Abstract
The solute carrier 2 family is composed of 14 transporters, which are members of the major facilitator superfamily. Despite their high physiological importance, there are still many open questions concerning their function and specificity, and in some cases, their physiological substrate is still unknown. To understand the determinants of the substrate and inhibitor specificity, we modeled all human glucose transport carriers (GLUTs) and simulated their interaction with known ligands. Comparative modeling was performed with the @TOME-2 pipeline, employing multiple templates and providing an ensemble of models for each GLUT. We analyzed models in both outward-occluded and inward-open conformations, to compare exofacial and endofacial binding sites throughout the family and understand differences in susceptibility of GLUTs to the inhibitor cytochalasin B. Finally, we employed molecular docking and bioinformatics to identify residues likely critical for recognition of myo-inositol by GLUT13 and urate by GLUT9. These results provide insights into the molecular basis for the specificity for these substrates. In addition, we suggested a potential recognition site of glucosamine by GLUT11 to be evaluated in future experiments.
Introduction
Glucose transport carrier (GLUT) transporters, described initially as glucose transporters, are essential proteins in homeostasis. In humans, there are 14 identified GLUT transporters, which compose the solute carrier (SLC)2 family and are members of the major facilitator superfamily. These proteins are composed of approximately 500 residues and 12 transmembrane helices. Their overall structure is characterized by two domains, containing six helices each, connected by an intracellular helical (ICH) domain.1
On the basis of the sequence identity, they are categorized into three classes: Class 1 (GLUT1–4, 14); Class 2 (GLUT5, 7, 9, and 11); and Class 3 (GLUT6, 8, 10, 12, and 13, alias Proton myo-inositol cotransporter proton myo-inositol cotransporter or proton myo-inositol cotransporter (MYCT)).2 They differ in their expression patterns and substrate specificity. Despite their high physiological importance, there are still many open questions concerning their function. Most GLUTs can transport glucose, but in some cases their physiological substrate is unknown. Additionally, some transporters recognize a wider range of substrates or present unique specificities.2 They are also important for other mechanisms, such as mediation of viral entry into cells, but those most likely involve outside regions of the receptors and fall beyond the scope of this study.3
Abnormal GLUT expression levels or mutant variants are related to several diseases, such as GLUT1 deficiency syndromes,4 renal hypouricemia,5,6 arterial tortuosity syndrome,7 or cancer.8 In the latter case, there is a clear interest in developing new GLUT inhibitors. However, it will be crucial to address and assess the specificity of such inhibitors.
Structural information should be of great value to understand both the determinants of the substrate and inhibitor specificity. Detailed examination of GLUT binding sites should help the development of new inhibitors that will specifically target defined member(s) of the GLUT/SLC2 family. Toward this goal, inhibitors selective for either GLUT19,10 or GLUT411,12 have been described as candidates for cancer treatment. A better understanding of molecular recognition within this family is also useful for other applications, such as the design of GLUT1 and GLUT5 ligands as molecular probes for breast cancer diagnostics.13−17
Although experimental difficulties have hampered determination of GLUT crystal structures, as generally observed for membrane proteins, significant progress has been achieved recently, particularly among Class 1 transporters. Thus, structure resolution has been reported for a close Escherichia coli homologue18 and human GLUT119,20 and GLUT3.21 The structures of rat and bovine GLUT5, a Class 2 transporter, have also been determined.22 Interestingly, these include complexes with sugars (glucose and maltose21) and with inhibitors, such as cytochalasin B (cytoB).20 Interestingly, the elucidated structures represent different transporter conformations, including outward-open, outward-occluded, and inward-open structures. Molecular dynamics simulation provide some more insights into the conformational changes associated with ligand binding and transport.23−25 All of these results provide a clearer picture of the mechanisms that govern substrate transport, on the basis of alternating accessibility of substrate-binding sites, from the extracellular (ECL) and intracellular (ICL) sides of the membrane.1
Thanks to these recent findings, it is now conceivable to build comparative models of all GLUT family members, as they harbor between 24 and 95% sequence identity to known crystal structures. Therefore, with the aim to improve our understanding of substrate and inhibitor recognition by GLUTs, we modeled all human GLUTs and simulated their interaction with ligands. For Class 1 GLUTs, modeling and comparative docking is straightforward and the functioning is highly similar according to the high overall sequence identity. Here, we focused our analysis mostly on Class 2 and 3 transporters, which have lower sequence identity with the available templates and have been much less studied than Class 1 GLUTs. To the best of our knowledge, this is the first time that models have been proposed for the whole GLUT family. This structural information, combined with bioinformatic techniques, allowed us to study ligand selectivity of GLUT family members that have major physiological functions.
Results
Comparative Modeling
We built comparative models for all GLUT family members through the @TOME-2 pipeline.26 The automated @TOME-2 procedure allowed us to use templates crystallized in apo or ligand-bound forms and in various conformations: outward-facing partially occluded, outward open, outward occluded, and inward-open (Table 1). For each GLUT, up to 20 models were selected based on the top ranking structural alignments for each sequence.
Table 1. Templates Used with @TOME-2 for Comparative Modeling of GLUT Family Members.
| PDB code | resolution (Å) | conformation | ligand | protein | references |
|---|---|---|---|---|---|
| 4GC0 | 2.6 | outward-facing, partially occluded | 6-bromo-6-deoxy-d-glucose | XylE E. coli | (18) |
| 4GBY | 2.8 | d-xylose | |||
| 4GBZ | 2.9 | d-glucose | |||
| 4ZW9 | 1.5 | outward-occluded | d-glucose | GLUT3 Homo sapiens | (21) |
| 4ZWC | 2.6 | outward-open | maltose | ||
| 4ZWB | 2.4 | outward-occluded | maltose | ||
| 4PYP | 3.2 | inward-open | apo | GLUT1 H. sapiens | (19) |
| 5EQG | 2.9 | inward-open | phenylamide 1 | GLUT1 H. sapiens | (20) |
| 5EQH | 3.0 | phenylamide 2 | |||
| 5EQI | 3.0 | cytochalasin B |
For several GLUTs (e.g., Class 1 GLUTs), there is enough sequence identity to allow accurate automated sequence alignment and comparative modeling. However, GLUT6 and 8 and 10–13 share a lower sequence identity (<35%) with the available templates. To improve the quality of models generated for these proteins, a manual process was also employed to generate additional models. First, templates for comparative modeling were selected on the basis of their resolution, structure quality, protein conformation, and cocrystallization in the presence of ligands. Binding sites undergo important changes in the transition from the outward to inward conformation, and some inhibitors are known to bind specifically only to a given conformation. Therefore, we built models both in the inward-open and outward-occluded conformations, respectively, based on templates of GLUT1 in complex with an inhibitor, cytoB (PDB 5EQI,20 3.0 Å resolution), and GLUT3 in complex with glucose (PDB 4ZW9,21 1.5 Å resolution). These models were validated based on Tito,27 Q-Mean,28 and Verify 3D29 scores, within the @TOME-2 pipeline.
Overall Structure
We were able to align to templates the 12 transmembrane (TM) regions, for all transporters of the GLUT family (Supporting Information Figure S1) and therefore able to build corresponding models. We also modeled short extracellular (ECL) and intracellular (ICL) loops, whereas long loops (ECL1 connecting TM1 and TM2 in Class 1 and Class 2 GLUTs and ECL5 connecting TM9 and TM10 in Class 3) were not included. Similarly, GLUT8 and GLUT10 contain a significant deletion in ICL3, between TM6 and TM7, the region which forms most of the ICH domain. Whereas this region is composed of 62 residues in GLUT1 (residues 207–268), it contains 52 residues in GLUT8 (202–253) and 43 residues in GLUT10 (185–228). Due to the lack of reliable alignment in this region and the fact that the deletion does not allow conservation of the secondary structure elements observed in GLUT1 and GLUT3 crystal structures, this region was not modeled either.
We based the validation of our models on the structural superposition of highly conserved residues, such as those involved in molecular gates21 and substrate-binding sites. The 11 residues previously described as necessary for molecular gates (G91, R92, E145, R153, G154, E(D)329, G332, R333, R(K)334, E393, and R400, following GLUT1 numbering) are found in all GLUTs and were structurally aligned in all models.
A further validation of our models consisted in the mapping of disease-related mutations (DRMs). Several DRMs have been described for the SLC2 family, including GLUT1, 2, 4, 9, and 10. Previous studies revealed that these DRMs form three clusters in GLUT1, one around the substrate-binding site and two in the regions of the extracellular and intracellular gates.19 We focused our analysis on GLUT9, for Class 2 GLUTs, and GLUT10, for Class 3 GLUTs, which show 34 and 24% sequence identity with the template used for building their models (PDB 4ZW9, Supporting Information Table S1), respectively.
