Multicomponent biomaterials using a high-throughput system regulate hyaline or hypertrophic chondrogenesis of hMSCs.
Abstract
Despite great progress in biomaterial design strategies for replacing damaged articular cartilage, prevention of stem cell-derived chondrocyte hypertrophy and resulting inferior tissue formation is still a critical challenge. Here, by using engineered biomaterials and a high-throughput system for screening of combinatorial cues in cartilage microenvironments, we demonstrate that biomaterial cross-linking density that regulates matrix degradation and stiffness—together with defined presentation of growth factors, mechanical stimulation, and arginine-glycine-aspartic acid (RGD) peptides—can guide human mesenchymal stem cell (hMSC) differentiation into articular or hypertrophic cartilage phenotypes. Faster-degrading, soft matrices promoted articular cartilage tissue formation of hMSCs by inducing their proliferation and maturation, while slower-degrading, stiff matrices promoted cells to differentiate into hypertrophic chondrocytes through Yes-associated protein (YAP)–dependent mechanotransduction. in vitro and in vivo chondrogenesis studies also suggest that down-regulation of the Wingless and INT-1 (WNT) signaling pathway is required for better quality articular cartilage-like tissue production.
INTRODUCTION
Cartilage defects are a common cause of pain and disability, and can progress to osteoarthritis (1, 2). Damaged articular cartilage cannot regenerate or repair due to its avascular nature and the limited quantity and mobility of chondrocytes, resulting in progressive total joint destruction (3). There are several clinical treatment options, including microfracture, mosaicplasty, and autologous chondrocyte implantation for patients with focal defects, and total joint arthroplasty for end-stage osteoarthritis. These treatments are extremely expensive, and it is still challenging to restore total functionality in damaged or diseased articular cartilage (4–6).
Stem cell–based strategies, alternative therapeutic approaches that may serve as long-term clinical solutions for cartilage regeneration, hold great promise for the treatment of damaged articular cartilage (7, 8). Since human mesenchymal stem cells (hMSCs) can differentiate into mesoderm-derived lineages, including osteoblasts and chondrocytes, hMSCs are an attractive stem cell source for cartilage repair and regeneration (9). However, maintenance of a stable articular cartilage phenotype formed by hMSCs is difficult due to the capacity of cartilage formed by hMSCs to undergo endochondral ossification (10). Emerging evidence suggests that identification of appropriate model systems that recapitulate the physical and biochemical signals present in complex cellular microenvironments (11), such as those from surrounding cell populations (12), the extracellular matrix (ECM) (e.g., cell adhesion ligands, matrix mechanics, and topography) (13, 14), mechanical stimulation (15), and soluble factors (e.g., growth factors, cytokines) (16), is critical to guide articular chondrogenesis of hMSCs. However, the majority of research efforts has focused on changing only one or two parameters while investigating their influence on cell behavior (12, 14–17), which makes it difficult to understand the interplay between all these important factors during articular chondrogenesis. Therefore, since precise control over cellular differentiation may not be possible without fully understanding this signal interplay (18), systems capable of deciphering the combinatorial effects of multiple cues on articular cartilage formation in a high-throughput manner would be valuable to elucidate critical therapeutic design parameters to prevent chondrocyte hypertrophy when engineering stable hyaline cartilage.
The mechanotransduction process, mediated extracellularly by the mechanics, structure, and composition of the microenvironment, influences stem cell fate decision and their terminal differentiation into chondrocytes (19, 20). The transcriptional cofactor Yes-associated protein (YAP) is known as a key mediator in the regulation of stem cell function through the nuclear transduction of physical cues (21, 22). For example, Karystinou et al. (23) demonstrated that YAP is down-regulated during chondrogenesis of hMSCs in vitro. Deng et al. (24) revealed the important role of YAP1 in inhibiting chondrocyte maturation by suppression of collagen type X alpha 1 chain (COL10A1) expression via interaction with runt-related transcription factor 2 (Runx2), a major regulator of chondrocyte hypertrophy. Furthermore, temporally dynamic changes in the cellular microenvironment can affect cell fate and maturation. For instance, a study by Yang et al. (21) demonstrated that YAP and Runx2 expressions in hMSCs were tunable by dynamically varying the substrate stiffness. In this study, we used a high-throughput system to explore how engineered biomaterials with a range of specific combinatorial physical and biochemical cues, in concert with defined compressive mechanical stimulation, affect articular and hypertrophic chondrogenesis of hMSCs (Fig. 1A).
Fig. 1. Stem cell–based cartilage repair strategy is based on engineered multicomponent biomaterials and a combinatorial system for screening of combinatorial physical and biochemical cues in cartilage microenvironments.
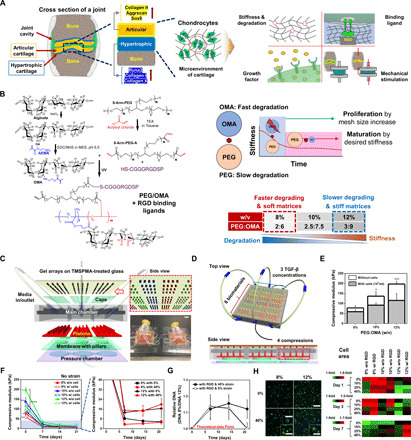
(A) Schematic depicting key factors guiding chondrogenic differentiation of hMSCs. (B) Chemical synthesis scheme of hybrid of photo–cross-linkable PEG/OMA hydrogels. (C) Schematic showing six layers (i.e., TMSPA-treated glass slides with hydrogel arrays, caps to maintain gel height, main chamber, pistons with different heights, membrane with pillars, and pressure chamber) comprising the combinatorial high-throughput system. A representative photograph showing a combinatorial system for screening combinatorial cues (Photo credit: J.L., University of California, Los Angeles) (please also see fig. S1, F and G). Scale bar, 20 mm. (D) Schematic illustrations of cross-sectional view of the combinatorial system. (E) Compressive modulus of PEG/OMA hydrogels (8, 10, and 12% PEG/OMA) with or without cells on day 0 (n = 5) (One-way ANOVA with Tukey’s significant difference post hoc test; *P < 0.05 and ***P < 0.005 compared with 8% without cells). (F) Time profile of hydrogel degradation without compression for 21 days (n = 5). (One-way ANOVA with Tukey’s significant difference post hoc test; *P < 0.05 compared with 10% without cell group, **P < 0.05 compared with 12% with cell group, ***P < 0.005 compared with 8% with cells, and ****P < 0.005 compared with 12% without cell group at day 0.) (G) Relative DNA content of cells in PEG/OMA hydrogels with compositions of 8 and 12%, TGF-β1 (10 ng/ml), and RGD conjugation under 0 or 40% cyclic compression (n = 6). (H) Representative DAPI/F-actin images of hMSCs cultured for 7 days in 8 and 12% PEG/OMA gels with or without 40% cyclic compression and quantification of cell spreading at days 1, 3, and 7 (n = 3). Scale bar, 300 μm (inset: 100 μm).
RESULTS AND DISCUSSION
Stem cell–based cartilage repair strategy using engineered multicomponent biomaterials and a combinatorial system
Although biomaterial degradation and changes in stiffness are critical design variables and interrelated, few studies have focused on the interplay between these important variables and their effect on cell behavior due to the complexity of considering the two phenomena, which typically occur simultaneously (25, 26). Since multicomponent hydrogels provide the potential for tunable material properties (e.g., stiffness and degradation) and biological functionality (27), in this study, eight-arm polyethylene glycol acrylates (PEG) and oxidized methacrylated alginate (OMA) were used to synthesize artificial ECMs. OMA is biodegradable and biocompatible and exhibits increased cell adhesion interactions compared with non–cell-adhesive, nonoxidized alginate (28). PEG is a synthetic biomaterial that is highly elastic (29), and for this study, we used a biodegradable and photo–cross-linkable PEG. By varying the composition of multicomponent hydrogels composed of PEG and OMA, we anticipate that PEG/OMA hydrogels could be produced that are highly elastic under cyclic mechanical loading and have controllable degradation rate and stiffness. Here, we used this biomaterial system to investigate (i) the effect of different levels of degradation on early MSC proliferation and (ii) the role of hydrogel stiffness changes and degradation profile in hMSC maturation into articular or hypertrophic chondrocytes (Fig. 1B). The degradation mechanism of the PEG/OMA hydrogels is based on hydrolysis of acrylate esters in the cross-linked hydrogel networks. In addition, OMA is further vulnerable to hydrolysis due to the oxidization process before methacrylation, which changes the uronate residue conformations to an open-chain adduct as previously reported (28). For this reason, the OMA in the PEG/OMA hydrogels is degraded faster than PEG, leading to the sudden decrease in mechanical properties of the system at the early time point. Moreover, the degree of degradation could possibly be controlled by varying the amount of OMA and PEG as well as by varying the levels of oxidation and methacrylation in the system. To explore how the cross-linking density could control both degradation and stiffness, PEG/OMA hydrogels (PEG:OMA = 1:3) were prepared with various final macromer concentrations (8, 10, and 12 % w/v). While hydrolysis is the only mechanism for degradation of the hydrogel network, the different susceptibilities of PEG and OMA to hydrolysis could regulate the mechanical properties of the PEG/OMA system over 3 weeks.
