics (CF and Orthopaedic/Plastics clinics respectively) at Sydney Children’s Hospital Randwick, Australia. Children with CF were matched to HC for gender and age (as closely as possible). We included children: (i) aged 0 to 18 years; (ii) with CF diagnosed according to the United States Cystic Fibrosis Foundation consensus criteria [51]; (iii) exocrine pancreatic insufficiency based on the 72-hour faecal fat and/or faecal elastase-1 [52, 53]; (iv) as healthy controls if they did not have CF or any gastrointestinal disease. We excluded children: (i) requiring antibiotic therapy for a pulmonary exacerbation [54] or intravenous antibiotics in the preceding four weeks before sampling; (ii) with gastroenteritis, on oral corticosteroids, probiotics and/or non-steroidal anti-inflammatory drugs in the preceding two weeks before sampling. We did not exclude children with CF on prophylactic oral or inhaled antibiotic therapy.
This study was approved by the South Eastern Sydney Area Health Service, Human Research Ethics Committee, Sydney Australia (HREC/10/240). Written informed consent was obtained from each subject or caregiver(s) and the study was carried out in accordance with the approved guidelines.
Sample and data collection
Stool samples were collected to provide a non-invasive representation of the intestinal viral communities and environment. Specifically, stool samples reflect the luminal contents of the large intestine, however for the purpose of this manuscript, will be referred to as a representation of the intestine. A single stool sample from each subject was collected and stored immediately at –80°C, or stored at –20°C (home freezer) until transport to the laboratory for storage at –80°C. Thawing of the sample during transport did not occur. At the time of sample collection, demographic and anthropometric data (height, weight and BMI z-scores) was recorded.
Virome pipeline
Sample preparation to enrich for faecal viruses followed an adjusted NetoVIR (Novel Enrichment Technique Of VIRomes) protocol [55]. Details are provided in S1 Methods. Briefly, sample processing involved: (i) stool being suspended in sterile phosphate-buffered solution, homogenised and centrifuged at 17,000g (ii) supernatant being filtered through a 0.8 μm PES centrifugal filter; (iii) filtered supernatant being treated with nuclease buffer and micrococcal nuclease; (iv) viral nucleic acids being extracted using the QIAamp Viral RNA Mini Kit (Qiagen) according to manufacturer’s instructions for the extraction of viral RNA and DNA, with the exception of centrifugation speed lowered from 20,000 × g to 17,000 × g; (v) nucleic acids being eluted to RNA storage solution (Ambion) and storage at -80°C; (vi) random amplification of nucleic acids using the Whole Transcriptome Amplification Kit 2 according to the manufacturer’s instructions (Sigma Aldrich); (vii) amplified products being purified using the Wizard® PCR Cleanup Kit (Promega); and (viii) library preparation using the Nextera XT kit and a repeat purification step. A DNA size selection step was omitted before sequencing as both solid phase reversible immobilisation beads and manual excision from agarose gel did not achieve desired results. Samples were sequenced using the Illumina MiSeq 500 platform (2 × 150bp).
Taxonomic assignment of virome reads was performed using the Vipie platform [56]. Paired-end sequences were uploaded (https://binf.uta.fi/vipie/) using the default parameters with the exception of: (i) read length of 150 base pairs; (ii) subsample of 0.80; (iii) Kmer length of 31; (iv) remapping of hits PER 1,000,000 and (v) minimum total matches of 2. Further details are provided in S2 Methods. The output included a search hit table of all viruses for each sample, which were combined for further analyses.
Functional annotation was performed using the Virsorter pipeline [33] to generate predictions which were searched against the KEGG (34), COG (35) and Pfam [36] databases.
Inflammatory biomarkers
Calprotectin was extracted and measured as described in a previous study [7] using the PhiCal kit (Calpro, San Diego, CA, US). The lower limit of detection for the assay was 19.5 mg/kg. Calprotectin > 50 mg/kg is considered elevated. Faecal M2-PK was extracted and measured as described in a previous study [11] using the ScheBo Tumour M2-PK kit (ScheBo Biotech, Giessen, Germany). The lower limit of detection of the assay was 1 U/mL.
