Abstract
Neurons can respond to decreased network activity with a homeostatic increase in the amplitudes of miniature EPSCs (mEPSCs). The prevailing view is that mEPSC amplitudes are uniformly multiplied by a single factor, termed “synaptic scaling.” Deviations from purely multiplicative scaling have been attributed to biological differences, or to a distortion imposed by a detection threshold limit. Here, we demonstrate in neurons dissociated from cortices of male and female mice that the shift in mEPSC amplitudes observed in the experimental data cannot be reproduced by simulation of uniform multiplicative scaling, with or without the distortion caused by applying a detection threshold. Furthermore, we demonstrate explicitly that the scaling factor is not uniform but is close to 1 for small mEPSCs, and increases with increasing mEPSC amplitude across a substantial portion of the data. This pattern was also observed for previously published data from dissociated mouse hippocampal neurons and dissociated rat cortical neurons. The finding of “divergent scaling” shifts the current view of homeostatic plasticity as a process that alters all synapses on a neuron equally to one that must accommodate the differential effect observed for small versus large mEPSCs. Divergent scaling still accomplishes the essential homeostatic task of modifying synaptic strengths in the opposite direction of the activity change, but the consequences are greatest for those synapses which individually are more likely to bring a neuron to threshold.
SIGNIFICANCE STATEMENT In homeostatic plasticity, the responses to chronic increases or decreases in network activity act in the opposite direction to restore normal activity levels. Homeostatic plasticity is likely to play a role in diseases associated with long-term changes in brain function, such as epilepsy and neuropsychiatric illnesses. One homeostatic response is the increase in synaptic strength following a chronic block of activity. Research is focused on finding a globally expressed signaling pathway, because it has been proposed that the plasticity is uniformly expressed across all synapses. Here, we show that the plasticity is not uniform. Our work suggests that homeostatic signaling molecules are likely to be differentially expressed across synapses.
Introduction
Synaptic plasticity occurs in multiple varieties, which have opposing effects on synaptic physiology (Vitureira and Goda, 2013; Zenke and Gerstner, 2017). Hebbian plasticity, including long-term potentiation (LTP) and long-term depression (LTD), involves a rapid induction and long-lasting change in synaptic strength at individual synapses in the direction of the induction–potentiation for strong stimulation; depression for weak stimulation (Bliss and Lomo, 1973; Bliss and Collingridge, 1993; Malenka and Bear, 2004; Collingridge et al., 2010). If Hebbian plasticity is left unchecked, it could promote network instability due to runaway excitation or complete silencing. Homeostatic synaptic plasticity of miniature EPSCs (mEPSCs) is a population-level effect induced by chronic perturbations to network activity and the resulting changes are in the opposite direction: increases following activity blockade and reductions following activity enhancement (O'Brien et al., 1998; Turrigiano et al., 1998; Turrigiano, 2012). Although initial studies of homeostatic plasticity used a nonphysiological paradigm in which pre- and postsynaptic action potentials are inhibited by tetrodotoxin (TTX) in dissociated neuronal cultures, subsequent studies demonstrated the phenomenon is also induced in visual cortex neurons by monocular deprivation or dark rearing (Desai et al., 2002; Goel and Lee, 2007).
Turrigiano and colleagues proposed that in homeostatic plasticity, mEPSCs are multiplied by a uniform factor following chronic changes in activity, and termed the process “synaptic scaling.” This conclusion was based on their finding that a rank-ordering of mEPSC amplitude distributions from control and chronically silenced cells plotted against each other produced a straight line. The parameters from the linear regression model fit to that line were used to scale the treated mEPSC distribution toward the control distribution. That the scaled distribution was statistically indistinguishable from the control distribution was taken as evidence that the transformation was uniformly multiplicative (Turrigiano et al., 1998). This conclusion led to the hypothesis that, because a single factor is applied to all synapses on a neuron, there must be a global signal generated in the cell body (Turrigiano, 2008, 2012; Keck et al., 2013; Vitureira and Goda, 2013). The impact of the multiplicative scaling hypothesis is manifest in the adoption of the term synaptic scaling, which appears in the title or abstract of over 250 articles since 2000. Although approximately half of the articles since 2018 use the term synaptic scaling without also employing the term “multiplicative,” the original concept of uniform multiplicative scaling remains influential. If it turns out that homeostatic plasticity of mEPSCs is non-uniform, it would have profound implications in the field, redirecting the focus from globally expressed mechanisms to developing an understanding of why the plasticity is non-uniform.
It has been noted that the original study establishing synaptic scaling (Turrigiano et al., 1998) included an intercept term in the linear equation used for the scaling transformation, which suggests that the transformation of mEPSCs might be more complex than multiplication by a uniform factor (Kim et al., 2012). The requirement for an intercept term in the linear regression fit has been attributed to the need to account for mEPSCs in the control distribution that fall below the detection threshold and disrupt the alignment of mEPSCs from untreated and activity-altered cultures (Turrigiano et al., 1998; Blackman et al., 2012). Kim and colleagues attempted to account for these unseen mEPSCs by using an iterative process to test potential scaling factors (Kim et al., 2012). The iterative process applies a range of multiplicative factors to downscale the distribution from activity-blocked cultures. The potential mismatch due to undetected control mEPSCs is addressed by discarding downscaled mEPSCS that fall below the detection threshold.
We recorded AMPA mEPSCs from a large number of cells in dissociated cultures of primary mouse cortical neurons, 13–14 days in vitro (DIV), chronically silenced with a 48-h treatment of 500 nm TTX. Parameters from the rank-order process (Turrigiano et al., 1998), or the iterative process (Kim et al., 2012), were unable to scale the data from activity-blocked cultures to match the control data, based on a Kolmogorov–Smirnov (K–S) test. That led us to reexamine the idea that homeostatic plasticity is a uniform multiplicative process.
Materials and Methods
Primary culture of mouse cortical neurons
Dissociated cultures of mixed neuronal and glial populations were prepared from unsexed P0-P1 neonatal mice. These mice are wild-type mice from a colony of Rab3A+/− mice housed at Wright State University. The Rab3A+/− mice are derived from C57BL/6J, 129 Sv-Rab3a^(tm1Sud) from The Jackson Laboratory, originally crossed three times to C57BL/6J (IMSR catalog #JAX:002443, RRID:IMSR_JAX:002443); we subsequently backcrossed the line 11 times to wild-type mice from a Rab3A+/Ebd colony. Rab3AEbd/Ebd are mice with a point mutation in Rab3A (Kapfhamer et al., 2002), identified in an ENU-mutagenesis screen of C57BL/6J, crossed once to C3H/HeJ for mapping, and backcrossed three times to C57BL/6J (MGI catalog #2388809, RRID:MGI:2388809). Pups were euthanized by rapid decapitation with sharp scissors, as approved by the Wright State University Institutional Animal Care and Use Committee. Brains were removed, and cortices were collected after removal and discarding of hippocampi. Tissue digestion was conducted by the addition of papain (Worthington Biochemical) at 20 units/ml in Neurobasal-A media (Invitrogen), osmolarity adjusted to 270 mOsm, and incubated in a 37°C H2O bath for 20 min with gentle stirring. After digestion, cortices were gently triturated with a fire-polished Pasteur pipette, filtered through a 100-μm cell strainer (Corning Falcon), and transferred to a sterile 15-ml conical tube. The cell suspension was centrifuged at 1100 rpm for 2 min, supernatant was removed, and the pellet re-suspended in room temperature Neurobasal-A media (osmolarity adjusted to 270 mOsm), supplemented with 5% fetal bovine serum (FBS; promotes glial growth), 2% B-27 supplement to promote neuronal growth, L-glutamine (2 mm), and gentamicin (0.01 mg/ml; all from Invitrogen). Neurons were plated onto 12-mm coverslips pre-coated with poly-L-lysine (Corning) at a density of 0.15 × 106 cells/coverslip. The culture media for the first day (0 DIV) was Neurobasal-A media supplemented with FBS, B-27, L-glutamine, and gentamicin, and was switched at 24 h (1 DIV) to media consisting of Neurobasal-A (270 mOsm), 2% B-27, and L-glutamine, without FBS. FBS was excluded to avoid its toxic effects on neuronal viability and health (Stellwagen and Malenka, 2006). Cells were maintained in culture for 13–14 DIV in a humidified 5% CO2 incubator at 37°C. Culture media was changed twice weekly by replacing half with freshly prepared media. Two days before experiments, culture dishes containing four 12-mm coverslips were randomly chosen to receive TTX (500 nm; Tocris) to chronically silence all network activity and induce homeostatic synaptic plasticity mechanisms, while sister cultures not receiving TTX treatment served as untreated controls.
Whole-cell voltage clamp to record mEPSCs
At 13–14 DIV, mEPSCs from TTX-treated and control sister cultures were recorded via whole-cell voltage clamp. Recordings were taken from pyramidal neurons, identified visually as those cells exhibiting a prominent apical dendrite and a triangular cell body (Fig. 1A). Cells were continuously perfused with a solution consisting of the following: 115 mm NaCl, 5 mm KCl, 2.5 mm CaCl2, 1.3 mm MgCl2, 23 mm dextrose, 26 mm sucrose, and 4.2 mm HEPES; pH 7.2 (Stellwagen and Malenka, 2006). The osmolarity of the media from the cultures was measured (normally 285–295 mOsm) and the perfusate osmolarity was adjusted using mannitol to match the culture osmolarity to prevent osmotic shock to the neurons. To isolate glutamatergic mEPSCs, TTX (500 nm) and picrotoxin (50 μm) were included in the perfusion solution to block action potentials and GABAergic currents, respectively. APV was not included in the perfusate because all mEPSCs were blocked with CNQX and picrotoxin, leaving no APV-sensitive events (data not shown). Patch electrodes were filled with an internal solution containing the following: 128 mm K-gluconate, 10 mm NaCl, 1 mm EGTA, 0.132 mm CaCl2, 2 mm MgCl2, and 10 mm HEPES; pH 7.2. Pipette solution osmolarity was adjusted with mannitol to 10 mOsm less than the perfusion solution osmolarity. Neurons were voltage clamped at −60 mV using an Axopatch 200B patch-clamp (Molecular Devices) and recorded with Clampex 10.0 (Molecular Devices) for 2–5 min using a sampling rate of 20 kHz and filtering at 5 kHz. All recordings were performed at room temperature. Pipette resistances were 3.5–5 MΩ. Recordings with holding currents >20 pA or unstable, rapidly fluctuating holding currents were discarded before analysis, as were recordings requiring >20-MΩ series resistance compensation (values usually between 6 and 12 MΩ).
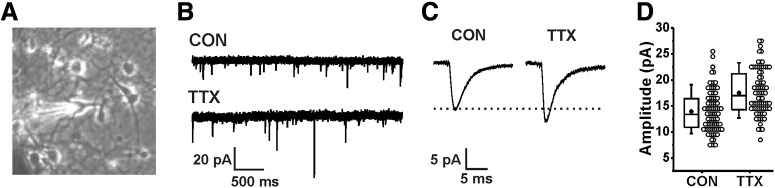
Figure 1.
Increased amplitude of mEPSCs in dissociated cultures of mouse cortical neurons after inhibition of action potentials for 48 h by treatment with 500 nm TTX. A, Phase contrast image of high-density cortical neuron culture with a pyramidal-shaped neuron chosen for recording. B, Representative example current traces of mEPSCs recorded in an untreated (CON) neuron and a TTX-treated (TTX) neuron. C, Average mEPSC current traces from the same recordings shown in B. D, Box and whisker plot of amplitude means for 86 untreated (14.0 ± 0.4 pA) and 77 TTX-treated neurons (17.5 ± 0.5 pA), from 23 cultures. p = 5.8 × 10−7, Kruskal–Wallis test. Box range, 25th–75th percentile; whisker, 10th–90th percentile; dot = mean; line = median.
