Abstract
Cancer development has been ascribed with diverse genetic variations which are identified in both mitochondrial and nuclear genomes. Mitochondrial DNA (mtDNA) alterations have been detected in several tumours which include lung, colorectal, renal, pancreatic and breast cancer. Several studies have explored the breast tumour-specific mtDNA alteration mainly in Western population. This study aims to identify mtDNA alterations of 20 breast cancer patients in Malaysia by next generation sequencing analysis. Twenty matched tumours with corresponding normal breast tissues were obtained from female breast cancer patients who underwent mastectomy. Total DNA was extracted from all samples and the entire mtDNA (16.6kb) was amplified using long range PCR amplification. The amplified PCR products were sequenced using mtDNA next-generation sequencing (NGS) on an Illumina Miseq platform. Sequencing involves the entire mtDNA (16.6kb) from all pairs of samples with high-coverage (~ 9,544 reads per base). MtDNA variants were called and annotated using mtDNA-Server, a web server. A total of 18 of 20 patients had at least one somatic mtDNA mutation in their tumour samples. Overall, 65 somatic mutations were identified, with 30 novel mutations. The majority (59%) of the somatic mutations were in the coding region, whereas only 11% of the mutations occurred in the D-loop. Notably, somatic mutations in protein-coding regions were non-synonymous (49%) in which 15.4% of them are potentially deleterious. A total of 753 germline mutations were identified and four of which were novel mutations. Compared to somatic alterations, less than 1% of germline missense mutations are harmful. The findings of this study may enhance the current knowledge of mtDNA alterations in breast cancer. To date, the catalogue of mutations identified in this study is the first evidence of mtDNA alterations in Malaysian female breast cancer patients.
Introduction
Breast cancer is the most common malignancy in women and one of the leading causes of cancer-related deaths in women worldwide, with nearly 2.1 million new cases estimated and responsible for the deaths of 629,679 women in 2018 [1]. Breast cancer incidences in Malaysia are estimated to be 7,593 (32.7%) new cases in 2018, with 2,894 deaths [1]. Despite various emerging treatment strategies and novel therapies in treating breast cancer, it is estimated that one in 30 Malaysian women is at risk of breast cancer in her lifetime [2]. Studies involving analysis of large samples and controls led to identification of genetic factors involved in predisposition to breast cancer [3, 4]. Albeit numerous studies reported the association of DNA mutations in cancer mainly in Western population, limited research exploring the effects of mitochondrial DNA (mtDNA) alterations specifically in Asian population were documented.
Mitochondria are the energy synthesising organelles in the cells. They are responsible for the generation of adenosine triphosphate (ATP) molecules through oxidative phosphorylation (OXPHOS). Each mitochondrion contains genetically compact circular double-stranded mtDNA loops with an average of 100–10,000 copies per cell at a high copy number. The mtDNA copy number per cell varies widely in different human tissues and is maintained to meet the energy requirement to sustain normal physiological functions of the cell [5]. mtDNA consists of 16,569 base pairs which contains 37 essential genes that encode for 13 respiratory chain subunits essential for the OXPHOS system, 22 transfer RNAs (tRNAs) and two ribosomal RNAs (rRNAs), namely 12S and 16S that are required for the transcription and translation of mitochondrial proteins [5].
It is increasingly apparent that mitochondria play a pivotal role in modulating oncogenesis by virtue of their key functions in energy production, synthesis of building blocks for tumour anabolism, generation of reactive oxygen species (ROS), regulation of cell death and immunosurveillance [6, 7]. The mtDNA plays an important role in regulating mitochondrial functions and are highly susceptible to damage. MtDNA is maternally inherited in which the integrity of all copies is maintained in the same sequence, a state known as “homoplasmy” [8]. However, these mtDNAs may undergo sequence variations and each cell may contain different proportions of mutant and normal (wild-type) mtDNAs during mitotic division, a state known as “heteroplasmy” [9]. The damaging effects of ROS radicals produced in the electron transport chain (ETC), replication cycle defects and lack of protective DNA repair mechanisms are known to contribute to the variation of the mtDNA gene sequence [10–12].
Various types of mtDNA mutations have been detected in breast cancers [13, 14]. Common genetic changes in mtDNA are germline and somatic mutations which include gene deletions, missense mutations, frame-shift mutations and insertions [15]. Both germline and somatic mutations are implicated in breast tumour formation. Germline mutations were shown as risk factor for invasive breast cancer whilst most of the mutations identified in breast cancers were somatic mutations [15, 16]. Mutations in mtDNA are known to perturb the OXPHOS system in various cancer cells [17]. Downregulation of OXPHOS activity was shown to be involved in the early stages of carcinogenesis as well as in the metastases of breast neoplastic cells [18, 19]. Interestingly, A10398G polymorphism of complex I subunit ND3 of the OXPHOS system has been associated with an increased risk of breast cancer in Malaysia [20]. Although the prevalence of this polymorphism is significant, there are very limited studies exploring the entire mtDNA genome of breast cancer patients in Malaysia.
In the current study, the entire mtDNA of adjacent normal-tumour pairs of 20 patients with breast cancer was screened using in-depth coverage provided by next generation sequencing (NGS) to catalogue the mtDNA mutations. Understanding the mtDNA alterations may add to the current knowledge of mitochondrial impairment in breast cancer. The relationship of mutations burden and clinical variables of the patients were also assessed. The outcome of this first preliminary study involving Malaysian breast cancer patients will potentially serve as a reference for future studies involving the entire mtDNA in breast cancer.
Materials and methods
Human breast cancer tissue samples collection
This study was approved by the Medical Research & Ethics Committee (MREC) of the National Institutes of Health Malaysia [NMRR ID-15-2085-28181 (IIR)] and Ethic Committee for Research Involving Human Subjects of Universiti Putra Malaysia (JKEUPM). Written informed consent was obtained from all participants and the study was conducted according to the ethical guidelines of the Helsinki Declaration. A total of 20 female breast cancer patients were recruited between April 2016 and December 2016 from Hospital Kuala Lumpur, National Cancer Institute and Hospital Putrajaya in Peninsular Malaysia. Breast tissue samples were obtained during surgery whilst the demographic and clinical data were extracted from the medical records. Women who were more than 18 years old, pathologically confirmed with primary breast carcinoma and underwent mastectomy surgery for breast cancer were included. Patients receiving neoadjuvant chemotherapy/radiotherapy, with serious infection or concomitant disease and absence of informed consent were excluded from this study. Tumour tissue and its matched control were obtained during mastectomy surgery. Normal breast tissue samples were located in the different quadrant and at a sufficient distance from the tumour. All samples were immediately immersed in liquid nitrogen and stored until further use. The demographic details of patients and clinical features of the samples are summarised in S1 Table.