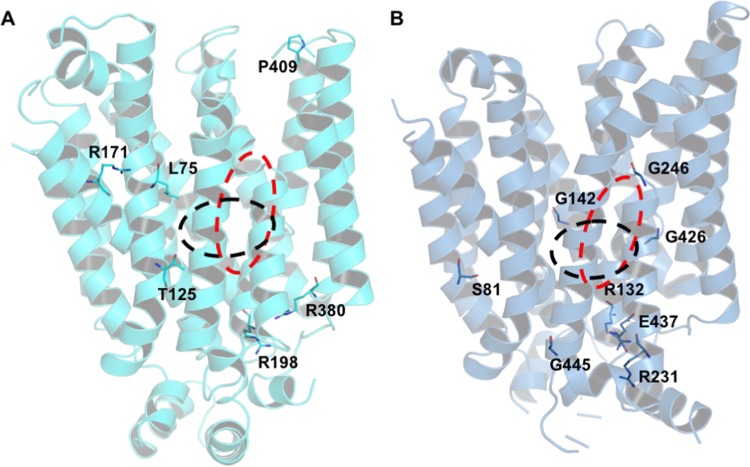
Five mutations in GLUT9 have been related to renal hypouricemia 2,5,6 and eight mutations in GLUT10, to arterial tortuosity syndrome (Figure 1, Table 2). Among these DRMs, only L75 and T125 in GLUT9 are found within the substrate-binding sites, as predicted by comparative docking (defined as all residues within 8 Å from ligand atoms), with L75 being in direct contact with the bound ligand, whereas T125 lies in a second shell surrounding the binding site. Interestingly, there is evidence for the functional or structural importance of most other DRMs of these transporters, as also illustrated by our modeling. Analysis of the environment surrounding these residues in our models unveiled their role in stabilization of the protein structure, as the predicted interactions with these residues could not be reproduced by the corresponding mutated residues. This can be exemplified by mutations that substituted neutral residues for wild-type positively charged residues, as seen in the R171C, R198C, and R380M changes in GLUT9 and in R132W in GLUT10. The corresponding arginines are either involved in salt bridges or hydrogen network, which is important for protein stability. Similarly, mutation E437K in GLUT10 eliminated the favorable salt bridge established with R296, resulting in charge repulsion in the corresponding DRM. In the case of glycines, we also observed that G246 and G426 in GLUT10 adopt backbone conformations not allowed to most other residues (Table 2). Similarly, mutation P412R eliminates the stabilizing N-capping of TM10 provided by the constrained proline present in wild-type GLUT9.
Figure 1.
Disease-related mutations mapped to GLUT9 (A) and GLUT10 (B) models. Approximate locations of endofacial and exofacial binding sites are indicated with black and red dashed lines, respectively.
Table 2. Analysis of Disease-Related Mutations in GLUT9 and GLUT10 Models.
| protein | mutation | location in protein | structural observations from model | reported information for aligned residuesa |
|---|---|---|---|---|
| GLUT9 | L75R | TM1 | direct contact with urate | no information found |
| T125M | TM2, near TM11 | buried, second shell from binding site | model by Dinour indicates residue towards channel | |
| R171C | TM4 | stabilizes local distortion in TM1 and TM2; H-bond to N74 backbone carbonyl in TM1 | R126 mutations in GLUT1 are related to GLUT1DS1; R133C, R113H, and R133L in XylE abolished xylose transport; model by Dinour indicated residues toward the channel | |
| R198C | TM6, near ICH domain | involved in salt bridge with E376 and H-bond with R505 backbone carbonyl | gate residue R153 in GLUT1; R153H in GLUT1 is related to GLUT1DS2; R160L in E.coli XylE failed to yield well-behaved recombinant protein | |
| R380W, R380M | TM9, near ICH domain | buried residue interacting with TM8 and TM10, through salt bridges with E376 and E440, and H-bond with I375 | gate residue R333 in GLUT1; R333Q in GLUT1 is related to GLUT1DS2; R341W in XylE abolished xylose transport | |
| P412R | TM10 | N-capping residue in TM10, conserved in GLUT1 (P362) | no information found | |
| GLUT10 | S81R | TM3 | surrounded by hydrophobic environment | no information found |
| R132W | TM6, near ICH domain | involved in salt bridge with E437 and H-bond with Q503 backbone carbonyl | see observations for GLUT9 R198C (equivalent residues) | |
| G142V | TM6 | close contact with TM1 (L20) | 100% conserved in GLUTs | |
| R231Q | TM7, near ICH domain | solvent exposed residue | no information found | |
| G246E | kink between TM7a/b | αL ϕ/Ψ angles; located close to binding sites | 100% conserved in GLUTs; involved in conformation change during transport | |
| G426W | kink between TM10a/b | ϕ/Ψ angles approximately (−120°, −90°); buried; located close to binding sites | 100% conserved in GLUTs; involved in conformation change during transport | |
| E437K | TM10, near ICH domain | salt bridge with R296 | gate residue E393 in GLUT1 | |
| G445E | TM11, near ICH domain | helical conformation | no information found |
Further evidence for the importance of these residues was found in the literature. Several of them map either to positions also described as DRM in GLUT1 (leading to GLUT1 deficiency syndrome 2—GLUT1DS2) or as gate residues in GLUT3.21 Site-directed mutagenesis in equivalent positions in XylE have also been shown to abolish xylose transport.18 For GLUT10, several mutations altering the presence of glycines that are 100% conserved in the family have been observed as DRM, including G142V in TM6, and G246E and G426W, which are located in the kinks of the discontinuous TM7 and TM10 helices. These glycines are located in the vicinity of the binding sites and are important in allowing the conformational changes of those regions that shift the most during gating movements that lead to substrate transport.21,22
Obtaining models for all GLUTs also provided an opportunity to compare the two binding sites (endofacial and exofacial ones) for each receptor, which are alternatively open during the transport cycle. A detailed discussion is presented below.
Endofacial Binding Site Comparison
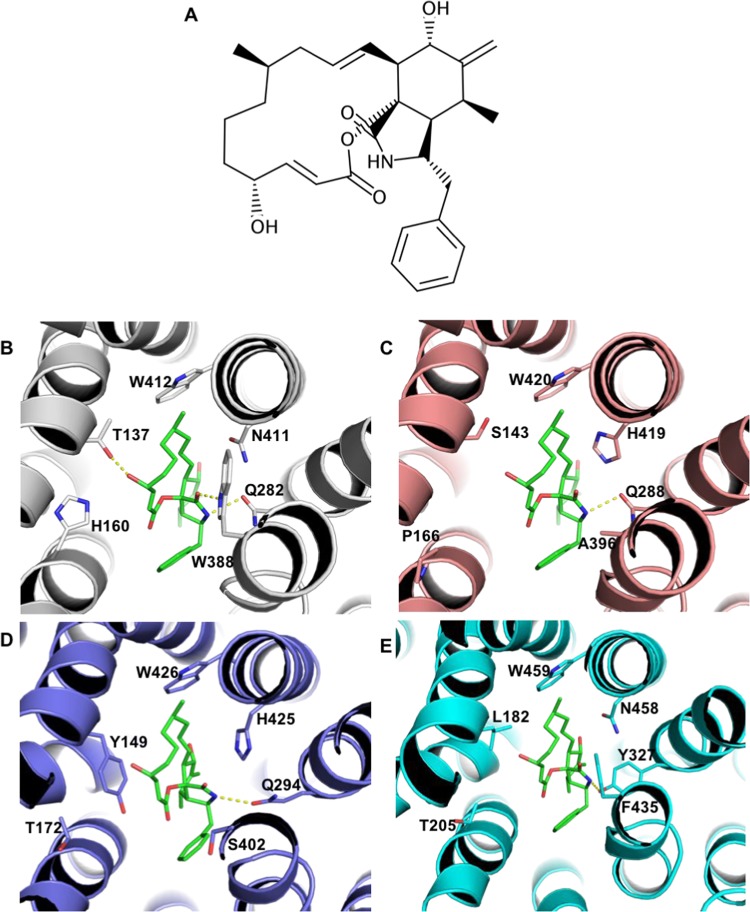
To compare endofacial binding sites throughout GLUT1–14, we superposed models in the inward-open conformation, built on the basis of GLUT1 templates (PDB 5EQG, 5EQH, or 5EQI). This conformation is known to bind to cytoB, a compound which inhibits several GLUTs, such as GLUT1, 3, 4, 8, and 13. On the other hand, other GLUTs such as the Class 2 transporters GLUT5, 7, and 9 are not inhibited by cytoB. Previously reported site-directed mutagenesis studies revealed that W388L and W412L impact GLUT1 photolabeling and reduced cytoB binding and are therefore important for molecular recognition of this compound.31 More recently, determination of the complex structure of GLUT1 bound to cytoB (PDB 5EQI) provided an understanding of the key residues involved in recognition and direct insights on the exclusive binding of the ligand to the inward-open conformation. Superposition of our inward-open conformation models to PDB 5EQI revealed several substitutions in positions surrounding cytoB (Figure 2). More specifically, residues Q282, W388, N411, and W412 are highly conserved among the GLUTs that are inhibited by cytoB (Table 3), whereas substitutions in at least one of these positions are observed for all receptors that are not inhibited (GLUT5, 7, and 9). In the complex with GLUT1, residues Q282, W388, and N411 are directly involved in interactions with the ligand. To investigate possible interactions between cytoB and GLUT5, 7, and 9, we performed comparative docking, transferring the ligand to each receptor on the basis of its binding mode to GLUT1. Docking results (Figure 2C–E) indicate that several of the interactions observed in the GLUT1–cytoB complex cannot be formed in the complexes with Class 2 transporters. These results suggest that in all Class 2 transporters exist substitutions expected to be detrimental to cytoB binding. Moreover, in the case of GLUT7, Y149 substitutes for a small residue (threonine or serine in the transporters inhibited by cytoB). The most favorable Y149 rotamers would clash with either ligand or with other binding site residues, and this residue would have to adopt an outlier rotamer to accommodate cytoB. Therefore, particularly in the case of GLUT7, we identified two features that were detrimental to inhibition by cytoB: substitution for residues that are essential for ligand recognition (corresponding to W388 and N411 in GLUT1) or presence of residues that would sterically hinder ligand binding, such as Y149.