Statistical analysis
Statistical analysis was performed in R v3.4.4. Alpha diversity indices (richness and Shannon index) were calculated. For viral taxa, relative abundance was calculated as the proportion of all contigs taxonomically assigned. For functions, relative abundance was calculated as the proportion of all assigned predictions. Bray-Curtis dissimilarities based on relative abundance and presence-absence data were calculated using the vegan package [57] and used to generate non-metric multidimensional scaling (NMDS) plots. Permutational multivariate analysis of variance (PERMANOVA) tests (permutations = 1000) were utilised to test if beta diversity significantly differed among groups (‘CF vs HC’ and ‘male vs female’) and age using the vegan function adonis [57]. A significant difference in abundance of viruses or protein functions between CF and HC groups was assessed using the ANCOM package v1.1–3, which uses non-parametric Kruskal-Wallis tests (Benjamini & Hochberg correction for multiple comparisons, q<0.05) [49]. Continuous variables were compared using a t-test or Wilcoxon signed-rank test for parametric and non-parametric data, respectively (p<0.05 considered statistically significant). Generalised linear models (glm function; using a Gaussian distribution) were constructed to control for age when comparing continuous variables between cohorts. Correlations between continuous variables were assessed using Spearman correlations (Benjamini & Hochberg correction, q<0.05) [58]. Graphs were generated using ggplot2 in R [59].
Supporting information
The number of viruses given the number of samples (A & B) and the number of viral sequences per stool sample (C & D).
(TIFF)
(A-E) Relative abundance of all viruses at each taxonomic rank: (A) group; (B) order; (C) family (top 20 most abundant); (D) genus (top 20 most abundant); (E) species (top 20 most abundant). CF and HC subjects ordered in increasing age (from left to right).
(TIF)
(A) Myoviridae was present in 6/8 (75%) CF and 7/8 (87.5%) HC samples. (B) Unclassified Gokushovirinae was present in 4/8 (50%) CF and 8/8 (100%) HC samples. (C) Faecalibacterium phage FP Taranis was not present in CF samples (0%), however, was present in 6/8 (75%) HC samples. No statistically significant differences (q<0.05) were identified at the group and order ranks.
(PDF)
COG (A) and Pfam (B) terms with a significantly different abundance between CF and HC cohorts using ANCOM analysis (q<0.05). KEGG (C) terms with a different abundance between CF and HC cohorts which was close to significance (q<0.1). COG3409, peptidoglycan-binding (PGRP) domain of peptidoglycan hydrolases; K08640, zinc D-Ala-D-Ala carboxypeptidase; PF02305.16, capsid protein (F protein).
(PDF)
Scatterplots of the relative abundance of: (A-C) viruses (family, genus, species); (D) a COG terms, (E) a Pfam terms, and (F) a KEGG term, against age in CF and HC cohorts. Cohort mean and 95% confidence intervals are constructed from generalised linear models and presented as lines and shaded regions, respectively.
(TIFF)
Calprotectin (A) was significantly elevated in children with CF compared to HC (98.9 mg/kg (50.1–104.9) vs. 19.5 mg/kg (19.5–30.5), respectively, p = 0.047). M2-PK (B) was significantly elevated in children with CF compared to HC (7.4 U/ml (6.3–22.6) vs. 1.0 U/ml (1.0–1.0), respectively, p = 0.005).
(TIFF)
*Spearman correlations (q<0.05) considered statistically significant, with the remaining Spearman correlations (q<0.1) considered close to significance. Analysed virus highlighted in bold, and multiple bold viruses in a single row indicates consistent results across taxonomic ranks. Calpro, calprotectin.
(DOCX)
*Spearman correlations (q<0.05) considered statistically significant, with the remaining Spearman correlations (q<0.1) considered close to significance.