The combinatorial system was fabricated with a main chamber and pistons composed of methacrylated polyester resin that was cross-linked in a three-dimensional (3D) printer, poly(methyl methacrylate) (PMMA) caps, and a polydimethylsiloxane (PDMS) membrane with pillars and pressure chamber; all parts were assembled using silicon glue (Fig. 1C and fig. S1A). The combinatorial system can control all of the important microenvironmental parameters described above [three compositions (with varying cross-linking density) × two adhesion ligand states [with and without the ubiquitous RGD (arginine-glycine-aspartic acid) peptide sequence coupled to the hydrogel backbone] × three concentrations of potent chondrogenic protein transforming growth factor–β1 (TGF-β1) (0, 5, and 10 ng/ml) × four dynamic compressive strain levels (0, 10, 25, and 40%)] at a frequency of 1 Hz × four replicates: a synergistic effect of all the parameters including three compositions, two RGD states, three TGF-β1 concentrations, and four dynamic mechanical strains with four replicates = array of 288 hydrogels. Six different biomaterial conditions (three compositions × two RGD states) were separated using a PMMA structure, and hydrogel arrays (1 mm in diameter and height) were cross-linked via ultraviolet (UV) exposure through a patterned photomask (fig. S1B). Three divided media chambers separated three TGF-β1 concentrations, and different heights of pistons (2.2, 2.5, 2.65, and 2.8 mm) permitted application of four different compressive strains (Measured actual compression: 0, 11, 25, and 41% strain) (fig. S1, C to G). The combinatorial system can identify combinations of cues to understand resulting 3D cellular responses during chondrogenesis (Fig. 1D).
To determine the weight % composition of PEG/OMA hydrogels that will facilitate the study of chondrogenic differentiation of encapsulated hMSCs, degradation of 4 to 12 % w/v hydrogels with or without cells and glycosaminoglycan (GAG) deposition were examined for 21 days in the chondrogenic differentiation media (fig. S2, A and B). Complete degradation was observed when cells were incorporated in PEG/OMA hydrogels with compositions of 4 and 6 % w/v within 21 days of culture, which indicated these compositions are not suitable for screening of chondrogenic differentiation of hMSCs over this time period. Therefore, a higher macromer concentration was chosen with three different PEG/OMA concentrations (8, 10, and 12 % w/v representing 2:6, 2.5:7.5, and 3:9 PEG/OMA, respectively) to identify the combinatorial effect of PEG/OMA’s degradation rate and stiffness on cartilage synthesis.
The mechanical stiffness of the PEG/OMA hydrogels increased with higher concentrations, but it was also noted that their stiffness decreased with the encapsulation of cells likely due to the disruption of cross-linking in the presence of encapsulated cells resulting in a decreased cross-linking density (Fig. 1E). The stiffness change of the PEG/OMA hydrogels was measured under different compressive strains for 21 days (Fig. 1F and fig. S2C). The stiffness of PEG/OMA hydrogels rapidly decreased over the first 7 days. However, a similar rate of decrease in stiffness was observed over 21 days regardless of hydrogel composition, and there was no significant change in compressive modulus after 7 days between two compositions (~4.5 kPa for 8% and ~15.2 kPa for 12% PEG/OMA). The swelling and mass loss of PEG/OMA hydrogels were measured for 21 days (fig. S2, D and E). As the hydrogels degraded, the swelling of all hydrogels continuously increased for 21 days. The 8% PEG/OMA hydrogel exhibited more extensive swelling and faster mass loss kinetics compared with the other groups. However, the trends in hydrogel swelling were different from those of the moduli. This may be because the sudden decrease in moduli is caused by the rapid degradation of OMA, but slowly degrading PEG could maintain the hydrogel shape, leading to a continuous increase in swelling over 3 weeks. Analysis of cell proliferation indirectly through DNA quantification over time demonstrated that faster degrading and soft matrices (8% PEG/OMA) induced a higher level of cell proliferation (0% strain: ~1.14-fold and ~1.13-fold, on days 7 and 14, respectively; 40% strain: ~1.12-fold, ~1.16-fold, and ~1.12-fold on days 7, 14, and 21, respectively) compared with slower degrading and stiff ones (12% PEG/OMA) (Fig. 1G). Increased levels of proliferation were observed for cells cultured in 8% PEG/OMA with and without compressive strain on days 7 and 14, but the maintained level was only observed for those cultured over 21 days under 40% strain. The encapsulated hMSCs exhibited high cell viability (>80%) in the PEG/OMA hydrogels (fig. S3A), indicating the macromers, photoencapsulation process, PEG/OMA hydrogels and their degradation byproducts, and the mechanical compression had minimal effect on hMSC survival. It was also examined how the polymer composition regulated cell spreading and proliferation (Fig. 1H). hMSCs photoencapsulated in 8% PEG/OMA showed higher cell area at early time points than in 12% PEG/OMA, followed by increased proliferation but their area decreased likely due to the larger number of multicellular clusters that formed in the 8% hydrogels compared with the 12% hydrogels (fig. S3B).
Cross-linking density of PEG/OMA that could guide different levels of both degradation and stiffness regulating articular or hypertrophic chondrogenesis of hMSCs
The combinatorial system allows for simultaneous combinatorial screening of the role of biomaterial composition, soluble signaling, and compressive mechanical stimulation on chondrogenesis of hMSCs. After 21 days in culture, the ability of these different parameters to direct hMSCs down the chondrogenic lineage was examined using immunohistochemistry against widely used chondrogenic markers, such as collagen type II and aggrecan. A synergistic effect of all the parameters (three compositions, two RGD states, three TGF-β1 concentrations, and four dynamic mechanical strains) on hMSC chondrogenesis was observed (Fig. 2, A and B, and fig. S4, A and B). Specifically, TGF-β1 plays a bigger role in promoting the chondrogenic phenotype of hMSCs than other cues used in this study, while biomaterial composition plays a key role in determining chondrogenic fate of hMSCs in the presence of the growth factor, mechanical compression, and RGD (Fig. 2C and fig. S4C). We observed that chondrogenic marker expression of hMSCs was significantly increased in 8% PEG/OMA hydrogels when synergized with the other factors used. However, hMSCs encapsulated in 12% PEG/OMA hydrogels exhibited a higher expression level of Runx2, a representative hypertrophy marker (Fig. 2, D to F, and fig. S4D). This is also supported by the staining results (costaining with aggrecan and Runx2) from cells cultured for 21 days in the combinatorial system, showing distinct phenotypes between cells cultured in 8 and 12% PEG/OMA with the presence of RGD (Fig. 2G and fig. S4E). Moreover, by using the combinatorial system, it can be explored how individual variables regulate the chondrogenesis of hMSCs. For instance, when only compositions and compressive strains are plotted as shown in Fig. 2G [TGF-β1 concentration (10 ng/ml) and RGD presence are fixed], the cross-linking density that regulates both degradation and stiffness plays a decisive role in directing articular (cells cultured in 8%) or hypertrophic (cells cultured in 12%) hMSC chondrogenesis regardless of applied cyclic compression level (0, 10, 25, and 40%).
Fig. 2. Cross-linking density of PEG/OMA that could guide different levels of both degradation and stiffness regulates expression level of markers associated with articular or hypertrophic chondrogenesis of hMSCs.
