A new approach connects synthetic biology with microelectronics via engineered microbial population dynamics.
Abstract
While there has been impressive progress connecting bacterial behavior with electrodes, an attractive observation to facilitate advances in synthetic biology is that the growth of a bacterial colony can be determined from impedance changes over time. Here, we interface synthetic biology with microelectronics through engineered population dynamics that regulate the accumulation of charged metabolites. We demonstrate electrical detection of the bacterial response to heavy metals via a population control circuit. We then implement this approach to a synchronized genetic oscillator where we obtain an oscillatory impedance profile from engineered bacteria. We lastly miniaturize an array of electrodes to form “bacterial integrated circuits” and demonstrate its applicability as an interface with genetic circuits. This approach paves the way for new advances in synthetic biology, analytical chemistry, and microelectronic technologies.
INTRODUCTION
Our ability to investigate gene expression through various monitoring methods plays an essential role in advancing our understanding of biology (1, 2). Synthetic biology is a field where these methods are of critical importance for characterizing genetic circuit behavior and applications where the output of circuit expression is used to accomplish a certain task, such as sensing. Most methods rely on fluorescent reporter genes, which can convert a biological response into a detectable signal (3, 4). These methods generally require sophisticated optics and may have some intrinsic challenges such as the presence of background signal, toxicity, or the need for substrates.
Meanwhile, advances in microelectronics have provided a promising alternative in the progress of smart analytical tools (5, 6). In particular, the properties of biological molecules have been leveraged in microelectronic devices for sensing (7, 8) and mechanical actuation applications (9). In these approaches, optical and electrochemical transducers have been coupled with enzymes, antibodies, and aptamers (10, 11). Despite the high selectivity of these technologies, they are limited by their detection capabilities due to the saturation after several measurements, the requirement of expensive purification procedures, and complex immobilization or labeling steps (12).
Connecting bacterial gene expression to electrodes is an appealing approach to interface genetic circuits with microelectronics for multiple applications. General progress in this direction has been focused on energy generation applications from bacterial cultures where electrons are transferred via extracellular pathways (13–19). In addition, redox species have been included or produced in the cell cultures to serve as a molecular connection between electronics and bacterial colonies (20–24). Although linking redox molecules to engineered expression allows a simple electronic output, these molecules are usually toxic at high levels and do not provide a general platform for tracking gene expression (25). A complementary focus is the creation of a general interface between cells and electronics that does not rely on electron transfer from bacteria to electrodes or redox intermediaries. Such a strategy would serve to facilitate advances in synthetic biology where the output of different kinds of genetic circuits may be easily connected to an electrical measure.
During bacterial growth, charged ions are naturally released because of metabolic processes (26–28). As a result, the surrounding medium becomes more conductive, which decreases the impedance to electrical current (29, 30). To leverage this phenomenon for the measurement of gene circuit behavior, we engineered the expression of a bacterial killing gene as the output. The resulting behavior can be conceptualized as a bacterial population decreasing in culture impedance over time due to the release of charged ions, after which cell death decreases ion generation and increases impedance (Fig. 1A). We monitor electrochemical impedance spectroscopy of the bacterial culture in a device that includes electrodes and a turbidity sensor as a secondary output measurement. We have developed a set of applications including a heavy metal sensor via a population control circuit and a dynamic genetic oscillator, which is synchronized to lyse periodically. We lastly pair an array of miniaturized electrodes to form “bacterial integrated circuits” (bICs) to demonstrate a portable interface with genetic circuits for new science advances in synthetic biology, analytical chemistry, and microelectronic technologies.
Fig. 1. Interfacing genetic circuits with electrical measurement.
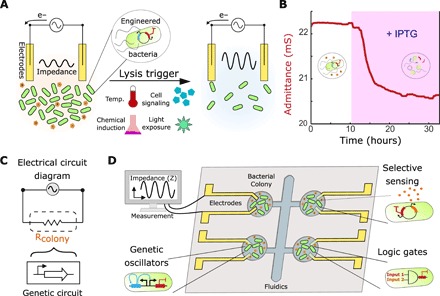
(A) Schematic of the approach using a culture of bacteria with a killing gene as the circuit output in contact with inert gold electrodes. The impedance of the culture reduces during growth (left) and increases upon the induction of bacterial death due to the clearance of charged metabolites (right). (B) Profile of the admittance (red), which is the inverse of impedance, using an IPTG-inducible lysis construct (pE35GFP) (42) in an electrochemostat device. The pink shaded region represents induction of lysis with 1 mM IPTG in the medium. (C) Schematic of the equivalent electrical circuit for our strategy using an alternating input voltage. The bacterial population is simplified to a resistor, which is controlled by a genetic circuit. (D) Schematic showing a microelectronic platform to interface between engineered bacteria and electronics. Several chambers may contain unique genetic circuits, connected via electrodes to an impedance output system.