(DOCX)
(DOCX)
(DOCX)
(CSV)
(CSV)
(CSV)
(CSV)
Acknowledgments
The authors thank the parents and families of our children with CF and our healthy participants. We thank the CF Clinic at Sydney Children’s Hospital Randwick (Dr Yvonne Belessis, Dr Penny Field, Dr John Widger, the late Dr John Morton, Rhonda Bell, Rebecca McDonald and Amanda Thomsen). We also thank Lily Nahidi (Sydney Children’s Hospital Clinical Research Centre), Roxanne Strachan, Jung M. Lee, Tamara Pang and Joanna Michalowski for their assistance in recruitment and sample collection. We thank the Virology Research Laboratory staff at Prince of Wales Hospital (Ms Susanne Booth, Dr Ki Wook Kim, Jessica Horton, Sonia Isaacs).
Data Availability
All data generated or analysed during this study are included in this published article and its supplementary information files. Raw sequence files are available from the Sequence Read Archive (http://www.ncbi.nlm.nih.gov/sra) under BioProject PRJNA547643.
Funding Statement
MJC was supported by the Sydney Children’s Hospital Foundation and the Australian Government Department of Education and Training (Australian Postgraduate Award). TT was supported by the Australian Research Council.
References
- 1.Ooi CY, Pang T, Leach ST, Katz T, Day AS, Jaffe A. Fecal Human beta-Defensin 2 in Children with Cystic Fibrosis: Is There a Diminished Intestinal Innate Immune Response? Digestive diseases and sciences. 2015;60(10):2946–52. 10.1007/s10620-015-3842-2 [DOI] [PubMed] [Google Scholar]
- 2.Ooi CY, Durie PR. Cystic fibrosis from the gastroenterologist's perspective. Nature reviews Gastroenterology & hepatology. 2016;13(3):175–85. [DOI] [PubMed] [Google Scholar]
- 3.Sutherland R, Katz T, Liu V, Quintano J, Brunner R, Tong CW, et al. Dietary intake of energy-dense, nutrient-poor and nutrient-dense food sources in children with cystic fibrosis. Journal of cystic fibrosis: official journal of the European Cystic Fibrosis Society. 2018;17(6):804–10. [DOI] [PubMed] [Google Scholar]
- 4.Nielsen S, Needham B, Leach ST, Day AS, Jaffe A, Thomas T, et al. Disrupted progression of the intestinal microbiota with age in children with cystic fibrosis. Scientific reports. 2016;6:24857 10.1038/srep24857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Debyser G, Mesuere B, Clement L, Van de Weygaert J, Van Hecke P, Duytschaever G, et al. Faecal proteomics: A tool to investigate dysbiosis and inflammation in patients with cystic fibrosis. Journal of cystic fibrosis: official journal of the European Cystic Fibrosis Society. 2016;15(2):242–50. [DOI] [PubMed] [Google Scholar]
- 6.Manor O, Levy R, Pope CE, Hayden HS, Brittnacher MJ, Carr R, et al. Metagenomic evidence for taxonomic dysbiosis and functional imbalance in the gastrointestinal tracts of children with cystic fibrosis. Scientific reports. 2016;6:22493 10.1038/srep22493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garg M, Leach ST, Coffey MJ, Katz T, Strachan R, Pang T, et al. Age-dependent variation of fecal calprotectin in cystic fibrosis and healthy children. Journal of cystic fibrosis: official journal of the European Cystic Fibrosis Society. 2017;16(5):631–6. [DOI] [PubMed] [Google Scholar]
- 8.Vernocchi P, Del Chierico F, Russo A, Majo F, Rossitto M, Valerio M, et al. Gut microbiota signatures in cystic fibrosis: Loss of host CFTR function drives the microbiota enterophenotype. PloS one. 2018;13(12):e0208171 10.1371/journal.pone.0208171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dhaliwal J, Leach S, Katz T, Nahidi L, Pang T, Lee JM, et al. Intestinal inflammation and impact on growth in children with cystic fibrosis. Journal of pediatric gastroenterology and nutrition. 2015;60(4):521–6. 10.1097/MPG.0000000000000683 [DOI] [PubMed] [Google Scholar]