MiniAnalysis to detect mEPSCs
mEPSCs were manually selected using MiniAnalysis software (Synaptosoft). A low-pass Butterworth filter of 2 kHz was applied before analyzing records; other parameters included using three points to average for peak and using 10,000 μs to average for baseline. Records were analyzed without reviewing the treatment group from which they came. In the earliest recordings, the detection threshold was set to 5 pA, as was done in previous studies (Turrigiano et al., 1998; Desai et al., 2002). Because the program would not accept some of the smaller visually identified events, the detection threshold was ultimately set to 3 pA. A separate analysis before and after the threshold change showed only modest alterations in the rank-order parameters, likely because mEPSCs below 5 pA were relatively rare. mEPSCs below 5 pA were observed in only 50% of the recordings from untreated cells, and 43% of those from TTX-treated cells. Of the recordings that contained 1 or more mEPSCs <5 pA, the average percent of mEPSCs below 5 pA was 1.6% and 0.92% for untreated and TTX-treated cells, respectively. Noise histograms were generated by saving 15 block segments of current traces displaying no mEPSCs, importing the traces to Origin 8, subtracting off the holding current, and combining multiple traces; ultimately >50,000 points were analyzed for each noise amplitude frequency histogram in Figure 2.
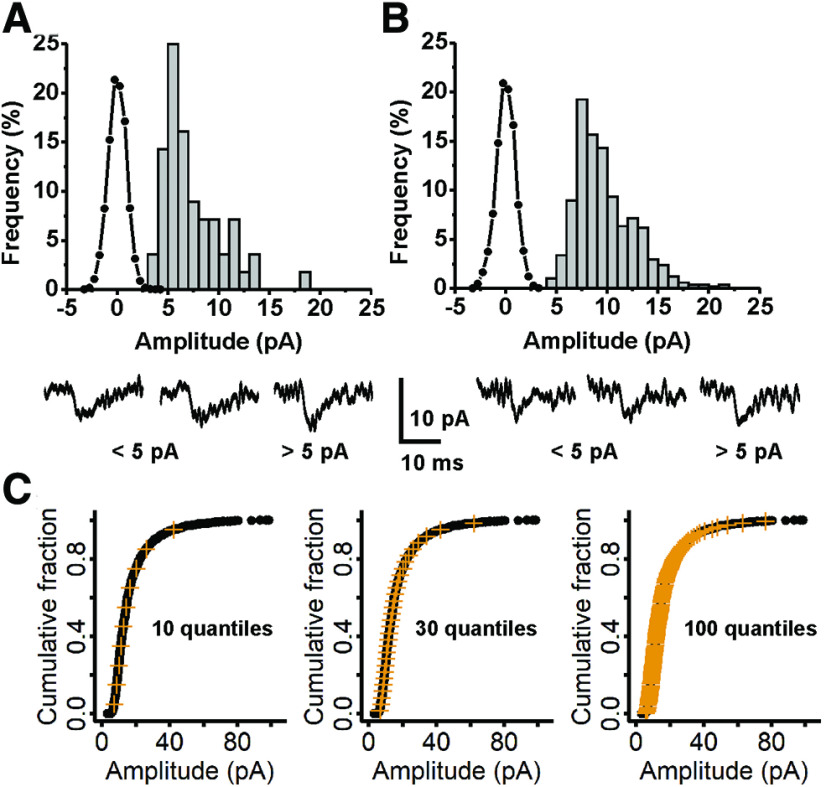
Figure 2.
Analysis of noise, detection of mEPSCs <5 pA, and demonstration of quantile sampling. A, Data from cell C0220000, mean amplitude 7.4 pA, 56 events. This cell had the highest percent of mEPSCS under 5 pA (10/56, 17.9%). The frequency histogram for currents in regions of the recording without mEPSCs (line and black circles) does not significantly overlap with the frequency histogram of mEPSCs detected by MiniAnalysis (gray bars). Below, current traces for three selected mEPSCs, two of which were below 5 pA and one which was slightly above. The mEPSCs have the following characteristics, left to right: 4.40 pA, 1.6-ms rise time, 5.5-ms decay; 4.15 pA, 1.5-ms rise time, 8.9-ms decay; 6.23 pA, 1.5-ms rise time, 4.1-ms decay. B, Data from cell D0512005, mean amplitude 9.7 pA, 504 events. Approximatley 1% of mEPSCs were below 5 pA (5/504). The noise histogram (line and black circles) is very similar to that of C0220000, although the two recordings were made more than a year apart; it does not overlap with the frequency histogram for mEPSCs detected by MiniAnalysis. Below, three selected mEPSCs, two below 5 pA and one above, with the following characteristics, left to right: 4.52 pA, 1.0-ms rise time, 2.4-ms decay; 4.88 pA, 2.3-ms rise time, 2.9-ms decay; 6.29 pA, 1.5-ms rise time, 2.8-ms decay. C, The cumulative distribution function (CDF) for amplitudes of mEPSCs recorded from cell E0324002 is shown as black circles, overlaid with 10 quantiles, 30 quantiles, and 100 quantiles, shown as vermilion crosses.
Sampling
Because the number of mEPSCs recorded varies by cell, the data must be sampled to ensure that each cell contributes a representative subset of equal size. Initially, a random subset of events with n = 30 was chosen without replacement from each cell; the events from untreated neurons were pooled to form the control distribution, and events from TTX-treated neurons were pooled to form the TTX distribution. A sampling approach based on random selection has the potential for sampling error—the generation of a non-representative sample due to random chance. To eliminate this possibility, we also used a quantile-based approach. Quantiles are values that describe a frequency distribution by dividing it into equal groups, such that each value represents the same fraction of the total data. For example, the median divides a distribution into two halves and represents the midpoint, or 50th percentile. We performed quantile-sampling by computing 30 evenly spaced quantile values from every cell, starting at the 1.67th percentile and ending at the 98.33rd percentile, with a step size of 3.33% (1/30). These sampling values were chosen to avoid including the maximum mEPSC amplitudes, which were the most variable. The choice of 30 quantiles is somewhat arbitrary, but is within the range of random samples that others have chosen when examining the distribution of mEPSC amplitudes, from 20 (Echegoyen et al., 2007) to 100 (Kim et al., 2012). To avoid excessive interpolation, we discarded recordings that contained fewer mEPSCs than the number of quantiles, so the choice of 30 quantiles meant that only 1 recording was discarded (giving 86 instead of 87 recordings from control cultures). Finally, the quantiles from untreated neurons were pooled to form the control distribution, and the quantiles from TTX-treated neurons were pooled to form the TTX distribution. Note that the actual number of mEPSCs recorded in untreated cells ranged from 46 to 2500 (average 391) and from 37 to 5029 for TTX-treated cells (average 500); in some recordings, the MiniAnalysis detection process was stopped when the number of mEPSCs reached 500, rather than after analyzing the entire record.
Simulated scaling
An empirical simulation of homeostatic plasticity was created based on the prevailing hypothesis of uniform synaptic scaling. Two non-identical samples of control data were generated by randomly sampling 30 mEPSCs from each control cell, pooling the data into a cumulative distribution function (CDF), and repeating that process until two CDFs were obtained that were different enough to give a p < 0.05 with a K–S test. One of the two datasets was multiplied by 1.25, to produce the simulated TTX dataset. Detection thresholds of 5 and 7 pA were simulated by discarding mEPSCs amplitudes below 5 or 7 pA from the CDFs of simulated control and simulated TTX data.
Rank-order process
In the rank-order process (Turrigiano et al., 1998) the control and TTX mEPSCs are ranked from smallest to largest and then plotted against each other, requiring the sample sizes to be identical. When the pooled samples have been created by randomly sampling an equal number of mEPSCs from each cell, the only ways to accomplish this are to randomly discard either mEPSCs or cells, from the distribution with the larger number of cells. However, in quantile sampling, the number of quantiles can be adjusted to make the quantity (number of quantiles times the number of cells) equal for the two distributions. For example, in the mouse cortical neuron data, 77 quantiles were sampled from each of the 86 control cells, and 86 quantiles were sampled from each of the 77 TTX cells. After plotting TTX versus control mEPSCs, the resulting relationship is fit with a linear regression model y = mx + b, using control as the predictor (x) and TTX as the response (y) to obtain slope (m) and intercept (b). The original pooled sample of TTX mEPSCs (created by sampling 30 quantiles per cell) is downscaled by the parameters from the linear regression model using the equation (TTX-b)/m, and the downscaled TTX data are compared with the original pooled sample of control mEPSCs (30 quantiles per cell) with a K–S test for the equivalence of distributions, and the test statistic and p value reported for comparison purposes (for details, see below, Experimental design and statistical analysis).
Iterative process
The iterative process to determine the scaling factor was proposed by Kim et al. (2012) as an alternative to the rank-order method for testing whether homeostatic plasticity causes multiplicative scaling. This method repeatedly downscales the pooled TTX data by an arbitrary factor, discards any downscaled amplitudes smaller than the smallest value in the control data, and compares the fit of the downscaled TTX data to the control data with a K–S test for equivalent distributions. The process is repeated for different values of scaling factor, and the factor that produces the best fit, as judged by the largest p value, is chosen. We applied the process both as described and without the removal of events that fell below the detection threshold after downscaling.
Experimental design and statistical analysis
The new results reported for dissociated mouse cortical neurons were obtained from 23 cultures, 86 recordings from untreated cells, and 77 recordings from TTX-treated cells. Data were included from a recording day only if there was at least 1 cell from each treatment group. The number of cells per day ranged from 1 to 10 [average 3.5 ± 2.1 (SD)]. Each culture is prepared from combining the cortical tissue from two mouse pups. We did not sex the pups so cannot address whether any of our results are sex specific. While it is not statistically correct to use n as the number of cells (Fig. 1) or number of mEPSCs (all other figures), we have continued to do so to compare our results to previous studies. The question we are asking is not whether activity blockade has a statistically significant effect, but whether activity blockade-induced homeostatic plasticity involves uniform multiplication of the control mEPSC distribution.
All analyses, including statistical tests, empirical modeling, and implementation of the rank-order and iterative tests, were performed using the R system for statistical computation (www.R-project.org). The K–S test for the equality of distributions has previously been used to analyze the similarity of two groups of mEPSC amplitudes. However, because multiple mEPSC samples are included per cell, such data are not independent and identically distributed (iid), which is an assumption of the K–S test (Mood et al., 1973; Raghavachari, 1973; Tygert, 2010). The K–S test expects every value in the sample to be independent, but here, any mEPSC is related to the other mEPSCs from the same recording; these are more likely to be similar to each other than mEPSCs from another recording. Violation of a statistical test's assumptions means that the distribution of the test statistic, and thus the critical value, is unknown, and the p values are therefore unreliable. Furthermore, the power of the K–S test increases with sample size, meaning that larger samples will, on average, yield smaller p values than smaller samples. When we include 30 values from each cell, the N that the K–S test sees is 30 × the number of independent samples, an inflated value. For these reasons, we cannot conclude that the true probability of obtaining a result in a K–S test is the p value we report. We believe the magnitude of the test statistic (which quantitates the difference between the two CDFs) and accompanying p value (which takes into account the sample size) indicate whether a comparison between two CDFs is a better or worse match than a comparison between two other CDFs if (1) the sampling has been performed in the same way, e.g., multiple samples included per cell; and (2) the sample sizes are comparable. Unless otherwise indicated, means are presented ±SEM.