Total DNA extraction
Total DNA was extracted from 50 mg of malignant and corresponding normal tissue samples using the QIAamp DNA Mini kit (Qiagen, Germany) according to the manufacturer’s instructions [21]. An ultraviolet-visible spectrophotometer (Thermo Scientific Nanodrop 2000, Thermo Scientific, United States) was used to quantify and assess the purity of DNA samples. All DNA samples were stored at -20°C until further use.
PCR amplification of mtDNA
Long-range polymerase chain reaction (LR-PCR) was performed to purify mtDNA from isolated total DNA. LR-PCR was performed in two separate reactions to amplify two large overlapping amplicons (9.1kb and 11.2kb) using two sets of primer pairs MTL-F1 (5’ AAA GCA CAT ACC AAG GCC AC 3’); R1 (5’ TTG GCT CTC CTT GCA AAG TT 3’) and MTL-F2 (5’ TAT CCG CCA TCC CAT ACA TT 3’); R2 (5’ AAT GTT GAG CCG TAG ATG CC 3’) as previously described [22]. TaKaRa LA Taq kit (Takara Clontech, Mountain View CA, USA) was used for amplification. The 50 μL of PCR reaction conditions were: 1 x LA PCR buffer, 0.4 mM dNTP mixture, 0.4 μM primer pairs, 2.5 U TaKaRa LA Taq, and 2 ng of genomic template DNA. Thermal cycling parameters were: initial denaturation at 95°C for 3 minutes, then 30 cycles at 95°C for 30 s, 68°C for 10 s, 60°C for 15 s and 68°C for 11 minutes, followed by a final extension at 72°C for 10 minutes. Agarose gel electrophoresis was performed to validate the size of amplified PCR products. PCR products were quantified using the Qubit dsDNA HS Assay Kit (Life Technologies Corporation, Oregon USA) with the Quantus Fluorometer (Promega, USA).
Library building and paired-end sequencing on Miseq
Indexed paired-end mtDNA libraries were prepared with the Nextera XT DNA Sample Prep Kit and the Nextera XT Index Kit (Illumina, USA) according to the manufacturer’s guidelines [23]. An input of 1ng/μl mtDNA amplicons from each of the two amplicon sets was pooled and diluted further for tagmentation on thermal cycler. Tagmentation involves fragmenting the mtDNA amplicons and tagging with adapter sequences. This is followed by amplification of tagmented mtDNA using a limited-cycle PCR program as described by the manufacturer [23]. The amplified library was purified using AMPure XP beads (Beckman Coulter, USA) to remove short library fragments. The purified library was quantified using Qubit dsDNA HS Assay Kit (Life Technologies Corporation, Oregon USA) and ran on Agilent Technology 2100 Bioanalyzer using a High Sensitivity DNA chip (Agilent, USA) to check for size distribution. An undiluted library mtDNA was normalized to the average library size (600–750 bp) for every 660 g/mol of mtDNA to produce 4 nM library mtDNA. This is followed by pooling of an equal volume of libraries of all tumour-normal sample pairs of 20 patients. Pooled libraries were denatured and diluted to 10 pM sequencing input with Miseq Reagent Kit v2 (Illumina, USA) as described by the manufacturer [24]. Illumina PhiX Control, derived from phiX174 (RF1) bacteriophage of 5386 bp circular genome was used as an Illumina sequencing positive control [25]. The sequencing reactions were performed on the Miseq V2 (2 x 250 bp) platform (Illumina, USA) in compliance with the manufacturer’s preparation guides for paired end runs [26].
Variant calling and annotation
The Illumina Miseq paired-end FASTQ files of all 20 tumour and matched normal samples were analysed with mtDNA-Server Version 1.0.6. [27]. Server default parameters were used to generate sequence data. FASTQ input sample files were aligned to rCRS and pair reads mapped before BAM files were generated. Several filters were applied to detect homoplasmic and heteroplasmic variants from the resulting BAM files. Only reads with a mapping quality score >20, alignment quality >30 and base quality >20 were used for variants detection and 1% minor component threshold for heteroplasmies. Indels were manually classified using similar filters with the raw data file. All samples were also checked for contamination by the server.
Variants detected in both tumour and corresponding normal tissue samples of a patient were classified as germline mutations as described in previous studies [28, 29]. Variant differences between tumour and matched normal mtDNA were classified as somatic mutations [28, 29]. For somatic mutations, only variants with allele frequency <1% in the normal and ≥1% in the matched tumour samples were analysed. Germline heteroplasmies were called for alleles present in normal tissue at level ≥1%. Variants were further searched in MITOMAP [30] and dbSNP [31] to identify novel variants and to determine the clinical significance of the reported variants.
The predicted functional effects of variants were determined using precomputed values of the Polyphen2, SIFT, CADD and APOGEE algorithms, collected within the MitImpact 3D database [32] and MutPred [33] within mtDNA-Server.
Statistical analysis and data visualization
The shift in heteroplasmy levels of heteroplasmic point mutations was analysed using a paired t-test. The distribution of germline mutations (all homoplasmic, heteroplasmic and indel mutations) and somatic mutations were assessed by dividing the mitochondrial genome into 18 regions (D-loop, other non-coding region, tRNA (all tRNA-coding genes), 12s rRNA, 16s rRNA, ATP8, ATP6, COX I, COX II, COX III, CYB, ND1, ND2, ND3, ND4,ND4L, ND5 and ND6). The proportions of mtDNA genes were calculated by dividing the total number of base pairs per gene by the number of base pairs of the entire mtDNA. The proportions of germline mutations and somatic mutations were calculated by dividing the number of mutations in a certain gene region with a total number of mutations. Testing differences in the distribution of mutations across the mtDNA genome between germline and somatic mutations were assessed using a two-sided Fisher’s exact test with rows indicating germline and somatic categories, and columns indicating regions of mutations. The association between the numbers of mutations in relation to gene size were tested for independency with a Pearson correlation analysis. Statistical test was performed using IBM SPSS version 23. A value of P<0.05 was considered significant, whereas values of P<0.01 and P<0.001 were considered highly significant. Tables were generated using Microsoft Excel 2010. Figures were generated using Microsoft Excel 2010 and Adobe Illustrator CS3.