RESULTS
To transduce gene circuit output to an electronic signal, we engineered bacterial circuits capable of controlling the release of ionic species via cell growth and death. We used inert gold electrodes to measure the AC impedance when applying a small sinusoidal voltage to the bacterial population without externally added redox mediators (fig. S1A). This measurement naturally includes resistive and capacitive effects (31, 32). These can be decoupled by using different input potential frequencies explored between 1 Hz to 100 kHz (fig. S1B). The correspondent Bode plots (representing impedance over frequency) indicate that at high frequencies, the AC current and voltage signals are in phase with each other. At these frequencies, the phase is virtually 0, and therefore, the circuit is simplified to an active bacterial media resistance. Since the impedance resulting from these resistive effects (fig. S1C) was generally more robust to changes in the input frequency than the one at lower frequencies (fig. S1D), we elected to track the culture resistance. In addition, this optimized measurement was found to accurately correlate with the population density (monitored via turbidity) as opposed to measurements at lower frequencies (fig. S2 and Supplementary Text). To demonstrate the electrochemical interface with a minimal expression system in bacteria, we tested a previously described construct pE35GFP containing a bacterial lysis gene. The engineered bacteria grew to a stable density in the continuous culture before triggering the lysis gene upon chemical induction with isopropyl β-d-1-thiogalactopyranoside or IPTG. We observed a decrease of more than half in culture density upon lysis induction, corresponding to an admittance (inverse of the impedance) change of approximately 2 millisiemens (mS; Fig. 1B). This demonstrates that the induction of bacterial lysis was sufficient to cause a marked change in the conductive properties of the culture.
In addition, we found that the impedance was lower in a heat-killed culture than in an intact one (fig. S1C). We also investigated the contribution of the bacterial cells and their metabolic by-products to the culture resistance (fig. S1, C to E). We found that the resistive effects were dependent on the metabolic by-products from the bacterial growth and not the bacterial cells (fig. S1E). Since the engineered bacteria are essentially modulating the flow of electrical current in this circuit, they can be viewed as a bacterial “conductor” (or “resistor”), where the conductance or admittance can be modulated by genetic modifications that control the population dynamics (Fig. 1C). A platform interfacing this property with genetic circuits can be envisioned, where bacterial colonies with unique circuit behaviors (sensors, logic gates, or oscillators) are directly connected to electrodes (Fig. 1D).
Interfacing to engineered biosensors
The dynamics of gene circuits have primarily been investigated using well-defined submillimeter-scale bacterial colonies (33–35). Interfacing synthetic biology with electrodes requires robust population behavior at larger scales (36), where dynamical behaviors are less characterized. To study the dynamic behavior in a macroscale continuous culturing system, we fabricated a customized milliliter scale chemostat with disposable gold electrodes in contact with the culture (Fig. 2A and fig. S3A). In addition, an in-house turbidity sensor provides real-time data and direct correlation between impedance and culture density, resulting in an “electrochemostat” system (fig. S3, B and C).
Fig. 2. Interfacing bacterial circuits for impedimetric sensing.
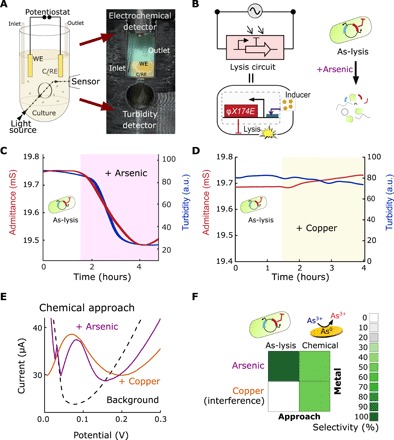
(A) Schematic and picture of electrochemostat components included in a 3D-printed holder containing a culture tube, in which the disposable electrode consisting of gold counter/reference (C/RE) and working (WE) electrodes are immersed, and an external turbidimeter detector. (B) Equivalent electrical circuit for a bacterial population engineered with an inducible promoter driving the expression of the lysis gene, E, and schematic of engineered bacterial (As-lysis) death upon arsenic induction. (C) Profiles of the admittance (red) and turbidity (blue) using an arsenic-sensitive strain in an electrochemostat device. The shaded region represents the duration of 250-ppb arsenic induction. (D) Profiles for the As-lysis strain induced with 250-ppb copper. a.u., arbitrary units. (E) Square-wave voltammetry profiles for 250-ppb arsenic (purple) and 250 ppb copper (orange). Dashed line shows the signal of the HNO3 buffer (background). The oxidation potential at the maximum current intensity versus background signal is indicative of the presence of the metal. (F) Heat maps of selectivity to arsenic versus copper using our bacterial lysis and chemical approaches. Photo credit: M. O. Din, UCSD.