Results
To characterize homeostatic plasticity of mEPSCs, we chose the well-studied paradigm of treating dissociated neurons with TTX. Cortical neurons were obtained from newborn mice and cultured for 13–14 DIV. Recordings of mEPSCs were performed after blocking firing activity with 500 nm TTX for 48 h. As has been shown in other studies, mEPSC amplitudes were significantly increased in neurons treated with TTX compared with untreated control neurons. Figure 1A shows a typical pyramidal-shaped neuron that was chosen for recording. Individual current traces recorded from an untreated and a TTX-treated cell are shown in Figure 1B, and the average mEPSCs waveforms from these recordings are shown in Figure 1C. The mean mEPSC amplitudes from 86 control and 77 TTX-treated cells were not normally distributed (Fig. 1D), and were significantly different based on a non-parametric Kruskal–Wallis test (control, 14.0 ± 0.4 pA; TTX, 17.5 ± 0.5 pA, p = 5.8 × 10−7).
Our dataset differs from those of previous studies. First, we put our cutoff for accepting mEPSCs at 3 pA instead of 5 pA (Turrigiano et al., 1998; Desai et al., 2002), because we noticed there were events that resembled mEPSCs in every way, but which could not be manually selected with a 5-pA cutoff. Examples of such events for two cells with small mean mEPSC amplitudes are shown in Figure 2, below graphs of the frequency histograms for noise amplitudes (Fig. 2A,B, line and symbol) and detected mEPSC amplitudes (Fig. 2A,B, gray bars) in these cells. The two traces on the left show mEPSCs with amplitudes <5 pA, and the one on the right shows an mEPSC with amplitude >5 pA. It is clear from these traces that mEPSCs below 5 pA occur and are similar to mEPSCs above 5 pA. The root mean square (RMS) noise was 1.44 ± 0.18 (SD) for the untreated cells (N = 86) and 1.43 ± 0.18 (SD) for TTX-treated cells (N = 77), justifying a cutoff of 4.32 (3 × RMS). We set the threshold below this calculated value, but manually selected mEPSCs, avoiding large fluctuations in noise or glitches that might be selected with the automated setting.
The second difference in our dataset concerns the way we generated the CDFs of mEPSC amplitudes. CDFs have been generated in previous studies by randomly selecting mEPSCs to keep the number of mEPSCs per cell the same. To avoid the random variability of this approach, we generated CDFs of mEPSC amplitudes by computing 30 quantiles per cell. Quantiles divide the distribution into equally spaced pieces; the median divides it into two halves; 30 quantiles divides it into 30 segments. We settled on 30 quantiles to be within the range of samples per cell used in previous studies and to not over- or under-sample the distributions (Fig. 2C; see Materials and Methods). Another characteristic of quantile sampling is that it provides a deterministic representation of each cell, whereas multiple random samples from the same cell will necessarily differ from each other. As expected, quantile sampling produced a pooled distribution with less variability than the distribution obtained by random sampling, but did not alter the overall appearance of the CDFs (Fig. 3, compare A, random sampling, B, quantile sampling).
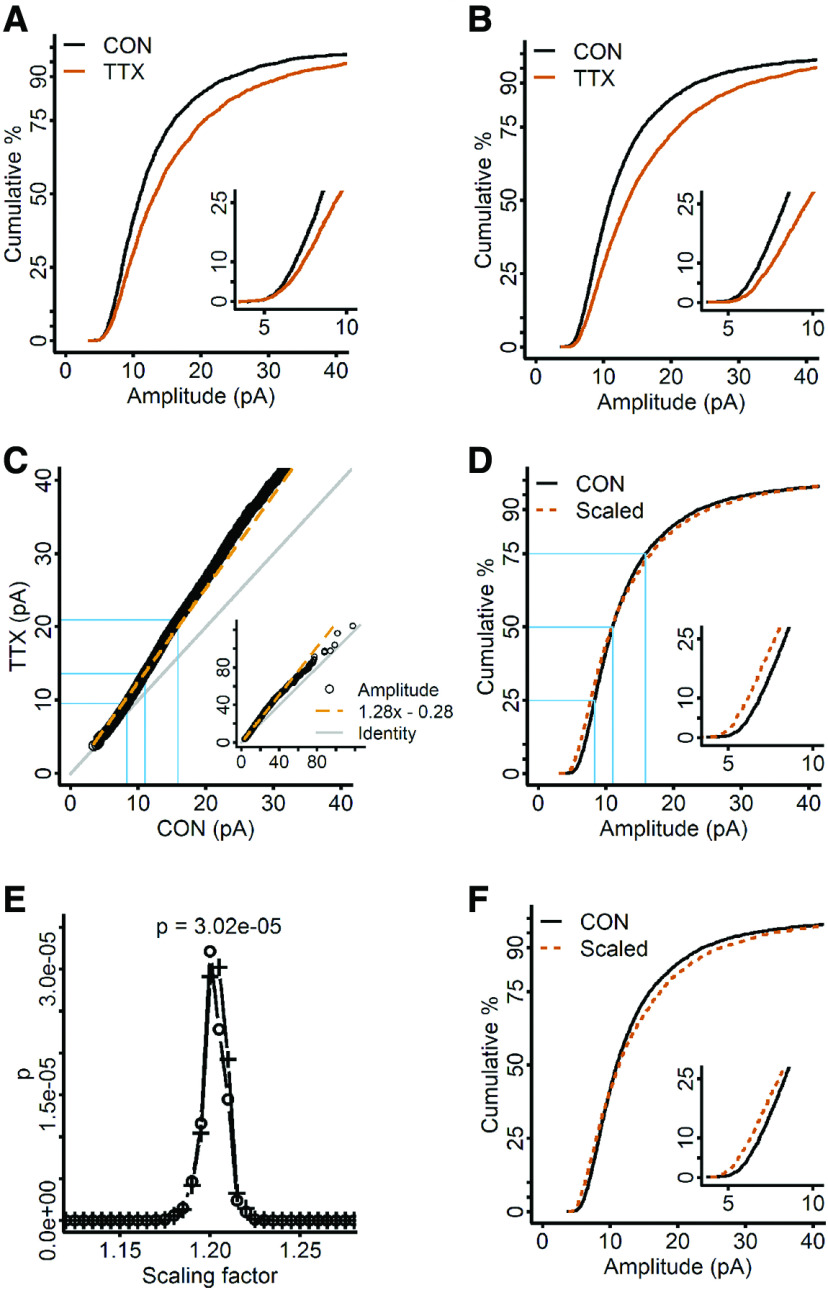
Figure 3.
Homeostatic plasticity in dissociated mouse cortical neurons is not well-described by the parameters obtained from the rank-order process, nor the single multiplication factor obtained by an iterative process. A, CDFs produced by randomly sampling 30 mEPSCs from each untreated (CON, black) and TTX-treated cell (TTX, vermilion). B, CDFs produced by computing 30 quantiles from the mEPSC distributions of each control (black) and TTX-treated cell (vermilion). C, Rank-order plot. To create identical size datasets as required, 77 quantiles were computed from each control cell, and 86 quantiles were computed from each TTX cell; the rank-order plot was obtained by sorting from smallest to largest amplitude in control and TTX data and plotting them against each other. A linear regression fit is shown in long vermilion dashes. D, Control CDF compared with TTX CDF after scaling (short dashes) using the parameters obtained from the linear regression fit of the rank-order plot in C; n, control = 2580; TTX = 2310; test statistic = 0.070, p = 1.3 × 10−5, K–S test. E, Scaling factors were chosen iteratively, used to downscale the TTX distribution, followed by discarding subthreshold amplitudes below 4.01 pA (smallest control mEPSC quantile), and the scaled distribution was compared with control with a K–S test to produce the corresponding p value. Crosses, Below-threshold events were discarded. Circles, Below threshold events were not discarded. Test statistic = 0.053, p = 3.0 × 10−5, K–S test. F, The scaling factor that produced the largest p value in E, 1.19, was used to downscale the TTX distribution (dashed vermilion curve) to the control (black curve); two events were discarded. Insets in A, B, D, F, Blow up of the low end of the data. Inset in C, Full range of the data. Line of identity is indicated in gray. Blue lines in C, D indicate the 25th, 50th, and 75th percentiles of the data.
The prevailing view of homeostatic plasticity is that it transforms amplitudes of mEPSCs via a uniform multiplicative factor (Turrigiano et al., 1998; Kim et al., 2012; Turrigiano, 2012). The rank-order process was introduced in the Turrigiano et al., study and continues to be a commonly used method for analyzing homeostatic plasticity data. In this process, the control and TTX amplitude data are ranked, plotted against each other, and fit with a linear regression model, yielding a slope and an interceptcoefficient that are used to quantify the mathematical transformation induced by homeostatic plasticity on the mEPSC amplitudes in TTX-treated neurons. If these coefficients accurately describe the effects of homeostatic plasticity, then using them to mathematically downscale the TTX distribution should reverse those effects and produce a scaled distribution almost identical to the control distribution. Figure 3C shows our rank-order data, which were fit with a linear regression model with a slope of 1.28 and an intercept of −0.28. We found that using the parameters from the regression model produced a scaled TTX distribution that had a different shape from the control distribution and gave a p ≪ 0.05 (Fig. 3D, dashed vermilion curve; test statistic = 0.070, p = 1.3 × 10−5, K–S test). Note that we cannot conclude significance or non-significance in these analyses, because the inclusion of multiple samples per cell violates the requirement of the K–S test for iid samples.
It has been suggested previously (Kim et al., 2012) that the existence of control mEPSCs that fall below the detection threshold could produce a mismatch in the rank-order data, since this population may be shifted into the detectable range after chronic silencing but would not have corresponding mEPSCs from the untreated dataset. Such a mismatch could distort the rank-order relationship, leading to parameters that do not accurately scale the TTX CDF to the control CDF. This issue has been addressed by Kim et al. (2012), who have developed an approach distinct from that of the rank-order process. They use a series of multiplicative factors to downscale the mEPSC amplitudes from TTX-treated cells, and discard any downscaled amplitudes that fall below the detection threshold (defined as the smallest observed mEPSC amplitude in the control data). If the deviation from multiplicative scaling is solely due to a mismatch caused by undetectable control mEPSCs, this process should be able to find a scaling factor that downscales the TTX data to a close match to the untreated data (small test statistic, large p value). We applied the iterative process to the quantile-sampled experimental data, obtaining a scaling factor of 1.19 (Fig. 3E, circles), and found it produced a downscaled TTX distribution (Fig. 3F, dashed vermilion curve) that gave a very small p value (Fig. 3F, black curve; test statistic = 0.053, p = 3.0 × 10−5, K–S test). This result suggests the data are not undergoing a uniform multiplicative transformation following chronic silencing. The fit was not noticeably changed by leaving out the correction that is supposed to address loss of mEPSCs due to the detection threshold (Fig. 3E, crosses). This lack of effect may be because of the low detection threshold we set, at 3 pA.