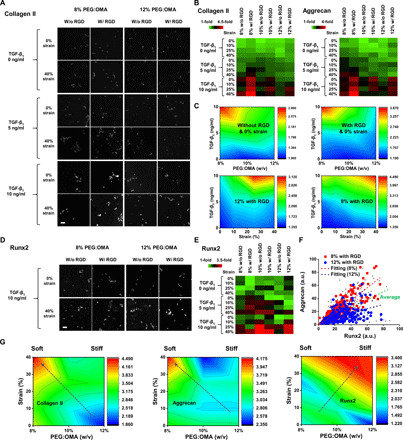
(A) Representative 3D confocal images of hMSCs stained with collagen II in different combinations of parameters. (B) Quantified heat maps of collagen II and aggrecan markers for cells encapsulated in the hydrogel microarrays cultured with combinations of all the factors for 21 days (n = 4). All data were normalized by the condition, 10% PEG/OMA without the presence of RGD, compressive stain, and TGF-β1 supplement. (C) Surface plot displaying the effect of the two variables on chondrogenic marker expression (collagen II) of hMSCs when other two factors are fixed. (D) Representative 3D confocal images of hMSCs stained with representative osteogenic marker (Runx2) in different combinations of parameters. (E) Quantified heat maps of Runx2 for cells encapsulated in the hydrogel microarrays cultured with combinations of all the factors for 21 days (n = 4). (F) Plot of measured immunofluorescence intensity data to define the role of Runx2 in lineage specification of hMSCs. Data were selected randomly (n = 1000). a.u., arbitrary unit. (G) Surface plot displaying the effect of the variables composition and strain on collagen II, aggrecan, Runx2 expression of hMSCs with other factors (with RGD and 10 ng/ml TGF-β1 supplement) fixed. Scale bars, 100 μm.
Engineered multicomponent biomaterials controlling hypertrophic chondrogenesis of hMSCs
To gain insight into how the mechanotransduction process mediated by the matrix degradation and mechanics influences the chondrogenesis of hMSCs, we studied the level of YAP activity for hMSCs cultured in the combinatorial system. The expression of nuclear YAP for hMSCs grown in different conditions was time dependent. Encapsulated hMSCs in PEG/OMA hydrogels showed a low level of nuclear YAP expression regardless of the composition for the first 3 days, while the expression level of nuclear YAP increased for cells at 7 days of culture in a composition-dependent manner. Increased nuclear YAP expression was evident in cells, especially those encapsulated in 12% PEG/OMA at day 7 (Fig. 3, A and B, and fig. S5A), and a similar trend was also observed at day 21 (Fig. 3C). Quantification of the nuclear/cytoplasmic ratio of YAP demonstrated that the condition for hypertrophic chondrogenesis (12% PEG/OMA together with other cues) induced a higher level of the ratio for cells cultured for 3 and 7 days compared with the condition for articular chondrogenesis (8% PEG/OMA together with other cues) (fig. S5B). In agreement with previous studies that have reported culturing hMSCs in chondrogenic culture medium for 7 days was enough to promote prechondrogenic differentiation (30, 31), we also observed elevated expression level of YAP for cells cultured in 12% conditions for 7 days, indicating that YAP-dependent mechanotransduction may play a crucial role in promoting the hypertrophic transition of prechondrogenic hMSCs. These trends in the YAP nuclear expression were further verified with immunofluorescence costaining of collagen II and YAP, which corresponds to the trends in expression (fig. S5C). To decipher differential roles of multiple parameters in different expression levels of markers, we obtained linear regression (LR) coefficients describing the correlation between variables and the response; a positive value indicates that as a variable increases, the response also increases, and vice versa (Fig. 3D). The coefficients predict that the levels of both collagen II and aggrecan are highly correlated to the RGD presence, higher TGF-β1 concentration, and 8% of PEG/OMA composition, while the levels of both Runx2 and YAP are negatively correlated to 8% of PEG/OMA composition. Moreover, all the combinations of different cues in the study were ranked on the basis of the normalized expression levels of positive (collagen II and aggrecan) and negative (Runx2 and YAP) markers, demonstrating that engineered multicomponent biomaterials play a key role in the regulation of articular and hypertrophic chondrogenesis of hMSCs (Fig. 3E and fig. S6).
Fig. 3. Engineered multicomponent biomaterials regulate hypertrophic chondrogenesis of hMSCs through YAP-dependent mechanotransduction and WNT signaling pathway.
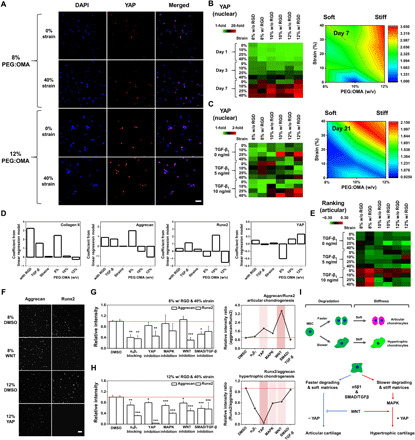
(A) Representative 3D confocal images of hMSCs cultured in different combinations of parameters for 7 days and stained with YAP. (B) Quantified heat map and surface plot of measured immunofluorescence nuclear intensity of YAP at days 1, 3, and 7 (n = 4). (C) Quantified heat map and surface plot of YAP for cells encapsulated in the hydrogel microarrays cultured with combinations of all the factors for 21 days (n = 4). (D) Plots of LR model coefficients across the parameters regulating expression levels of articular (collagen II and aggrecan) and hypertrophic (Runx2 and YAP) cartilage markers. (E) Quantified heat map of scores based on positive correlation of articular (collagen II and aggrecan) and negative correlation of hypertrophic (Runx2 and YAP) cartilage marker expression levels for the best chondrogenic metrics on day 21. (F) Representative 3D confocal images of cells stained with aggrecan and Runx2 when treated with an inhibitor of the WNT signaling pathway for the 8% PEG/OMA condition or of the YAP signaling pathway for the 12% PEG/OMA condition. Relative expression of aggrecan and Runx2 markers for cells cultured in (G) 8% PEG/OMA hydrogels with or without WNT inhibitor or (H) 12% PEG/OMA hydrogels with or without YAP inhibitor, demonstrating possible pathways that promote hypertrophy in articular-like or hypertrophic-like chondrogenesis (n = 4). (P values were obtained on the basis of one-way ANOVA with Tukey’s post hoc testing *P < 0.05, **P < 0.005, and ***P < 0.0005 compared with the DMSO condition.) (I) Proposed pathway for mechanical stimuli guiding articular or hypertrophic chondrogenesis of hMSCs. Scale bars, 100 μm.
To identify possible mechanotransduction pathways that regulate the articular and hypertrophic chondrogenesis of hMSCs, cells were treated with blocking antibodies against integrin α5β1 and inhibitors for YAP, mitogen-activated protein kinases (MAPKs), WNT signaling, or SMAD/TGF-β. Addition of dimethyl sulfoxide (DMSO) led to negligible changes in the expression of aggrecan but a notable increase in the Runx2 expression, especially for the 12% condition (fig. S7A). Since the pathway inhibitors were supplemented with DMSO (1 μl/ml), we also supplemented all other cultures including controls and blocking antibodies with the same concentration of DMSO to accurately compare between the conditions. Addition of the WNT inhibitor led to a significant decrease in the expression of Runx2 for cells cultured in the condition promoting articular chondrogenesis with a slight increase in the aggrecan expression, leading to an elevated level of aggrecan/Runx2 ratio (Fig. 3, F and G). This suggests down-regulation of the WNT signaling pathway may promote articular-like cartilage formation by cells cultured in faster degrading and soft matrices. However, supplementing the cultures in the condition guiding hypertrophic chondrogenesis with YAP inhibitor significantly attenuated the expression of Runx2 with a relatively subtle decrease in aggrecan expression, resulting in a huge decrease in the level of the Runx2/aggrecan ratio in slower degrading and stiff matrices (Fig. 3H). This result supports the theory that the cells differentiate into hypertrophic chondrocytes through YAP-dependent mechanotransduction in slower degrading and stiff matrices. Together, these results suggest that cross-linking density that could guide different levels of both degradation and stiffness can modulate different mechanotransduction and signaling pathways to regulate articular and hypertrophic chondrogenesis of hMSCs (Fig. 3I).