Figure 2.
Molecular basis for cytochalasin B selectivity within the GLUT family. (A) Cytochalasin B (cytoB) chemical structure. (B) Binding mode of cytoB to GLUT1 (PDB 5EQI). (C–E) Comparative docking to GLUT5, 7, and 9, respectively, highlighting residue substitutions. Hydrogen bonds (distance cutoff = 3.5 Å) are shown as yellow dashed lines.
Table 3. Substitutions Observed in Selected Residues in the Cytochalasin B Binding Site.
| inhibition by cytoB | transporter | residue | |||||
|---|---|---|---|---|---|---|---|
| yes | GLUT1 | T137 | H160 | Q282 | W388 | N411 | W412 |
| GLUT3 | T135 | N158 | Q280 | W386 | N409 | W410 | |
| GLUT4 | S153 | N176 | Q298 | W404 | N427 | W428 | |
| GLUT8 | S138 | V161 | Q267 | W394 | N417 | W418 | |
| GLUT13 | S189 | N212 | Q336 | W529 | N552 | W553 | |
| GLUT14 | T159 | N182 | Q304 | W410 | N433 | W434 | |
| no | GLUT5 | S143 | P166 | Q288 | A396 | H419 | W420 |
| GLUT7 | Y149 | T172 | Q294 | S402 | H425 | W426 | |
| GLUT9 | L182 | T205 | Y327 | F435 | N458 | W459 | |
Exofacial Binding Site Comparison
To compare models in the outward-occluded conformation, we superposed the best models built based on GLUT3 bound to glucose as template (PDB 4ZW9). This superposition allowed us to compare the exofacial glucose binding sites throughout the family members. We examined the conservation of the seven residues found to hydrogen-bond to glucose (Table 4). Within the five Class 1 GLUTs, all of these residues were strictly conserved, whereas for all other GLUTs, at least two substitutions were observed at these positions. The most conserved residues are (using GLUT3 numbering) Q280, Q281, N286, and W386, all of which are present in at least 10 family members (conservation >70%). On the other hand, position E378 was conserved only in Class 1 transporters.
Table 4. Conservation throughout the GLUT Family of Selected Residues in the Glucose Binding Site.
| residues
that H-bond to glucose in PDB 4ZW9 |
||||||||
|---|---|---|---|---|---|---|---|---|
| protein | # conserved residues (out of 7) | Q159 | Q280 | Q281 | N286 | N315 | E378 | W386 |
| GLUT1 | 7 | Q161 | Q282 | Q283 | N288 | N317 | E380 | W388 |
| GLUT2 | 7 | Q193 | Q314 | Q315 | N320 | N349 | E412 | W420 |
| GLUT3 | 7 | Q159 | Q280 | Q281 | N286 | N315 | E378 | W386 |
| GLUT4 | 7 | Q177 | Q298 | Q299 | N304 | N333 | E396 | W404 |
| GLUT14 | 7 | Q183 | Q304 | Q305 | N310 | N339 | E402 | W410 |
| GLUT5 | 5 | Q167 | Q288 | Q289 | N294 | N325 | A380 | A396 |
| GLUT7 | 4 | E173 | Q294 | Q295 | N300 | N331 | S394 | S402 |
| GLUT9 | 2 | A206 | Y327 | Q328 | N333 | E364 | C427 | F435 |
| GLUT11 | 0 | A163 | M284 | E285 | D290 | E321 | G384 | G392 |
| GLUT6 | 4 | Q174 | Q286 | Q287 | T292 | R322 | A410 | W418 |
| GLUT8 | 5 | Q162 | Q267 | Q268 | N273 | Q302 | A386 | W394 |
| GLUT10 | 3 | E140 | Q242 | Q243 | P248 | K280 | S424 | W432 |
| GLUT12 | 2 | E172 | V289 | Q290 | P295 | K327 | S476 | W484 |
| GLUT13 | 5 | T213 | Q336 | Q337 | N342 | N374 | A521 | W529 |
| residue conservation | 8 | 11 | 13 | 10 | 8 | 5 | 10 | |
| % conservation | 57 | 79 | 93 | 71 | 57 | 36 | 71 | |
Notably, we observed conservation of only three or fewer of the residues analyzed in the four receptors. This was the case for GLUT9–12, all of which have been reported to transport glucose (Supporting Information Table S2). Receptors GLUT9 and GLUT11, which showed the lowest exofacial site conservation, as well as GLUT13, which presents distinctive particularities, will be discussed in more detail in separate sections. The remaining two, GLUT10 and GLUT12, are known to transport glucose and galactose, which is rather intriguing with regard to their low conservation in the glucose binding site. To verify if, in each GLUT, the residues highlighted in Table 4 are likely functionally important, we first analyzed their conservation among the 250 closest orthologues (as well as some homologues) (Supporting Information Tables S3 and S4). Overall, these residues were highly conserved in homologous sequences: all residues had percentages of conservation higher than 97%, with the exception of W484 in GLUT12, which is still 88% conserved. Among the former are two charged residues E140/E172 and K280/K327 (in GLUT10/12) that are in a rather close vicinity and might form a buried salt bridge. They can still interact favorably with a polar compound such as glucose. A more pronounced variation comes from the mutation of an asparagine common in other GLUTS but herein replaced by proline (P248/P295). Interestingly, the asparagine is positioned near a kink at the beginning of a helical segment. Accordingly, the proline should not destabilize the local backbone conformation but its hydrophobic side chain will disallow any hydrogen bond. We then asked whether GLUT10 and GLUT12 could recognize and stabilize glucose, despite the differences observed in the putative binding site. Analysis of the best scoring binding modes predicted for glucose to these receptors revealed, in both cases, that two of the hydroxyls could still be involved in hydrogen bonds. In GLUT10, the glucose C2 hydroxyl is predicted to interact with Q242 and Q243, whereas the C4 hydroxyl would H-bond to E140 and W432. In the case of GLUT12, glucose C2 and C4 hydroxyls are predicted to hydrogen-bond to Q290 and N507. These results indicated partial glucose stabilization, but this would be significantly less than that observed in Class 1 receptors.
Specificity of Myo-inositol Transport by GLUT13
GLUT13 is an atypical SLC2 transporter: it is the only H+-coupled transporter in the family, and it is also characterized by its unique specificity as the only documented myo-inositol transporter.1,32 Like other members of the Class 3 GLUTs, GLUT13 has low sequence identity (between 26 and 31%) to Class 1 transporters. However, high residue conservation was observed within the exofacial binding sites (Table 4). This provides an interesting case of specificity, which we sought to better understand with our models.