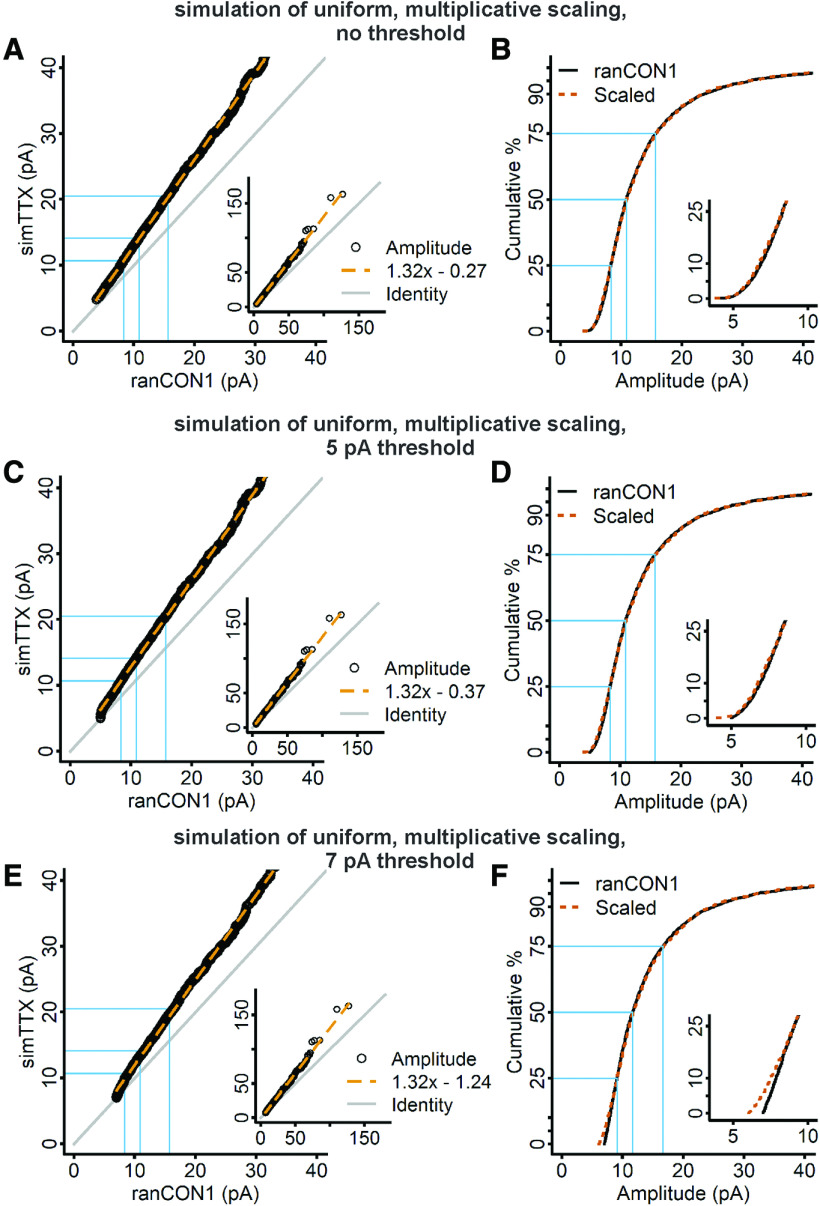
The differences between the scaled and control CDFs for both the rank-order and the iterative processes applied to the experimental data are extremely important. If the scaling process represents the true transformation conferred by homeostatic plasticity, it should by definition exactly match the downscaled treated distribution to the control distribution. That we do not get an exact match means that our assumption of a uniform multiplicative transformation must be incorrect. Because our data fail to support uniform multiplicative scaling, we wanted to simulate an example of uniform multiplicative scaling and observe how the resulting data behave when subjected to the rank-order and iterative processes. In particular, we wanted to observe what happens when data that we know are scaled uniformly by a multiplicative factor are subjected to an arbitrary detection threshold, and whether the existence of undetected control mEPSCs affect the ability of the rank-order and iterative processes to scale the data from TTX-treated cultures. Our idea was to simply multiply the distribution of control events (CON) by 1.25. To simulate expected biological variability, we did not quantile sample but created two non-identical samples of control data by randomly sampling 30 mEPSCs from each cell, repeatedly producing pooled datasets until we obtained two control CDFs that gave a p < 0.05 with a K–S test (Fig. 4A, black curve, ranCON1, vs vermilion dashed curve, ranCON2, p = 0.04). We then multiplied the second randomly sampled control dataset (ranCON2) by 1.25, to simulate uniform multiplicative scaling in data with realistic variability (simTTX; Fig. 4A, vermilion curve). Note that, because of the variability we introduced between the two random control samples, the second randomly sampled control dataset is larger than the first by a factor of 1.06. Therefore, the actual scaling factor between ranCON1 and simTTX is 1.31 instead of 1.25. We were surprised, based on the experimental data in Figure 3, our own previous work (Altimimi and Stellwagen, 2013; Garcia-Bereguiain et al., 2013; Fong et al., 2015), and others' previous studies (Turrigiano et al., 1998; Desai et al., 2002; Thiagarajan et al., 2005; Ibata et al., 2008; Blackman et al., 2012; Gerkin et al., 2013), that the CDF produced by multiplication with a single factor is larger than the control CDF from the smallest mEPSCs in the distribution (Fig. 4A, inset). For all of the studies cited here, and for our experimental data, the TTX CDF runs very close to the control CDF in the small amplitude ranges, and separates only as amplitude increases.
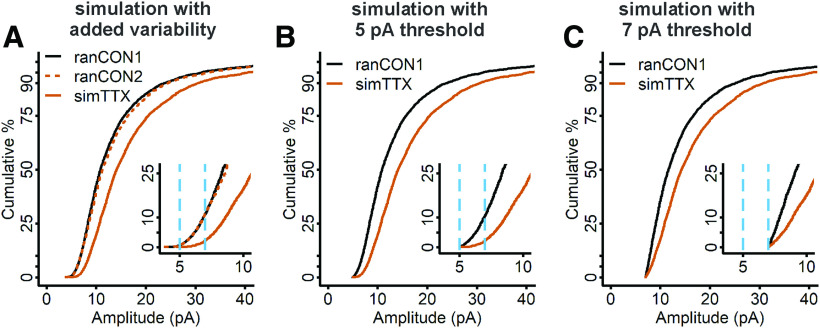
Figure 4.
Empirical simulations of uniform multiplicative scaling show that mEPSCs of TTX-treated cells are larger than those of control cells from the lowest percentiles of the distributions; application of a detection threshold causes a small overlap. A, Two distinct control distributions (ranCON1, black curve; and ranCON2, dashed vermilion curve) were obtained by random sampling and differed on a K–S test with a test statistic = 0.039, p = 0.04. The second control distribution was multiplied by 1.25 to simulate uniform multiplicative scaling (simTTX, solid vermilion curve). B, A detection threshold of 5 pA was applied to the ranCON1 and simTTX data, meaning that all mEPSCs below 5 pA were removed from both distributions. C, A detection threshold of 7 pA was applied to the ranCON1 and simTTX data; all data below 7 pA were removed from both distributions. Insets, Blow up of the low end of the distribution. Dashed blue lines indicate the location of 5- and 7-pA cutoffs, which represent ∼2% and 10% of the control dataset, respectively.
To address whether the deviation of the experimental data from simulated uniform multiplicative scaling could be caused by the existence of an undetected population of control mEPSCs, we used the data shown in Figure 4A to simulate detection thresholds of 5 and 7 pA (Fig. 4B,C, respectively). To simulate these thresholds, we discarded all mEPSC amplitudes below 5 pA or 7 pA from both the ranCON1 and simTTX distributions. The detection thresholds are shown with the resulting truncated distributions on an expanded scale in Figure 4B,C, insets, and can be compared with the original distributions in Figure 4A. Both detection thresholds eliminate a greater proportion of events from ranCON1 than simTTX, just as we imagine occurs in an electrophysiological experiment, although fewer events are removed by the 5-pA detection threshold. Following application of a 7-pA detection threshold, ranCON1 and simTTX CDFs are no longer separated at the beginning of the distributions, supporting the possibility that a mismatch caused by a detection threshold could lead to the appearance of non-uniform scaling such as we observe in the experimental data. However, the simulation of a 7-pA threshold only causes an overlap of the first pair of data points, whereas the CDFs from the experimental data overlap for multiple points, and in addition, run closely together for >10% of the distributions.
Once we had successfully simulated the mismatch caused by a detection threshold, we examined the datasets with the simulated 5- and 7-pA detection thresholds using the rank-order process, and compared their behaviors to that of the untruncated data from the original simulation. A linear regression model applied to the rank-order data from the original simulation produced a slope coefficient close to the expected value (Fig. 5A; slope = 1.316, expected value 1.31), and an intercept close to zero (−0.27). The simTTX distribution scaled by these parameters closely matched the ranCON1 CDF (Fig. 5B; test statistic = 0.018, p = 0.77). The 5-pA detection threshold simulation data were fit with a slope coefficient close to the expected value of 1.31 (Fig. 5C; slope = 1.320), and an intercept only slightly larger than that of the original untruncated data (−0.37). These parameters again were able to scale simTTX to be a close match of the ranCON1 CDF (Fig. 5D; test statistic = 0.018, p = 0.63). Finally, the 7-pA detection threshold data were fit with a slope coefficient of 1.320 and an intercept of −1.24 (Fig. 5E), parameters which scaled the simTTX to closely match the upper 75% of ranCON1, but the lower 25% were misaligned (Fig. 5F), the test statistic was 0.047 and p = 0.012. This result suggests that the mismatch caused by a detection threshold can lead to a nonzero intercept term and a p < 0.05. However, the intercept term did not increase in proportion with the cutoff value, and larger intercepts have been observed for previously published experimental data (−12.2, −2.6, and −4.9 for Turrigiano et al., 1998; Blackman et al., 2012; Kim et al., 2012, respectively). In summary, these results show that the rank-order process is capable of finding a nearly perfect fit in uniformly scaled data, and is only modestly disrupted by a large detection threshold. Although the p values cannot be used to conclude statistical significance, the simulated dataset has the identical sampling issues as the experimental dataset, yet the p values are much larger for the simulated dataset. Therefore, the inability of the rank-order process to scale the experimental TTX CDF to the control CDF reflects a true deviation from uniform multiplicative scaling.
Figure 5.
The scaling transformation computed by the rank-order process detects the correct scaling factor and produces a nearly perfect fit of data simulated with multiplication by a uniform factor, and is only slightly disrupted when data have been truncated with a large detection threshold. A, RanCON1 and simTTX mEPSC amplitudes were sorted from smallest to largest and plotted against each other, and fit with a linear regression model (dashed vermilion line). B, The parameters from the linear regression fit shown in A were used to downscale the data from the TTX simulation, and produced a close match between the scaled distribution (dashed vermilion curve) and the ranCON1 data (black curve; ranCON1, n = 2610; simTTX, n = 2610; test statistic = 0.018, p = 0.77, K–S test). C, Rank-ordered mEPSCs of ranCON1 and simulated TTX cells with a detection threshold set at 5 pA, plotted against each other and fit with a linear regression model (dashed vermilion line). After removing mEPSC amplitudes < 5 pA from ranCON1 and simTTX, the samples sizes had to be rematched to be identical for the rank-order process, which was accomplished by randomly discarding mEPSCs from the now larger simTTX dataset (new n = 2594 for both control and TTX). D, The parameters from the linear regression fit shown in C were used to downscale simTTX (test statistic = 0.018, p = 0.63, K–S test). E, Rank-ordered mEPSCs of ranCON1 and simulated TTX data with a detection threshold of 7 pA, plotted against each other and fit with a linear regression model (dashed vermilion line). After removing mEPSC amplitudes < 7 pA, the sample sizes were rematched by randomly discarding from the now larger simTTX dataset (new n = 2321 for both control and TTX). F, The parameters from the linear regression fit in F were used to scale the simulated TTX data down to the ranCON1 data (test statistic = 0.047, p = 0.012, K–S test). Gray lines in A, C, E indicate the line of identity. Blue lines indicate the 25th, 50th, and 75th percentiles of the data. Insets in A, C, E, View of the entire data range. Insets in B, D, F, Blow up of the initial part of the distribution.