We explored the capabilities of interfacing electrochemistry with synthetic biology using a construct capable of inducing lysis in the presence of a heavy metal toxin, probing the concept with an arsenic-sensitive strain (As-lysis; Fig. 2B). We observed, after steady population state, that a fast triggering of lysis resulted from the induction with arsenic, in both admittance and turbidity (Fig. 2C). Good reproducibility was found for the drop in signal with relative standard deviations (RSDs) of 7.2 and 2.1% for turbidity readouts, respectively, in n = 3 experiments. On the other hand, this bacterial construct did not lyse in response to a related toxin such as copper (Fig. 2D). Furthermore, the absence of sensitive bacteria showed a negligible increase in conductivity and no change in turbidity when induced with arsenic (fig. S4A). This arsenic biosensor was compared with a chemical sensing methodology, stripping voltammetry. This approach relies on the reduction of the arsenic ions on the gold electrode and next stripping and study of the arsenic oxidation at different potentials (37, 38). Arsenic and copper can be individually detected at +0.20 V versus Ag/AgCl (Fig. 2E); however, the similarity in oxidation potential causes copper to interfere in the chemical approach for arsenic analysis but not for the bacterial approach (Fig. 2F). A further control using a copper-sensitive strain (Cu-lysis) demonstrated the lysis triggering in the presence of copper (fig. S4B). Thus, using our approach, we may discriminate between two related heavy metal toxins compared to the standard electrochemical assay for this application.
Electrochemical population dynamics
Apart from the analytical capabilities, we also sought to implement this strategy to oscillatory synthetic circuits such as the synchronized lysis circuit (SLC; Fig. 3A). This bacterial circuit was previously shown to generate oscillatory population dynamics in microfluidic devices and in animal models, which occurs via cycles of growth and lysis (35). The genetic circuit contains a common promoter (pLuxI) that drives the expression of LuxI, which produces the quorum sensing molecule acyl homoserine lactone (AHL) that further activates the promoter by binding the activator LuxR. The promoter also drives the expression of the lysis gene from the phage phiX174, E. AHL provides an intercellular synchronization mechanism: after reaching a threshold level, synchronized lysis at the population level ensues. A remaining population of bacteria yields new AHL where the cycle repeats, resulting in oscillatory growth and lysis behavior. When measuring the SLC strain with our electrochemostat, we observe oscillatory culture density and corresponding behavior in the admittance signal over time (Fig. 3B). Good correlation of the periodical lysis events were found between the admittance and turbidity signals, with a period of 3.2 ± 0.6 hours. It is likely that the long-term bacterial population oscillations result in accumulation of bacterial debris overtime, leading to increasing baseline in the turbidity signal. Compared to common variable resistors, which usually require manual manipulation or computer coding to modify the resistance, the SLC allows a bacterial population to exhibit autonomous oscillatory resistance variations over time.
Fig. 3. Interfacing genetic oscillators.
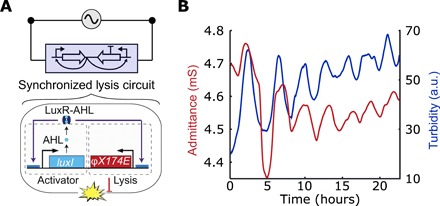
(A) Schematic of the equivalent electrical circuit when using a SLC connected to the potential source. The circuit is composed of an AHL-based quorum sensing system driving a phage lysis gene (35). The protein LuxI regulates the synthesis of autoinducer, AHL, which binds to LuxR and activates lysis after a threshold population level. (B) Profiles of the admittance (red line) and turbidity (blue line) using this strain in an electrochemostat device. The SLC is harbored by a strain Salmonella enterica subsp. enterica serovar Typhimurium (see Materials and Methods).
Bacterial integrated circuits
One substantial challenge in synthetic biology is minimally tracking gene expression without the need for fluorescent proteins and associated complex imaging equipment. Given recent efforts in the development of electrochemical platforms (8, 36) and the minimal nature of impedance measurement for circuit dynamics by directly connecting bacteria with electrodes, we sought to develop a miniature device to demonstrate the utility of our approach. To accomplish this, we fabricated a fluidic chip with multiple milliscale growth chambers, where each chamber contained electrodes (Fig. 4A). The electrodes were composed of conductive interlayers of chrome and gold, which made up the reference/counter and working electrodes (fig. S5, A, B, and E). Each of the electrode-containing growth chambers was seeded with engineered bacteria, which were referred to as bICs, since each one connects genetic circuit output to electrochemical measurement. Multiple bICs are connected in parallel to a single potentiostat and a multiplexing module, allowing for the measurement of multiple strains (Fig. 4B). We found that the resistive effects can be measured in the growth chambers of this device over a wide range of input frequencies (fig. S5C). We then showed that these electrodes can detect changes in environmental ion concentration via admittance measurement (fig. S5D). Thus, the bIC chip displayed potential to detect gene circuit behavior via impedance detection of ions in solution.
Fig. 4. bICs in millifluidic devices.