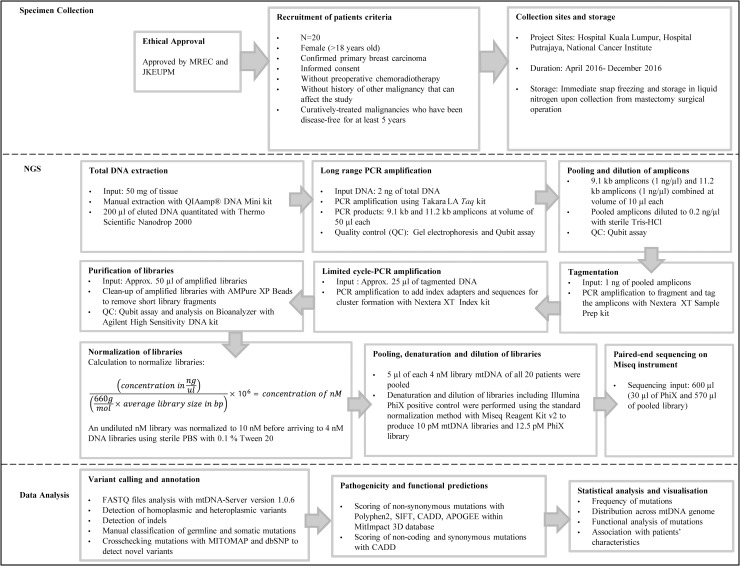
Study flowchart
The mtDNA mutation study was conducted at the Integrative Pharmacogenomics Institute (iPROMISE) in Universiti Teknologi MARA Puncak Alam Campus in Malaysia. A flow chart summarizing the methodology is shown in Fig 1.
Fig 1. Flow chart describing the mtDNA mutation study.
Results
Patient characteristics
The corresponding normal breast tissue specimens act as controls. The mean age of patients at the time of initial surgery was 57.45 years (range 38–78 years) (S1 Table). Briefly, all tumours were invasive breast carcinomas and six of the tumours were confirmed to be triple-negative. Ten of the patients were diagnosed with stage 2, nine with stage 3 and one with stage 4 breast cancer. NGS was performed to sequence the entire mtDNA for all 20 tumours paired with normal breast tissue.
Annotation of variants
Lists of homoplasmic and heteroplasmic variants identified by mtDNA-server for all 20 tumour-normal sample pairs are presented in Table 1 of S2 Table. Inspection of the variants led to the exclusion of mitochondrial hotspot 3017 according to the rCRS (Tables 2–5 in S2 Table). The average read depth of entire mtDNA genome for all samples was around 9500x (mean ± SEM = 9544x ± 350x). The homoplasmic and heteroplasmic variants including indels identified by the mtDNA-server were further classified as germline or somatic mutations using a variant allele fraction (VAF) threshold ≥ 1%. Variants detected in both tumour and corresponding normal tissue samples of a patient were classified as germline mutations as previously described [28, 29]. Variant differences between tumour and matched normal mtDNA were classified as somatic mutations [28, 29].
Germline mtDNA mutations
All patients (100%) carried germline mtDNA mutations. In total, 753 germline mtDNA mutations were identified with an average of 37.7 mutations per patient. Among the germline mutations identified, 632 (83.9%) were homoplasmic single nucleotide variants (S3 Table), 63 (8.4%) were heteroplasmic single nucleotide variants (S4 Table) and 58 (7.7%) were insertions and deletions (indels) (S5 Table).
Of the homoplasmic mutations, 630 (99.7%) were known variants reported in MITOMAP [30] database, whereas two mutations (0.32%) were novel. Patients were found sharing similar homoplasmic mutations in their germline: variants A73G, A263G, A750G, A1438G, A2706G, A4769G, C7028T, A8860G, G11719A, C14766T and A15326G were detected in all patients (100%); variant T16519C was found in 70% of the population; variants C12705T and C16223T was found in 50% of the population; variants T489C, A8701G, T9540C, C10400T, T10873C, T14783C, G15043A and G15301A were found in 40% of the population; variant T16362C was found in 30% of the population; variants A827G, T6392C and G13928C were found in 20% of the population; variants G499A, C3970T, G4820A, G10310A, G12372A, G13590A, C15535T, T16136C, A16183C, T16189C, T16189C, T16217C, T16249C, C16292T and T16304C accounts for 15% of the population. The rest of the germline variant accounts for 5–10% of patient population. Of note, the current study observed A10398G germline mutation in 45% of tested population. Prevalence of A10398G polymorphism was previously reported in invasive breast cancer Malay patients in peninsular Malaysia [20]. However, the observed population consisted of Malay (30%), Chinese (20%) and Indian (5%) (S1 Fig).
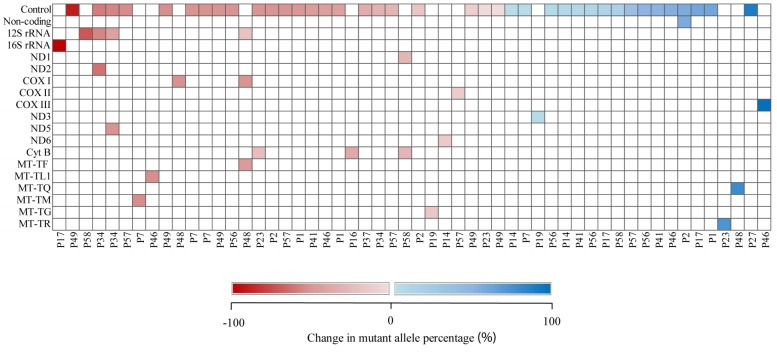
A total of 63 (8.4%) germline heteroplasmies at 46 distinct nucleotide positions were detected in 18 individual breast cancer patients (S4 Table). Seven of the heteroplasmies were present in multiple individuals with A189G in 44% of the population, T16189C in 22% of the population, T204C and T16093C in 16.7% of the population, G207A, G16129A and A16183C in 11.1% of the population. Of the heteroplasmic mutations, 61 (96.8%) were known variants reported in MITOMAP [30] and two others (3.2%) were novel. A mixture of reference and non-reference alleles in the same base position is regarded as heteroplasmies. The abundance of non-reference alleles ranged between 1.02% and 98.96% (median 2.72%) in normal tissue samples, whereas non-reference alleles ranged between 0.03% and 99.7% (median 1.26%) in corresponding tumour tissue samples. In overall, there was a decreasing trend towards loss of non-reference alleles or mutant alleles in the corresponding tumour samples (P = 0.13, t = 1.53) (Fig 2).
Fig 2. Shifts in the variant allele percentage of heteroplasmic germline variants.
Each coloured rectangle corresponds to a heteroplasmic germline mutation (white = no heteroplasmy present, red = decreases in mutant allele percentage from normal to tumour, blue = increases in mutant allele percentage from normal to tumour). The shifts in heteroplasmy are presented in tabular format in S4 Table. Adapted from [34].