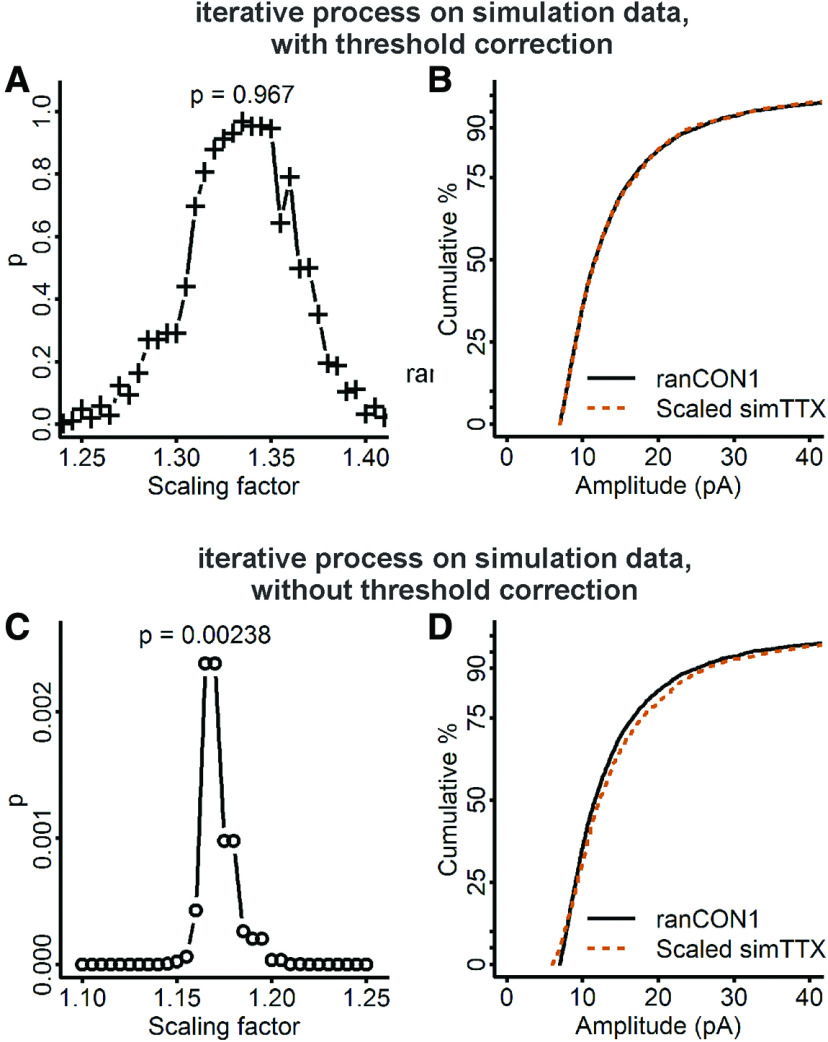
We next applied the iterative process of Kim et al. (2012) to the simulation data with the 7-pA threshold, since this process should be able to take even this large threshold into account. As expected, the iterative process produced an almost perfect fit between the scaled simTTX and ranCON1 distributions with a factor of 1.335 (Fig. 6A,B; test statistic = 0.016, p = 0.97, K–S test). The iterative process consists of two distinct steps. The first is to move through a range of factors to downscale the TTX dataset, this is an unbiased optimization process. The second is to discard scaled TTX data that fall below the detection threshold, this step addresses the potential mismatch caused by a detection threshold. To verify that the goodness of the fit was not simply due to using an unbiased optimization process, we repeated the iterative process but did not discard subthreshold events from the downscaled simTTX data. This version of the process selected a factor of 1.165, no longer close to the expected value of 1.31, and gave a much smaller p value (Fig. 6C,D; test statistic = 0.056, p = 2.4 × 10−3, K–S test), confirming that the removal of subthreshold events is essential when there is a detection threshold truncating the dataset. These results strongly demonstrate that the iterative process should be able to produce an almost perfect fit if the only issue is the inability to detect below threshold mEPSCs. Therefore, the large test statistic and small p value obtained when the iterative process is applied to our experimental data cannot be attributed to a mismatch caused by undetected, subthreshold control mEPSCs.
Figure 6.
Iterative selection of the scaling factor detects the expected factor with an almost perfect fit in data simulated with multiplication by a single factor and truncated by a large detection threshold. A, Each scaling factor was used to downscale the simulated TTX distribution, subthreshold events (<7 pA) were discarded from the downscaled data, and the scaled distribution was compared with simulated control with a K–S test to produce the corresponding p value. A factor of 1.335 produced a nearly perfect fit (test statistic = 0.016, p = 0.967, K–S test). B, Cumulative distributions of ranCON1 data (black curve) and scaled simTTX (downscaled by 1.335, 13 subthreshold simTTX mEPSCs removed, 5.6% of the data; dashed vermilion curve). C, When subthreshold mEPSCs were not removed, the scaling factor that produced the best fit of scaled simTTX to ranCON1 was 1.165, but the p value was much smaller than when subthreshold values were removed (test statistic = 0.056, p = 2.4 × 10−3, K–S test). D, Cumulative distributions of ranCON1 (black curve) and scaled simTTX (downscaled by 1.165, no events discarded; dashed vermilion curve).
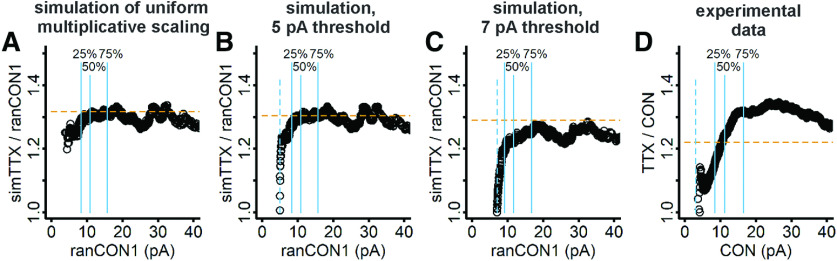
Thus far, we have shown that simulation of uniform multiplicative scaling, even with a large detection threshold imposed, does not reproduce the behavior of our experimental data. In order to more directly observe the magnitude of the homeostatically-induced scaling factor across mEPSC amplitudes, we plotted the ranked mEPSC data as the ratio of TTX/control against control amplitudes for each pair of ranked amplitudes. In data transformed by uniform multiplication, the ratio should be constant across amplitudes. Because a detection threshold causes the smallest TTX mEPSC amplitudes to overlap with control mEPSC amplitudes, we predict that a detection threshold will manifest as a reduced ratio at the smallest amplitudes, and the extent of the threshold's effect on the data will be visible as the magnitude and range of this reduction. We plotted the data from the simulation with no detection threshold, which confirmed our expectation that the ratio plot yields an approximately constant value very close to the expected multiplication factor of 1.31 (Fig. 7A). The fluctuations of data in this plot are attributable to the variability introduced by random sampling (Fig. 4A). The ratio plot of the simulation data with a 5-pA detection threshold also exhibited a near constant ratio close to the expected multiplication factor, but the ratio was underestimated for approximately the smallest 10% of the data (Fig. 7B). In the simulation of the 7-pA detection threshold, the ratio was underestimated for the bottom 25% of the data, then approached the expected multiplication factor, but ultimately was systematically slightly smaller than the expected value (Fig. 7C). In dramatic contrast to the behaviors exhibited by data from all three simulations, the ratio calculated for the experimental data increased gradually over >60% of the data before reaching an apparent plateau at a value which was substantially greater than the other estimates we have of the factor from the rank-order and iterative processes (Fig. 7D). These results clearly demonstrate that the scaling induced in our data by homeostatic plasticity is not the result of multiplication by a uniform factor. Since the scaling factor is smallest for small amplitude mEPSCs, and increases with increasing amplitude, we propose a new name for this type of homeostatic plasticity, “divergent scaling,” to distinguish it from uniform multiplicative scaling. To illustrate the effect of divergent scaling, a 10 pA mEPSC will be 11 pA after scaling (10% increase); a 20 pA mEPSC will be 26 pA after scaling (30% increase).
Figure 7.
After rank-ordering the control and TTX data, a plot of TTX mEPSC/control mEPSC as a function of control mEPSC shows a roughly constant ratio for simulated uniform multiplicative scaling, a predictable deviation over the initial portion of the data for simulation of a detection threshold, but a non-constant ratio over a large portion of the data range for the experimental data. A, For simTTX/ranCON1, the ratio is close to the expected value of 1.31 (dashed vermilion line). B, For data truncated by a 5-pA detection threshold, the ratio simTTX/ranCON1 is underestimated for a small range of data in the range of the 5-pA threshold, but is otherwise close to the expected value (dashed vermilion line). C, For data truncated by a 7-pA detection threshold, the ratio is underestimated for the bottom 25% of the data, then reaches a plateau; the plateau systematically underestimates the expected value thereafter (dashed vermilion line). D, For experimental data the ratio increases smoothly until reaching a plateau halfway between the 50th and 75th percentiles; the ratio is first under and then surpasses the slope factor estimated from linear regression of the rank-ordered data (dashed vermilion line). Solid blue lines indicate 25th, 50th, and 75th percentiles. Dashed blue lines indicate the simulated 5-pA (B) and 7-pA (C) thresholds, and the 3-pA detection threshold used for experimental data (D).