Down-regulation of WNT signaling pathway required for production of better quality of articular cartilage–like tissue in vivo
To examine whether the finding obtained at the small scale using the combinatorial system may be translated to tissue-relevant constructs, the hydrogels (small-scale gels with 1-mm diameter to large-scale gels with 8-mm diameter) were scaled up and chondrogenic differentiation of encapsulated cells was characterized. Different dynamic strains (0 or 40%) at 1 Hz were applied to the 8 or 12% PEG/OMA modified with RGD peptide in the presence of TGF-β1 (10 ng/ml) for 21 days. Similar to the immunohistochemistry results from the microgels (Fig. 2, B and E), total amount or normalized amount to the DNA content of GAG, a representative articular cartilage marker, or alkaline phosphatase (ALP) activity, a representative osteogenic marker, increased when cells were cultured in 8 or 12% PEG/OMA under dynamic compression, respectively, at days 7, 14, and 21 (Fig. 4A and fig. S7B). After 21 days of culture, hematoxylin and eosin (H&E) staining and staining with markers of articular chondrogenesis (Safranin O and Toluidine blue O) were performed to further confirm the composition-based articular chondrogenic differentiation (Fig. 4B and figs. S7C and S8). The intensity of the Toluidine blue O staining signals of the 8% PEG/OMA group was greater than that of the 12% PEG/OMA group (Fig. 4B). We also performed gene expression analysis using real-time quantitative reverse transcription polymerase chain reaction (qRT-PCR) of a panel of markers associated with articular [collagen type II a1 (Col2a1), aggrecan (ACAN), and Sox9] and hypertrophic [ALP, collagen type I a1 (Col1a1), collagen type X a1 (Col10a1), osteocalcin (OCN), and osterix (OSX)] (Fig. 4C and fig. S9, A and B). After 21 days of culture, a higher degree of articular chondrogenic transcripts for cells cultured in the condition promoting articular chondrogenesis (8% PEG/OMA with RGD at 40% strain stimulation) and a higher expression of hypertrophic transcripts for cells cultured in the condition for hypertrophy (12% PEG/OMA with RGD at 40% strain stimulation) were observed, which was consistent with the immunofluorescence (Fig. 2, B and E) and histology (Fig. 4B) results. Cells cultured in the condition promoting articular chondrogenesis showed a ~2.3-fold enhanced expression of key articular marker gene Col2a1 compared with cells cultured in the hypertrophic chondrogenesis condition. The same trend was also observed in the expression of ACAN. (~1.6-fold) and Sox9 (~1.3-fold). In contrast, the hypertrophic condition resulted in ~4.9-fold, ~13.8-fold, ~12.4-fold, ~1.3-fold, and ~1.4-fold increases in ALP, Col1a1, Col10a1, OCN, and OSX expression, respectively. These results demonstrate clear differences in gene expression linked to the degree of matrix degradation and stiffness that support the immunofluorescence findings at the small scale in the combinatorial system.
Fig. 4. hMSCs cultured in high-ranked conditions for the prevention of hypertrophic chondrogenesis show higher potency for cartilage repair in vitro and in vivo.
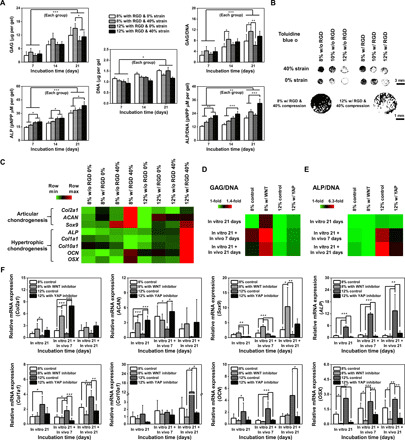
(A) Quantification of GAG/DNA and ALP/DNA for 21 days in culture of hMSCs in RGD-conjugated PEG/OMA gels supplemented with TGF-β1 under 40% compression compared with those cultured without compression (n = 6). (B) Toluidine blue O staining after treatment with threshold-based imaging analysis. (C) Heat map of real-time PCR gene expression results associated with articular or hypertrophic chondrogenic differentiation for cells cultured in the combinatorial bioreactor with modulation of mechanical stimuli for 21 days (n = 6 for Col2a1, ACAN, Sox9, and ALP & n = 3 for Col1a1, Col10a1, OCN, and OSX). Quantification of (D) GAG/DNA and (E) ALP/DNA for cells cultured for 21 days with or without WNT and YAP inhibitors and transplanted in vivo for 7 or 21 days after the in vitro pretreatment (n = 6). (F) Real-time PCR quantification of gene expression associated with chondrogenic (Col2a1, ACAN, and Sox9) or osteogenic (ALP, Col1a1, Col10a1, OCN, and OSX) differentiation for cells before and after transplantation in vivo (n = 3 to 5, please see fig. S9D). (*P < 0.05, **P < 0.005, and ***P < 0.0005 based on one-way ANOVA with Tukey’s post hoc testing). Scale bar, 100 μm.
We speculated that the signaling pathways promoting elevated expression of hypertrophic markers (WNT for 8% and YAP for 12%) could influence the fate of articular or hypertrophic phenotypes after transplantation in vivo. To verify this, encapsulated hMSCs were pretreated for 21 days with WNT (for 8% articular phenotype promoting hydrogels) or YAP (for 12% hypertrophic phenotype promoting hydrogels) inhibitors to prevent them from progressing down the hypertrophic pathways, followed by subcutaneous transplantation into 6- to 8-week-old C57BL/6 mice. The transplanted constructs were collected at days 7 and 21 to compare the degree of articular and hypertrophic chondrogenesis of hMSCs with those before implantation that had been cultured for 21 days in vitro. Both GAG and ALP normalized to DNA content, and qRT-PCR analysis revealed that cells cultured in the condition promoting articular chondrogenesis kept their phenotypes even after 21 days in culture in vivo (Fig. 4, D to F, and fig. S9, C and D). In addition, both the conditions supplemented with WNT inhibitor (8% PEG/OMA) and YAP inhibitor (12% PEG/OMA) showed attenuated expression of markers associated with hypertrophic chondrogenesis compared with the controls without a supplement of inhibitors, which show elevated expression of hypertrophic markers. Despite suppression of the hypertrophic chondrogenic markers with WNT and YAP inhibitions, the trend of H&E and Toluidine blue O staining results did not follow the qRT-PCR results (fig. S9E). Together, these results reveal that a combination strategy of engineered biomaterials with cross-linking density that could guide different levels of both degradation and stiffness while blocking hypertrophic pathways may be a promising strategy for generating articular phenotypes for cartilage regeneration.
Our results demonstrate that the design of engineered microenvironments and biomaterials is critical for regulating cell behavior and the resulting chondrogenic fate of hMSCs and cartilage phenotype. However, the majority of current research is focused on understanding the role of individual factors affecting cell fate decisions. Thus, screening combinatorial cues is important to decipher the interacting roles of signals existing in the complex microenvironment. In this regard, we uncovered that cross-linking density that could guide different levels of both degradation and stiffness in concert with other factors known for promoting chondrogenesis (i.e., cell-biomaterial interactions, TGF-β, and cyclic compression) enhances articular chondrogenesis while suppressing hypertrophic chondrogenesis. Although we revealed that faster degrading and soft matrices could promote articular cartilage tissue formation of hMSCs by inducing their proliferation and maturation, we acknowledge that decoupling of degradation and stiffness in our system may lead to additional valuable insights into how these factors may interact during hMSC chondrogenesis. Future studies could be conducted with mixtures of degradable and nondegradable biomaterials, which may permit decoupling of the two parameters. In addition, since we used chondrogenic differentiation media in culture, hMSCs across the conditions underwent prechondrogenic differentiation. hMSCs cultured in both 8 and 12% hydrogels underwent prechondrogenic differentiation in culture as evidenced by (i) the amount of GAG measured increasing regardless of the conditions for cells cultured over 3 weeks as shown in Fig. 4A and (ii) the high level of Toluidine blue O staining observed regardless of the conditions, although the intensity of the Toluidine blue O staining of the 8% PEG/OMA group was greater than that of the 12% PEG/OMA group (Fig. 4B). However, the differentiation cocktail was not the only factor influencing differentiation down the chondrogenic lineage; the PEG/OMA composition was a decisive factor that could regulate articular or hypertrophic chondrogenesis of hMSCs together with soluble cues known to support hMSC chondrogenesis. Therefore, these findings emphasize the importance of tailored microenvironments and biomaterial design in tissue engineering and, specifically, may play a pivotal role in positively augmenting strategies to regenerate functional cartilage tissue.