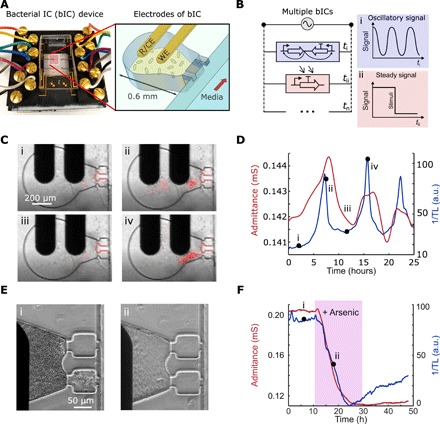
(A) Photograph of the bIC device (left) and schematic showing the design of the growth chamber and adjoining gold electrodes. Each bIC integrates lithography-fabricated electrodes, including a reference/counter electrode (R/CE) and a WE electrode, on 0.6-mm-diameter traps where bacteria form 3D continuous cultures. (B) Schematic of the equivalent electrical circuit illustrating multiple parallel interconnecting bICs measured with a single potentiostat. The dynamics in each bIC is measured sequentially in time intervals (Δt). A device containing n unique bICs may output an oscillatory signal at ti, a steady signal at tii, and other distinct signals up to tn. (C) Images of a bIC, containing the Salmonella SLC strain, were taken using transmitted light (TL). The bacteria begin at a low cell density (i) from which they reach the quorum threshold and lyse (ii and iii), and repeat the process (iv) cell growth in red is superimposed with the original time lapses in movie S1. (D) Profiles of admittance (red line) and inverse of the TL (blue line) for the strain in (C). (E) Images of the response of the bIC with the arsenic-inducible construct in E. coli MG1655 showing the steady growth state (i) and after 250-ppb arsenic induction (ii). (F) Profiles of admittance (red line) and inverse of the TL (blue line) for the strain in (E). Photo credit: A. Martin, UCSD.
We next characterized the capabilities of the electrochemical platform using the miniaturized device. We first investigated the SLC strain, using both impedance and transmitted light (TL) measurements to confirm that the impedimetric output corresponded to population dynamics. We observed bacterial growth oscillations in both TL and admittance, showing that we can achieve oscillatory impedance output with this circuit as a bIC (Fig. 4, C and D, and movie S1). Periodical lysis events were found every 7.6 ± 1.2 hours for both admittance and turbidity signals. In comparison with the electrochemostat, the admittance values in this device are lower, likely due to the lower electrode surface area. The period discrepancy is related to the differences between approaches and culture sizes. We also explored the sensing capabilities of the device using a construct capable of inducing lysis in the presence of arsenic. We observed that the bacterial population reaches a steady state in both TL and admittance before induction with arsenic. Subsequent lysis resulted in the admittance sharply decreasing to another steady state within 1 hour (Fig. 4, E and F, and movie S2). The TL exhibited a slight drift after the lysis event, likely due to a small portion of the bacterial population exhibiting both growth and lysis in the presence of arsenic, but where the population was small enough not to be detected by electrochemical measurement. Reproducibility between traps indicated RSDs of 10.8 and 13.0% (n = 4) for TL and admittance, respectively. These results demonstrate the functionality of the bIC device as a miniature platform for electrochemically measuring genetic circuit output. More generally, the electrochemical monitoring of engineered bacteria provides a simple, label-free means for the real-time collection of expression data, in contrast to standard optical modalities.
DISCUSSION
The development of synthetic biology has been motivated by ideas from electrical engineering, with gene circuits built to act as logic gates, switches, clocks, sensors, and actuators. Assessment of these gene circuit functionalities has been predominately reliant on detection of specific fluorescent or colorimetric proteins. A largely unexplored area is the direct integration of synthetic biology with electronic circuitry. A platform for direct interfacing of engineered microorganisms with microelectronics would provide a framework for a new class of hybrid biological electronic devices and biosensors where cellular “logic” informs electronic output. We have demonstrated that engineered bacterial circuits can be interfaced with microelectronics with simple impedance readouts via population lysis. Although changes in admittance with population growth have been demonstrated (27), the biochemical basis still requires further studies. The rise of conductivity of the solution is generally due to energy metabolism or transportation of ions through the cell membrane, where uncharged or weakly charged substrates are transformed into highly charged smaller ions (31). Direct interfacing of electrodes with the cell culture represents the most simplified format with which we can connect bacteria to an electrical output. This was performed without the requirement of additional substrates (6) or redox probes (20). Consequently, the bacterial population dynamics are not affected by other external inputs rather than the targets of interest. In this sense, bacterial lysis, engineered or otherwise induced, are proposed here as a general interface for electronics.