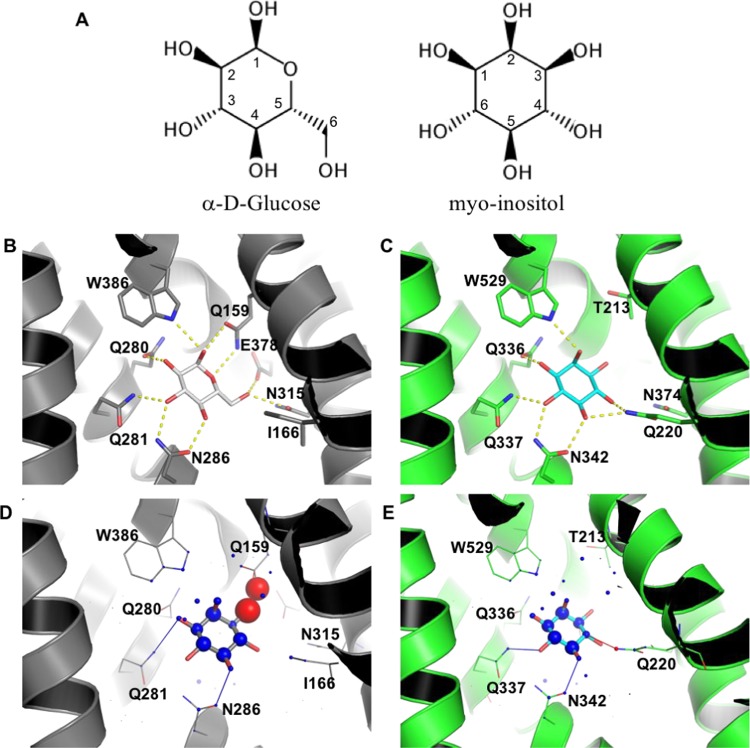
Myo-inositol is a small polyhydroxylated cyclic compound that differs from the carbohydrate glucose in two positions: the ring oxygen is replaced by a carbon attached to a hydroxyl group; the hydroxymethyl at C5 is also replaced by a hydroxyl (Figure 3A). Despite the high similarity between these molecules, glucose is transported by most GLUT receptors, whereas only GLUT13 transports myo-inositol. To understand the molecular bases for this specificity, we conducted docking studies and binding site comparisons. Before docking, reference complexes were assembled in @TOME-2 by transferring ligands from templates to the models built. This allowed calculation of protein–ligand interaction profiles and subsequent application of pharmacophoric restraints when docking with PLANTS,33,34 while favoring poses similar to the position and interaction profile of anchor compounds. We took into account both global and local protein flexibility during docking, by employing models in several conformations and allowing side-chain flexibility.
Figure 3.
Molecular bases for the specific binding of myo-inositol to GLUT13. (A) Chemical structures of α-d-glucose and myo-inositol. (B) Glucose binding mode to GLUT3 (PDB 4ZW9). (C) myo-inositol binding mode to GLUT13; prediction based on docking with PLANTS. Highlighted residues (shown as sticks) are predicted to be important for ligand binding, either by their involvement in hydrogen bonds or through improvement of sterical complementarity. Hydrogen bonds (distance cutoff = 3.5 Å) are shown as yellow dashed lines. Visualization of myo-inositol docking to GLUT3 (D) and GLUT13 (E) with DSX online. Favorable and unfavorable potentials are shown as blue and red spheres, respectively. Favorable and unfavorable distances are shown as blue and red lines, respectively.
The best scoring binding mode of myo-inositol to GLUT13 was obtained against a model in the outward-open conformation (template PDB 4ZW9), with glucose as the anchor. Remarkably, this was the best scoring pose according to all scoring functions (PLANTS, Medusa, XScore, and DSX). This binding mode is highly similar to the one described for glucose against GLUT3, with most hydroxyls stabilized by hydrogen-bond interactions to the protein (Figure 3B,C). These include residues that are highly conserved in GLUTs (present in at least 10 receptors), such as Q336, Q337, N342, and W529, positions which were also observed to H-bond to glucose. As expected from the similarity between glucose and myo-inositol and the respective binding sites in GLUT3 and GLUT13, the conserved region in these small molecules is predicted to hydrogen-bond precisely to the same residues (Figure 3B,C). Additionally, GLUT13 residue Q220, which is not conserved, is predicted to H-bond to the C4 hydroxyl. The only other receptor containing a polar residue (Ser) at this position is GLUT6. The smaller side chain of serine places its hydroxyl too far to direct hydrogen bonding to the ligand, but it would allow a water molecule to fill in the gap and stabilize their interaction. In all other GLUTs, there is either an isoleucine or a valine residue at this position and therefore no other GLUT harbors the C4 hydroxyl that could be favorably accommodated.
Myo-inositol specific binding to GLUT13 is also supported by its unique ability to accommodate the hydroxyl at C3. GLUT13 contains a threonine (T213) at a position that is usually occupied by a bulkier residue, which would sterically clash with the hydroxyl (glutamine or glutamate in most GLUTs, except for GLUT9 and GLUT11, which bear an alanine at this position). This is captured by DSX as unfavorable potentials for both ligand and protein in the GLUT3–myo-inositol complex (Figure 3D), whereas in the same region, good potentials are observed for the GLUT13/myo-inositol complex (Figure 3E). Therefore, our results suggested that two residues that are uniquely present in GLUT13 seemed critical for myo-inositol recognition: T213 and Q220 (Supporting Information Table S5). Analysis of homologous protein sequences revealed that these residues are, respectively, 96.4 and 99.6% conserved in the 250 closest GLUT13 homologues, further supporting our observation.
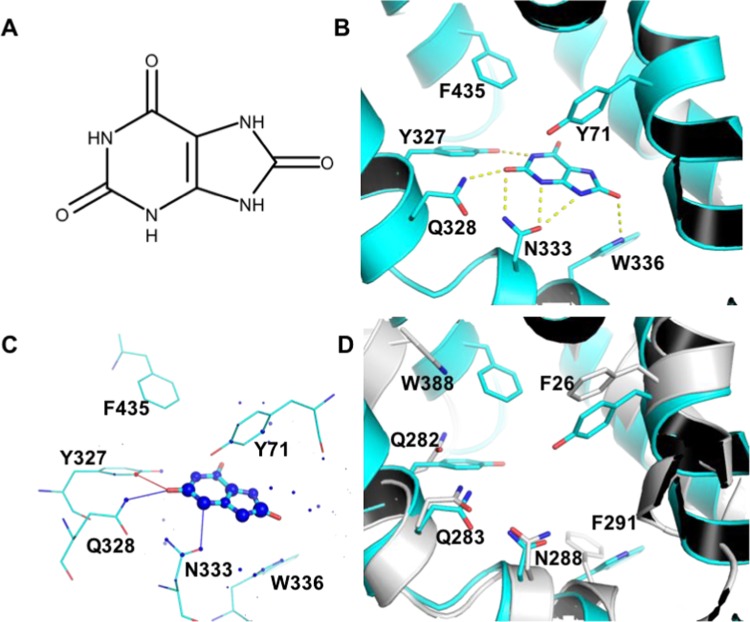
Specificity of GLUT9 to Urate
GLUT9 represents another interesting case of substrate specificity as the only known urate transporter in the family. We therefore employed docking studies to obtain insights into the molecular bases for this specificity. The best scoring binding mode for urate to GLUT9, according to the estimated affinity deduced by @TOME-2, was obtained when docking into a outward-facing model, in a partially occluded conformation (PDB 4GC0).18 In this pose, the ligand hydrogen-bonds to residues Y327, Q328, N333, and W336 (Figure 4A,B). Evaluation by DSX online indicated positive contributions from most ligand atoms and several surrounding residues (Figure 4C). In agreement with exclusive urate transport by GLUT9, docking urate to other GLUTs did not reveal binding modes similar to the one proposed for GLUT9. This result is also coherent with the urate binding site and binding mode proposed by docking. Among residues predicted to hydrogen-bond with urate, Q328 and N333 were highly conserved in the family, whereas Y327 (Q282 in GLUT1) and W336 (F291 in GLUT1) were not observed in any other GLUT (Supporting Information Table S6). Therefore, the former residues are likely to be crucial for the specificity of GLUT9 for urate recognition and transport. Other distinctive residues in the active site are Y71, C427, and F435 (respectively, F26, E380, and W388 in GLUT1, Figure 4D), among which Y71 is predicted to interact with urate through edge-face pi-stacking. To document further the importance of these residues, we analyzed their conservation within 250 closest homologues found through a BLASTp search. The sequences we retrieved had a global sequence identity of at least 78.9% to our query and encompassed a wide phylogenetic range of vertebrates, including birds, bats, insectivores, carnivores, rodents, and primates. Overall, the least conserved residue in the binding site was Y71 (96% conservation), underlining the evolutionary high conservation of the structural properties for this site.
Figure 4.
Molecular bases for urate specific binding of GLUT9. (A) Urate chemical structure. (B) urate binding mode to GLUT9, predicted by docking with PLANTS. Residues predicted to hydrogen-bond with the ligand or which differ from most GLUTs are highlighted residues shown as sticks. Hydrogen bonds (distance cutoff = 3.5 Å) are shown as yellow dashed lines. (C) Visualization of the docking result with DSX online. Favorable and unfavorable potentials are shown as blue and red spheres, respectively. Favorable and unfavorable distances are shown as blue and red lines, respectively. (D) superposition of GLUT9 (cyan) and GLUT1 (white) binding sites.