MATERIALS AND METHODS
PEG/OMA gel preparation and binding ligand immobilization
PEG was synthesized by conjugating acryloyl chloride (Sigma-Aldrich, St. Louis, MO) to the hydroxyl groups of eight-arm polyethylene glycol (10 kDa; JenKem Technology USA, Allen, TX) in the presence of triethylamine (Sigma-Aldrich), as previously described (32). PEG was collected by precipitating the reaction into a 2:1 mixture of diethyl ether/hexane. The polymer was then rehydrated in ultrapure deionized water (diH2O) followed by dialysis against diH2O using a 3500-Da cutoff membrane for 3 days at 4°C. The dialyzed solution was filtered (0.22-μm filter), frozen, and lyophilized until dry. To characterize the resulting polymer, PEG was dissolved in deuterium oxide (D2O), and the 1H nuclear magnetic resonance (NMR) spectrum was recorded on a Varian Unity 300 (300 MHz) NMR spectrometer (Varian Inc.) using 3-(trimethylsilyl)propionic acid-d4 sodium salt (0.05 % w/v) as an internal standard.
The oxidized alginate was prepared by reacting sodium alginate (Protanal LF 20/40, FMC BioPolymer) with sodium periodate (Sigma-Aldrich) using a modification of a previously described method (28). Briefly, sodium alginate (10 g) was dissolved in ultrapure diH2O (900 ml) overnight. Sodium periodate (1.08 g) was dissolved in 100 ml of diH2O and added to alginate solution under stirring in the dark at room temperature (RT) for 24 hours. The OMA macromer was prepared by reacting oxidized alginate (OA) with 2-aminoethyl methacrylate (AEMA: Polysciences Inc.) (28). To synthesize OMA, 2-morpholinoethanesulfonic acid (19.52 g; Sigma-Aldrich) and NaCl (17.53 g) were directly added to an OA solution (1 liter), and the pH was adjusted to 6.5. N-hydroxysuccinimide (NHS, 1.45 g; Sigma-Aldrich) and 1-ethyl-3-(3-dimethylaminopropyl)-carbodiimide hydrochloride (EDC, 4.84 g; Sigma-Aldrich) (molar ratio of NHS:EDC = 1:2) were added to the mixture to activate 20% of the carboxylic acid groups of the alginate. After 5 min, AEMA (2.09 g) (molar ratio of NHS:EDC:AEMA = 1:2:1) was added to the product, and the reaction was maintained in the dark at RT for 24 hours. The reaction mixture was precipitated with the addition of excess of acetone, dried in a fume hood, and rehydrated to a 1% w/v solution in diH2O for further purification. The OMA was purified by dialysis against diH2O [molecular weight cutoff (MWCO), 3500; Spectrum Laboratories Inc.] for 3 days, treated with activated charcoal (5 g/liter, 50 to 200 mesh: Thermo Fisher Scientific) for 30 min, filtered (0.22-μm filter), and lyophilized. To determine the levels of alginate oxidation and methacrylation, the OMA was dissolved in D2O to 2 % w/v, and the 1H NMR spectrum was recorded on a Varian Unity 300 (300 MHz) NMR spectrometer using 3-(trimethylsilyl)propionic acid-d4 sodium salt (0.05 % w/v) as an internal standard.
PEG and OMA [2:6 (8%), 2.5:7.5 (10%), and 3:9 (12%) weight ratio] were dissolved in Dulbecco’s modified Eagle’s medium (DMEM) with a photo initiator [2-hydroxy-4′-(2-hydroxyethoxy)-2-methylpropiophenone, 0.05 % w/v, Sigma-Aldrich] at pH 7.4. Specifically, CGGGRGDSP peptide (20 mg peptide per gram macromer) was mixed with macromer solutions for 1 hour at RT. To form PEG/OMA hydrogels, PEG/OMA macromer solutions were photo–cross-linked with 365-nm UV light (OmniCure S1000, EXFO Photonic Solutions Inc.) at 20 mW/cm2 for 1 min.
Characterization of the PEG/OMA hydrogels
The photo–cross-linked PEG/OMA hydrogels were lyophilized, and dry weights (Wi) were measured. Dried samples were immersed in 15 ml of DMEM and incubated at 37°C, and the DMEM was replaced after 7 days. At predetermined time points, samples were removed, rinsed with fresh DMEM, and the swollen hydrogel sample weights (Ws) were measured. The swelling ratio (Q) was calculated by Q = Ws/Wi. After weighing the swollen hydrogel samples, the samples were lyophilized and weighed (Wd). The percent mass loss was calculated by (Wi – Wd)/Wi × 100.
The elastic moduli of the PEG/OMA hydrogels were determined by performing uniaxial, unconfined constant strain rate compression tests at RT using a constant crosshead speed of 1 mm/min on a mechanical testing machine (Instron, 5542, USA) equipped with a 100-N load cell. Elastic moduli were calculated from the first nonzero linear slope of the stress versus strain plots within 0 to 10% strain with at least four experimental replicates.
hMSC isolation and culture
To isolate bone marrow-derived hMSCs, bone marrow aspirates were obtained from the posterior iliac crest under a protocol approved by the University Hospitals of Cleveland Institutional Review Board and processed as previously described (33). Briefly, the aspirates were washed with growth medium composed of low-glucose DMEM (DMEM-LG; Sigma-Aldrich) with 10% prescreened fetal bovine serum (FBS; Gibco). Mononuclear cells were isolated by centrifugation in a Percoll (Sigma-Aldrich) density gradient, and the isolated cells were plated at 1.8 × 105 cells/cm2 in DMEM-LG containing 10% FBS, 1% penicillin/streptomycin (P/S; Thermo Fisher Scientific), and fibroblast growth factor–2 (FGF-2) (10 ng/ml; R&D Systems) in an incubator at 37°C and 5% CO2. After 4 days of culture, nonadherent cells were removed, and adherent cells were maintained in DMEM-LG containing 10% FBS, 1% P/S, and FGF-2 (10 ng/ml) with media changes every 3 days. After 14 days of culture, the cells were passaged at a density of 5 × 103 cells/cm2, cultured for an additional 14 days, and then stored in cryopreservation media in liquid nitrogen until use.
hMSCs were cultured in DMEM-LG media (10 mg/ml; Gibco, no. 11885084) supplemented with 10% FBS (Thermo Fisher Scientific, no. 10438026), 1% PS, and recombinant human FGF basic (10 ng/ml; R&D Systems, 146aa) protein. To chondrogenically differentiate hMSCs, cells were cultured in DMEM high-glucose medium (4.5 mg/ml; Gibco, no. 11965092) supplemented with 1% PS, 1% ITS [insulin and transferrin (6.25 μg/ml), bovine serum albumin (6.25 ng/ml), and linoleic acid (5.35 μg/ml)] (Sigma-Aldrich no. l3146), 100 nM dexamethasone (Sigma-Aldrich, D4902), 100 mM sodium pyruvate (GE Healthcare Life Sciences, SH3023901), 100 μM nonessential amino acids (Gibco, no. 11140-076), ascorbic acid-2 phosphate (37.5 μg/ml; Wako USA, no. 013-12061), and recombinant human TGF- β1 [10 ng/ml; human embryonic kidney (HEK) 293 derived; no. 100-21). Passage 3 hMSCs were used at a cell density of around 107 cells/ml.