- 10.Ooi CY, Syed SA, Rossi L, Garg M, Needham B, Avolio J, et al. Impact of CFTR modulation with Ivacaftor on Gut Microbiota and Intestinal Inflammation. Scientific reports. 2018;8(1):17834 10.1038/s41598-018-36364-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garg M, Leach ST, Pang T, Needham B, Coffey MJ, Katz T, et al. Age-related levels of fecal M2-pyruvate kinase in children with cystic fibrosis and healthy children 0 to 10years old. Journal of cystic fibrosis: official journal of the European Cystic Fibrosis Society. 2018;17(1):109–13. [DOI] [PubMed] [Google Scholar]
- 12.Antosca KM, Chernikova DA, Price CE, Ruoff KL, Li K, Guill MF, et al. Altered Stool Microbiota of Infants with Cystic Fibrosis Shows a Reduction in Genera Associated with Immune Programming from Birth. Journal of bacteriology. 2019;201(16). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Lisle RC. Decreased Expression of Enterocyte Nutrient Assimilation Genes and Proteins in the Small Intestine of Cystic Fibrosis Mouse. Journal of pediatric gastroenterology and nutrition. 2016;62(4):627–34. 10.1097/MPG.0000000000001030 [DOI] [PubMed] [Google Scholar]
- 14.Visca A, Bishop CT, Hilton S, Hudson VM. Oral reduced L-glutathione improves growth in pediatric cystic fibrosis patients. Journal of pediatric gastroenterology and nutrition. 2015;60(6):802–10. 10.1097/MPG.0000000000000738 [DOI] [PubMed] [Google Scholar]
- 15.Duytschaever G, Huys G, Boulanger L, De Boeck K, Vandamme P. Amoxicillin-clavulanic acid resistance in fecal Enterobacteriaceae from patients with cystic fibrosis and healthy siblings. Journal of cystic fibrosis: official journal of the European Cystic Fibrosis Society. 2013;12(6):780–3. [DOI] [PubMed] [Google Scholar]
- 16.Reyes A, Haynes M, Hanson N, Angly FE, Heath AC, Rohwer F, et al. Viruses in the faecal microbiota of monozygotic twins and their mothers. Nature. 2010;466(7304):334–8. 10.1038/nature09199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Minot S, Bryson A, Chehoud C, Wu GD, Lewis JD, Bushman FD. Rapid evolution of the human gut virome. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(30):12450–5. 10.1073/pnas.1300833110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Waller AS, Yamada T, Kristensen DM, Kultima JR, Sunagawa S, Koonin EV, et al. Classification and quantification of bacteriophage taxa in human gut metagenomes. The ISME journal. 2014;8(7):1391–402. 10.1038/ismej.2014.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zuo T, Lu XJ, Zhang Y, Cheung CP, Lam S, Zhang F, et al. Gut mucosal virome alterations in ulcerative colitis. Gut. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barr JJ, Auro R, Furlan M, Whiteson KL, Erb ML, Pogliano J, et al. Bacteriophage adhering to mucus provide a non-host-derived immunity. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(26):10771–6. 10.1073/pnas.1305923110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Minot S, Sinha R, Chen J, Li H, Keilbaugh SA, Wu GD, et al. The human gut virome: inter-individual variation and dynamic response to diet. Genome research. 2011;21(10):1616–25. 10.1101/gr.122705.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rodriguez-Brito B, Li L, Wegley L, Furlan M, Angly F, Breitbart M, et al. Viral and microbial community dynamics in four aquatic environments. The ISME journal. 2010;4(6):739–51. 10.1038/ismej.2010.1 [DOI] [PubMed] [Google Scholar]
- 23.Norman JM, Handley SA, Baldridge MT, Droit L, Liu CY, Keller BC, et al. Disease-specific alterations in the enteric virome in inflammatory bowel disease. Cell. 2015;160(3):447–60. 10.1016/j.cell.2015.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wat D, Gelder C, Hibbitts S, Cafferty F, Bowler I, Pierrepoint M, et al. The role of respiratory viruses in cystic fibrosis. Journal of cystic fibrosis: official journal of the European Cystic Fibrosis Society. 2008;7(4):320–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fothergill JL, Mowat E, Walshaw MJ, Ledson MJ, James CE, Winstanley C. Effect of antibiotic treatment on bacteriophage production by a cystic fibrosis epidemic strain of Pseudomonas aeruginosa. Antimicrobial agents and chemotherapy. 