The use of engineered population control circuits with electrochemical measurement may lead to new opportunities for simple and cost-effective sensing, bypassing optical requirements. As an alternative to conventional optics, the impedance measurement allows exploring more complex nonoptically transparent materials and samples. Compared to other electrochemical approaches such as stripping voltammetry, these genetic circuits can provide the selectivity expected from a biosensor by connecting genetic sensing elements naturally present in cells to impedimetric output via lysis. By connecting the bacterial sensing output directly to electrodes, isolation and preparation steps are not required to build a biochemical sensor. In addition, compared to common molecular-based biosensors, which use enzymes or antibodies, the proposed lysis triggering of bacteria enables dynamic sensing because cells can be grown in continuous environmental conditions.
One of the main challenges of our approach is to reduce the response time. For example, the sensor responds in 40 ± 10 min after triggering (Fig. 2C). The response is likely dependent on the translation of the proteins. Thus, further efforts in the analysis and new gene circuits may decrease these times. Another challenge in measuring bacterial cultures with electrodes is the long-term stability due to biofouling (39). Some of these processes could be a reason for the overtime divergence between turbidity and admittance (Fig. 3B). To mitigate these challenges, each experiment used a new disposable screen-printed or lithography-fabricated electrode. In addition, the population control and the interfacing of genetic lysis circuits with electronics provide an inherent antibiofouling mechanism because of the continuous clearing of the chamber after certain periods of time or after certain triggering conditions.
Here, we have included arsenic, copper, and IPTG as manual triggering factors. Other triggering factors may include light, temperature, other chemicals, and bacteriophages (Fig. 1A). Further analytical efforts will be focused on improving bICs, which may include isolating the electrodes from the bacterial culture using permeable membranes (40) or using microfluidic chambers that exclusively allow the supernatant to be measured. Three-dimensional (3D) layering could be possible with the use of electrodes compared to conventional imaging methods, as well as in vivo depth tissue measurements using implantable electronics for applications where bacteria reside in vivo.
Last, the capabilities of multiplexing have been evaluated using microelectronics integrated to a microfluidic device. The compartmentalization of bacterial colonies in the bIC paves the way for high-throughput monitoring of large libraries. The presented approach may be broadly applicable to readily connect microelectronics with a variety genetic circuits such as logic gates, switches, oscillators, sensors, and more sophisticated microbial communities (41). We envision that these strategies may lead to the construction of hybrid computational devices, where an array of bacterial colonies process external information through genetic circuitry, which is then transferred via impedance, to an electronic device for analysis. The realization of this platform may lead to bIC-based devices that sense environmentally relevant molecules or enable unexplored investigations of engineered bacterial populations in the context of synthetic biology.
MATERIALS AND METHODS
Bacterial circuit strains
All of our circuits used plasmids constructed by the circular polymerase extension, standard restriction digest/ligation, or Gibson assembly cloning methods. Attenuated Salmonella enterica subsp. enterica serovar typhimurium, SL1344 strain ELH1301, and Escherichia coli MG1655 were used as the bacterial hosts for transformation and electroporation of the experimental plasmids. For the IPTG-induced lysis experiments, we used the plasmid pE35GFP, which was provided by R. Young (42). This plasmid contains a lac promoter that transcribes gene E after IPTG induction, which causes lysis of the host. The arsenic-induced lysis experiments were performed with the As-lysis plasmid. This plasmid contains an arsenic-sensitive promoter driving the expression of its repressor (ArsR, from E. coli plasmid R773) (43) and the lysis gene, E. The control with copper-sensitive (Cu-lysis) strain contains a copper-sensitive promoter driving the expression of its repressor (CueR, from E. coli plasmid) (44) and the lysis gene, E. For the SLC, we used the strains designated MOD46A and MOD101, previously described by our group (35).
Electrochemical measurements
Impedance characterization was performed using a two-electrode format using inert gold material. All electrochemical measurements were performed with a commercial potentiostat (PalmSens4 with an impedance module, Enschede, Holland) at a 10-mV voltage amplitude and zero DC bias to avoid possible electrochemical reactions. We performed the measurements in the range of frequencies starting from 1 MHz to 1 Hz. Corresponding impedance versus frequency plots were analyzed using the Randles model as the electrical equivalent circuit in a non-Faradaic system (32). Under these conditions, the total impedance could be expressed by the medium resistance (Rs) and the double-layer capacitance (Cdl) in series. By treating the impedance as a vector, the total impedance is divided into real and imaginary parts (45). Thus, when the phase is zero, the real impedance is the main component of the total impedance and can be represented as the medium resistance.
Stripping voltammetry measurements (Fig. 2E) were performed using commercial screen-printed gold electrodes using Ag/AgCl as the reference electrode (C220AT, DropSens-Metrohm, Riverview, FL, USA) and a commercial potentiostat (PalmSens4, Enschede, Holland). The arsenic and copper solutions were prepared in 0.2 M HNO3 from standard solutions at 10 ppm (ERA Waters, Golden, CO, USA). Deposition of metals on the working electrode was performed at −0.6 V for 300 s. Square-wave voltammetry profiles were obtained between 0 to +0.3 V versus Ag/AgCl, at 25 Hz, 5 mVs−1, and 40 mV, applying a cleaning step at +0.3 V for 60 s between measurements.