Indels were found in all patients (100%) accounting for a total of 58 indels at 7 distinct nucleotide positions (S5 Table). All germline indels have occurred in non-coding regions and have been previously reported. The majority of indels were heteroplasmic (89.7%) found at nucleotide regions of 309, 315, 514–517 and 8271–8279 base pairs. The remaining (10.3%) were homoplasmic deletions occurred in nucleotide region 249, 514–515 and 8270–8278 base pairs. Percentage of population carrying the indels is 315 ins C (95%), 309 ins C (80%), 309 ins CC (30%), 514–515 CA (35%), 249 del A (20%), 8271–8279 del (20%), 514–517 del (5%) and 8270–8278 del (5%).
Somatic mtDNA mutations
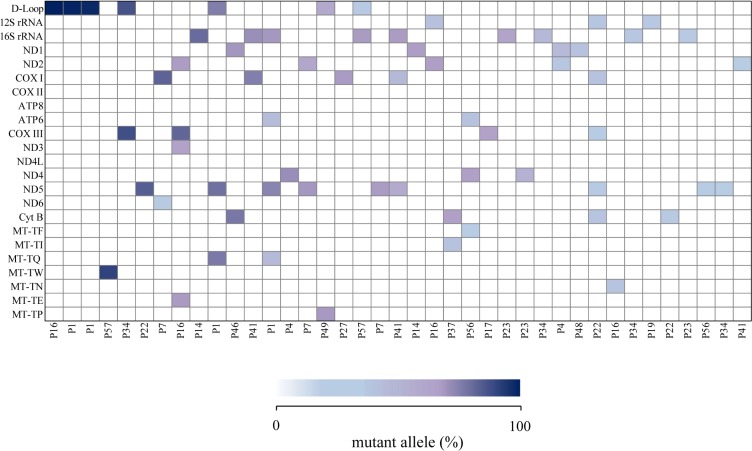
Somatic mutations were detected in 18 of 20 patients (90%). A total of 65 somatic mutations were detected, with a mean of 3.25 mutations per tumour sample. Of the 65 somatic mutations, 30 (46.2%) were novel and 35 (53.8%) were known variants in MITOMAP and dbSNP databases (Table 1). All somatic mutations were heteroplasmic in the tumour samples, 44 (67.7%) of which were low-level heteroplasmies (<10%) while 21 (32.3%) were high-level heteroplasmies (≥10%). Mutant allele abundance in the tumour samples ranged between 1.01% and 85.7% (median = 3.9%) (Fig 3). Each somatic mutation occurred only once, unique to each individual patient, and 13 (20%) of the mutations were associated with disease in previous studies (S6 Table).
Table 1. Somatic mtDNA mutations in breast cancer.
| Patient ID | Gene | rCRS | Mutation | N > T | Variants % | AAC | Pathogenicity predictions | |||
|---|---|---|---|---|---|---|---|---|---|---|
| CADD | Polyphen2 | SIFT | APOGEE | |||||||
| P001 | D-loop | C | C150T | Homo > Hetero | 80.51 | Neutral | ||||
| D-loop | A | A214G | Homo > Hetero | 83.32 | Neutral | |||||
| 16S rRNA | T | T2171Ca | Homo > Hetero | 10.25 | Neutral | |||||
| TI | G | G4309A | Homo > Hetero | 2.24 | Neutral | |||||
| TQ | A | A4343G | Homo > Hetero | 28.95 | Neutral | |||||
| ATP6 | T | T8547C | Homo > Hetero | 2.07 | L7P | Deleterious | Probably damaging | Neutral | Neutral | |
| ND5 | G | G12979Aa | Homo > Hetero | 34.34 | G215S | Deleterious | Benign | Neutral | Neutral | |
| ND5 | G | G13138Aa | Homo > Hetero | 22.19 | E268K | Deleterious | Probably damaging | Neutral | Neutral | |
| D-loop | A | A16233G | Homo > Hetero | 26.18 | Neutral | |||||
| P004 | ND1 | C | C3738T | Homo > Hetero | 2.26 | V144V | Neutral | |||
| ND2 | A | A4767G | Homo > Hetero | 1.35 | M100V | Neutral | Benign | Neutral | Neutral | |
| ND4 | T | T10908C | Homo > Hetero | 15.6 | F50S | Neutral | Benign | Neutral | Neutral | |
| P007 | ND2 | G | G4665Aa | Homo > Hetero | 3.6 | A66T | Deleterious | Probably damaging | Neutral | Neutral |
| CO1 | G | G6517Aa | Homo > Hetero | 43.05 | G205D | Deleterious | Probably damaging | Deleterious | Pathogenic | |
| ND5 | T | T12365Ca | Homo > Hetero | 10.09 | L10P | Deleterious | unknown | Neutral | Neutral | |
| ND5 | T | T14102Ca | Homo > Hetero | 7.01 | L589P | Deleterious | Probably damaging | Neutral | Neutral | |
| ND6 | G | G14453A | Homo > Hetero | 1.23 | A74V | Deleterious | Probably damaging | Neutral | Pathogenic | |
| P014 | 16S rRNA | G | G2701A | Homo > Hetero | 37.97 | Neutral | ||||
| ND1 | G | G3380A | Homo > Hetero | 5.63 | R25Q | Deleterious | Probably damaging | Neutral | Pathogenic | |
| P016 | D-loop | C | C186T | Homo > Hetero | 85.65 | Neutral | ||||
| 12S rRNA | G | G951A | Homo > Hetero | 1.86 | Neutral | |||||
| ND2 | A | A4870Ta | Homo > Hetero | 6.28 | Q134L | Deleterious | Probably damaging | Neutral | Neutral | |
| ND2 | T | T4911C | Homo > Hetero | 5.15 | S148P | Deleterious | Benign | Neutral | Neutral | |
| TN | A | A5711G | Homo > Hetero | 1.62 | Neutral | |||||
| CO3 | T | T9280Ca | Homo > Hetero | 41.59 | L25P | Deleterious | Probably damaging | Neutral | Neutral | |
| ND3 | T | T10076C | Homo > Hetero | 3.88 | I6I | Neutral | ||||
| TE | G | G14698Aa | Homo > Hetero | 7.19 | Neutral | |||||
| P017 | CO3 | G | G9838Aa | Homo > Hetero | 3.88 | G211D | Deleterious | Probably damaging | Neutral | Pathogenic |
| P019 | 12S rRNA | A | A1528Ga | Hetero > Hetero | 1.29 | Neutral | ||||
| P022 | 12S rRNA | G | G945Aa | Homo > Hetero | 1.46 | Neutral | ||||
| CO1 | G | G6810Aa | Homo > Hetero | 1.68 | A303T | Deleterious | Probably damaging | Neutral | Neutral | |
| CO3 | G | G9820A | Homo > Hetero | 1.08 | G205E | Deleterious | Probably damaging | Neutral | Pathogenic | |
| ND5 | A | A12612G | Homo > Hetero | 45.94 | V92V | Neutral | ||||
| ND5 | A | A13183G | Homo > Hetero | 1.16 | I283V | Deleterious | Benign | Neutral | Neutral | |
| CYB | G | G14869A | Homo > Hetero | 1.25 | L41L | Neutral | ||||
| CYB | G | G15699Aa | Homo > Hetero | 1.6 | R318H | Deleterious | Probably damaging | Neutral | Neutral | |
| P023 | 16S rRNA | T | T2233Ca | Homo > Hetero | 3.85 | Neutral | ||||
| 16S rRNA | G | G2732Aa | Homo > Hetero | 1.14 | Neutral | |||||
| ND4 | G | G11226Aa | Homo > Hetero | 2.86 | G156D | Deleterious | Probably damaging | Deleterious | Pathogenic | |
| P027 | CO1 | G | G5991A | Homo > Hetero | 7.72 | G30S | Deleterious | Probably damaging | Deleterious | Pathogenic |