GLUT11 Binding Site and Potential Ligands
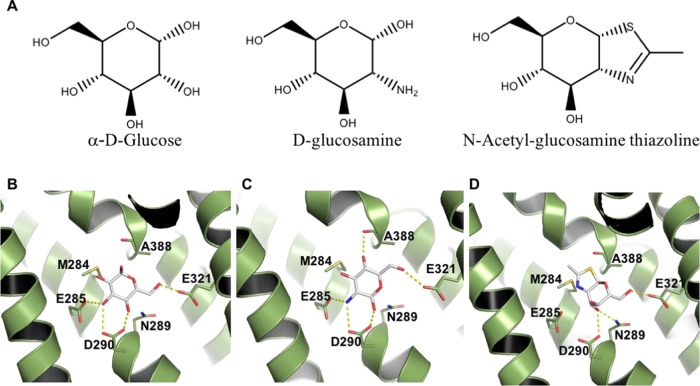
Finally, we examined GLUT11, one of the least studied transporters in the SLC2 family and the one with the most divergent exofacial binding site. Compared to Class 1 receptors, there is no residue conservation in the positions that hydrogen-bond to glucose, even though GLUT11 has been shown to transport glucose in reconstituted membranes.35−37 These results initially seemed rather in contradiction and incited us to perform further modeling analyses of this GLUT family member. As for the other transporters, we first investigated whether the predicted GLUT11 binding site was phylogenetically conserved. Among the 250 closest homologues, we observed that the corresponding seven residues (highlighted in Table 4) are highly conserved (with at least 88% conservation), suggesting a conserved functional binding site (Supporting Information Table S7). Of note, when compared to Class 1 GLUTs, three residue differences were rather conservative although they consisted in the change of a neutral residue in GLUT11 (asparagine or glutamine) to the isosteric but negatively charged residue (aspartate or glutamate; E285, D290, and E321). Most other differences involved the smallest residues, such as alanine and glycine, at three other positions. This should allow some water molecules to fill in the extra space made available and form some hydrogen bond with the polar substrate. The only difference, when compared to GLUT1, that is predicted to be detrimental is M284 in GLUT11, which is hydrophobic and as large as the glutamine in GLUT1 (Q282). Next, we evaluated whether glucose could still be accommodated in GLUT11 exofacial binding site. Docking studies were performed on our models that we built based on templates with 33– 35% sequence identity (to GLUT3 and GLUT1, respectively). Despite the lack of residue conservation, docking studies revealed glucose poses that were stabilized by up to five hydrogen bonds. Assuming a conserved orientation when compared to the glucose binding mode of GLUT3, hydroxyls at C2–C4 could be stabilized by four hydrogen bonds with GLUT11 residues E285, D290, and E321 (Figure 5B). Therefore, even though glucose recognition does not seem as favorable as for Class 1 transporters, the GLUT11 binding site still appeared compatible with glucose transport, as also experimentally observed.35−37
Figure 5.
Predicted binding mode for putative GLUT11 substrates. (A) Chemical structures of docked ligands. Docking predicted poses for: (B) α-d-glucose, (C) d-glucosamine, and (D) N-acetyl-glucosamine thiazoline. GLUT11 is shown as a cartoon, with selected residues and ligands highlighted as sticks and colored by atom. Salt bridges and hydrogen bonds (distance cutoff = 3.5 Å) are shown as yellow dashed lines.
Nevertheless, our model indicated that glucose is unlikely the primary substrate for GLUT11, which has a bigger and more negatively charged exofacial binding site than that of Class 1 GLUTs. These differences suggested transport of either a positively charged substrate or a larger molecule. We therefore investigated the binding mode of putative substrates by docking. d-glucosamine was evaluated as an example of a putatively relevant and positively charged sugar. The best scoring binding mode obtained (best pKd according to @TOME) is characterized by positioning the positively charged nitrogen among two negatively charged residues (E285 and D290), at 3.1 Å from each, and additional hydrogen bonds to the side chains of D290, E321 and the backbone carbonyl from A388 (Figure 5C).
To identify further putative ligands, we searched GLUT11 annotations in the STITCH database.38 Among those, we found N-acetyl-glucosamine thiazoline (NAG-thiazoline), a d-glucosamine derivative. NAG-thiazoline predicted binding mode suggested that this ligand could fit in the GLUT11 binding site, since its polar region is stabilized by hydrogen bonds to N289, D290, and E321, whereas the thiazoline ring would occupy a more hydrophobic region of the binding site, close to residues A388 and M284 (Figure 5D). The latter positions are occupied by polar residues in most of the other GLUTs (Table 4).
Discussion
In this study, we employed recently obtained structural information from members of the SLC2 family to provide comparative models for the whole family. This approach allowed us to obtain insights into previously described trends studying substrate and inhibitor specificity. Our first contribution is to provide models for all family members and this, in multiple conformations and together with their validation data. These models are made readily available to the community through the @TOME-2 platform. To the best of our knowledge, no comparative models have been published either for any Class 3 transporters or some Class 2 transporters (GLUT7 and GLUT11).
Sequence identity between the GLUTs to be modeled and available templates varied widely, from 24 to 95%. Therefore, whereas the model building was trivial for some transporters, in some cases, the automatic alignment could be locally inaccurate, especially around insertions and deletions, and a more precise procedure was employed. This involved incorporating information from secondary structure prediction, manually checking for alignment of conserved residues and use of ViTO,39 an interface that easily allows the visualization of the correspondence between sequence alignment and protein structure. Such procedures resulted in models for the transmembrane regions and smalls loops of all GLUTs, whereas we opted not to model flexible regions without proper templates, such as long loops connecting transmembrane helices and intracellular N- and C-terminal sequences. Even though these regions, which we did not include in our modeling, are indeed biologically relevant, we reasoned that the transmembrane core that contains ligand binding sites was the most critical region for the analyses reported here.
Obtaining accurate models for the core regions of all GLUTs allowed us to compare their endofacial and exofacial binding sites, highlighting conserved residues and allowing structure-based hypothesis that supported substrate distinctive specificities, as observed within the GLUT family members. Understanding how selectivity might be achieved within the SLC2 family is critical from a medicinal chemistry standpoint. Several diseases have been associated with aberrant expression of members of this family, and these proteins have been therefore targeted for the development of inhibitors. Several GLUT mutations have also been associated to deficient expression and/or transport. Despite the high binding site conservation observed in Class 1 GLUTs, inhibitors selective for either GLUT19,10 or GLUT411,12 have been described, in response to a strong motivation to develop selective inhibitors as therapeutic agents. CytoB is a well-studied inhibitor which binds to several Class 1 and Class 3 transporters, with no inhibition documented for Class 2 proteins. Two tryptophans (W388 and W412 in GLUT1) have been previously described as essential for cytoB binding.31 On the basis of comparative docking analyses, here we describe additional substitutions in the cytoB binding site, which may also explain the lack of inhibition of this compound for some of the GLUTs.
Understanding substrate specificity within the GLUT family is still a fundamental question to be answered in the field.1 Here, we provide contributions toward this direction by predicting binding modes for urate and myo-inositol, formulating a hypothesis of critical residues for substrate recognition and suggesting possible GLUT11 substrates. Such hypotheses were based on docking studies, binding site comparisons, and conservation among homologues from different species.
GLUT9 is the only urate transporter known in the GLUT family. To the best of our knowledge, here we provide for the first time a putative binding mode for urate in GLUT9. Comparative models for this transporter have been previously proposed, both by Dinour et al.5 and Clemençon et al.40 However, neither study included a detailed description of binding sites. Here, we considered multiple templates for comparative modeling and docking, including structures with higher resolution and obtained from sequences with higher sequence identity. This approach allowed us to provide a hypothesis for urate binding mode and to propose residues Y327 and W336 as keys for recognition and specificity. Additional bioinformatics analysis supported this hypothesis: No other protein in the family contains these residues at the structurally aligned positions, which is coherent with the exclusive transport by GLUT9, and these residues were 100% conserved in homologues from varied species. Finally, no polymorphism has yet been reported at these positions in humans. GLUT9 is expressed as two splice variants, GLUT9a and GLUT9b, which differ in their amino-terminal cytoplasmic tails.41 Although this difference is important for protein localization,41 it does not interfere with the affinity for urate.42 Even though we focused our studies on GLUT9a, the longest variant (540 amino acid long), and the more widely expressed of the two splice variants, our analysis should be valid for both variants.
Another family member with distinct substrate specificity is GLUT13. No polymorphism has been described for this transporter. The GLUT13 glucose binding site is very similar to that observed for Class 1 transporters. However, we have identified key modifications that may be related to selectivity. In the case of both GLUT9 and GLUT13, formal confirmation of the importance of the residues proposed here should be provided by experimental assessment. Here, we employed various bioinformatic techniques to document our hypotheses and orientate further experimentation on GLUT biochemistry and biology.