Electrochemostat setup
The macroscale bacterial characterization was performed using an electrochemostat composed of a custom-built 3D printed housing where the electrochemical and turbidity detectors are enclosed (fig. S3). This assembly was placed in a 37°C incubator for experiments. The 3-ml bacterial culture, confined in an autoclavable beaker, is stirred at 700 rpm. Fresh LB media (containing 0.075% Tween-20; BD Difco, LB broth, Miller) is continuously supplied with a P625/10K.143 peristaltic pump (Instech, Plymouth Meeting, PA, USA) at a typical flow rate of 25 μl/min. A removable lid holds in place inlet and outlet tubings and the disposable screen-printed electrochemical sensor consisting of a 3-mm diameter gold working electrode and a 4-mm gold counter/reference electrode (C220AT, DropSens-Metrohm, Riverview, FL, USA). The lid also has an extra opening for aerobic conditions. A new electrode is used for each experiment and connected to a type B screen–printed electrode adapter (IO Rodeo, Pasadena, CA, USA) interfaced to the potentiostat. The optimal potential frequency for impedance measurement in these experiments was 100 kHz. Turbidity and impedance values were taken every 30 s and transmitted via USB to a PC.
bIC device
Photolithography was used to pattern the multi-bICs on a 75 mm by 50 mm glass slide. Interconnected layers of Cr (20 nm)/Au (200 nm) were deposited (Denton Discovery 18 Sputter System) and chemically etched to form the eight pairs of electrodes. A second layer of polyimide (PI-2545, HD Microsystems, USA) as electrical insulation was coated to yield a 1.2-μm film. This top layer was etched by reactive ion etching (Plasmalab Oxford P80), exposing only the electrodes and connection pads. All the contact pads are aligned to a 3D-printed holder, which enables easy alignment with the microscope and connection using conductive screws interconnected to a flat ribbon cable attached to the MUX8-R2 multiplexer (fig. S5E). This module is controlled by the potentiostat that measures the eight electrodes with a 30-s delay between measurements. Optical and impedance measurements were taken every 4 and 5 min, respectively, for every bIC. A potential frequency of 1 MHz was used for the impedance measurements. Our millifluidic devices were constructed from polydimethylsiloxane (Dow Corning, Sylgard 184), which was molded and baked on a silicon master with customized features (more details about flow conditions, bacterial seeding microscope features, and data analysis are included in the Supplementary Materials).
Supplementary Material
Acknowledgments
We would like to thank A. Miano for her help with the microfluidic device fabrication, and J. Rowan and S. Hasty for technical assistance. We also acknowledge M. Ferry for advice with the electrochemostat and microfluidic designs and W. Mather for help processing data. We would also like to thank R. Young for providing the pE35GFP plasmid used in this study and T. J. Dawkins for useful scientific discussions. Funding: This work was supported by DARPA. Author contributions: All the authors have contributed extensively to the work presented in this paper. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/21/eaaz8344/DC1
REFERENCES AND NOTES
- 1.Specht A. E., Braselmann E., Palmer A. E., A critical and comparative review of fluorescent tools for live-cell imaging. Annu. Rev. Physiol. 79, 93–117 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tsien R. Y., Imagining imaging's future. Nat. Rev. Mol. Cell Biol. 2003, SS16–SS21 (2003). [PubMed] [Google Scholar]
- 3.Mimee M., Nadeau P., Hayward A., Carim S., Flanagan S., Jerger L., Collins J., McDonnell S., Swartwout R., Citorik R. J., Bulovic V., Langer R., Traverso G., Chandrakasan A. P., Lu T. K., An ingestible bacterial-electronic system to monitor gastrointestinal health. Science 360, 915–918 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ostrov N., Jimenez M., Billerbeck S., Brisbois J., Matragrano J., Ager A., Cornish V. W., A modular yeast biosensor for low-cost point-of-care pathogen detection. Sci. Adv. 3, e1603221 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vaughan O., Better sensing with bacteria. Nat. Electron. 1, 376 (2018). [Google Scholar]
- 6.Adkins J. A., Boehle K., Friend C., Chamberlain B., Bisha B., Henry C. S., Colorimetric and electrochemical bacteria detection using printed paper- and transparency-based analytic devices. Anal. Chem. 89, 3613–3621 (2017). [DOI] [PubMed] [Google Scholar]