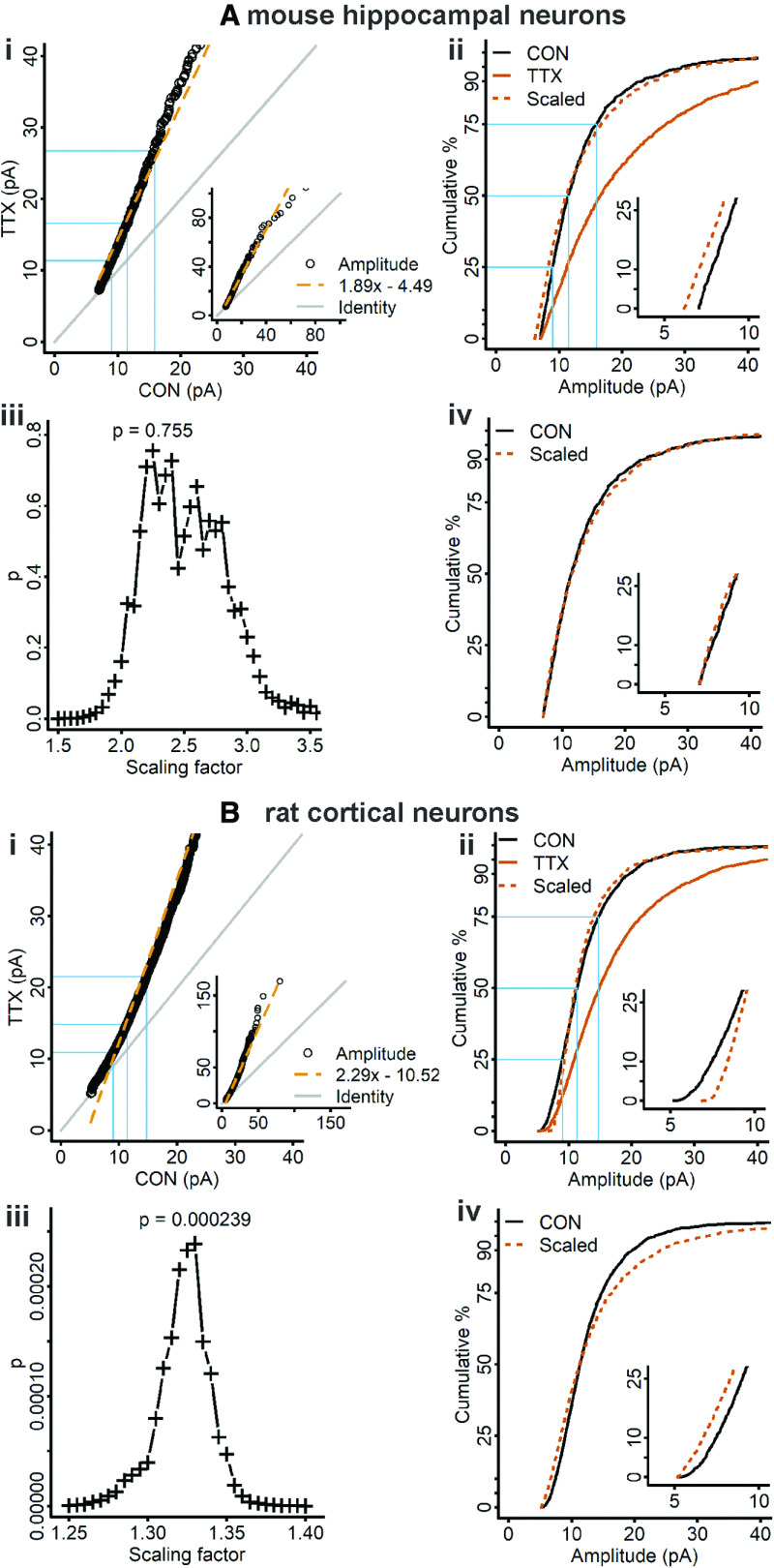
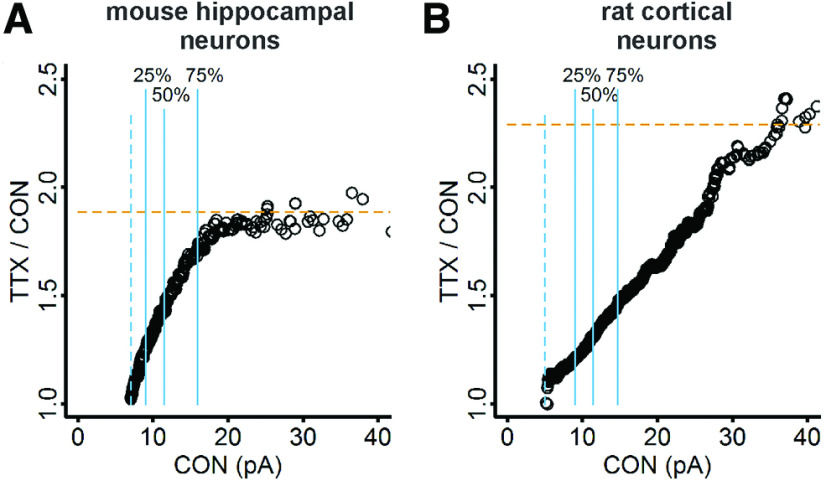
To determine whether the presence of divergent scaling is unique to the current experimental conditions, relatively mature (13–14 DIV) dissociated mouse cortical cultures, we examined previously published data from our laboratories: 12–15 DIV dissociated mouse hippocampal cultures treated with TTX for48 h (Altimimi and Stellwagen, 2013) and 10–12 DIV dissociated rat cortical cultures treated with TTX for 24 h (Fong et al., 2015). We applied a linear regression model to the rank-order data for both preparations (Fig. 8Ai,Bi), then used the parameters of the fits to scale the CDFs from TTX-treated cells to the CDFs of control cells (Fig. 8Aii,Bii). Note that the CDFs were created from quantile sampling whereas previously, random sampling was used, and the new plots differ slightly from the previously published plots (Altimimi and Stellwagen, 2013, their Fig. 3, compare A, B and D, E; Fong et al., 2015, their Supplementary Fig. 3A). Scaling of the TTX-treated data produced a CDF that closely matched the control CDF in the mouse hippocampal and rat cortical data (Fig. 8Aii,Bii, dashed vermilion curves), but the intercepts were far from zero (−4.49 and −10.52, respectively), and the K–S test returned small p values (7.7 × 10−3 and 2.9 × 10−7, respectively). Although the scaled data from mouse hippocampal neurons deviate to the left of control (Fig. 8Aii, inset) while the scaled data from rat cortical neurons deviate to the right (Fig. 8Bii, inset), these differences may not reflect biological differences but only that the linear regression model is not an accurate representation of the data (Fig. 8Ai,Bi). Going back to the results of the rank-order process on our simulation data, we expect an intercept close to zero and a nearly perfect match of scaled data to control data, if the scaling relationship is truly uniformly multiplicative (Fig. 5). The results of the rank-order process call into question whether these data are uniformly scaled.
Figure 8.
Rank-order and iterative processes applied to previously published data from dissociated mouse hippocampal neurons and rat cortical neurons. Ai, Rank-order plot of mouse hippocampal neuron data, with linear regression fit (dashed vermilion line). To match the sample sizes, 20 quantiles were computed for each control cell and 18 quantiles for each TTX cell (n = 360 for both groups). Aii, Cumulative distributions of mEPSC amplitudes from control (black curve) and TTX-treated mouse hippocampal neurons (vermilion curve). Slope and intercept parameters from the linear regression fit in Ai were used to downscale the TTX data (dashed vermilion curve; control, 30 quantiles for 18 cells, n = 540; TTX, 30 quantiles for 20 cells, n = 600; test statistic = 0.099, p = 7.7 × 10−3, K–S test). Aiii, For the mouse hippocampal neuron data, the scaling factor that produced the best fit of scaled TTX to control data after discarding downscaled mEPSCs below 7 pA is 2.25 (test statistic = 0.037, p = 0.755, K–S test), but > 40% of the data were discarded after downscaling (n after downscaling = 318; original n = 600). Aiv, Cumulative distributions of scaled TTX (dashed vermilion curve) downscaled by 2.25 and subthreshold mEPSCs removed, and control mEPSCs from mouse hippocampal neurons (black curve). Bi, Rank-order plot of rat cortical neuron data, with linear regression fit (dashed vermilion line). To match the sample sizes, 58 quantiles were computed for each control cell and 47 quantiles were computed for each TTX cell, n = 2726 for both groups. Bii, Cumulative distributions of mEPSC amplitudes for control (black curve) and TTX-treated rat cortical neurons (vermilion curve). Slope and intercept parameters from the linear regression fit in Bi were used to downscale the TTX data (dashed vermilion curve; control, 30 quantiles for 47 cells, n = 1410; TTX, 30 quantiles for 58 cells, n = 1740; test statistic = 0.101, p = 2.9 × 10−7, K–S test). Biii, For the rat cortical neuron data, the scaling factor that produced the best fit of scaled TTX to control data after discarding downscaled mEPSCs below 5 pA is 1.33 (test statistic = 0.079, p = 2.4 × 10−4, K–S test); only 25 mEPSCs were discarded after downscaling. Biv, Cumulative distributions of scaled TTX (dashed vermilion curve), downscaled by 1.33 and subthreshold mEPSCs removed, and control data from rat cortical neurons (black curve). Gray lines indicate the line of identity. Blue lines indicate the 25th, 50th, and 75th percentiles. Insets in Ai, Bi, View of the entire data range. Insets in Aii, Aiv, Bii, Biv, Blow up of the initial part of the distribution.
To further examine whether the previously published data show uniform multiplicative scaling, we applied the iterative process of Kim et al. (2012). The iterative process applied to the mouse hippocampal culture data selected a multiplicative factor that scaled the TTX CDF to the control CDF with a large p value (Fig. 8Aiii,iv; test statistic = 0.037, p = 0.755), suggesting these data may be uniformly scaled. However, the large magnitude of the scaling factors applied resulted in a substantial portion of the data (>40%) being discarded as subthreshold following downscaling. The large fraction of data being discarded may also contribute to the ability of a wide range of factors to fit the data with large p values (scaling factors from 2.0 to 3.0), as a smaller sample size leads to larger p values in the K–S test. For cultures of rat cortical neurons, the iterative process selected a multiplicative factor that gave a small p value when the scaled TTX CDF was compared with the control CDF (Fig. 8Biii,iv; test statistic = 0.079, p = 2.4 × 10−4). For these data, as well as the mouse cortical neurons, <2% of the mEPSC amplitudes were discarded in the downscaling step. The differences in amount of data discarded are difficult to interpret, but these results clearly show that in rat and mouse cortical cultures, scaling is non-uniform.
We generated plots of the ratio of TTX/CON amplitudes as a function of control amplitudes for the mouse hippocampal and rat cortical neurons, to see whether the ratios were uniform. Because the iterative process gave a large p value when comparing scaled TTX data to control data for mouse hippocampal cultures, we expected the ratio to exhibit a constant value, with minor deviations due to the detection threshold. Surprisingly, the ratios from the mouse hippocampal neurons show the divergent scaling pattern, with the ratio increasing across roughly 75% of the data, a larger proportion than for the data from mouse cortical neurons (compare Figs. 9A, 7D). Despite being most closely related to the canonical preparation (Turrigiano et al., 1998), rat cortical neurons show the most divergent scaling of all, with the scaling ratio continuing to increase far past the 75th percentile (Fig. 9B). Whether a plateau is reached is inconclusive due to the presence of very few mEPSC amplitudes in that range. In contrast to the mouse cortical neurons, where the difference between the smallest and largest scaling factor is a modest 25%, for mouse hippocampal and rat cortical neurons, the differences are 80% and 125%, respectively. To illustrate the impact of this effect, in a rat cortical neuron, a 10 pA mEPSC will be 11.8 pA after divergent scaling (18%) but a 20 pA mEPSC will be 33 pA (65%). From these findings, we conclude that (1) divergent scaling cannot be detected by the rank-order or iterative processes; (2) the quantitative difference between uniform multiplicative scaling and divergent scaling can be substantial; and (3) divergent scaling is not limited to particular experimental conditions but may be a common outcome of homeostatic plasticity.
Figure 9.
Ratio plots show divergent scaling in dissociated mouse hippocampal and rat cortical neurons. A, For mouse hippocampal data, the ratio of TTX mEPSC/control mEPSC as a function of control mEPSC increases smoothly until reaching an approximately constant value above the 75th percentile. B, For rat cortical data, the ratio increases continuously over most of the data range. Solid blue lines indicate 25th, 50th, and 75th percentiles. Dashed blue lines show thresholds used in acquisition: 7 pA for mouse hippocampal data and 5 pA for rat cortical data. Dashed horizontal vermilion lines indicate the expected scaling factor determined from the slope of linear regression of the rank-order data.