Currently, GLUT11 is among the least studied GLUTs. It is expressed mainly in heart and skeletal muscles,35 although different isoforms appear to be expressed in kidney, adipose tissues, placenta, and pancreas.37 The three GLUT11 isoforms can transport glucose and fructose but not galactose.37 Not much information has been reported concerning substrates or inhibitors for this transporter. Here, analysis of the GLUT11 binding sites revealed a strikingly low level of similarity to other GLUTs, characterized by an exofacial binding site more negatively charged and larger than the corresponding site in Class 1 transporters. Taking into account these characteristics and annotations in the STITCH database, d-glucosamine and N-acetyl-glucosamine thiazoline were successfully docked into GLUT11. Further experiments are needed to assess whether the latter compounds are potential ligands for this receptor. Alternatively, another hypothesis is that the GLUT11 binding site may favor glucose transport in more acidic conditions, as found in active muscles, due to protonation of the acidic side chains.
Conclusions
Through a combination of bioinformatic tools, we obtained models for GLUT1–14, compared their exofacial and endofacial binding sites, and studied possible determinants for inhibitor and substrate specificity within this family. We identified residues likely critical for recognition and transport by GLUTs that present unique specificity (Y317 and W336 for urate transport by GLUT9 and T213 and Q220 for myo-inositol transport by GLUT13), providing hypotheses to be confirmed by mutagenesis and functional studies. Additionally, models provided a basis for understanding selectivity trends for cytoB, a widely employed GLUT inhibitor.
Experimental Section
Automated Comparative Modeling with the @TOME-2 Pipeline
For each GLUT, models were generated in an automated way by the @TOME-2 pipeline.26 Briefly, starting from each query sequence, template detection was performed with Fugue,43 HHSearch,44 and PSI-BLAST.45 Models were then built with TiTO27 and Scwrl.46 Structural alignments obtained through these programs were evaluated with different 1D, 2D, and 3D tools and scored, taking into account the percentage sequence identity, 3D-Coffee sequence alignment score, 3DJury rank, Tito Score, Verify 3D,29 and QMEAN28 evaluation scores. Long cytoplasmatic or extracellular loops (Class 1: L2–3, Class 3: L9–10) and the ICH domain from GLUT8 and GLUT10 were not modeled, due to the lack of appropriate templates. For each GLUT, models were generated from all templates detected, and up to 20 models were selected on the basis of the top ranking structural alignments for each sequence. Results are available at http://atome3.cbs.cnrs.fr/htbin-post/AT23/SUPERATOME/aff_study_stat_base.cgi?&WD=AT23/GLUT_HUMAN.
Sequence Alignment and Comparative Modeling
Human GLUT6, 8 and 10–13 have sequence identity to the available GLUT templates (GLUT3 and GLUT1) that is lower than 35%. For these, additional models were generated from a nonautomated procedure. Sequences were obtained from the UniProt database30 (GLUT1: P11166; GLUT2: P11168; GLUT3: P11169; GLUT4: P14672; GLUT5: P22732; GLUT6: Q9UGQ3; GLUT7: Q6PXP3; GLUT8: Q9NY64; GLUT9: Q9NRM0; GLUT10: O95528; GLUT11: Q9BYW1; GLUT12: Q8TD20; GLUT13 (MYCT): Q96QE2; GLUT14: Q8TDB8). Sequence alignment was manually checked in ViTO,39 taking into account secondary structure prediction by the PSIPRED server.47 Models were then built based on templates PDB 4ZW9 (GLUT3) and PDB 5EQI (GLUT1) with Tito and Swrcl and uploaded into the @TOME-2 platform for validation and docking studies. We opted not to use the rat and bovine GLUT5 structures as templates, given their low resolution (3.27 and 3.2 Å, respectively) and the fact that the human GLUT1 and GLUT3 structures available had higher resolution, were in complex with ligands, and in the same conformation (inward open and outward open, respectively) as the GLUT5 structures.
Ligand File Preparation
Isomeric SMILES were retrieved from PubChem (https://pubchem.ncbi.nlm.nih.gov/). OpenBabel48 was employed to convert SMILES into MOL2, which were then prepared for docking by reprotonation with SPORES49 and addition of Gasteiger charges with OpenBabel.
Molecular Docking
All docking procedures were performed in the @TOME-2 platform. Before docking, complexes were built by transferring ligands from experimental structures to each model, in a process called comparative docking. For each complex, the binding constant (pKd) is calculated as an average of four scoring functions: MedusaScore,50 Xscore,51 PLANTS,33,34 and DSX.52 On the basis of the calculated pKd, a subset of these complexes was then selected as reference complexes, for which protein–ligand interaction profiles were built, to allow anchoring of new ligands during docking with PLANTS.
For cytoB, we only performed the comparative docking procedure in @TOME-2, in which the crystallographic ligand coordinates were transferred to the models of several GLUTs, in all cases, built on the basis of PDB 5EQI(20) as the template. Unconserved side chains in the ligand binding site were optimized by SCWRL after ligand transfer. In the case of GLUT7, the complex obtained was minimized with the YASARA force field, through the YASARA web server.53
For other ligands, docking was performed with PLANTS, through the @TOME-2 platform, with the following parameters: up to 20 modeled receptors were considered, with 1 ligand-anchor/model, chosen on the basis of anchor/candidate similarity, as measured by the Tanimoto coefficient. The binding site size was set to anchor radius plus 8 Å, the quality/speed ratio for PLANTS software set to speed 2, and side chains of the binding site were recalculated by PLANTS. Ligand reposition/anchor constraint for Plants software was usually set to −3 and in the case of urate, calculations were also run with no constraints (parameter set to 0). Docking results were examined in Pymol. All docking results discussed in the manuscript are listed in Supporting Information Table S8 and available for download through the link: http://atome4.cbs.cnrs.fr/AT23/GLUT_HUMAN/DBINFO/Inf/Table-S1.html.
DSX Online
Selected docking results were evaluated with the online version of the DSX scoring function,52 available at http://pc1664.pharmazie.uni-marburg.de/drugscore/. Protein PDB and ligand MOL2 files were obtained from the best scoring results generated by @TOME-2. Complexes were evaluated with CSD potentials. Torsion potentials and solvent accessible surface were taken into account during scoring. A Pymol script for visualization of per-atom contributions was generated by the server and examined in Pymol.
Sequence Conservation in Orthologues
For the most functionally and structurally divergent GLUTs, namely, GLUT9–13, sequence conservation among orthologues was carefully surveyed. To search for homologous genes, a Blastp search was performed with standard parameters (word size: 6; expected value: 10; gap costs: existence = 11, extension = 1; Matrix: BLOSUM62), except for the number of hits, which was set to 250. Query sequences, obtained from UniProt, were Q9NRM0 for human GLUT9, O95528 for GLUT10, Q9BYW1 for human GLUT11, Q8TD20 for GLUT12, and Q96QE2 for human GLUT13. The sequences obtained were then aligned with Clustal Omega and the resulting file was visualized and analyzed with Jalview, to determine conservation and occupancy of specific residues within putative binding sites.
Stitch Database
Annotated ligands for GLUT11 were retrieved from the Stitch database (http://stitch.embl.de/),38 on the basis of a search with the term “GLUT11” and selecting “Homo sapiens” as the organism.
Acknowledgments
R.S.F. holds a CNPq researcher fellowship (Bolsa de Produtividade em Pesquisa, nível 2) and has received the L’Oréal-UNESCO-ABC “For Women in Science Award, Category Chemistry, Brazil, 2017” and L’Oréal-UNESCO “International Rising Talent Award 2018”. We thank Marc Stibon, Jean-François Guichou, Martin Cohen-Gonsaud, Muriel Felin, and Corinne Lionne for critically reading and providing suggestions on this manuscript.
Glossary
Abbreviations
- SLC
solute carrier
- GLUT
glucose transport carrier
- ICH
intracellular helical
- MYCT
proton myo-inositol cotransporter
- TM
transmembrane
- DRM
disease-related mutations
- GLUT1DS2
GLUT1 deficiency syndrome 2
- NAG-thiazoline
N-acetyl-glucosamine thiazoline
- cytoB
cytochalasin B
- pKd
–log of the dissociation constant
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsomega.8b03447.
Sequence alignment of human GLUT1–14; reported substrates for GLUT1–14; conservation of GLUT10, GLUT11, and GLU12 exofacial binding site in homologous proteins; myo-inositol binding site residues and their conservation within GLUTs and GLUT13 homologues; urate binding site residues and their conservation within GLUTs and GLUT9 homologues; list of reported docking results, with a link to access file coordinates; authors will release the atomic coordinates and experimental data upon article publication (PDF)
Author Contributions
J.-L.P., R.S.F., and G.L. performed comparative modeling, R.S.F. prepared ligands for docking and performed all docking studies and performed bioinformatic studies, R.S.F. and G.L. designed the study, and R.S.F. wrote the manuscript, with contributions from J.L.P. and G.L.
This study was funded by the ANR grant 10-BINF-0003 and by Coordenação de Aperfeiçoamento de Pessoal de Nível Superior—Brasil (CAPES). R.S.F. acknowledges funding from Foundation pour la Recherche Médicale (FRM, France) and CAPES fellowship (Professor Visitante no Exterior, Processo 88881.170377/2018-01).