- 7.Turner A. P. F., Biosensors--sense and sensitivity. Science 290, 1315–1317 (2000). [DOI] [PubMed] [Google Scholar]
- 8.Kim J., Campbell A., Esteban-Fernandez B., Wang J., Wearable biosensors for healthcare monitoring. Nat. Biotechnol. 37, 389–406 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cao Y., Feng Y., Ryser M. D., Zhu K., Herschlag G., Cao C., Marusak K., Zauscher S., You L., Programmable assembly of pressure sensors using pattern-forming bacteria. Nat. Biotech. 35, 1087–1093 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roggo C., van der Meer J. R., Miniaturized and integrated whole cell living bacterial sensors in field applicable autonomous devices. Curr. Opin. Biotech. 45, 24–33 (2017). [DOI] [PubMed] [Google Scholar]
- 11.Zamora-Gálvez A., Morales-Narváez E., Mayorga-Martinez C. C., Merkoçi A., Nanomaterials connected to antibodies and molecularly imprinted polymers as bio/receptors for bio/sensor applications. Appl. Mater. Today 9, 387–401 (2017). [Google Scholar]
- 12.Liu Q., Wu C., Cai H., Hu N., Zhou J., Wang P., Cell-based biosensors and their application in biomedicine. Chem. Rev. 114, 6423–6461 (2014). [DOI] [PubMed] [Google Scholar]
- 13.TerAvest M. A., Ajo-Franklin C. M., Transforming exoelectrogens for biotechnology using synthetic biology. Biotechnol. Bioeng. 113, 687–697 (2016). [DOI] [PubMed] [Google Scholar]
- 14.Zeng J., Banerjee A., Kim J., Deng Y., Chapman T. W., Daniel R., Sarpeshkar R., A novel bioelectronic reporter system in living cells tested with a synthetic biological comparator. Sci. Rep. 9, 7275 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kracke F., Lai B., Yu S., Krömer J. O., Balancing cellular redox metabolism in microbial electrosynthesis and electro fermentation-A chance for metabolic engineering. Metab. Eng. 45, 109–120 (2018). [DOI] [PubMed] [Google Scholar]
- 16.Pankratova G., Gorton L., Electrochemical communication between living cells and conductive surfaces. Curr. Opin. Electrochem. 5, 193–202 (2017). [Google Scholar]
- 17.Kumar A., Hsu L. H.-H., Kavanagh P., Barrière F., Lens P. N. L., Lapinsonnière L., Lienhard J. H. V, Schröder U., Jiang X., Leech D., The ins and outs of microorganism-electrode electron transfer reactions. Nat. Rev. Chem. 1, 0024 (2017). [Google Scholar]
- 18.Ding M., Shiu H.-Y., Li S.-L., Lee C. K., Wang G., Wu H., Weiss N. O., Young T. D., Weiss P. S., Wong G. C. L., Nealson K. H., Huang Y., Duan X., Nanoelectronic investigation reveals the electrochemical basis of electrical conductivity in shewanella and geobacter. ACS Nano 10, 9919–9926 (2016). [DOI] [PubMed] [Google Scholar]
- 19.Logan B. E., Exoelectrogenic bacteria that power microbial fuel cells. Nat. Rev. Microbiol. 7, 375–381 (2009). [DOI] [PubMed] [Google Scholar]
- 20.Tschirhart T., Kim E., McKay R., Ueda H., Wu H.-C., Pottash A. E., Zargar A., Negrete A., Shiloach J., Payne G. F., Bentley W. E., Electronic control of gene expression and cell behaviour in Escherichia coli through redox signalling. Nat. Commun. 8, 14030 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bellin D. L., Sakhtah H., Rosenstein J. K., Levine P. M., Thimot J., Emmett K., Dietrich L. E. P., Shepard K. L., Integrated circuit-based electrochemical sensor for spatially resolved detection of redox-active metabolites in biofilms. Nat. Commun. 5, 3256 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jensen H. M., Albers A. E., Malley K. R., Londer Y. Y., Cohen B. E., Helms B. A., Weigele P., Groves J. T., Ajo-Franklin C. M., Engineering of a synthetic electron conduit in living cells. Proc. Natl. Acad. Sci. U.S.A. 107, 19213–19218 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Buzid A., Shang F., Reen F. J., Muimhneacháin E. Ó., Clarke S. L., Zhou L., Luong J. H. T., O’Gara F., McGlacken G. P., Glennon J. D., Molecular signature of Pseudomonas aeruginosa with simultaneous nanomolar detection of quorum sensing signaling molecules at a boron-doped diamond electrode. Sci. Rep. 6, 30001 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tschirhart T., Zhou X. Y., Ueda H., Tsao C.-Y., Kim E., Payne G. F., Bentley W. E., Electrochemical measurement of the β-galactosidase reporter from live cells: A comparison to the Miller assay. ACS Synth. Biol. 5, 28–35 (2016). [DOI] [PubMed] [Google Scholar]