2011;55(1):426–8. 10.1128/AAC.01257-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rolain JM, Francois P, Hernandez D, Bittar F, Richet H, Fournous G, et al. Genomic analysis of an emerging multiresistant Staphylococcus aureus strain rapidly spreading in cystic fibrosis patients revealed the presence of an antibiotic inducible bacteriophage. Biology direct. 2009;4:1 10.1186/1745-6150-4-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Willner D, Furlan M, Haynes M, Schmieder R, Angly FE, Silva J, et al. Metagenomic analysis of respiratory tract DNA viral communities in cystic fibrosis and non-cystic fibrosis individuals. PLoS ONE [Electronic Resource]. 2009;4(10):e7370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lim YW, Schmieder R, Haynes M, Willner D, Furlan M, Youle M, et al. Metagenomics and metatranscriptomics: windows on CF-associated viral and microbial communities. Journal of cystic fibrosis: official journal of the European Cystic Fibrosis Society. 2013;12(2):154–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Quinn RA, Lim YW, Maughan H, Conrad D, Rohwer F, Whiteson KL. Biogeochemical forces shape the composition and physiology of polymicrobial communities in the cystic fibrosis lung. MBio. 2014;5(2):e00956–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moran Losada P, Chouvarine P, Dorda M, Hedtfeld S, Mielke S, Schulz A, et al. The cystic fibrosis lower airways microbial metagenome. ERJ open research. 2016;2(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cherney B, Guan E, Rosenberg A. Background Information for Adventitious Agent Safety Control of Creon, a Pancreatic Enzyme Produce Derived from Porcine Tissue. Presentation to the FDA, Antiviral Drugs Advisory Committee. December, 2008.
- 32.Caruso C, Gobbi E, Biosa T, Andra M, Cavallazzi U, Masoero L. Evaluation of viral inactivation of pseudorabies virus, encephalomyocarditis virus, bovine viral diarrhea virus and porcine parvovirus in pancreatin of porcine origin. Journal of virological methods. 2014;208:79–84. 10.1016/j.jviromet.2014.07.032 [DOI] [PubMed] [Google Scholar]
- 33.Roux S, Enault F, Hurwitz BL, Sullivan MB. VirSorter: mining viral signal from microbial genomic data. PeerJ. 2015;3:e985 10.7717/peerj.985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic acids research. 2000;28(1):27–30. 10.1093/nar/28.1.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tatusov RL, Galperin MY, Natale DA, Koonin EV. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic acids research. 2000;28(1):33–6. 10.1093/nar/28.1.33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.El-Gebali S, Mistry J, Bateman A, Eddy SR, Luciani A, Potter SC, et al. The Pfam protein families database in 2019. Nucleic acids research. 2019;47(D1):D427–d32. 10.1093/nar/gky995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Coffey MJ, Garg M, Homaira N, Jaffe A, Ooi CY. Probiotics for people with cystic fibrosis. Cochrane Database of Systematic Reviews. 2020(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brentlinger KL, Hafenstein S, Novak CR, Fane BA, Borgon R, McKenna R, et al. Microviridae, a family divided: isolation, characterization, and genome sequence of phiMH2K, a bacteriophage of the obligate intracellular parasitic bacterium Bdellovibrio bacteriovorus. Journal of bacteriology. 2002;184(4):1089–94. 10.1128/jb.184.4.1089-1094.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cornuault JK, Petit MA, Mariadassou M, Benevides L, Moncaut E, Langella P, et al. Phages infecting Faecalibacterium prausnitzii belong to novel viral genera that help to decipher intestinal viromes. Microbiome. 2018;6(1):65 10.1186/s40168-018-0452-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Quevrain E, Maubert MA, Michon C, Chain F, Marquant R, Tailhades J, et al. Identification of an anti-inflammatory protein from Faecalibacterium prausnitzii, a commensal bacterium deficient in Crohn's disease. Gut. 2016;65(3):415–25. 