The authors declare no competing financial interest.
Supplementary Material
References
- Yan N. A Glimpse of Membrane Transport through Structures—Advances in the Structural Biology of the GLUT Glucose Transporters. J. Mol. Biol. 2017, 429, 2710–2725. 10.1016/j.jmb.2017.07.009. [DOI] [PubMed] [Google Scholar]
- Mueckler M.; Thorens B. The SLC2 (GLUT) Family of Membrane Transporters. Mol. Aspects Med. 2013, 34, 121–138. 10.1016/j.mam.2012.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manel N.; Battini J.-L.; Sitbon M. Human T Cell Leukemia Virus Envelope Binding and Virus Entry Are Mediated by Distinct Domains of the Glucose Transporter GLUT1. J. Biol. Chem. 2005, 280, 29025–29029. 10.1074/jbc.M504549200. [DOI] [PubMed] [Google Scholar]
- Pascual J.; Wang D.; Lecumberri B.; Yang H.; Mao X.; Yang R.; De Vivo D. GLUT1 Deficiency and Other Glucose Transporter Diseases. Eur. J. Endocrinol. 2004, 150, 627–633. 10.1530/eje.0.1500627. [DOI] [PubMed] [Google Scholar]
- Dinour D.; Gray N. K.; Ganon L.; Knox A. J. S.; Shalev H.; Sela B. A.; Campbell S.; Sawyer L.; Shu X.; Valsamidou E.; et al. Two Novel Homozygous SLC2A9 Mutations Cause Renal Hypouricemia Type 2. Nephrol., Dial., Transplant. 2012, 27, 1035–1041. 10.1093/ndt/gfr419. [DOI] [PubMed] [Google Scholar]
- Claverie-Martin F.; Trujillo-Suarez J.; Gonzalez-Acosta H.; Aparicio C.; Justa Roldan M. L.; Stiburkova B.; Ichida K.; Martín-Gomez M. A.; Herrero Goñi M.; Carrasco Hidalgo-Barquero M.; et al. URAT1 and GLUT9 Mutations in Spanish Patients with Renal Hypouricemia. Clin. Chim. Acta 2018, 481, 83–89. 10.1016/j.cca.2018.02.030. [DOI] [PubMed] [Google Scholar]
- Németh C. E.; Marcolongo P.; Gamberucci A.; Fulceri R.; Benedetti A.; Zoppi N.; Ritelli M.; Chiarelli N.; Colombi M.; Willaert A.; et al. Glucose Transporter Type 10-Lacking in Arterial Tortuosity Syndrome-Facilitates Dehydroascorbic Acid Transport. FEBS Lett. 2016, 590, 1630–1640. 10.1002/1873-3468.12204. [DOI] [PubMed] [Google Scholar]
- Ancey P.-B.; Contat C.; Meylan E. Glucose Transporters in Cancer - from Tumor Cells to the Tumor Microenvironment. FEBS J. 2018, 285, 2926–2943. 10.1111/febs.14577. [DOI] [PubMed] [Google Scholar]
- Siebeneicher H.; Cleve A.; Rehwinkel H.; Neuhaus R.; Heisler I.; Müller T.; Bauser M.; Buchmann B. Identification and Optimization of the First Highly Selective GLUT1 Inhibitor BAY-876. ChemMedChem 2016, 11, 2261–2271. 10.1002/cmdc.201600276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebeneicher H.; Bauser M.; Buchmann B.; Heisler I.; Müller T.; Neuhaus R.; Rehwinkel H.; Telser J.; Zorn L. Identification of Novel GLUT Inhibitors. Bioorg. Med. Chem. Lett. 2016, 26, 1732–1737. 10.1016/j.bmcl.2016.02.050. [DOI] [PubMed] [Google Scholar]
- Wei C.; Bajpai R.; Sharma H.; Heitmeier M.; Jain A. D.; Matulis S. M.; Nooka A. K.; Mishra R. K.; Hruz P. W.; Schiltz G. E.; et al. Development of GLUT4-Selective Antagonists for Multiple Myeloma Therapy. Eur. J. Med. Chem. 2017, 139, 573–586. 10.1016/j.ejmech.2017.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hresko R. C.; Hruz P. W. HIV Protease Inhibitors Act as Competitive Inhibitors of the Cytoplasmic Glucose Binding Site of GLUTs with Differing Affinities for GLUT1 and GLUT4. PLoS One 2011, 6, e25237 10.1371/journal.pone.0025237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soueidan O.-M.; Scully T. W.; Kaur J.; Panigrahi R.; Belovodskiy A.; Do V.; Matier C. D.; Lemieux M. J.; Wuest F.; Cheeseman C.; et al. Fluorescent Hexose Conjugates Establish Stringent Stereochemical Requirement by GLUT5 for Recognition and Transport of Monosaccharides. ACS Chem. Biol. 2017, 12, 1087–1094. 10.1021/acschembio.6b01101. [DOI] [PubMed] [Google Scholar]
- Wuest M.; Hamann I.; Bouvet V.; Glubrecht D.; Marshall A.; Trayner B.; Soueidan O.-M.; Krys D.; Wagner M.; Cheeseman C.; et al. Molecular Imaging of GLUT1 and GLUT5 in Breast Cancer: A Multitracer Positron Emission Tomography Imaging Study in Mice. Mol. Pharmacol. 2018, 93, 79–89. 10.1124/mol.117.110007. [DOI] [PubMed] [Google Scholar]
- Otsuka Y.; Sasaki A.; Teshima T.; Yamada K.; Yamamoto T. Syntheses of D-Glucose Derivatives Emitting Blue Fluorescence through Pd-Catalyzed C–N Coupling. Org. Lett. 2016, 18, 1338–1341. 10.1021/acs.orglett.6b00280. [DOI] [PubMed] [Google Scholar]
- Begoyan V. V.; Weseliński Ł.J.; Xia S.; Fedie J.; Kannan S.; Ferrier A.; Rao S.; Tanasova M. Multicolor GLUT5-Permeable Fluorescent Probes for Fructose Transport Analysis. Chem. Commun. 2018, 54, 3855–3858. 10.1039/C7CC09809J. [DOI] [PubMed] [Google Scholar]
- Kannan S.; Begoyan V.; Fedie J.; Xia S.; Weseliński Ł.; Tanasova M.; Rao S. Metabolism-Driven High-Throughput Cancer Identification with GLUT5-Specific Molecular Probes. Biosensors 2018, 8, 39 10.3390/bios8020039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L.; Zeng X.; Yan C.; Sun X.; Gong X.; Rao Y.; Yan N. Crystal Structure of a Bacterial Homologue of Glucose Transporters GLUT1–4. Nature 2012, 490, 361–366. 10.1038/nature11524. [DOI] [PubMed] [Google Scholar]
- Deng D.; Xu C.; Sun P.; Wu J.; Yan C.; Hu M.; Yan N. Crystal Structure of the Human Glucose Transporter GLUT1. Nature 2014, 510, 121–125. 10.1038/nature13306. [DOI] [PubMed] [Google Scholar]
- Kapoor K.; Finer-Moore J. S.; Pedersen B. P.; Caboni L.; Waight A.; Hillig R. C.; Bringmann P.; Heisler I.; Müller T.; Siebeneicher H.; et al. Mechanism of Inhibition of Human Glucose Transporter GLUT1 Is Conserved between Cytochalasin B and Phenylalanine Amides. Proc. Natl. Acad. Sci. U.S.A. 2016, 113, 4711–4716. 10.1073/pnas.1603735113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng D.; Sun P.; Yan C.; Ke M.; Jiang X.; Xiong L.; Ren W.; Hirata K.; Yamamoto M.; Fan S.; et al. Molecular Basis of Ligand Recognition and Transport by Glucose Transporters. Nature 2015, 526, 391–396. 10.1038/nature14655. [DOI] [PubMed] [Google Scholar]
- Nomura N.; Verdon G.; Kang H. J.; Shimamura T.; Nomura Y.; Sonoda Y.; Hussien S. A.; Qureshi A. A.; Coincon M.; Sato Y.; et al. Structure and Mechanism of the Mammalian Fructose Transporter GLUT5. Nature 2015, 526, 397–401. 10.1038/nature14909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X.; Zhang G.; Liu R.; Wei J.; Zhang-Negrerie D.; Jian X.; Gao Q. Mechanistic Study of Human Glucose Transport Mediated by GLUT1. J. Chem. Inf. Model. 2016, 56, 517–526. 10.1021/acs.jcim.5b00597. [DOI] [PubMed] [Google Scholar]
- Mohan S.; Sheena A.; Poulose N.; Anilkumar G. Molecular Dynamics Simulation Studies of GLUT4: Substrate-Free and Substrate-Induced Dynamics and ATP-Mediated Glucose Transport Inhibition. PLoS One 2010, 5, e14217 10.1371/journal.pone.0014217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ainsley J.; Chaturvedi S. S.; Karabencheva-Christova T. G.; Tanasova M.; Christov C. Z. Integrating Molecular Probes and Molecular Dynamics to Reveal Binding Modes of GLUT5 Activatory and Inhibitory Ligands. Chem. Commun. 2018, 54, 9917–9920. 10.1039/C8CC04843F. [DOI] [PubMed] [Google Scholar]