Fabrication of combinatorial system
The combinatorial system was fabricated by a combination of 3D-printed main chamber and pistons using methacrylated polyester resin cross-linked in a 3D printer (FormLabs 2 3D printer with biocompatible dental SG resin), PMMA caps (width, 63.5 mm; wide, 2.2 mm; height, 0.5 mm) cut by laser cutter, and PDMS (Sylgard 184, Dow Corning, MI) membrane with pillars and pressure chamber. Specifically, 3D-printed constructs (main chamber and pistons) were designed by AutoCAD and printed by the 3D printer, followed by washing with isopropyl alcohol for 10 min, UV after curing (~20 mW/cm2 on a 70°C hotplate for 10 min each side), and then autoclaving according to the manufacturer’s instructions. PMMA molds to form PDMS N2 pressure chamber were obtained using a laser cutter. The mold for the pressure chamber layer consisted of the inverse of the cylindrical chamber with the diameter of 14 and 1-mm height that was divided into two sections to apply even pressure to whole chambers, and each section was connected with 3-mm-wide connecting channels to allow the application of pressure from a single gas inlet. The corresponding PDMS layers (10:1 w/w mixture of PDMS base and curing agent) fabricated onto the PMMA molds were cured at 80°C for 1 hour. PDMS membranes with pillars were obtained with the same process for the PDMS chamber using the dimension of the PMMA mold with the cylindrical chamber (diameter, 8 mm; height, 1.5 mm) and membrane (thickness, 0.5 mm). These PDMS membranes with pillars and pressure chamber were plasma bonded by oxygen plasma treatment (200 mTorr, 80 W for 120 s). The main chamber (width, 70 mm; length, 54 mm; and height, 5.5 mm) designed through AutoCAD and fabricated by the 3D printer was divided into three different media chambers to separate three different growth factor conditions, and pistons with different heights (2.2, 2.5, 2.65, and 2.8 mm) were fabricated to apply different compressional strains (0, 11, 25, and 41% strain) at 1 Hz for 21 days (for 24 hours a day) onto the cross-linked gels (1-mm diameter and height) sandwiched between 3-(trimethoxysilyl)propyl methacrylate (TMSPMA)–treated glass slides and PMMA caps.
Cell encapsulation and culture in the combinatorial system
Cells were trypsinized from the cell culture flask and suspended in PEG/OMA prepolymer solutions with 0.05% photo initiator at a cell density of around 107 cells/ml. To create a hydrogel microarray (1-mm diameter and height), the cell suspended PEG/OMA prepolymer solutions were sandwiched between TMSPMA (Sigma-Aldrich, no. 440159)–treated glass slides and hydrophobically treated glass slides; 1-mm spacers were used to make the gel’s height 1 mm. A photomask (288 hydrogel cylinders with 1-mm diameter) was aligned on the top of the TMSPMA-treated glass slides to selectively polymerize the hydrogel onto the post array. To separate PEG/OMA prepolymer solutions with six different biomaterial conditions (three different hydrogel compositions × two with or without RGD), a PMMA-based structure consisting of eight separate bridges (two at each end for placing a spacer and six for hydrogel separation) was fabricated by CO2 laser cutting. Spacers (1 mm thick) were located on the first and last bridges to make 1-mm-tall hydrogel arrays. TMSPMA-coated glass was placed on top of the bridge structure with separated hydrogel prepolymer solution and subsequently cross-linked by UV exposure (25 mW/cm2 for 180 s) through the mask with 288 holes with 1-mm diameter. The uncross-linked prepolymer solution was removed, and the cross-linked gels were washed with phosphate-buffered saline (PBS) twice. The TMSPMA-treated glass slides containing PEG/OMA hydrogel array were aligned onto the combinatorial system and sealed with Si glue (Fig. 1C). The combinatorial system with hydrogel array was filled with expansion hMSC culture media with recombinant human FGF basic protein (10 ng/ml) or chondrogenic differentiation media. All cell culture experiments were conducted in a humidified incubator with 5% CO2 at 37°C. A WAGO controller and a custom-designed MATLAB program were used to apply cyclic compressions with actuation pressure of around 14 kPa and cyclic frequency of 1 Hz (N2 gas) to the cross-linked hydrogels within the combinatorial system during 21 days of culture.
Cell culture in macroscale gels with a bioreactor
To evaluate the effect of mechanical stimulation on the chondrogenic differentiation of hMSCs encapsulated in macroscale PEG/OMA hydrogels, macromer solutions (8, 10, and 12% weight ratio) with CGGGRGDSP peptide (20 mg peptide per gram macromer) were prepared as described above, and then hMSCs (passage number 3, 5 × 106 cells/ml) were suspended in the macromer solutions. hMSC suspended macromer solutions were placed between quartz (top) and glass (bottom) plates separated by 0.75-mm spacers and then photo–cross-linked with UV light to form hydrogels as described above. hMSC-laden hydrogel construct disks were created using an 8-mm-diameter biopsy punch and placed in wells of 24-well tissue culture plates with 0.5 ml of chondrogenic differentiation media [10% ITS + Premix, 100 nM dexamethasone, l-ascorbic acid-2-phosphate (37.5 μg/ml), 1 mM sodium pyruvate, 100 μM nonessential amino acids, and TGF-β1 (10 ng/ml) in DMEM–high glucose]. The chondrogenic medium was changed every 2 days. The hydrogel constructs were subjected to strain-controlled, unconfined, dynamic mechanical compression using a BOSE bioreactor (ElectroForce BioDynamic Test Instrument, Bose) equipped with a 200-N load cell. Compressive mechanical stimulation was performed using a sine wave with a frequency of 0.5 Hz at 40% strain for 24 hours/day during the entire culture period.
Biochemical assay analysis
Cell viability was examined at days 1, 3, and 7 using a LIVE/DEAD Viability/Cytotoxicity Kit (Invitrogen) according to the manufacturer’s instructions. Cells were cultured in different conditions in different PEG/OMA compositions (8, 10, and 12%) with or without cyclic compression (0 or 40%) or RGD presence for 1, 3, or 7 days. The hydrogels were washed with PBS twice, followed by imaging using an inverted fluorescence microscope (Zeiss Axio Observer 5 microscope, Germany). The number of live/dead cells was counted manually for four samples of each group using ImageJ software.
To determine whether chondrogenic differentiation of hMSCs encapsulated in PEG/OMA hydrogels could be enhanced by mechanical stimulation in vitro, at predetermined time points, chondrogenically differentiated hMSC/hydrogel constructs were homogenized on ice at 35,000 rpm for 30 s using a TH homogenizer (Omni International) in 1 ml of papain buffer (pH 6.5) containing papain (25 μg/ml; Sigma-Aldrich), l-cysteine (2 × 10−3 M; Sigma-Aldrich), sodium phosphate (50 × 10−3 M), and EDTA (2 × 10−3 M). Half of the homogenate was papain digested at 65°C overnight. The following day, GAG content was measured using a dimethylmethylene blue assay in 96-well plates. In each well, 50 μl of supernatant was mixed with 250 μl of dye containing dimethylmethylene blue (16 μg/ml) and glycine (3.04 mg/ml, pH 1.5). The absorbance was read at 595 nm using a plate reader (FMAX, Molecular Devices). Chondroitin-6-sulfate (Sigma-Aldrich) from shark cartilage was used to construct the standard curve. DNA content in supernatant (100 μl) was measured using a Quant-iT Picogreen assay kit (Invitrogen) according to the manufacturer’s instructions. Fluorescence intensity of the dye-conjugated DNA solution was measured using a fluorescence plate reader (FMAX) set at 485-nm excitation and 538-nm emission. To measure the ALP activity, an equal volume of ALP lysis buffer (CelLytic M, Sigma-Aldrich) was added into the homogenate. After vigorous mixing for 1 min, the mixed solutions were centrifuged at 500g with a Sorvall Legend RT Plus Centrifuge (Thermo Fisher Scientific). For ALP activity measurements, supernatant (100 μl) was treated with p-nitrophenylphosphate (pNPP) ALP substrate (100 μl; Sigma-Aldrich) at 37°C for 30 min, and then 0.1 N NaOH (50 μl) was added to stop the reaction. The absorbance was measured at 405 nm using a plate reader (FMAX). A standard curve was made using the known concentrations of 4-nitrophenol (Sigma-Aldrich).
Immunofluorescence and histology
After culturing for 21 days, cells encapsulated in PEG/OMA hydrogels were washed with PBS and fixed with 4% PFA (Sigma-Aldrich) for 20 min. Cells were permeabilized in 0.1% Triton X-100 in PBS for 30 min, followed by blocking with 1% BSA for 15 min. Primary antibody labeling was performed in 1% BSA in PBS for 2 hours at RT (20°C) with antibodies listed in table S1A, followed by washing thrice with PBS. Secondary antibody labeling was performed in 2% goat serum containing 1% BSA in PBS for 30 min in a humid chamber (37°C) with secondary antibodies listed in table S1A. Immunofluorescence microscopy was conducted using a confocal microscope (BC LSM880) or an inverted fluorescence microscope (Zeiss Axio Observer 5 microscope, Germany) for the live/dead assay only.