- 25.Muller M., Glutathione modulates the toxicity of, but is not a biologically relevant reductant for, the Pseudomonas aeruginosa redox toxin pyocyanin. Free Radic. Biol. Med. 50, 971–977 (2011). [DOI] [PubMed] [Google Scholar]
- 26.Butler D., Goel N., Goodnight L., Tadigadapa S., Ebrahimi A., Detection of bacterial metabolism in lag-phase using impedance spectroscopy of agar-integrated 3D microelectrodes. Biosens. Bioelectron. 129, 269–276 (2019). [DOI] [PubMed] [Google Scholar]
- 27.Gomez-Sjoberg R., Morisette D. T., Bashir R., Impedance microbiology-on-a-chip: Microfluidic bioprocessor for rapid detection of bacterial metabolism. J. Microelectromech. S. 14, 829–838 (2005). [Google Scholar]
- 28.Ward A. C., Hannah A. J., Kendrick S. L., Tucker N. P., MacGregor G., Connolly P., Identification and characterisation of Staphylococcus aureus on low cost screen printed carbon electrodes using impedance spectroscopy. Biosens. Bioelectron. 110, 65–70 (2018). [DOI] [PubMed] [Google Scholar]
- 29.Cheng X., Liu Y.-s., Irimia D., Demirci U., Yang L., Zamir L., Rodriguez W. R., Toner M., Bashir R., Cell detection and counting through cell lysate impedance spectroscopy in microfluidic devices. Lab Chip 7, 746–755 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Silley P., Forsythe S., Impedance microbiology––a rapid change for microbiologists. J. Appl. Bacteriol. 80, 233–243 (1996). [DOI] [PubMed] [Google Scholar]
- 31.Yang L., Ruan C., Li Y., Detection of viable Salmonella typhimurium by impedance measurement of electrode capacitance and medium resistance. Biosens. Bioelectron. 19, 495–502 (2003). [DOI] [PubMed] [Google Scholar]
- 32.Yang L., Bashir R., Electrical/electrochemical impedance for rapid detection of foodborne pathogenic bacteria. Biotechnol. Adv. 26, 135–150 (2008). [DOI] [PubMed] [Google Scholar]
- 33.Bennett M. R., Hasty J., Microfluidic devices for measuring gene network dynamics in single cells. Nat. Rev. Genet. 10, 628–638 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Potvin-Trottier L., Luro S., Paulsson J., Microfluidics and single-cell microscopy to study stochastic processes in bacteria. Curr. Opin. Microbiol. 43, 186–192 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Din M. O., Danino T., Prindle A., Skalak M., Selimkhanov J., Allen K., Julio E., Atolia E., Tsimring L. S., Bhatia S. N., Hasty J., Synchronized cycles of bacterial lysis for in vivo delivery. Nature 536, 81–85 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chawla K., Bürgel S. C., Schmidt G. W., Kaltenbach H.-M., Rudolf F., Frey O., Hierlemann A., Integrating impedance-based growth-rate monitoring into a microfluidic cell culture platform for live-cell microscopy. Microsyst. Nanoeng. 4, 8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Majid E., Hrapovic S., Liu Y., Male K. B., Luong J. H. T., Electrochemical determination of arsenite using a gold nanoparticle modified glassy carbon electrode and flow analysis. Anal. Chem. 78, 762–769 (2006). [DOI] [PubMed] [Google Scholar]
- 38.Salaün P., van den Berg C. M. G., Voltammetric detection of mercury and copper in seawater using a gold microwire electrode. Anal. Chem. 78, 5052–5060 (2006). [DOI] [PubMed] [Google Scholar]
- 39.Campuzano S., Yañez-Sedeño P., Pingarron J. M., Reagentless and reusable electrochemical affinity biosensors for near real-time and/or continuous operation. Advances and prospects. Curr. Opin. Electrochem. 16, 35–41 (2019). [Google Scholar]
- 40.Trouillon R., Combs Z., Patel B. A., O’Hare D., Comparative study of the effect of various electrode membranes on biofouling and electrochemical measurements. Electrochem. Commun. 11, 1409–1413 (2009). [Google Scholar]
- 41.Tsoi R., Dai Z., You L., Emerging strategies for engineering microbial communities. Biotechnol. Adv. 37, 107372 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bernhardt T. G., Roof W. D., Young R., Genetic evidence that the bacteriophage φX174 lysis protein inhibits cell wall synthesis. Proc. Natl. Acad. Sci. U.S.A. 97, 4297–4302 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu J., Rosen B. P., Metalloregulated expression of the ars operon. J. Biol. Chem. 268, 52–58 (1993). [PubMed] [Google Scholar]
- 44.Yamamoto K., Ishihama A., Transcriptional response of Escherichia coli to external copper. Mol. Microbiol. 56, 215–227 (2005). [DOI] [PubMed] [Google Scholar]
- 45.A. J. Bard, L. R. Faulkner, Electrochemical methods: fundamentals and applications (Wiley, ed. 2, 1980), pp. 316–369.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/21/eaaz8344/DC1