Discussion
Synaptic scaling is not uniformly multiplicative
In a large dataset from mature mouse cortical cultures (13–14 DIV), blocking network activity with TTX resulted in an increase of mEPSC amplitudes, as shown in many previous studies in multiple preparations. However, we were surprised to find that our data were not well fit by the parameters from the linear fit to the rank-ordered data, nor by a factor chosen by an iterative process that takes the detection threshold into account. While the scaled distributions are close to the control distribution, the deviations are important, since we have already applied a scaling process to make the two distributions the same. The differences that remain after this scaling reveal that the scaling process does not fully capture the transformation due to homeostatic plasticity. We simulated uniform multiplicative scaling by multiplying a random sample of control data by a uniform factor, and found it did not reproduce the behavior of the experimental data: the rank-order and iterative processes almost perfectly scaled the simulation data, even when a detection threshold was applied. These findings make it highly unlikely that a population of unseen small control mEPSCs caused the failure of our data to show uniform multiplicative scaling. Finally, we found that the ratio of TTX/control amplitudes plotted as a function of control mEPSC amplitude is not uniform, but increases with control amplitudes over a large proportion of the data. We named this phenomenon divergent scaling, and show that it is not unique to dissociated mouse cortical neurons but extends to dissociated mouse hippocampal and rat cortical neurons.
While we are not the first to suggest that activity blockade can produce non-uniform homeostatic increases in mEPSC amplitudes (Thiagarajan et al., 2005; Echegoyen et al., 2007; Goel and Lee, 2007; Cingolani and Goda, 2008; Pozo and Goda, 2010), uniform multiplicative scaling has remained the dominant idea in the field (for review, see Watt and Desai, 2010; Turrigiano, 2012; Keck et al., 2013; Chen et al., 2014; Lee et al., 2014). Our point is not that we are reporting yet another example that does not follow the norm of uniform multiplicative scaling, but that uniform multiplicative scaling is not the norm. This conclusion is based on (1) our finding that simulated uniform multiplicative scaling is different from experimental data for three preparations; (2) that the CDFs for the control and scaled TTX data in simulated uniform multiplicative scaling are separated from each other starting with the smallest mEPSCs, in contrast to the CDFs of mEPSCs from control and activity-blocked rat cortical neurons in Turrigiano et al. (1998); (3) that the linear regression model applied to the rank-order data in Turrigiano et al. (1998) requires a large intercept term (−12.3); and finally (4) that we show simulation of a detection threshold does not produce deviations from uniform multiplicative scaling. We cannot rule out that there may be a set of conditions under which homeostatic plasticity causes a uniform multiplicative shift. It will be important to identify such cases using rigorous criteria (close to zero intercept in the rank-order process, flat ratio of treated/control amplitude across the range of control mEPSC amplitudes), because they are likely to employ a unique repertoire of biological mechanisms.
It must be acknowledged that simultaneously blocking action potentials of excitatory and inhibitory neurons in a network has no correlate in vivo. Homeostatic plasticity is also induced by more realistic visual and auditory deprivation paradigms (Desai et al., 2002; Goel and Lee, 2007; Teichert et al., 2017), but downregulation of synaptic strength during sleep is the only non-pathologic example that has been proposed (Diering et al., 2017). While the culture model is imperfect, we show for the first time a way to distinguish non-uniform from uniform homeostatic plasticity. The role of divergent and uniform homeostatic plasticity in normal nervous system function will be revealed once genetic models are identified that selectively lack one or the other.
Additional evidence for non-uniform scaling
There have been a few reports that directly demonstrate homeostatic plasticity leads to non-uniform scaling. In dissociated rat hippocampal neurons cultured for 21 d, homeostatic plasticity in response to increased or decreased network activity was restricted to the largest and most proximal synapses, from which arise the largest amplitude mEPSCs (Lee et al., 2013). In another study, GluA1 fluorescence increased significantly for large synapses but not for small synapses after blocking activity in dissociated mouse hippocampal cultures with NBQX (Thiagarajan et al., 2005). When changes in spine size in layer 5 cortical dendrites were followed for 48 h after monocular deprivation, spine size increased, but not in every dendrite; ∼50% showed no change (Barnes et al., 2017). The authors applied the iterative process of Kim et al. (2012) to scale the distribution of control mEPSC amplitudes to that of the monocularly-deprived neurons, and found that the fit was much improved when, based on their morphologic data, the authors selected a random subset of 50% of the mEPSCs to remain unscaled.
The increase in glutamate receptors following activity blockade is the most well-established molecular mechanism accompanying homeostatic plasticity of mEPSCs (O'Brien et al., 1998; Ju et al., 2004; Thiagarajan et al., 2005; Wierenga et al., 2005; Shepherd et al., 2006; Stellwagen and Malenka, 2006; Aoto et al., 2008; Hou et al., 2008; Ibata et al., 2008; Corrêa et al., 2012; Garcia-Bereguiain et al., 2013). However, only a few studies have examined how activity blockade affects the cumulative distribution of fluorescence intensity values (Wierenga et al., 2005; Hou et al., 2008; Ibata et al., 2008; Corrêa et al., 2012). In three out of four of these (Wierenga et al., 2005; Hou et al., 2008; Corrêa et al., 2012), a smaller effect of activity blockade on low intensity puncta, compared with high intensity puncta, can be discerned, consistent with our description of divergent scaling. For the fourth study, newly inserted receptor clusters with the lowest intensity values did not show an increase in intensity following activity blockade, but these insensitive clusters were excluded from the CDF analysis (Ibata et al., 2008). Together, these results suggest that postsynaptic receptor expression is differentially regulated at synapses with low versus high levels of receptors.
Biological implications
The power of the uniform multiplicative scaling hypothesis first proposed by Turrigiano and colleagues in 1998 is its mathematical simplicity, and its ability to explain how the rankings and the proportional relationships between synaptic strengths, established by other forms of activity-dependent plasticity, will be exactly as they were before (Turrigiano et al., 1998; Turrigiano and Nelson, 2004; Pozo and Goda, 2010; Watt and Desai, 2010; Turrigiano, 2012, 2017; Vitureira and Goda, 2013; Keck et al., 2017a,b; Yee et al., 2017). These characteristics of uniform multiplicative synaptic scaling have been readily incorporated into computational models of network behavior to stabilize synaptic weights in the face of Hebbian plasticity (Fröhlich et al., 2008; Tetzlaff et al., 2012; Zenke et al., 2013; Toyoizumi et al., 2014; Chistiakova et al., 2015; Effenberger et al., 2015; González et al., 2015; Zenke et al., 2017). It might seem that divergent scaling, in which weak synapses show the least increase in strength and strong synapses show the greatest increase, will lead to runaway growth of synaptic weights. However, the changes are still in the opposite direction of the overall network activity level, so addressing the strongest synapses may be the most efficient way to bring network activity back into balance. If such a mechanism holds true for the homeostatic response to excessive network activity (mimicked experimentally by treatment with bicuculline), the strongest synapses would have the most dramatic reduction, again, an efficient way to rapidly restore normal network behavior. Multiple studies suggest the effects of bicuculline are divergent: in the low range of mepscs, rank-order plots follow the line of identity (Qiu et al., 2012), and cumulative plots run close together (Turrigiano et al., 1998; Shepherd et al., 2006; Qiu et al., 2012; Xu and Pozzo-Miller, 2017). Furthermore, Turrigiano et al. (1998) report a non-zero intercept term (2.0) in the linear fit of rank-ordered bicuculline data. In future, computational models will need to examine the impact of divergent homeostatic plasticity on synaptic weights.
To our knowledge, no one has yet recorded mEPSCs at single synapses before and following activity blockade, and therefore the physiological behavior at this microscopic level eludes us. For example, we cannot determine whether high-activity synapses react differently than low activity synapses. Recently, fluorescently labeled receptors were imaged following a short period (4 h) of activity blockade; the majority of clusters showed an increase in fluorescence, but a small group showed a decrease (Wang et al., 2019). The smallest clusters showed larger increases, not smaller, than the overall average, but when all the data were pooled, the population behavior was identical to what we observe—the CDFs of receptor amount before and after treatment run closely together in the smallest values. Any computational model of homeostatic plasticity must explain how stochastic increases and decreases produce divergent (or uniform) plasticity at the population level (Statman et al., 2014; Shomar et al., 2017).
Footnotes
The authors declare no competing financial interests.
This work was supported by the National Institute of Neurological Disorders and Stroke (NINDS) Grant P01 NS057228 (to K.L.E.; project 4; Tim Cope, PI of PPG); the NINDS Grant R01NS065992 (to P.W.); Canadian Institutes of Health Research and the Natural Sciences and Engineering Research Council of Canada (D.S.); the National Science Foundation Graduate Research Fellowship GRFP 09-603 (to M.F.); and the Heart and Stroke Foundation, Canada (H.F.A.). We thank David Ladle and Mark Rich for comments on drafts of this manuscript, Steve Potter for providing access to data from rat cortical neurons, and Shuxia Sun for consultation on statistical matters.