| P034 | 16S rRNA | A | A2623G | Homo > Hetero | 2.38 | Neutral | ||||
| 16S rRNA | G | G2702A | Homo > Hetero | 1.35 | neutral | |||||
| CO3 | G | G9868A | Homo > Hetero | 56.27 | R221H | Deleterious | Possibly damaging | Neutral | Pathogenic | |
| ND5 | C | C12906T | Homo > Hetero | 1.08 | I190I | Neutral | ||||
| D-loop | A | A16165G | Hetero > Hetero | 53.49 | Neutral | |||||
| P037 | TI | G | G4282A | Homo > Hetero | 1.68 | Neutral | ||||
| CYB | C | C15702Ta | Homo > Hetero | 4.7 | P319L | Deleterious | Probably damaging | Neutral | Neutral | |
| P041 | 16S rRNA | G | G2333Aa | Homo > Hetero | 6.7 | Neutral | ||||
| 16S rRNA | G | G2815A | Homo > Hetero | 15.3 | Neutral | |||||
| ND2 | T | T5130Ca | Homo > Hetero | 1.04 | L221L | Neutral | ||||
| ND2 | T | T5200Ca | Homo > Hetero | 2.31 | I244T | Deleterious | Benign | Neutral | Neutral | |
| CO1 | G | G6028Aa | Homo > Hetero | 25.54 | G42D | Deleterious | Probably damaging | Deleterious | Neutral | |
| ND5 | G | G13099Ca | Homo > Hetero | 3.3 | A255P | Deleterious | Probably damaging | Neutral | Pathogenic | |
| P046 | ND1 | G | G4244Aa | Homo > Hetero | 10.81 | S313N | Neutral | Benign | Neutral | Neutral |
| CYB | G | G14963A | Homo > Hetero | 30.63 | V73M | Deleterious | Probably damaging | Neutral | Neutral | |
| P048 | ND1 | T | T4007Ca | Homo > Hetero | 1.71 | M234T | Deleterious | Probably damaging | Neutral | Pathogenic |
| P049 | TP | T | T15971Ca | Homo > Hetero | 8.83 | Neutral | ||||
| D-loop | C | C16295Ta | Homo > Hetero | 3.46 | Neutral | |||||
| P056 | TF | G | G627Aa | Homo > Hetero | 1.01 | Neutral | ||||
| ATP6 | G | G8854A | Homo > Hetero | 1.68 | A110T | Deleterious | Benign | Deleterious | Neutral | |
| ND4 | G | G10775A | Homo > Hetero | 4.59 | V6I | Neutral | Benign | Neutral | Neutral | |
| ND5 | G | G12835Aa | Homo > Hetero | 1.08 | A167T | Deleterious | Probably damaging | Neutral | Neutral | |
| P057 | D-loop | G | G513A | Homo > Hetero | 1.06 | Neutral | ||||
| 16S rRNA | G | G3022A | Homo > Hetero | 7.58 | Neutral | |||||
| TW | G | G5521A | Homo > Hetero | 37.04 | Neutral | |||||
a Novel variants; N>T, normal to tumour heteroplasmic pattern; AAC, amino acid change.
Fig 3. Somatic mtDNA mutations.
Each coloured rectangle corresponds to a somatic mtDNA mutation, with a shading indicating mutant allele percentage (white = 0%, dark blue = 100%). Adapted from [34].
Distribution of germline and somatic mtDNA mutations across mtDNA genome
Overall, 245 (32.5%) of germline mutations were within the D-loop region, 399 (53.0%) were within protein-coding region, 88 (11.7%) were within rRNA region, 14 (1.9%) were within tRNA region and 7 (0.9%) were within other non-coding regions. A similar pattern of distribution was observed for somatic mutations whereby 7 (10.8%) of somatic mutations were within the D-loop region, 38 (58.5%) were within protein-coding region, 12 (18.5%) were within rRNA region and 8 (12.3%) were within tRNA region. The proportional distribution of germline and somatic mtDNA mutations across the mtDNA genome are depicted in Fig 4. The differences in distribution of mutations across the mtDNA genome between germline and somatic mutations is highly significant (Fisher’s P<0.001). Additionally, among the protein-coding genes, the proportional number of somatic mutations correlated strongly with the proportional transcript size of the gene in relation to the entire mitochondrial genome (Pearson r = 0.84; P = 0.01). This suggests that somatic mutations in the protein coding genes occur in respect to the length of gene in the mtDNA genome of breast cancer patients. When the number of mutations was normalized to the size of the gene region, it was found that the D-loop region showed 2× and 6× susceptibility to somatic and germline events in breast cancer, respectively.
Fig 4. Distribution of mutations across the mtDNA genome in breast cancer patients.
(A) The proportion of the 16.6kb mtDNA genome. (B) The distribution of germline mutations (Total = 753) across mtDNA genome. (C) The distribution of somatic mutations (Total = 65) across mtDNA genome.
Given the role of protein-coding genes in the OXPHOS for the generation of cellular energy, the distribution of somatic and germline mutations within the OXPHOS cascade protein complexes was studied. Table 2 shows the distribution of somatic and germline mutations among OXPHOS components (complexes I, III, IV and V) encoded by mtDNA. There are no major variations in mutation distribution between the germline and somatic mutations (Fisher P = 0.159). The Complex I genes, however have the largest number of somatic mutations (36.9%) and germline mutations (25.5%). Interestingly, ND5 gene of Complex I harboured the largest number of somatic (37.5%) and germline mutations (26.0%) (Fig 4), consistent with a previous breast cancer study [34]. Complex III had (6.2%) somatic mutations and (12.4%) germline mutations, while Complex IV had (12.3%) somatic and (9.6%) germline mutations. The least number of mutations was observed in complex V genes, with (3.1%) of somatic mutations and (5.6%) of germline mutations.