Histomorphometric analysis was performed to evaluate the chondrogenic differentiation of hMSCs encapsulated in PEG/OMA hydrogels after 21 days of culture. The histological specimens were fixed in formalin, embedded in paraffin, sectioned at a thickness of 10 μm, and examined with H&E staining. The GAG distribution in the PEG/OMA hydrogel constructs was visualized via Safranin O (Acros Organics) staining with a Fast Green (Fisher Chemical) counter stain. Stained sections were imaged using an Olympus BS61VS microscope (Olympus) equipped with a Pike F-505 camera (Allied Vision Technologies). Intensities were compared between different groups with thresholds after subtracting backgrounds by using ImageJ software.
Combinatorics analysis
This involved outlier removal (Tukey’s rule) followed by calculating LR coefficients. Then, the best (top 10) combinations for optimal chondrogenicity were computed, indicated by the lowest values of YAP and Runx2 and the highest values of collagen and aggrecan chondrogenic markers. Figure S6 shows the top 10 combinations and the corresponding heat map.
RNA isolation and RT-PCR
Cells were cultured in PEG/OMA hydrogels for 21 days. The hydrogels were homogenized and lysed in TRIzol reagent (Invitrogen), and total RNA was isolated by chloroform extraction and ethanol precipitation. The total RNA was amplified using TargetAmp 1-Round aRNA Amplification Kit 103 (Epicentre Biotechnologies) according to the manufacturer’s instructions, and then it was reverse transcribed using Superscript III First-Strand Synthesis System for RT-PCR (Invitrogen). SYBR Green Real-Time PCR Master Mix (Invitrogen) was used to perform RT-PCR linearly by cycle number of each primer set. Primer sequences are shown in table S1B.
Inhibition assays
YAP (verteporfin: 3 μM; MedChemExpress, no. HY-B0146), MAPK (PD98059: 10 μM; Selleck Chemicals Llc.), WNTs (ETC-159: 100 nM; MedChemExpress, no. ETC-159), and SMAD/TGF-βR [Galunisertib (LY2157299): 1 μM, Selleckchem, no. S2230] inhibitions were performed by adding media supplemented with these inhibitors after combinatorial system fabrication and with each media change. Integrin-blocking antibodies, especially for α5β1 (Millipore, no. MAB1969), were added to cells in media before encapsulation and with each media change at 1 μg/ml.
Ethics statement
All experiments using live animals followed animal welfare ethical regulations by the U.S. Department of Health and Human Services’ Office of Research Integrity. The experiments were approved by the Institutional Animal Care and Use Committee (IACUC) before experimentation.
In vivo mice study
hMSCs encapsulated PEG/OMA hydrogel construct disks [8 and 12% weight ratio with CGGGRGDSP peptide (20 mg peptide per gram macromer)] were prepared as described above and then placed in wells of 24-well tissue culture plates with 0.5 ml of chondrogenic differentiation media [10% ITS + Premix, 100 nM dexamethasone, l-ascorbic acid-2-phosphate (37.5 μg/ml), 1 mM sodium pyruvate, 100 μM nonessential amino acids, and TGF-β1 (10 ng/ml) in DMEM–high glucose]. To examine whether the characteristics of articular cartilage tissues formed in vitro could be maintained by preventing hypertrophic progression in vivo, WNT inhibitor (ETC-159: 100 nM) and YAP inhibitor (verteporfin: 3 μM) were added into the chondrogenic differentiation media containing 8 and 12% PEG/OMA macroscale hydrogel constructs, respectively. The hydrogel constructs were subjected to strain controlled, unconfined, dynamic mechanical compression using a BOSE bioreactor (ElectroForce BioDynamic Test Instrument, Bose) equipped with a 200-N load cell. Compressive mechanical stimulation was performed using a sine wave with a frequency of 0.5 Hz at 40% strain for 24 hours/day during the entire culture period. The chondrogenic medium was changed every 2 days. After 21 days of culture, to evaluate the maintenance of articular cartilage phenotype, chondrogenically differentiated hMSCs encapsulated in PEG/OMA hydrogel constructs were subcutaneously implanted into mice. The surgical procedures used in this study were conducted according to a protocol approved by the IACUC of Case Western Reserve University, which adhered to the National Institutes of Health Guide for the Care and Use of Laboratory Animals. Six- to 8-week-old athymic mice (SCID) were anesthetized with 3% isoflurane, and chondrogenically differentiated hydrogel constructs were implanted subcutaneously on the dorsa of mice (two constructs per mouse). The incisions were closed, and the mice were administered buprenorphine (0.1 mg/kg) after surgery. hMSC/hydrogel constructs that were differentiated without an inhibitor were implanted into the left side of the subcutaneous pouch, and hMSC/hydrogel constructs differentiated with an inhibitor were implanted into the right side of the subcutaneous pouch. At predetermined time points, the mice were euthanized, and the implants were retrieved. Histomorphometric analysis, biochemical assay, and qRT-PCR were performed as described above.
Microscopy data analysis
Immunofluorescence images were analyzed using ImageJ software. At least five different focal planes were imaged for each single hydrogel pillar, and four different hydrogel replicates were used. Fluorescence intensities of single cells cultured in PEG/OMA hydrogels were used to compare marker expression (aggrecan and collagen II). Using CellProfiler5, an analysis pipeline to identify and segment cell nuclei [4′,6-diamidino-2-phenylindole (DAPI)] and cell bodies was used. A variety of fluorescent marker parameters were then calculated in the cell nuclei and bodies across all the different conditions studied for comparison. For Runx2, average integrated intensity in the nucleus was presented. For YAP, the average ratio of nuclear to cytoplasmic fluorescence intensity was presented. All data were normalized by the lowest value in each set. Analysis pipelines are available upon request.
Statistical analysis
Data were obtained from at least three experimental replicates and repeated at least two times to confirm the trend. The mean and SD were presented unless otherwise specified. One-way analysis of variance (ANOVA) with Tukey’s significant difference post hoc test was used for statistical comparisons between groups. Differences were considered significant at P < 0.05.