10.1136/gutjnl-2014-307649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hoffman LR, Pope CE, Hayden HS, Heltshe S, Levy R, McNamara S, et al. Escherichia coli dysbiosis correlates with gastrointestinal dysfunction in children with cystic fibrosis. Clinical infectious diseases: an official publication of the Infectious Diseases Society of America. 2014;58(3):396–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roux S, Krupovic M, Poulet A, Debroas D, Enault F. Evolution and diversity of the Microviridae viral family through a collection of 81 new complete genomes assembled from virome reads. PloS one. 2012;7(7):e40418 10.1371/journal.pone.0040418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chung-Faye G, Hayee B, Maestranzi S, Donaldson N, Forgacs I, Sherwood R. Fecal M2-pyruvate kinase (M2-PK): a novel marker of intestinal inflammation. Inflammatory bowel diseases. 2007;13(11):1374–8. 10.1002/ibd.20214 [DOI] [PubMed] [Google Scholar]
- 44.de Freitas MB, Moreira EAM, Tomio C, Moreno YMF, Daltoe FP, Barbosa E, et al. Altered intestinal microbiota composition, antibiotic therapy and intestinal inflammation in children and adolescents with cystic fibrosis. PloS one. 2018;13(6):e0198457 10.1371/journal.pone.0198457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Eppinga H, Sperna Weiland CJ, Thio HB, van der Woude CJ, Nijsten TE, Peppelenbosch MP, et al. Similar Depletion of Protective Faecalibacterium prausnitzii in Psoriasis and Inflammatory Bowel Disease, but not in Hidradenitis Suppurativa. Journal of Crohn's & colitis. 2016;10(9):1067–75. [DOI] [PubMed] [Google Scholar]
- 46.Karuppannan AK, Opriessnig T. Possible risks posed by single-stranded DNA viruses of pigs associated with xenotransplantation. Xenotransplantation. 2018;25(4):e12453 10.1111/xen.12453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Denner J, Mankertz A. Porcine Circoviruses and Xenotransplantation. Viruses. 2017;9(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shkoporov AN, Clooney AG, Sutton TDS, Ryan FJ, Daly KM, Nolan JA, et al. The Human Gut Virome Is Highly Diverse, Stable, and Individual Specific. Cell host & microbe. 2019;26(4):527–41.e5. [DOI] [PubMed] [Google Scholar]
- 49.Mandal S, Van Treuren W, White RA, Eggesbo M, Knight R, Peddada SD. Analysis of composition of microbiomes: a novel method for studying microbial composition. Microbial ecology in health and disease. 2015;26:27663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vemuri R, Shankar EM, Chieppa M, Eri R, Kavanagh K. Beyond Just Bacteria: Functional Biomes in the Gut Ecosystem Including Virome, Mycobiome, Archaeome and Helminths. Microorganisms. 2020;8(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Farrell PM, Rosenstein BJ, White TB, Accurso FJ, Castellani C, Cutting GR, et al. Guidelines for diagnosis of cystic fibrosis in newborns through older adults: Cystic Fibrosis Foundation consensus report. The Journal of pediatrics. 2008;153(2):S4–S14. 10.1016/j.jpeds.2008.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jeejeebhoy KN, Ahmad S, Kozak G. Determination of fecal fats containing both medium and long chain triglycerides and fatty acids. Clinical biochemistry. 1970;3(2):157–63. [PubMed] [Google Scholar]
- 53.Loser C, Mollgaard A, Folsch UR. Faecal elastase 1: a novel, highly sensitive, and specific tubeless pancreatic function test. Gut. 1996;39(4):580–6. 10.1136/gut.39.4.580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fuchs HJ, Borowitz DS, Christiansen DH, Morris EM, Nash ML, Ramsey BW, et al. Effect of aerosolized recombinant human DNase on exacerbations of respiratory symptoms and on pulmonary function in patients with cystic fibrosis. The Pulmozyme Study Group. The New England journal of medicine. 1994;331(10):637–42. 10.1056/NEJM199409083311003 [DOI] [PubMed] [Google Scholar]