- Pons J.-L.; Labesse G. @TOME-2: A New Pipeline for Comparative Modeling of Protein-Ligand Complexes. Nucleic Acids Res. 2009, 37, W485–W491. 10.1093/nar/gkp368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labesse G.; Mornon J. Incremental Threading Optimization (TITO) to Help Alignment and Modelling of Remote Homologues. Bioinformatics 1998, 14, 206–211. 10.1093/bioinformatics/14.2.206. [DOI] [PubMed] [Google Scholar]
- Benkert P.; Biasini M.; Schwede T. Toward the Estimation of the Absolute Quality of Individual Protein Structure Models. Bioinformatics 2011, 27, 343–350. 10.1093/bioinformatics/btq662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg D.; Lüthy R.; Bowie J. U. VERIFY3D: Assessment of Protein Models with Three-Dimensional Profiles. Methods Enzymol. 1997, 277, 396–404. 10.1016/S0076-6879(97)77022-8. [DOI] [PubMed] [Google Scholar]
- Consortium T. U. UniProt: The Universal Protein Knowledgebase. Nucleic Acids Res. 2017, 45, D158–D169. 10.1093/nar/gkw1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inukai K.; Asano T.; Katagiri H.; Anai M.; Funaki M.; Ishihara H.; Tsukuda K.; Kikuchi M.; Yazaki Y.; Oka Y. Replacement of Both Tryptophan Residues at 388 and 412 Completely Abolished Cytochalasin B Photolabelling of the GLUT1 Glucose Transporter. Biochem. J. 1994, 302, 355–361. 10.1042/bj3020355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uldry M.; Ibberson M.; Horisberger J.; et al. Identification of a Mammalian H + -Myo-Inositol Symporter Expressed Predominantly in the Brain. EMBO J. 2001, 20, 4467–4477. 10.1093/emboj/20.16.4467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korb O.; Stützle T.; Exner T. E. An Ant Colony Optimization Approach to Flexible Protein–Ligand Docking. Swarm Intell. 2007, 1, 115–134. 10.1007/s11721-007-0006-9. [DOI] [Google Scholar]
- Korb O.; Stützle T.; Exner T. E. Empirical Scoring Functions for Advanced Protein-Ligand Docking with PLANTS. J. Chem. Inf. Model. 2009, 49, 84–96. 10.1021/ci800298z. [DOI] [PubMed] [Google Scholar]
- Doege H.; Bocianski A.; Scheepers A.; Axer H.; Eckel J.; Joost H. G.; Schürmann A. Characterization of Human Glucose Transporter (GLUT) 11 (Encoded by SLC2A11), a Novel Sugar-Transport Facilitator Specifically Expressed in Heart and Skeletal Muscle. Biochem. J. 2001, 359, 443–449. 10.1042/bj3590443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X.; Li W.; Sharma V.; Godzik A.; Freeze H. H. Cloning and Characterization of Glucose Transporter 11, a Novel Sugar Transporter That Is Alternatively Spliced in Various Tissues. Mol. Genet. Metab. 2002, 76, 37–45. 10.1016/S1096-7192(02)00018-5. [DOI] [PubMed] [Google Scholar]
- Scheepers A.; Schmidt S.; Manolescu A.; Cheeseman C. I.; Bell A.; Zahn C.; Joost H. G.; Schürmann A. Characterization of the Human SLC2A11 (GLUT11) Gene: Alternative Promoter Usage, Function, Expression, and Subcellular Distribution of Three Isoforms, and Lack of Mouse Orthologue. Mol. Membr. Biol. 2005, 22, 339–351. 10.1080/09687860500166143. [DOI] [PubMed] [Google Scholar]
- Kuhn M.; von Mering C.; Campillos M.; Jensen L. J.; Bork P. STITCH: Interaction Networks of Chemicals and Proteins. Nucleic Acids Res. 2007, 36, D684–D688. 10.1093/nar/gkm795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catherinot V.; Labesse G. ViTO: Tool for Refinement of Protein Sequence-Structure Alignments. Bioinformatics 2004, 20, 3694–3696. 10.1093/bioinformatics/bth429. [DOI] [PubMed] [Google Scholar]
- Clémençon B.; Lüscher B. P.; Fine M.; Baumann M. U.; Surbek D. V.; Bonny O.; Hediger M. A. Expression, Purification, and Structural Insights for the Human Uric Acid Transporter, GLUT9, Using the Xenopus Laevis Oocytes System. PLoS One 2014, 9, e108852 10.1371/journal.pone.0108852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augustin R.; Carayannopoulos M. O.; Dowd L. O.; Phay J. E.; Moley J. F.; Moley K. H. Identification and Characterization of Human Glucose Transporter-like Protein-9 (GLUT9): Alternative Splicing Alters Trafficking. J. Biol. Chem. 2004, 279, 16229–16236. 10.1074/jbc.M312226200. [DOI] [PubMed] [Google Scholar]
- Caulfield M. J.; Munroe P. B.; O’Neill D.; Witkowska K.; Charchar F. J.; Doblado M.; Evans S.; Eyheramendy S.; Onipinla A.; Howard P.; et al. SLC2A9 Is a High-Capacity Urate Transporter in Humans. PLoS Med. 2008, 5, 1509–1523. 10.1371/journal.pmed.0050197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J.; Blundell T. L.; Mizuguchi K. FUGUE: Sequence-Structure Homology Recognition Using Environment-Specific Substitution Tables and Structure-Dependent Gap Penalties. J. Mol. Biol. 2001, 310, 243–257. 10.1006/jmbi.2001.4762. [DOI] [PubMed] [Google Scholar]
- Söding J. Protein Homology Detection by HMM-HMM Comparison. Bioinformatics 2005, 21, 951–960. 10.1093/bioinformatics/bti125. [DOI] [PubMed] [Google Scholar]
- Altschul S. F.; Madden T. L.; Schäffer A. A.; Zhang J.; Zhang Z.; Miller W.; Lipman D. J. Gapped BLAST and PSI-BLAST: A New Generation of Protein Database Search Programs. Nucleic Acids Res. 1997, 25, 3389–3402. 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q.; Canutescu A. A.; Dunbrack R. L. SCWRL and MolIDE: Computer Programs for Side-Chain Conformation Prediction and Homology Modeling. Nat. Protoc. 2008, 3, 1832–1847. 10.1038/nprot.2008.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan D. W. A.; Minneci F.; Nugent T. C. O.; Bryson K.; Jones D. T. Scalable Web Services for the PSIPRED Protein Analysis Workbench. Nucleic Acids Res. 2013, 41, W349–W357. 10.1093/nar/gkt381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Boyle N. M.; Banck M.; James C. A.; Morley C.; Vandermeersch T.; Hutchison G. R. Open Babel: An Open Chemical Toolbox. J. Cheminf. 2011, 3, 33 10.1186/1758-2946-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ten Brink T.; Exner T. E. Influence of Protonation, Tautomeric, and Stereoisomeric States on Protein-Ligand Docking Results. J. Chem. Inf. Model. 2009, 49, 1535–1546. 10.1021/ci800420z. [DOI] [PubMed] [Google Scholar]
- Yin S.; Biedermannova L.; Vondrasek J.; Dokholyan N. V. MedusaScore: An Accurate Force Field-Based Scoring Function for Virtual Drug Screening. J. Chem. Inf. Model. 2008, 48, 1656–1662. 10.1021/ci8001167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R.; Lai L.; Wang S. Further Development and Validation of Empirical Scoring Functions for Structure-Based Binding Affinity Prediction. J. Comput.-Aided Mol. Des. 2002, 16, 11–26. 10.1023/A:1016357811882. [DOI] [PubMed] [Google Scholar]
- Neudert G.; Klebe G. DSX: A Knowledge-Based Scoring Function for the Assessment of Protein-Ligand Complexes. J. Chem. Inf. Model. 2011, 51, 2731–2745. 10.1021/ci200274q. [DOI] [PubMed] [Google Scholar]
- Krieger E.; Joo K.; Lee J.; Lee J.; Raman S.; Thompson J.; Tyka M.; Baker D.; Karplus K. Improving Physical Realism, Stereochemistry, and Side-Chain Accuracy in Homology Modeling: Four Approaches That Performed Well in CASP8. Proteins: Struct., Funct., Bioinf. 2009, 77, 114–122. 10.1002/prot.22570. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.