Table 2. Distribution of somatic and germline mtDNA mutations in the OXPHOS components.
| Region | Germline Mutation N (%) | Somatic Mutation N (%) |
|---|---|---|
| Complex I | 192 (25.5) | 24 (36.9) |
| Complex III | 93 (12.4) | 4 (6.2) |
| Complex IV | 72 (9.6) | 8 (12.3) |
| Complex V | 42 (5.6) | 2 (3.1) |
Impact of mtDNA mutations
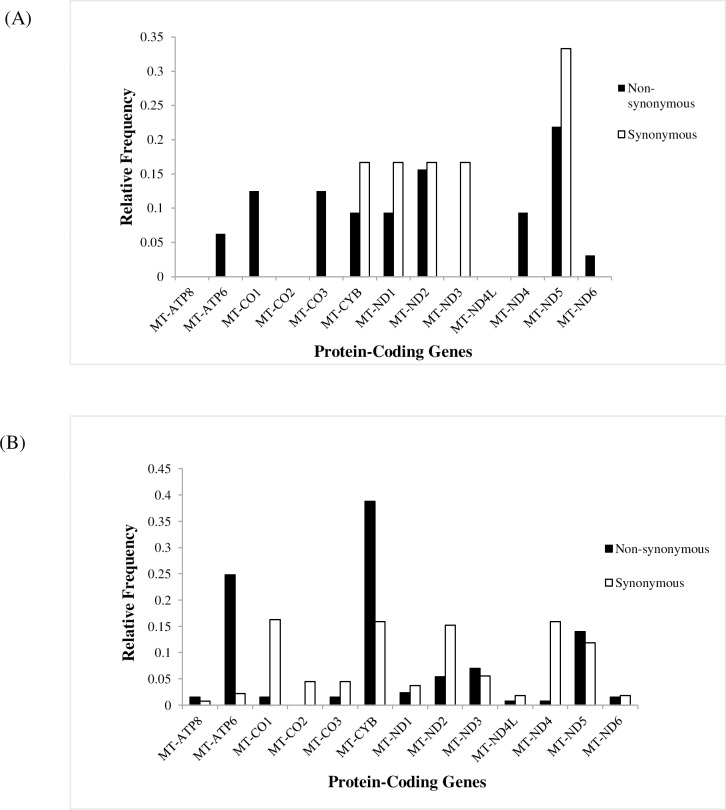
Of the 38 somatic mutations observed in protein-coding regions, 32 (84.2%) were non-synonymous while 6 (15.8%) were synonymous (Fig 5A). In contrast, of the 399 germline mutations observed in the protein-coding regions, 129 (32.3%) were non-synonymous while 270 (67.7%) were synonymous (Fig 5B). There was a significant enrichment of non-synonymous somatic mutations as compared with germline mutations (Fisher P<0.0001). Based on the overall deleterious and functional prediction scores through CADD, Polyphen2, SIFT and APOGEE, 10 missense somatic mutations were potentially damaging (see Table 1). Mutations occurred predominantly in Complex I (15.6%) and Complex IV (15.6%) genes. Comparatively, there are only five homoplasmic germline mutations known to cause deleterious protein function (S3 Table). The frequency of these mutations within the tested population was M1T (5%), L237M (10%), L17F (10%), F26S (5%) and N333I (5%). Mutations L237M and L17F were found to occur in two cases, P017 and P058. Interestingly, patients P017 and P058 were found to share the major haplogroup D. Based on Mitomap, the two mutations have been previously associated to other diseases but not with cancer. In addition, less severe missense mutations T333A and I78T, as well as neutral mutations P183P, P36P and M2M, were found in the same two patients with major haplogroup D. Further study in a larger population is required to understand the significance of haplogroup D mutations as potential biomarkers in breast cancer. The identity of the mtDNA haplogroup of all patients can be found listed in Table 1 of S2 Table. A venn diagram of tested population with the major haplogroups is listed in S2 Fig.
Fig 5. Relative frequencies of non-synonymous and synonymous mutations across mtDNA protein-coding genes.
(A) Somatic mtDNA mutations; (B) Germline mtDNA mutations.
In finding the possible role of tRNA mutations found in the current study, somatic mutations G4309A, G4282A and G5521A were classified as pathogenic when assessed with MitoTIP [35]. Each of these mutations occurred in 5% of the tested population. Eight (1.3%) germline homoplasmic tRNA mutations and 6 (22.2%) germline heteroplasmic tRNA mutations were mostly identified as benign mutations (92.9%) in the MitoTIP [35]. Each of these germline tRNA mutations was found to occur in 5% of the population studied.
The functional impact of mutations in the rRNA genes is currently unknown. However, differences in the occurrence of somatic and germline mutations in rRNA genes in the current study is worthy of future investigation. It was found that among 12 (18.5%) somatic mutations in the rRNA genes, a predominant (13.8%) of mutations occurred in the 16S rRNA gene region. Whereas, among 88 (12.7%) germline mutations identified in the rRNA genes (65.9%) were predominantly found in the 12S rRNA gene region.
As reported earlier, the D-loop contains a substantial number of somatic (10.8%) and germline mutations (31.9%) in comparison to other genes despite its small size and non-coding properties. The somatic D-loop mutations in 25% of the population, most commonly occurred in the hypervariable region 1 (HVR-1) (4.6%) and HVR-2 (4.6%). Similarly, germline D-loop mutations in all patients (100%) occurred largely in the HVR-1 (54.7%) and HVR-2 (37.0%). Somatic mutations in the HVR-2 especially C150T, C186T and A214G as well as germline mutations from nucleotide positions 146–374 are likely to affect heavy-strand synthesis as they were located at the origin of heavy-strand replication (OHR) sites.
Association with clinicopathological parameters
The association between the frequency of somatic and germline mtDNA mutations with patients’ age, ethnicity, tumour grades and stages, nodal stages and hormone receptors in breast cancer were investigated. Significant association was found between frequency of somatic mutations in the rRNA gene region and tumour grades (P = 0.01) and tumour stages (P = 0.022). Breast tumour grade III (73%) appeared to express more rRNA somatic mutation than the grade II tumours (11%), while stage III breast tumours (78%) carried more rRNA somatic mutation than the other tumour stages. In addition, the frequency of somatic mutations in the protein coding genes was significantly different between different nodal stages in breast cancer (P = 0.016) compared to other gene regions. An overall lack of associations with various other clinicopathological variables may be attributed to the limited sample size for the current study. S7 Table describes the mtDNA mutation status and its association with patients’ characteristics.
Discussion
MtDNA mutations occur frequently in cancer and have recently emerged as non-invasive cancer biomarkers for evaluating the risk and prognosis of the disease [36, 37]. Various mutations in the coding and non-coding regions of mtDNA are associated with an increased risk of breast cancer [38]. To date, the current study is the first evidence of germline and somatic mtDNA genome mutations in Malaysian breast cancer patients.