- 55.Conceicao-Neto N, Zeller M, Lefrere H, De Bruyn P, Beller L, Deboutte W, et al. Modular approach to customise sample preparation procedures for viral metagenomics: a reproducible protocol for virome analysis. Scientific reports. 2015;5:16532 10.1038/srep16532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lin J, Kramna L, Autio R, Hyoty H, Nykter M, Cinek O. Vipie: web pipeline for parallel characterization of viral populations from multiple NGS samples. BMC genomics. 2017;18(1):378 10.1186/s12864-017-3721-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Anderson JL, Miles C, Tierney AC. Effect of probiotics on respiratory, gastrointestinal and nutritional outcomes in patients with cystic fibrosis: A systematic review. Journal of cystic fibrosis: official journal of the European Cystic Fibrosis Society. 2017;16(2):186–97. [DOI] [PubMed] [Google Scholar]
- 58.Torondel B, Ensink JH, Gundogdu O, Ijaz UZ, Parkhill J, Abdelahi F, et al. Assessment of the influence of intrinsic environmental and geographical factors on the bacterial ecology of pit latrines. Microbial biotechnology. 2016;9(2):209–23. 10.1111/1751-7915.12334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wickham H. ggplot2: Elegant Graphics for Data Analysis: Springer Publishing Company, Incorporated; 2009. 216 p.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The number of viruses given the number of samples (A & B) and the number of viral sequences per stool sample (C & D).
(TIFF)
(A-E) Relative abundance of all viruses at each taxonomic rank: (A) group; (B) order; (C) family (top 20 most abundant); (D) genus (top 20 most abundant); (E) species (top 20 most abundant). CF and HC subjects ordered in increasing age (from left to right).
(TIF)
(A) Myoviridae was present in 6/8 (75%) CF and 7/8 (87.5%) HC samples. (B) Unclassified Gokushovirinae was present in 4/8 (50%) CF and 8/8 (100%) HC samples. (C) Faecalibacterium phage FP Taranis was not present in CF samples (0%), however, was present in 6/8 (75%) HC samples. No statistically significant differences (q<0.05) were identified at the group and order ranks.
(PDF)
COG (A) and Pfam (B) terms with a significantly different abundance between CF and HC cohorts using ANCOM analysis (q<0.05). KEGG (C) terms with a different abundance between CF and HC cohorts which was close to significance (q<0.1). COG3409, peptidoglycan-binding (PGRP) domain of peptidoglycan hydrolases; K08640, zinc D-Ala-D-Ala carboxypeptidase; PF02305.16, capsid protein (F protein).
(PDF)
Scatterplots of the relative abundance of: (A-C) viruses (family, genus, species); (D) a COG terms, (E) a Pfam terms, and (F) a KEGG term, against age in CF and HC cohorts. Cohort mean and 95% confidence intervals are constructed from generalised linear models and presented as lines and shaded regions, respectively.
(TIFF)
Calprotectin (A) was significantly elevated in children with CF compared to HC (98.9 mg/kg (50.1–104.9) vs. 19.5 mg/kg (19.5–30.5), respectively, p = 0.047). M2-PK (B) was significantly elevated in children with CF compared to HC (7.4 U/ml (6.3–22.6) vs. 1.0 U/ml (1.0–1.0), respectively, p = 0.005).
(TIFF)
*Spearman correlations (q<0.05) considered statistically significant, with the remaining Spearman correlations (q<0.1) considered close to significance. Analysed virus highlighted in bold, and multiple bold viruses in a single row indicates consistent results across taxonomic ranks. Calpro, calprotectin.
(DOCX)
*Spearman correlations (q<0.05) considered statistically significant, with the remaining Spearman correlations (q<0.1) considered close to significance.
(DOCX)
(DOCX)
(DOCX)
(CSV)
(CSV)
(CSV)
(CSV)
Data Availability Statement
All data generated or analysed during this study are included in this published article and its supplementary information files. Raw sequence files are available from the Sequence Read Archive (http://www.ncbi.nlm.nih.gov/sra) under BioProject PRJNA547643.