References
- Altimimi HF, Stellwagen D (2013) Persistent synaptic scaling independent of AMPA receptor subunit composition. J Neurosci 33:11763–11767. 10.1523/JNEUROSCI.1102-13.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoto J, Nam CI, Poon MM, Ting P, Chen L (2008) Synaptic signaling by all-trans retinoic acid in homeostatic synaptic plasticity. Neuron 60:308–320. 10.1016/j.neuron.2008.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes SJ, Franzoni E, Jacobsen RI, Erdelyi F, Szabo G, Clopath C, Keller GB, Keck T (2017) Deprivation-induced homeostatic spine scaling in vivo is localized to dendritic branches that have undergone recent spine loss. Neuron 96:871.e5–882.e5. 10.1016/j.neuron.2017.09.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackman MP, Djukic B, Nelson SB, Turrigiano GG (2012) A critical and cell-autonomous role for MeCP2 in synaptic scaling up. J Neurosci 32:13529–13536. 10.1523/JNEUROSCI.3077-12.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bliss TV, Lomo T (1973) Long-lasting potentiation of synaptic transmission in the dentate area of the anaesthetized rabbit following stimulation of the perforant path. J Physiol 232:331–356. 10.1113/jphysiol.1973.sp010273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bliss TV, Collingridge GL (1993) A synaptic model of memory: long-term potentiation in the hippocampus. Nature 361:31–39. 10.1038/361031a0 [DOI] [PubMed] [Google Scholar]
- Chen L, Lau AG, Sarti F (2014) Synaptic retinoic acid signaling and homeostatic synaptic plasticity. Neuropharmacology 78:3–12. 10.1016/j.neuropharm.2012.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chistiakova M, Bannon NM, Chen JY, Bazhenov M, Volgushev M (2015) Homeostatic role of heterosynaptic plasticity: models and experiments. Front Comput Neurosci 9:89. 10.3389/fncom.2015.00089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cingolani LA, Goda Y (2008) Differential involvement of beta3 integrin in pre- and postsynaptic forms of adaptation to chronic activity deprivation. Neuron Glia Biol 4:179–187. 10.1017/S1740925X0999024X [DOI] [PubMed] [Google Scholar]
- Collingridge GL, Peineau S, Howland JG, Wang YT (2010) Long-term depression in the CNS. Nat Rev Neurosci 11:459–473. 10.1038/nrn2867 [DOI] [PubMed] [Google Scholar]
- Corrêa SA, Hunter CJ, Palygin O, Wauters SC, Martin KJ, McKenzie C, McKelvey K, Morris RG, Pankratov Y, Arthur JS, Frenguelli BG (2012) MSK1 regulates homeostatic and experience-dependent synaptic plasticity. J Neurosci 32:13039–13051. 10.1523/JNEUROSCI.0930-12.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai NS, Cudmore RH, Nelson SB, Turrigiano GG (2002) Critical periods for experience-dependent synaptic scaling in visual cortex. Nat Neurosci 5:783–789. 10.1038/nn878 [DOI] [PubMed] [Google Scholar]
- Diering GH, Nirujogi RS, Roth RH, Worley PF, Pandey A, Huganir RL (2017) Homer1a drives homeostatic scaling-down of excitatory synapses during sleep. Science 355:511–515. 10.1126/science.aai8355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echegoyen J, Neu A, Graber KD, Soltesz I (2007) Homeostatic plasticity studied using in vivo hippocampal activity-blockade: synaptic scaling, intrinsic plasticity and age-dependence. PLoS One 2:e700. 10.1371/journal.pone.0000700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Effenberger F, Jost J, Levina A (2015) Self-organization in balanced state networks by STDP and homeostatic plasticity. PLoS Comput Biol 11:e1004420. 10.1371/journal.pcbi.1004420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong MF, Newman JP, Potter SM, Wenner P (2015) Upward synaptic scaling is dependent on neurotransmission rather than spiking. Nat Commun 6:6339. 10.1038/ncomms7339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fröhlich F, Bazhenov M, Sejnowski TJ (2008) Pathological effect of homeostatic synaptic scaling on network dynamics in diseases of the cortex. J Neurosci 28:1709–1720. 10.1523/JNEUROSCI.4263-07.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Bereguiain MA, Gonzalez-Islas C, Lindsly C, Butler E, Hill AW, Wenner P (2013) In vivo synaptic scaling is mediated by GluA2-lacking AMPA receptors in the embryonic spinal cord. J Neurosci 33:6791–6799. 10.1523/JNEUROSCI.4025-12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerkin RC, Nauen DW, Xu F, Bi GQ (2013) Homeostatic regulation of spontaneous and evoked synaptic transmission in two steps. Mol Brain 6:38. 10.1186/1756-6606-6-38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goel A, Lee HK (2007) Persistence of experience-induced homeostatic synaptic plasticity through adulthood in superficial layers of mouse visual cortex. J Neurosci 27:6692–6700. 10.1523/JNEUROSCI.5038-06.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González OC, Krishnan GP, Chauvette S, Timofeev I, Sejnowski T, Bazhenov M (2015) Modeling of age-dependent epileptogenesis by differential homeostatic synaptic scaling. J Neurosci 35:13448–13462. 10.1523/JNEUROSCI.5038-14.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Q, Zhang D, Jarzylo L, Huganir RL, Man HY (2008) Homeostatic regulation of AMPA receptor expression at single hippocampal synapses. Proc Natl Acad Sci USA 105:775–780. 10.1073/pnas.0706447105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibata K, Sun Q, Turrigiano GG (2008) Rapid synaptic scaling induced by changes in postsynaptic firing. Neuron 57:819–826. 10.1016/j.neuron.2008.02.031 [DOI] [PubMed] [Google Scholar]
- Ju W, Morishita W, Tsui J, Gaietta G, Deerinck TJ, Adams SR, Garner CC, Tsien RY, Ellisman MH, Malenka RC (2004) Activity-dependent regulation of dendritic synthesis and trafficking of AMPA receptors. Nat Neurosci 7:244–253. 10.1038/nn1189 [DOI] [PubMed] [Google Scholar]
- Kapfhamer D, Valladares O, Sun Y, Nolan PM, Rux JJ, Arnold SE, Veasey SC, Bućan M (2002) Mutations in Rab3a alter circadian period and homeostatic response to sleep loss in the mouse. Nat Genet 32:290–295. 10.1038/ng991 [DOI] [PubMed] [Google Scholar]
- Keck T, Keller GB, Jacobsen RI, Eysel UT, Bonhoeffer T, Hübener M (2013) Synaptic scaling and homeostatic plasticity in the mouse visual cortex in vivo. Neuron 80:327–334. 10.1016/j.neuron.2013.08.018 [DOI] [PubMed] [Google Scholar]
- Keck T, Hübener M, Bonhoeffer T (2017a) Interactions between synaptic homeostatic mechanisms: an attempt to reconcile BCM theory, synaptic scaling, and changing excitation/inhibition balance. Curr Opin Neurobiol 43:87–93. 10.1016/j.conb.2017.02.003 [DOI] [PubMed] [Google Scholar]
- Keck T, Toyoizumi T, Chen L, Doiron B, Feldman DE, Fox K, Gerstner W, Haydon PG, Hubener M, Lee HK, Lisman JE, Rose T, Sengpiel F, Stellwagen D, Stryker MP, Turrigiano GG, van Rossum MC (2017b) Integrating Hebbian and homeostatic plasticity: the current state of the field and future research directions. Philos Trans R Soc Lond B Biol Sci 372:20160158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Tsien RW, Alger BE (2012) An improved test for detecting multiplicative homeostatic synaptic scaling. PLoS One 7:e37364. 10.1371/journal.pone.0037364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KF, Soares C, Béïque JC (2014) Tuning into diversity of homeostatic synaptic plasticity. Neuropharmacology 78:31–37. 10.1016/j.neuropharm.2013.03.016 [DOI] [PubMed] [Google Scholar]
- Lee KJ, Queenan BN, Rozeboom AM, Bellmore R, Lim ST, Vicini S, Pak DT (2013) Mossy fiber-CA3 synapses mediate homeostatic plasticity in mature hippocampal neurons. Neuron 77:99–114. 10.1016/j.neuron.2012.10.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malenka RC, Bear MF (2004) LTP and LTD: an embarrassment of riches. Neuron 44:5–21. 10.1016/j.neuron.2004.09.012 [DOI] [PubMed] [Google Scholar]
- Mood AM, Graybill FA, Boes DC (1973) Introduction to the theory of statistics, Ed 3 New York: McGraw-Hill. [Google Scholar]
- O'Brien RJ, Kamboj S, Ehlers MD, Rosen KR, Fischbach GD, Huganir RL (1998) Activity-dependent modulation of synaptic AMPA receptor accumulation. Neuron 21:1067–1078. 10.1016/S0896-6273(00)80624-8 [DOI] [PubMed] [Google Scholar]
- Pozo K, Goda Y (2010) Unraveling mechanisms of homeostatic synaptic plasticity. Neuron 66:337–351. 10.1016/j.neuron.2010.04.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Z, Sylwestrak EL, Lieberman DN, Zhang Y, Liu XY, Ghosh A (2012) The Rett syndrome protein MeCP2 regulates synaptic scaling. J Neurosci 32:989–994. 10.1523/JNEUROSCI.0175-11.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghavachari M. (1973) Limiting distributions of Kolmogorov-Smirnov type statistics under alternative. Ann Statist 1:67–73. 10.1214/aos/1193342382 [DOI] [Google Scholar]
- Shepherd JD, Rumbaugh G, Wu J, Chowdhury S, Plath N, Kuhl D, Huganir RL, Worley PF (2006) Arc/Arg3.1 mediates homeostatic synaptic scaling of AMPA receptors. Neuron 52:475–484. 10.1016/j.neuron.2006.08.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shomar A, Geyrhofer L, Ziv NE, Brenner N (2017) Cooperative stochastic binding and unbinding explain synaptic size dynamics and statistics. PLoS Comput Biol 13:e1005668. 10.1371/journal.pcbi.1005668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Statman A, Kaufman M, Minerbi A, Ziv NE, Brenner N (2014) Synaptic size dynamics as an effectively stochastic process. PLoS Comput Biol 10:e1003846. 10.1371/journal.pcbi.1003846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stellwagen D, Malenka RC (2006) Synaptic scaling mediated by glial TNF-alpha. Nature 440:1054–1059. 10.1038/nature04671 [DOI] [PubMed] [Google Scholar]
- Teichert M, Liebmann L, Hübner CA, Bolz J (2017) Homeostatic plasticity and synaptic scaling in the adult mouse auditory cortex. Sci Rep 7:17423. 10.1038/s41598-017-17711-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tetzlaff C, Kolodziejski C, Timme M, Wörgötter F (2012) Analysis of synaptic scaling in combination with Hebbian plasticity in several simple networks. Front Comput Neurosci 6:36. 10.3389/fncom.2012.00036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiagarajan TC, Lindskog M, Tsien RW (2005) Adaptation to synaptic inactivity in hippocampal neurons. Neuron 47:725–737. 10.1016/j.neuron.2005.06.037 [DOI] [PubMed] [Google Scholar]
- Toyoizumi T, Kaneko M, Stryker MP, Miller KD (2014) Modeling the dynamic interaction of Hebbian and homeostatic plasticity. Neuron 84:497–510. 10.1016/j.neuron.2014.09.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turrigiano GG. (2008) The self-tuning neuron: synaptic scaling of excitatory synapses. Cell 135:422–435. 10.1016/j.cell.2008.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turrigiano G. (2012) Homeostatic synaptic plasticity: local and global mechanisms for stabilizing neuronal function. Cold Spring Harb Perspect Biol 4:a005736. 10.1101/cshperspect.a005736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turrigiano GG. (2017) The dialectic of Hebb and homeostasis. Philos Trans R Soc Lond B Biol Sci 372:20160258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turrigiano GG, Nelson SB (2004) Homeostatic plasticity in the developing nervous system. Nat Rev Neurosci 5:97–107. 10.1038/nrn1327 [DOI] [PubMed] [Google Scholar]
- Turrigiano GG, Leslie KR, Desai NS, Rutherford LC, Nelson SB (1998) Activity-dependent scaling of quantal amplitude in neocortical neurons. Nature 391:892–896. 10.1038/36103 [DOI] [PubMed] [Google Scholar]
- Tygert M. (2010) Statistical tests for whether a given set of independent, identically distributed draws comes from a specified probability density. Proc Natl Acad Sci USA 107:16471–16476. 10.1073/pnas.1008446107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitureira N, Goda Y (2013) Cell biology in neuroscience: the interplay between Hebbian and homeostatic synaptic plasticity. J Cell Biol 203:175–186. 10.1083/jcb.201306030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Zhong J, Guttieres D, Man HY (2019) Non-scaling regulation of AMPA receptors in homeostatic synaptic plasticity. Neuropharmacology 158:107700. 10.1016/j.neuropharm.2019.107700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt AJ, Desai NS (2010) Homeostatic plasticity and STDP: keeping a neuron's cool in a fluctuating world. Front Synaptic Neurosci 2:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierenga CJ, Ibata K, Turrigiano GG (2005) Postsynaptic expression of homeostatic plasticity at neocortical synapses. J Neurosci 25:2895–2905. 10.1523/JNEUROSCI.5217-04.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Pozzo-Miller L (2017) EEA1 restores homeostatic synaptic plasticity in hippocampal neurons from Rett syndrome mice. J Physiol 595:5699–5712. 10.1113/JP274450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yee AX, Hsu YT, Chen L (2017) A metaplasticity view of the interaction between homeostatic and Hebbian plasticity. Philos Trans R Soc Lond B Biol Sci 372:20160155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenke F, Gerstner W (2017) Hebbian plasticity requires compensatory processes on multiple timescales. Philos Trans R Soc Lond B Biol Sci 372:20160259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenke F, Hennequin G, Gerstner W (2013) Synaptic plasticity in neural networks needs homeostasis with a fast rate detector. PLoS Comput Biol 9:e1003330. 10.1371/journal.pcbi.1003330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenke F, Gerstner W, Ganguli S (2017) The temporal paradox of Hebbian learning and homeostatic plasticity. Curr Opin Neurobiol 43:166–176. 10.1016/j.conb.2017.03.015 [DOI] [PubMed] [Google Scholar]