The D-loop region is the most studied mtDNA variants as it possesses high mutation rate, associated to later stages of cancer and poor prognosis in breast cancer [39]. Several somatic mtDNA mutations in breast cancer were reported to be accumulated in the hypervariable regions HVR1 and HVR2 within the D-loop region [28]. All indels in this study are germline mutations localized in the non-coding regions. D-loop showed higher susceptibility to both germline (6×) and somatic mutations (2×) compared to other regions. However, no associations were derived in relation to stages of cancer. The CADD scores in this study showed none of the D-loop mutations were found to be deleterious and it was noted both germline and somatic mutations in the D-loop occur preferentially in HVR-1 and HVR-2. This is consistent with a previous study that supports hypervariable sites in the mtDNA control region as mutational hotspots [40].
Analysis of mutations in the coding region showed that genes encoding for OXPHOS components that regulate energy production in cells were mostly affected. Complex I genes carried the highest number of mutations as described in Table 2. The substantial amount of somatic mutations in Complex I were previously reported with ND5 subunit among Complex I genes harbouring the most somatic mutations [31, 34]. Similarly, the current study observed highest frequencies of somatic mutations (37.5%) in the ND5 subunit alone. Among the ND5 somatic mutations, 29.2% were missense deleterious mutations (Table 1). Alterations in the ND5 subunit were postulated to disrupt the Complex I assembly leading to resistance in apoptosis and ultimately promoting tumour formation and growth [41]. The current evidence suggests that breast tumours may select for mutations within functional regions of the mitochondrial genome which modulates altered metabolism and support tumorigenesis. Nonetheless, further investigation is necessary to determine the effect of ND5 somatic mutations. In this study, germline mutation A10398G was identified as a non-synonymous mutation that is benign indicated by the low pathogenicity score (S3 Table). This mutation was found in Chinese (20%) and Indian (5%) patients with breast cancer. Interestingly, A10398G in ND3 subunit has been previously reported to be a significant polymorphism (OR = 2.29, P = 0.007) linked to increased risk of invasive breast cancer in the Malay population of Peninsular Malaysia [19]. The potential use of 10398G as a biomarker in invasive breast cancer among Malaysian population warrants for further investigation.
In conclusion, the screening of mtDNA in a small cohort of patients with breast cancer in Malaysia identified germline and somatic mtDNA mutations. Evaluation for pathogenicity and functionality suggest mtDNA alterations affect protein functions. It was noted somatic mtDNA mutations were enriched for nonsynonymous changes compared to germline mutations. The findings from this study will serve as a basis in understanding mitochondrial genome of breast cancer in Malaysian population.
Supporting information
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(PDF)
(PDF)
Acknowledgments
The authors are grateful to all the volunteered breast cancer patients. We thank the Director and Deputy Director of Integrative Pharmacogenomics Institute (iPROMISE), UiTM Puncak Alam Campus, Prof. Dato’ Dr. Mohd Zaki Salleh as well as the science officers and laboratory staff of iPROMISE, UiTM, Department of Human Anatomy and Department of Pathology, Faculty of Medicine and Health Sciences, UPM for experimental support and assistance.
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
RO was supported by Graduate Research Fellowship from University Putra Malaysia. The study was funded by Ministry of Education Malaysia (Programme Grant: Fundamental Research Grant Scheme FRGS/1/2015/SKK02/UPM/02/4). The URL of funder: http://mygrants.gov.my. The funder had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: a cancer journal for clinicians. 2018. November; 68(6):394–424. [DOI] [PubMed] [Google Scholar]
- 2.Azizah AM, Nor Saleha IT, Noor Hashimah A, Asmah ZA, Mastulu W. Malaysian national cancer registry report 2007–2011. Malaysia cancer statistics, data and figure Putrajaya: National Cancer Institute, Ministry of Health; 2016. [Google Scholar]
- 3.Yip CH, Bhoo Pathy N, Teo SH. A review of breast cancer research in Malaysia. Med J Malaysia. 2014. August; 69(Suppl A):8–22. [PubMed] [Google Scholar]
- 4.Ministry of Health and Academy of Medicine of Malaysia. Clinical Practice Guidelines for the Management of Breast Cancer 2nd Edition [Internet]. 2010 [cited 2020 March 9]. Available from: http://www.acadmed.org.my
- 5.Chinnery PF, Hudson G. Mitochondrial genetics. British medical bulletin. 2013. June 1; 106(1):135–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vakifahmetoglu-Norberg H, Ouchida AT, Norberg E. The role of mitochondria in metabolism and cell death. Biochemical and biophysical research communications. 2017. January 15;482(3):426–31. 10.1016/j.bbrc.2016.11.088 [DOI] [PubMed] [Google Scholar]
- 7.Porporato PE, Filigheddu N, Bravo-San Pedro JM, Kroemer G, Galluzzi L. Mitochondrial metabolism and cancer. Cell research. 2018. March;28(3):265–80. 10.1038/cr.2017.155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hagström E, Freyer C, Battersby BJ, Stewart JB, Larsson NG. No recombination of mtDNA after heteroplasmy for 50 generations in the mouse maternal germline. Nucleic acids research. 2013. October 25;42(2):1111–6. 10.1093/nar/gkt969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wallace DC, Chalkia D. Mitochondrial DNA genetics and the heteroplasmy conundrum in evolution and disease. Cold Spring Harbor perspectives in biology. 2013. November 1;5(11):a021220 10.1101/cshperspect.a021220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Penta JS, Johnson FM, Wachsman JT, Copeland WC. Mitochondrial DNA in human malignancy. Mutation Research/Reviews in Mutation Research. 2001. May 1;488(2):119–33. [DOI] [PubMed] [Google Scholar]
- 11.Lagouge M, Larsson NG. The role of mitochondrial DNA mutations and free radicals in disease and ageing. Journal of internal medicine. 2013. June;273(6):529–43. 10.1111/joim.12055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scheibye-Knudsen M, Fang EF, Croteau DL, Wilson DM III, Bohr VA. Protecting the mitochondrial powerhouse. Trends in cell biology. 2015. March 1;25(3):158–70. 10.1016/j.tcb.2014.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schon EA, DiMauro S, Hirano M. Human mitochondrial DNA: roles of inherited and somatic mutations. Nature Reviews Genetics. 2012. December;13(12):878 10.1038/nrg3275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duchen MR, Szabadkai G. Roles of mitochondria in human disease. Essays in biochemistry. 2010. June 14;47:115–37. 10.1042/bse0470115 [DOI] [PubMed] [Google Scholar]