Supplementary Material
Acknowledgments
Funding: We acknowledge funding from the NIH (AR066193). Author contributions: J.L., O.J, E.A., and A.K. conceived the ideas and designed the experiments. J.L., O.J., M.K, A.A.A., J.-Y.S., H.N.L., Y.B.L., W.S., P.B., D.S.A., K.L., H.-J.K., S.J.L., and S.R.S. conducted the experiments, and J.L., O.J., S.C. S.R.S., E.A., and A.K. analyzed the data. All authors interpreted the data and wrote the manuscript. Competing interests: E.B. is an inventor on a patent related to this work (no. 10512708, filed 7 March 2014, published 24 December 2019). E.B. and O.J. are inventors on the following patents related to this work: no. 8273373, filed 30 December 2009, published 25 September 2012 and no. 9642914, filed 31 August 2012, published 9 May 2017. E.B. and O.J. are inventors on a pending patent related to this work (No. 15/590,818, filed 9 May 2017). E.B., O.J., and J.S. are inventors on the following pending patents related to this work: no. 16/107,756, filed 21 August 2018; no. PCT/US2018/043643, filed 21 August 2018; and no. PCT/US2019/026678, filed 9 April 2019. The authors declare that they have no other competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/21/eaaz5913/DC1
REFERENCES AND NOTES
- 1.Huey D. J., Hu J. C., Athanasiou K. A., Unlike Bone, Cartilage regeneration remains elusive. Science 338, 917–921 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Felson D. T., Lawrence R. C., Dieppe P. A., Hirsch R., Helmick C. G., Jordan J. M., Kington R. S., Lane N. E., Nevitt M. C., Zhang Y., Sowers M., McAlindon T., Spector T. D., Poole A. R., Yanovski S. Z., Ateshian G., Sharma L., Buckwalter J. A., Brandt K. D., Fries J. F., Osteoarthritis: New insights. Part 1: The disease and its risk factors. Ann. Intern. Med. 133, 635–646 (2000). [DOI] [PubMed] [Google Scholar]
- 3.Zhang W., Ouyang H., Dass C. R., Xu J., Current research on pharmacologic and regenerative therapies for osteoarthritis. Bone Res. 4, 15040 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Iorio R., Robb W. J., Healy W. L., Berry D. J., Hozack W. J., Kyle R. F., Lewallen D. G., Trousdale R. T., Jiranek W. A., Stamos V. P., Parsley B. S., Orthopaedic surgeon workforce and volume assessment for total hip and knee replacement in the united states: Preparing for an Epidemic. J. Bone Joint Surg. Am. 90, 1598–1605 (2008). [DOI] [PubMed] [Google Scholar]
- 5.Ebert J. R., Robertson W. B., Woodhouse J., Fallon M., Zheng M. H., Ackland T., Wood D. J., Clinical and magnetic resonance imaging-based outcomes to 5 years after matrix-induced autologous chondrocyte implantation to address articular cartilage defects in the knee. Am. J. Sports Med. 39, 753–763 (2011). [DOI] [PubMed] [Google Scholar]
- 6.Makris E. A., Gomoll A. H., Malizos K. N., Hu J. C., Athanasiou K. A., Repair and tissue engineering techniques for articular cartilage. Nat. Rev. Rheumatol. 11, 21–34 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caplan A. I., Review: Mesenchymal stem cells: Cell–based reconstructive therapy in orthopedics. Tissue Eng. 11, 1198–1211 (2005). [DOI] [PubMed] [Google Scholar]
- 8.Redman S. N., Oldfield S. F., Archer C. W., Current strategies for articular cartilage repair. Eur. Cells Mater. 9, 23–32 (2005). [DOI] [PubMed] [Google Scholar]
- 9.Chen F. H., Rousche K. T., Tuan R. S., Technology insight: Adult stem cells in cartilage regeneration and tissue engineering. Nat. Clin. Pract. Rheumatol. 2, 373–382 (2006). [DOI] [PubMed] [Google Scholar]
- 10.Scotti C., Tonnarelli B., Papadimitropoulos A., Scherberich A., Schaeren S., Schauerte A., Lopez-Rios J., Zeller R., Barbero A., Martin I., Recapitulation of endochondral bone formation using human adult mesenchymal stem cells as a paradigm for developmental engineering. Proc. Natl. Acad. Sci. U.S.A. 107, 7251–7256 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alsberg E., von Recum H. A., Mahoney M. J., Environmental cues to guide stem cell fate decision for tissue engineering applications. Expert Opin. Biol. Ther. 6, 847–866 (2006). [DOI] [PubMed] [Google Scholar]
- 12.Hui T. Y., Cheung K. M. C., Cheung W. L., Chan D., Chan B. P., In vitro chondrogenic differentiation of human mesenchymal stem cells in collagen microspheres: Influence of cell seeding density and collagen concentration. Biomaterials 29, 3201–3212 (2008). [DOI] [PubMed] [Google Scholar]
- 13.Murphy W. L., Mcdevitt T. C., Engler A. J., Materials as stem cell regulators. Nat. Mater. 13, 547–557 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Salinas C. N., Anseth K. S., The enhancement of chondrogenic differentiation of human mesenchymal stem cells by enzymatically regulated RGD functionalities. Biomaterials 29, 2370–2377 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Horpe S. D., Buckley C. T., Vinardell T., O’Brien F. J., Campbell V. A., Kelly D. J., The response of bone marrow-derived mesenchymal stem cells to dynamic compression following TGF- β3 induced chondrogenic differentiation. Ann. Biomed. Eng. 38, 2896–2909 (2010). [DOI] [PubMed] [Google Scholar]
- 16.Awad H. A., Halvorsen Y.-D. C., Gimble J. M., Guilak F., Effects of transforming growth factor β1 and dexamethasone on the growth and chondrogenic differentiation of adipose-derived stromal cells. Tissue Eng. 9, 1301–1312 (2003). [DOI] [PubMed] [Google Scholar]
- 17.Bosnakovski D., Mizuno M., Kim G., Takagi S., Okumura M., Fujinaga T., Chondrogenic differentiation of bovine bone marrow mesenchymal stem cells (MSCs) in different hydrogels: Influence of collagen type II extracellular matrix on MSC chondrogenesis. Biotechnol. Bioeng. 93, 1152–1163 (2006). [DOI] [PubMed] [Google Scholar]
- 18.Lutolf M. P., Gilbert P. M., Blau H. M., Designing materials to direct stem-cell fate. Nature 462, 433–441 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Watt F. M., Huck W. T., Role of the extracellular matrix in regulating stem cell fate. Nat. Rev. Mol. Cell Biol. 14, 467–473 (2013). [DOI] [PubMed] [Google Scholar]
- 20.Zhong W., Tian K., Zheng X., Li L., Zhang W., Wang S., Qin J., Mesenchymal stem cell and chondrocyte fates in a multishear microdevice are regulated by yes-associated protein. Stem Cells Dev. 22, 2083–2093 (2013). [DOI] [PubMed] [Google Scholar]
- 21.Yang C., Tibbitt M. W., Basta L., Anseth K. S., Mechanical memory and dosing influence stem cell fate. Nat. Mater. 13, 645–652 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dupont S., Morsut L., Aragona M., Enzo E., Giulitti S., Cordenonsi M., Zanconato F., le Digabel J., Forcato M., Bicciato S., Elvassore N., Piccolo S., Role of YAP/TAZ in mechanotransduction. Nature 474, 179–183 (2011). [DOI] [PubMed] [Google Scholar]
- 23.Karystinou A., Roelofs A. J., Neve A., Cantatore F. P., Wackerhage H., de Bari C., Yes-associated protein (YAP) is a negative regulator of chondrogenesis in mesenchymal stem cells. Arthritis Res. Ther. 17, 147 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Deng Y., Wu A., Li P., Li G., Qin L., Song H., Mak K. K., Yap1 regulates multiple steps of chondrocyte differentiation during skeletal development and bone repair. Cell Rep. 14, 2224–2237 (2016). [DOI] [PubMed] [Google Scholar]
- 25.Madl C. M., LeSavage B. L., Dewi R. E., Dinh C. B., Stowers R. S., Khariton M., Lampe K. J., Nguyen D., Chaudhuri O., Enejder A., Heilshorn S. C., Maintenance of neural progenitor cell stemness in 3D hydrogels requires matrix remodelling. Nat. Mater. 16, 1233–1242 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peng Y., Liu Q. J., He T., Ye K., Yao X., Ding J., Degradation rate affords a dynamic cue to regulate stem cells beyond varied matrix stiffness. Biomaterials 178, 467–480 (2018). [DOI] [PubMed] [Google Scholar]
- 27.Jia X., Kiick K. L., Hybrid multicomponent hydrogels for tissue engineering. Macromol. Biosci. 9, 140–156 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jeon O., Alt D. S., Ahmed S. M., Alsberg E., The effect of oxidation on the degradation of photocrosslinkable alginate hydrogels. Biomaterials 33, 3503–3514 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McGann C. L., Akins R. E., Kiick K. L., Resilin-PEG hybrid hydrogels yield degradable elastomeric scaffolds with heterogeneous microstructure. Biomacromolecules 17, 128–140 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Herlofsen S. T., Küchler A. M., Melvik J. E., Brinchmann J. E., Chondrogenic differentiation of human bone marrow-derived mesenchymal stem cells in self-gelling alginate discs reveals novel chondrogenic signature gene clusters. Tissue Eng. Part A 17, 1003–1013 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang J., Tao Y., Zhou X., Li H., Liang C., Li F., Chen Q.-X., The potential of chondrogenic pre-differentiation of adipose-derived mesenchymal stem cells for regeneration in harsh nucleus pulposus microenvironment. Exp. Biol. Med. 241, 2104–2111 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nguyen M. K., Jeon O., Krebs M. D., Schapira D., Alsberg E., Sustained localized presentation of RNA interfering molecules from in situ forming hydrogels to guide stem cell osteogenic differentiation. Biomaterials 35, 6278–6286 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lennon D. P., Haynesworth S. E., Bruder S. P., Jaiswal N., Caplan A. I., Human and animal mesenchymal progenitor cells from bone marrow: Identification of serum for optimal selection and proliferation. Vitr. Cell. Dev. An. 32, 602–611 (1996). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/21/eaaz5913/DC1