- 15.Yadav N, Chandra D. Mitochondrial DNA mutations and breast tumorigenesis. Biochimica et Biophysica Acta (BBA)-Reviews on Cancer. 2013. December 1;1836(2):336–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li L, Chen L, Li J, Zhang W, Liao Y, Chen J, et al. Correlational study on mitochondrial DNA mutations as potential risk factors in breast cancer. Oncotarget. 2016. May 24;7(21):31270 10.18632/oncotarget.8892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ashton TM, McKenna WG, Kunz-Schughart LA, Higgins GS. Oxidative phosphorylation as an emerging target in cancer therapy. Clinical Cancer Research. 2018. June 1;24(11):2482–90. 10.1158/1078-0432.CCR-17-3070 [DOI] [PubMed] [Google Scholar]
- 18.He X, Zhou A, Lu H, Chen Y, Huang G, Yue X, et al. Suppression of mitochondrial complex I influences cell metastatic properties. PloS one. 2013. April 22;8(4):e61677 10.1371/journal.pone.0061677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Owens KM, Kulawiec M, Desouki MM, Vanniarajan A, Singh KK. Impaired OXPHOS complex III in breast cancer. PloS one. 2011. August 25;6(8):e23846 10.1371/journal.pone.0023846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baharudin NT, Jaafar H, Zainuddin Z. Association of mitochondrial DNA 10398 polymorphism in invasive breast cancer in malay population of peninsular malaysia. The Malaysian journal of medical sciences: MJMS. 2012. January;19(1):36 [PMC free article] [PubMed] [Google Scholar]
- 21.Qiagen. QIAamp DNA Mini Blood Mini Handbook-EN [Internet]. 2016 [cited 2018 April 10]. Available from: https://www.qiagen.com/us/resources/resourcedetail?id=62a200d6-faf4-469b-b50f-2b59cf738962&lang=en
- 22.Stawski H. Preparing whole genome human mitochondrial DNA libraries for next generation sequencing using Illumina Nextera XT. Western Carolina University; 2013. [cited 2018 April 10]. Available from: https://libres.uncg.edu/ir/wcu/f/Stawski2013.pdf [Google Scholar]
- 23.Illumina, Inc. Human mtDNA Genome [Internet]. 2016 [cited 2018 April 10]. Available from: https://support.illumina.com/downloads/human_mtdna_genome_guide_15037958.html
- 24.Illumina, Inc. Miseq System Denature and Dilute Libraries Guide [Internet]. 2016 [cited 2018 April 10]. Available from: https://support.illumina.com/downloads/prepare_libraries_for_sequencing_miseq_15039740.html
- 25.Kircher M, Stenzel U, Kelso J. Improved base calling for the Illumina Genome Analyzer using machine learning strategies. Genome biology. 2009. August 1;10(8):R83 10.1186/gb-2009-10-8-r83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Illumina, Inc. Miseq System Guide [Internet]. 2018 [cited 2018 April 10]. Available from: https://support.illumina.com/downloads/miseq_system_user_guide_15027617.html
- 27.Weissensteiner H, Forer L, Fuchsberger C, Schöpf B, Kloss-Brandstätter A, Specht G, et al. mtDNA-Server: next-generation sequencing data analysis of human mitochondrial DNA in the cloud. Nucleic acids research. 2016. April 15; 44(W1):W64–9. 10.1093/nar/gkw247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tan DJ, Bai RK, Wong LJ. Comprehensive scanning of somatic mitochondrial DNA mutations in breast cancer. Cancer research. 2002. February 15;62(4):972–6. [PubMed] [Google Scholar]
- 29.Zhu W, Qin W, Bradley P, Wessel A, Puckett CL, Sauter ER. Mitochondrial DNA mutations in breast cancer tissue and in matched nipple aspirate fluid. Carcinogenesis. 2005. January 1;26(1):145–52. 10.1093/carcin/bgh282 [DOI] [PubMed] [Google Scholar]
- 30.Lott MT, Leipzig JN, Derbeneva O, Xie HM, Chalkia D, Sarmady M, et al. mtDNA variation and analysis using MITOMAP and MITOMASTER. Current protocols in bioinformatics. 2013. December; 44(1):1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Variation Viewer [Internet]. National Centre for Biotechnology Information, U.S. National Library of Medicine; [cited 2018 April 10]. Available from: https://www.ncbi.nlm.nih.gov/variation/view/
- 32.Castellana S, Rónai J, Mazza T. MitImpact: an exhaustive collection of pre‐computed pathogenicity predictions of human mitochondrial non‐synonymous variants. Human mutation. 2015. February;36(2):E2413–22. 10.1002/humu.22720 [DOI] [PubMed] [Google Scholar]
- 33.Li B, Krishnan VG, Mort ME, Xin F, Kamati KK, Cooper DN, et al. Automated inference of molecular mechanisms of disease from amino acid substitutions. Bioinformatics. 2009. September 3;25(21):2744–50. 10.1093/bioinformatics/btp528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McMahon S, LaFramboise T. Mutational patterns in the breast cancer mitochondrial genome, with clinical correlates. Carcinogenesis. 2014. January 18;35(5):1046–54. 10.1093/carcin/bgu012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sonney S, Leipzig J, Lott MT, Zhang S, Procaccio V, Wallace DC, et al. Predicting the pathogenicity of novel variants in mitochondrial tRNA with MitoTIP. PLoS computational biology. 2017. December 11;13(12):e1005867 10.1371/journal.pcbi.1005867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kirches E. MtDNA as a cancer marker: a finally closed chapter?. Current genomics. 2017. June 1;18(3):255–67. 10.2174/1389202918666170105093635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.De Paepe B. Mitochondrial markers for cancer: relevance to diagnosis, therapy, and prognosis and general understanding of malignant disease mechanisms. ISRN Pathology. 2012. November 18;2012. 10.5402/2012/283670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jiménez-Morales S, Pérez-Amado CJ, Langley E, Hidalgo-Miranda A. Overview of mitochondrial germline variants and mutations in human disease: Focus on breast cancer. International journal of oncology. 2018. September 1;53(3):923–36. 10.3892/ijo.2018.4468 [DOI] [PubMed] [Google Scholar]
- 39.Hertweck KL, Dasgupta S. The landscape of mtDNA modifications in cancer: a tale of two cities. Frontiers in oncology. 2017. November 2;7:262 10.3389/fonc.2017.00262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stoneking M. Hypervariable sites in the mtDNA control region are mutational hotspots. The American Journal of Human Genetics. 2000. October 1;67(4):1029–32. 10.1086/303092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vartak R, Deng J, Fang H, Bai Y. Redefining the roles of mitochondrial DNA-encoded subunits in respiratory Complex I assembly. Biochimica et Biophysica Acta (BBA)-Molecular Basis of Disease. 2015. July 1;1852(7):1531–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(PDF)
(PDF)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.